Summary
SecM is an E. coli secretion monitor capable of stalling translation on the prokaryotic ribosome without co-factors. Biochemical and structural studies have demonstrated that the SecM nascent chain interacts with the 50S subunit exit tunnel to inhibit peptide bond formation. However, the timescales and pathways of stalling on a mRNA remain undefined. To provide a dynamic mechanism for stalling, we directly tracked the dynamics of elongation on ribosomes translating the SecM stall sequence (FSTPVWISQAQGIRAGP) using single-molecule fluorescence techniques. Within one minute, three peptide-ribosome interactions work cooperatively over the last 5 codons of the SecM sequence, leading to severely impaired elongation rates beginning from the terminal proline and lasting 4 codons. Our results suggest that stalling is tightly linked to the dynamics of elongation and underscore the roles that the exit tunnel and nascent chain play in controlling fundamental steps in translation.
Introduction
Proteins are synthesized by the ribosome by selecting the correct aminoacyl-tRNA, catalyzing peptide bond formation, and advancing one codon along the mRNA repetitively during translation elongation (Aitken et al., 2010; Chen et al., 2012a). Direct regulation of protein synthesis allows rapid adaptation to environmental changes within seconds to minutes. In addition to variable translation factors and tRNA abundance, the nascent polypeptide chain itself can modulate elongation (Tenson and Ehrenberg, 2002), indicating a dynamic interplay between the nascent chain and the ribosome. Stall sequences within nascent chains dramatically alter elongation, leading to a prolonged arrest of translation and controlling expression of co-transcribed genes (Ito and Chiba, 2013; Oliver et al., 1998).
The SecM stall sequence from E. coli relies solely upon peptide-ribosome interactions to stall elongation (Nakatogawa and Ito, 2001; Yap and Bernstein, 2009). In secretion-deficient conditions, SecM-induced stalling up regulates SecA expression, an ATPase secretion protein (McNicholas et al., 1997; Schmidt et al., 1988; Yap and Bernstein, 2011). However, when the cell is secretion competent, SecM stalled ribosomes are docked to the translocon machinery and the nascent chain “pulled” to relieve the stall (Butkus et al., 2003). The stability and simplicity of SecM has made it a tool to anchor the nascent peptide chain to the 50S subunit in bulk and single-molecule experiments (Evans et al., 2005; Uemura et al., 2008).
Bulk biochemical studies have identified a 17-amino-acid sequence 150FSTPVWISQAQGIRAGP166 near the C-terminus of SecM as the minimal stall sequence (Nakatogawa and Ito, 2002). It resides within the 50S subunit exit tunnel when stalling occurs. A growing body of evidence suggests that the exit tunnel, thought previously to be an inert passage way, interacts with the nascent peptide to arrest translation (Seidelt et al., 2009; Vazquez-Laslop et al., 2010; Vazquez-Laslop et al., 2008; Wilson and Beckmann, 2011). Arg163 and Pro166 are essential; mutations of either amino acid completely abolish stalling (Nakatogawa and Ito, 2002). Bulk fluorescence resonance energy transfer (FRET) measurements of peptide length within the tunnel revealed that the C-terminus is compacted, induced by interactions further upstream on the nascent chain and the constriction in the exit tunnel formed by the large subunit proteins L4 and L22 (Woolhead et al., 2006). A cryo-EM structure have suggested that the SecM peptide interacts with the tunnel entrance to remodel the geometry in the peptidyl transferase center (PTC) on the 50S subunit by moving the P-site tRNA away from the A-site tRNA (Bhushan et al., 2011). Specifically, Arg163 may interact with A2062 and U2585 of the 23S rRNA, which moves the CCA-end of the P-site tRNA away from the CCA-end of the A-site tRNA (Gumbart et al., 2012). The increased distance between the tRNAs slows peptide bond formation rate and the rigid structure of the terminal proline would then arrest translation. These elegant studies have thus identified the peptide sequence and portions of the exit tunnel anatomy necessary for stalling.
However, the proposed mechanisms implicitly assume that the final state captures stalling in its entirety. These previous studies isolated and observed stalled ribosomes hours after they had begun translation, whereas translation of the SecM sequence itself only requires a few minutes. Whether stalling abruptly stops the ribosome when all amino acids are moved into the exit tunnel or progressively changes the dynamics of elongation as the sequence is translated is not known. Previous studies have also focused on identifying a single stall site on the mRNA, inferring that the stalled state is uniform. If stalling involves a gradual buildup with subpopulations of varying stability, a distribution of stall sites is possible. Thus the dynamic pathways of stalling remain undetermined; the timescales for the ribosome to enter and exit the stalled state(s) and the spatial distribution of ribosomes on the SecM sequence have not been defined.
Single-molecule fluorescence methods can probe these mechanism with dynamics occurring over hundreds of milliseconds to hundreds of seconds (Petrov et al., 2012), observing less stable stalling locations along the SecM sequence masked by bulk averaging. Here we have leveraged our ability to track directly conformational and compositional dynamics of translating ribosomes to observe how SecM-mediated stalling unfolds. Our results show that stalling is a dynamic process where several key interactions work cooperatively within a defined time-window to reduce elongation rates over a distribution of positions along the SecM mRNA.
Results
Bulk translation experiments confirm that SecM reduces elongation rates
To confirm stalling by SecM, bulk translation rates were measured by the incorporation of tritium (3H) labeled amino acids. Two mRNAs were prepared: a stall-competent sequence (3H optimized SecM sequence) and readthrough sequence that has the critical arginine mutated (3H optimized R15A sequence)to abrogate stalling (Nakatogawa and Ito, 2002). Both sequences include unique codons at Phe2 (immediately after the start codon), Lys22 (3 codons after the stall sequence), and Leu30 (at the end of the coding sequence). After starting the elongation reaction by mixing 70S IC (initiation complex) with a factor mix containing charged total tRNA (30 μM) and elongation factors (EF-G at 1 μM), the reactions are quenched at predetermined time points (up to 10 minutes) in TCA (trichloroacetic acid) and trapped on glass fiber filters for scintillation counting (Pavlov and Ehrenberg, 1996).
The three amino acids are incorporated in order for both mRNAs: Phe first, followed by Lys, and finally Leu (Figure S1 A and B); however, translation on the SecM sequence shows significant delays between the three amino acids. The times required to incorporate the three amino acids for SecM, fitted to single-exponential distributions are: 100 ± 11 s (Phe), 150 ± 19 s (Lys), and 430 ± 90 s (Leu). This results in an average of 2.5 s per codon between Phe2-Lys22 and 35 s per codon between Lys22-Leu30. The delays are much shorter with the R15A sequence: 65 ± 7 s (Phe), 85 ± 12 s (Lys), and 100 ± 16 (Leu). The average incorporation times per codon are 1 s per codon for Phe2-Lys22 and 1.9 s per codon for Lys22-Leu30, close to the maximum translation rate observed in vitro of 0.2~1 s per codon (Underwood et al., 2005). These results confirm that the SecM sequence significantly reduces elongation rate whereas R15A abrogates stalling.
Intersubunit FRET signal allows for direct and continuous observation of an elongating ribosome
The bulk translation experiments identified a timescale of approximately 5 minutes for translational arrest on the SecM sequence. To observe translational dynamics directly, we observed translating ribosomes employing intersubunit FRET between specifically labeled E. coli 30S and 50S subunits (Dorywalska et al., 2005; Marshall et al., 2008) in zero-mode waveguides (ZMW) (Chen et al., 2013a; Uemura et al., 2010). The subunits have an inserted hairpin loop on surface-exposed portions of the rRNA (helix 44 for the 30S and helix 101 for the 50S subunit) that is complementary to a dye-labeled DNA oligonucleotide. The labeling sites are far from active sites and the dyes-oligonucleotides are exposed to solution, yielding longer continuous dye fluorescence until photobleaching or blinking.
Continuous observation of the intersubunit FRET signal between a Cy3B (donor dye) labeled 30S and BHQ2 (Black Hole Quencher 2) labeled 50S subunit monitors the conformational dynamics of a translating ribosome over multiple rounds of elongation (Aitken and Puglisi, 2010; Chen et al., 2012b). During each cycle of elongation, the ribosome undergoes two global conformational changes (Figure 1 A): (1) a 30S body rotation relative to the 50S subunit that follows peptide bond formation and (2) a reverse rotation upon EF-G catalyzed translocation. The initial non-rotated state has a higher FRET value, which corresponds to a lower Cy3B intensity. The rotated state has a lower FRET value, leading to a higher Cy3B intensity. Therefore the number of high-low-high FRET cycles reports on the number of codons translated, while the lifetimes of the FRET states report on the time to translate each codon. In all translation experiments, an aminoacylated mixture of total E. coli tRNAs was used to provide full coverage for all possible codons. Unless noted otherwise, experiments were performed at 2.5 μM total tRNA and 160 nM EF-G in the presence of 4 mM GTP.
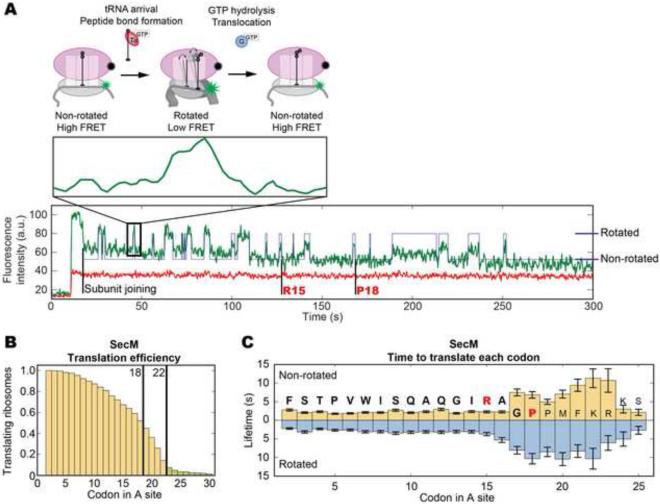
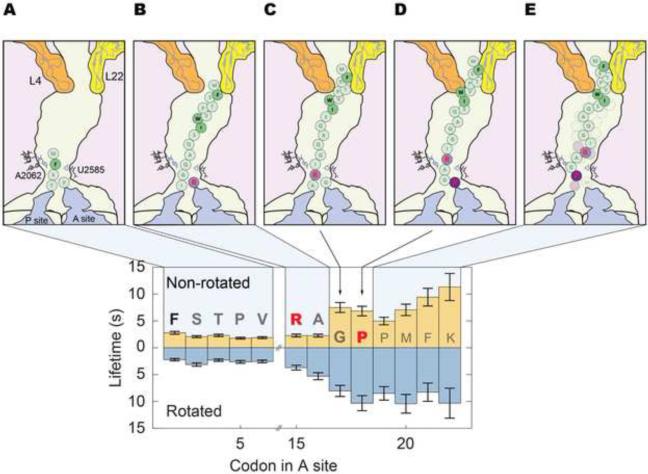
Figure 1. SecM inhibits elongation rates over specific codons.
(A) Cy3B (green) labeled 30S subunits are paired with a non-fluorescent FRET quencher BHQ2 (Black Hole Quencher 2, black)-labeled 50S subunits (Chen et al., 2012b). The ribosome begins elongation in the non-rotated state (low Cy3B intensity). Peptide bond formation rotates the subunits into the rotated (high Cy3B intensity) state. EF-G catalyzed translocation counter-rotates the subunits back to the non-rotated (low Cy3B intensity) state. Each cycle of low-high-low Cy3B intensity represents the translation of one codon. A representative trace from our SecM experiment is shown with state assignment overlaid (Arg15 and Pro18 shown for referecne). (B) Translation efficiency is plotted as the fraction of ribosomes (y-axis) still active over each codon (x-axis) within a 5 minute observation window. Translating the SecM mRNA (n = 347), 93% of the ribosomes do not proceed pass codon 22, with the sharpest drop between codons 18~22. 7% of ribosomes translating past codon 22 are colored light green. (C) The lifetimes of each state, when combined gives the time to translate a codon, are plotted the y-axis against the codon in the A site on the x-axis. The sequence translated is shown with the SecM sequence bolded and the critical amino acids in red. Before codon 15, each codon requires 4~5 s to translate at 2.5 μM total tRNA and 160 nM EF-G. Elongation rates slow down 3~5-fold over codons 16~23 to 15~20 s per codon. The non-rotated lifetimes peak over codons 17~18 and 20~23. The rotated lifetimes remain long between codons 16~22. Lifetimes are fitted to single-exponential distributions and error bars represent s.e.m. See Figure S1 for bulk translation and control experiments.
We first monitored translation on the gp32 mRNA (gene product 32 of the T4 bacteriophage) to validate application of the intersubunit FRET signal for a complex mRNA and as a standard for unhindered translation. As shown in Figure S1 C and D, more than 30% of the ribosomes reach beyond codon 30 within 5 minutes, at a constant rate of 3~4 s per codon (1.3~1.8 s in the non-rotated and 1.8~2.2 s in the rotated state). 100 μM of spectinomycin (Figure S1 E & F), a translocation blocker, increases the rotated state lifetime by 3-fold to 5.8~7.3 s while the non-rotated state remains unchanged, (Chen et al., 2013b). This slowdown compounds over each codon; only 10% of ribosomes reach codon 30 within five minutes. With 1 μM of erythromycin, a sharp drop in ribosomes translating past 10 codons is observed, with no ribosomes reaching codon 30 (Figure S1 G & H). This is consistent with previous observation that erythromycin binds in the exit tunnel and blocks peptide synthesis beyond 6~10 amino acids (Aitken and Puglisi, 2010; Tenson et al., 2003). These results confirm our ability to track elongation in real time on heteropolymeric mRNAs with codon resolution up to 20 codons, and observe specific inhibition of elongation induced by drugs.
Translation of SecM leads to codon-specific inhibitions of the elongation cycle
Applying this signal to the SecM stall sequence, a significantly different behavior compared to gp32 was observed. 93% of ribosomes do not translate past codon 22 (Figure 1 B) within five minutes. However, 45% of the ribosomes translate beyond the terminal proline (Pro18), with a sharp drop off in actively translating ribosomes between codons 18~22. Over the first 15 codons, translation proceeds efficiently at 4~5 s per codon, with short lifetimes in both the non-rotated (1.8~2.2 s) and rotated (2.2~2.9 s) states (Figure 1 C). Translation of the subsequent codons are 3~4 fold slower, at 15~20 s per codon. Beginning at codon 17, the non-rotated state lifetimes increase by 4-fold to 6.8~11 s and, beginning at codon 16, the rotated state lifetimes increase by 3-fold to 8.0~10 s. Two peaks are apparent in the non-rotated state lifetimes spanning codons 17~18 and 20~23. Conversely, the rotated state lifetimes remain consistently high between codons 16 and 23. For the 7% of ribosomes that proceeded beyond codon 22, translation rates recover after codon 22. These results suggest stalling requires 4~8 codons to fully unfold, with the process beginning before the entire SecM sequence has been translated. Additionally, SecM significantly slows elongation rates over codons 17~23 and not over a single stall site. Elongation rates of ribosomes that read through the stall eventually recover.
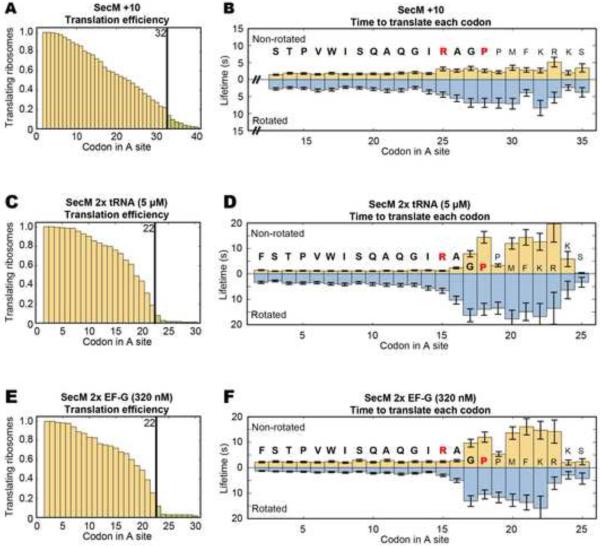
Adding 10 codons prior to the stall sequence (SecM +10 mRNA, Figure 2 A and B) results in a corresponding 10 codon shift in translational slowdown to codons 26~33. Doubling the concentration of total tRNA (2x tRNA, Figure 2 C and D) to 5 μM decreases the non-rotated lifetimes to 1.0~1.2 s (~4 s per codon) and doubling EF-G (2× EF-G, Figure 2 E and F) to 320 nM decreases the rotated lifetimes to 1.3~1.8 s (~3 s per codon). Nevertheless, their behavior around codons 16~23 remains unchanged compared to our standard conditions, suggesting that the decrease in elongation rate is not affected by changes in ligand concentrations, but rather involves intrinsic nascent-peptide/ribosome interactions.
Figure 2. SecM stalling is not affected by changes in the position of the stall sequence and the concentrations of tRNAs and EF-G.
(A) Stalling can be observed near codon 32 when SecM is shifted 10 codons downstream in our intersubunit FRET experiments (n = 242) with 2.5 μM charged total tRNA and 160 nM EF-G. (B) Increases in the non-rotated state lifetimes can be seen around codons 26~33 and increases in the rotated state lifetimes can be seen from codon 25 onwards. (C and D) Doubling charged total tRNA to 5 μM (n = 189), stalling still occurs, but there is a reduction of the non-rotated state lifetimes before codon 16. (E and F) Doubling EF-G to 320 nM, the translation profile (n = 173) remains comparable to SecM, but there is a reduction of the rotated state lifetimes before codon 15. Lifetimes are fitted to single-exponential distributions and error bars represent s.e.m.
Observing tRNA transit dynamics shows decreased elongation rates and stable incorporation of tRNA 3~4 codons beyond the terminal proline on SecM
Intersubunit FRET shows ribosomes translating up to 4 codons past the terminal Pro18; however, most ribosomes do no proceed past Phe21 or Lys22. To confirm the elongation dynamics within this region we applied an independent signal, tracking Cy5 labeled tRNAPhe transit through the ribosome as described in (Uemura et al., 2010). These experiments were performed with 200 nM of Cy5 labeled tRNAPhe in combination with 2.5 μM dark total tRNA (ΔPhe) and 160 nM EF-G. Phe tRNA binding to the ribosome results in a pulse of Cy5 fluorescence (Figure 3 A). The SecM mRNA and a truncated mRNA with all codons after Phe21 removed were tested. Both mRNAs encode Phe2 and Phe21. An intersubunit FRET experiment using the truncated mRNA shows that ribosomes have difficulty translating up to the last codon (Phe21) of the truncated mRNA within 5 minutes (Figure S2 A and B).
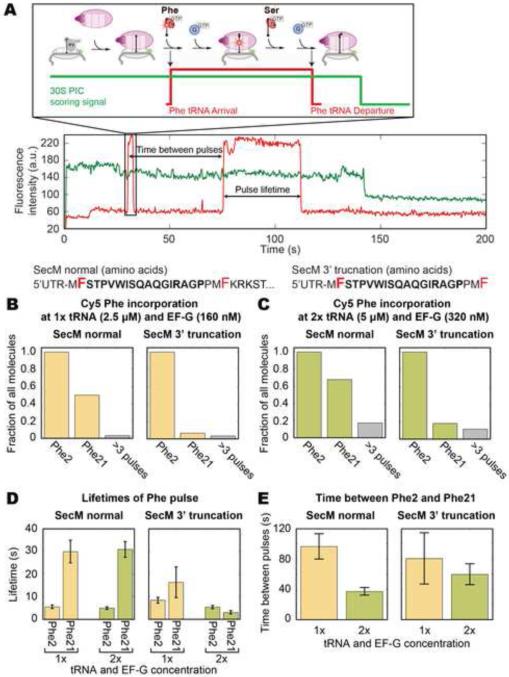
Figure 3. tRNA transit experiments show stable tRNA incorporation beyond the terminal proline and reduced elongation rates.
(A) Translating with 200 nM of Cy5 labeled tRNAPhe, accommodation of the tRNA can be tracked through stable Cy5 pulses colocalized to a 30S-Cy3B (Uemura et al., 2010). The tRNA remains on the ribosome for another round of elongation and dissociates once moved into the E site, whose dwell time thus measures the elongation rate. The SecM mRNA contains 32 codons after Phe21 whereas the SecM 3’ truncated mRNA ends exactly at Phe21. The Phe codons (2 and 21) are marked in red. (B) The fraction, normalized to ribosomes showing at least one labeled tRNA binding event, incorporating a tRNA at each Phe codon is shown. At 2.5 μM total tRNA and 160 nM EF-G (n = 220 for normal, n = 151 for truncated), 50% of ribosomes on normal SecM show tRNA binding over Phe21, which is suppressed on the truncated mRNA. (C) Doubling total tRNA to 5 μM and EF-G to 320 nM (n = 382 for normal, n = 288 for truncated) does not change the overall behavior, but increases the fraction of tRNA binding over Phe21. (D) The duration of binding is short over Phe2 but approaches the photobleaching time of the Cy5 dye over Phe21 on the normal SecM mRNA. (E) The time to translate from Phe2 to Phe21 is consistent with intersubunit FRET experiments and decreases when tRNA and EF-G concentrations are doubled. Lifetimes are fitted to single-exponential distributions and error bars represent s.e.m. See Figure S2 for controls and additional tRNA transit experiments.
50% of the ribosomes on the SecM mRNA show two pulses, suggesting the incorporation of both Phe2 and Phe21, whereas the truncated mRNA suppressed the second Cy5 pulse, commensurate with the intersubunit FRET experiment (Figure 3 B). A small fraction shows additional binding signals, which are likely unstable binding attempts of a labeled tRNA to the A site that did not result in successful tRNA accommodation. Doubling both the total tRNA and EF-G concentrations (Figure 3 C) yields a modest increase in molecules showing a second Phe pulse for both mRNAs. Compared to the short Phe2 pulse (5 s), the Phe21 pulse on the SecM mRNA is long (30 s) (Figure 3 D). This lifetime is close to the photobleaching lifetime of Cy5-labeled tRNAs, which provides a lower bound for stability of the tRNA on the ribosome. The lifetime of the Phe2 pulse on the truncated mRNA is similarly short at 8 s while the lifetime of the Phe21 pulse has a large error due to the low number of molecules. The time required to translate from Phe2 to Phe21 for the full-length SecM mRNA, calculated by the time between the two Cy5 pulses, is 100 s (Figure 3 E), consistent with intersubunit FRET experiments (130 s). Doubling the tRNA and EF-G concentration decreases the Phe2-Phe21 translation time to 40 s. The Phe2-Phe21 translation time for the truncated mRNA shows a large error due to the small number of ribosomes showing a second Phe pulse.
Additional tRNA transit experiments were conducted with a Cy2 labeled tRNALys on the SecM mRNA and the R15A readthrough mutant with 5 μM dark total tRNA (ΔPhe ΔLys), 200 nM each of Cy5 labeled tRNAPhe and Cy2 labeled tRNALys, and 320 nM EF-G. Most ribosomes on both mRNAs exhibit up to 2 pulses at Phe2 and Phe21 (55% for SecM and 68% for R15A), but only the R15A mutant shows significant Lys tRNA pulses at Lys22 and Lys24 (8% for SecM and 68% for R15A, Figure S2 C and D). Lifetimes of the labeled tRNA pulses on the SecM mRNA for both Phe21 (37 s) and Lys22 (20 s) are near the photobleaching lifetime of their respective dyes (Figure S2 E). For the R15A mRNA, all tRNA lifetimes are short, around 2~6 s. On the SecM mRNA, the times between Phe2-Phe21 (51 s) and Phe21-Lys22 (16 s) are consistent with the reagent concentrations and the distances between the codons (Figure S2 F). For R15A, the times between pulses are: 56 s for Phe2-Phe21, 7.2 s for Phe21-Lys22, and 15 s for Lys22-Lys24. These results show that some ribosomes can translate SecM mRNA up to Phe21, even Lys22. The long lifetimes of Phe21 and Lys22, as well as the longer Phe21-Lys22 time on SecM compared to the R15A mutant, suggest significantly decreased elongation rates at these codons, as seen in the intersubunit FRET measurements.
The terminal proline must specifically be at codon 18 to precipitate stalling
Changes to elongation dynamics begin 3 codons before Pro18. To specify the role of Pro18 in stalling, we performed intersubunit FRET experiments on a Pro18 to Ala mRNA (P18A). This mRNA retains Pro19; therefore, this experiment additionally probes if proline must be precisely at codon 18.
Despite retaining Pro19, stalling is abrogated in P18A (Figure 4 A), with 50% of ribosomes translating past codon 22. Similar lengthening in lifetimes of both the non-rotated and rotated state (Figure 4 B) to the wild-type SecM is observed up to Gly17. Nevertheless, the non-rotated state lifetimes return to ~2 s immediately thereafter. The rotated state lifetimes return to ~3 s by codon 20 and are shorter than the wild-type sequence (4.5~7.5 s vs. 5.3~10 s). Aze (azetidine-2-carboxylic acid), a proline analogue, has been shown to hinder stalling (Nakatogawa and Ito, 2001). Translating the wild-type SecM mRNA using total tRNA changed with Aze in place of Pro leads to a translation profile similar to P18A (Figure 4 C and D), both in the abrogation of stalling and the lifetimes over each codon. In contrast, mutating only Pro19 to Ala (P19A, Figure 4 E and F) does not abolish stalling and has no discernable differences compared to the wild-type sequence. Therefore, the positioning of proline within the A-site at exactly codon 18 is central for stalling. Its incorporation then leads to greatly reduced elongation rates between codons 18~23.
Figure 4. Proline must be positioned at codon 18 to precipitate stalling.
(A) The P18A (Pro18 to Ala) mutant (n = 231), despite retaining Pro19, allows 50% of ribosomes to translate past codon 22. (B) The lifetimes of each state is similar to the wild-type SecM up to codon 17, but the non-rotated state lifetimes quickly recover thereafter and lengthening of the rotated state lifetimes is less. (C) Translating SecM with total tRNA charged with Aze (azetidine-2-carboxylic acid) in place of Pro prevents stalling (n = 293). (D) The time to translate each codon is very similar to the P18A mutant. (E) The P19A mutant (n = 276) behaves exactly like the wild-type sequence, inducing stalling between codons 18~22. (F) The lifetimes per codon profile of P19A is similar to the wild-type SecM. Lifetimes are fitted to single-exponential distributions and error bars represent s.e.m.
Arg15 is responsible for increasing the non-rotated state lifetimes in advance of Pro18 arrival
Arginine at codon 15 is the other critical amino acid required for stalling. In all intersubunit FRET experiments described above, the non-rotated lifetimes increase when Arg15 enters the exit tunnel entrance (2 codons after incorporation). Thus Arg15 was mutated to Ala (R15A) to probe if Arg15 is responsible for these lifetime increases.
R15A mutation prevents stalling: nearly 60% of ribosomes translate past codon 22 (Figure 5 A). The non-rotated state lifetimes remain constant at 1.5~2.0 s (Figure 5 B). Lengthened rotated state lifetimes (4.6~7.5 s) are still seen between codons 16~19, but they are less severe than the wild-type (5.3~10 s). Replacing the positively charged Arg15 with a negatively charged Glu (R15E) (Figure S3 A and B) or positively charged Lys (R15K) (Figure S3 C and D) does not restore stalling and translation dynamics remains similar to R15A. These results suggest that Arg interacts with the exit tunnel entrance through its unique properties, such as its delocalized charge or ability to form specific hydrogen bonds. Mutating Arg15 removes increases in the non-rotated lifetimes over codons 17 and 18 persevered by the P18A mutant. Therefore, Arg15 interaction with the exit tunnel is likely responsible those effects, perhaps indicating difficulty in peptide bond formation.
Figure 5. Arg15 increases non-rotated state lifetimes and the N-terminal sequence lengthens rotated state lifetimes.
(A) The R15A mutant (n = 262) abolishes stalling, allowing 60% of ribosomes to translate past codon 22. (B) No increase in the non-rotated state lifetimes is seen with the R15A mutant and the increase in the rotated state lifetimes is less compared to the wild-type sequence. (C) Deleting the first 9 amino acids (Δ2-10, n = 234) of the SecM sequence abolishes stalling. (D) The lifetimes in each ribosome conformational state remain constant at 4~5 s per codon throughout the truncated sequence. Lifetimes are fitted to single-exponential distributions and error bars represent s.e.m. See Figure S3 for additional experiments on R15 and peptide bond formation.
SecM stalled ribosomes are resistant to peptide cleavage by puromycin
Prior work showed that SecM-stalled ribosomes are in conformational states resistant to peptide cleavage by the antibiotic puromycin (Muto et al., 2006). We probed peptide bond formation rates of our ribosome though puromycin release with our intersubunit FRET signal on the minimal SecM sequence or the 6(FK) mRNA. The 6(FK) mRNA codes for 6 repeating Phe-Lys codons, a stop codon, and 4 more Phe codons. Ribosomes are given 5 minutes to translate their respective sequences and either stall due to the SecM sequence or over the stop codon on 6(FK) due to a lack of release factors. Puromycin causes a permanent non-rotated to rotated (high FRET to low FRET) state transition as it cleaves off the peptide from the P-¬site tRNA (Marshall et al., 2008). In these experiments, the quencher on the 50S was replaced with Cy5 so that the FRET efficiency can be accurately quantified.
We delivered 500 μM of puromycin to 70S complexes stalled on either the 6(FK) mRNA or our SecM mRNA (Figure S3 E). On the 6(FK) mRNA, a high to low FRET transitions is clearly observed for each trace, with an average time of 20.4 ± 5.3 s (fitted to single-exponential distribution, error is s.e.m., n = 117). Ribosomes over SecM do not display any high to low FRET transitions. Instead, most traces end with the Cy5 signal photobleaching with an average lifetime of 69 ± 15 s (fitted to single-exponential distribution, error is s.e.m., n = 90), suggesting puromycin cannot attack the P-site tRNA and form a peptide bond in those ribosomes within 5 minutes. Thus the increased non-rotated state lifetimes likely stem from reduced peptide bond formation rates when the ribosome translates SecM.
The N-terminus of the SecM sequence is responsible for the increase in rotated state lifetimes in advance of both Arg15 and Pro18
The mutation experiments have demonstrated that while both Pro18 and Arg15 are essential for stalling, they are not responsible for the lengthening of the rotated state lifetimes around codons 16~19, perhaps suggesting barriers to translocation. The N-terminal region of the SecM nascent peptide likely interacts with the L4-L22 constriction point in the exit tunnel, leading to peptide compaction (Woolhead et al., 2006). To investigate if this region is responsible for the increased rotated state lifetimes, the first 9 codons of the SecM mRNA (Δ2-10) were deleted, while the rest of the sequence was retained.
Despite maintaining the critical Arg and Pro, Δ2-10 shows no stalling with close to 80% of ribosomes translating past codon 13 (Figure 5 C). Furthermore, the lifetimes of the two ribosomal conformational states are not increased, remaining at approximately 2 s for the non-rotated and 3 s for the rotated state (Figure 5 D). This suggests that the N-terminal region of SecM lengthens the rotated state between codons 16 to 19, without which Arg and Pro cannot exert their effects.
SecM-induced peptide compaction increases the energy barrier to translocation
As the rotated state ends with EF-G catalyzed translocation, we examined whether SecM interferes with EF-G binding or lengthens the lifetime through other means. An intersubunit FRET experiment using Cy5 labeled EF-G (160 nM) was performed, allowing cross-correlation of ribosome conformation to the binding of EF-G (Figure 6 A), as described (Chen et al., 2013b).
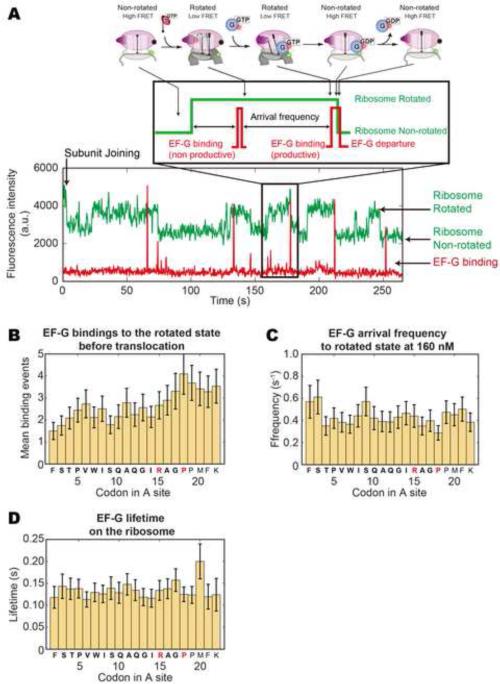
Figure 6. SecM increases the barrier to translocation but does not hinder EFG binding.
(A) Using Cy5 labeled EF-G and Cy3B/BHQ2 intersubunit FRET, EF-G binding attempts to the rotated state leading to a successful translocation can be tracked as a EF-G binding signal concurrent with a high to low Cy3B intensity transition (Chen et al., 2013b). (B) At 2.5 μM charged total tRNA and 160 nM EF-G-Cy5, the average number of EF-G binding attempts increases from 1.5~2.5 to 3.5~4 after codon 15 (n = 91), indicating an increased energy barrier to translocation. Error bars represent standard error. (C) EF-G binding frequency remains constant at 0.4~0.5 s−1, suggesting that EF-G binding to the rotated state is not inhibited. (D) EF-G dwell times on the ribosome remain constant at 0.10~0.15 s. Frequencies and lifetimes are fitted to single-exponential distributions and error bars represent s.e.m.
Translocation initially occurs within an average of 1.5~2 EF-G binding attempts to the rotated ribosome (Figure 6 B). Beyond codon 15, EF-G binding attempts increase, peaking at 4 attempts over codon 17. It remains elevated at ~3.5 attempts thereafter. However, the binding frequency of EF-G to the rotated ribosome (Figure 6 C) remains at 0.4~0.5 s−1. Likewise, the average lifetimes of EF-G on the rotated ribosome (Figure 6 D) is constant at 0.10~0.15 s. Thus the SecM sequence increases the barrier to translocation requiring increased EF-G sampling without inhibiting EF-G binding to the ribosome.
Discussion
Employing diverse single-molecule fluorescence signals, we observed translation of the SecM sequence with codon resolution over 5 minutes, a highly window to the process in vivo. We directly tracked stalling through ribosome conformational changes, tRNA binding, and EF-G binding, without extensive steps to isolate stalled ribosomes. In all of our signals, we observed clear effects of stalling. We propose that SecM stalls by significantly reducing translation rates, tightly linking stalling to elongation dynamics.
SecM induces severe slowdowns in elongation dynamics through three precisely positioned interactions with the ribosome exit tunnel
Translating the minimal SecM sequence requires at most a few minutes. Several precisely positioned interactions between the nascent peptide and the ribosome exit tunnel must take place within this window; disruption to any of these interactions abrogates stalling. Specifically, we investigated the kinetic roles of three critical elements in order to form a dynamic mechanism: the N-terminal that induce peptide compaction (Woolhead et al., 2006), Arg15 that interacts with the tunnel entrance, and Pro18 near the C-terminus (Nakatogawa and Ito, 2002). Our results lead to the model outlined below.
When the ribosome encounters the SecM sequence, it translates the first 15 codons efficiently (Figure 7 A). The nascent peptide then compacts due to interactions with the constriction point of the 50S exit tunnel formed by L4 and L22 (Figure 7 B). The mechanical stress of compaction is transduced through the nascent chain and/or the tunnel, leading to an increased energy barrier to translocation, manifested as lengthened rotated state lifetimes and increased EF-G binding attempts before translocation.
Figure 7. SecM induced stalling is a dynamic phenomenon that leads to a region of significantly reduced elongation rates.
(A) The ribosome translates the SecM sequence normally over the first 13 codons. (B) The peptide then interacts with the constriction point in the exit tunnel, formed by the large subunit proteins L4 and L22. This compacts and increases the mechanical stress on the peptide. The effect propagates downstream via the peptide or the tunnel, leading to reduced translocation rates. (C) Compaction of the peptide positions Arg15 to interact with the exit tunnel entrance to remodel tRNA geometry in the peptidyl transferase center (PTC), slowing down peptide bond formation. (D) This opens a window for Pro18 to lock the SecM peptide in conformations that induces a high level of mechanical stress leading to a heavily remodeled PTC geometry. (E) Elongation rates are greatly slowed over the next 4~5 codons past the terminal proline, leading to stalling that is stable up to an hour.
This stress limits the conformational freedom of the nascent chain, constraining Arg15 (Figure 7 C) to interact with rRNA near the tunnel entrance, which includes A2062 and U2585 (Bhushan et al., 2011; Gumbart et al., 2012). This alters the geometry of the tRNAs in the peptidyl transferase center (PTC), leading to the lengthened non-rotated state lifetime starting from codon 15. This specifically requires Arg and likely stems from its delocalized positive charge (for stacking and charge-charge interactions) and its ability to form two hydrogen bonds.
Pro18 then precipitates stalling, through the intrinsically slower peptide bond formation rates of Pro (Muto et al., 2006; Pavlov et al., 2009) and/or the rigidity that Pro imposes on the nascent chain backbone (Figure 7 D). That mutating Pro18 but not Pro19 abolished stalling, and that translating SecM with Aze, also containing a secondary amine group, abolishes stalling, suggests that stalling arises from the specific conformational properties of Pro18. The result is a heavily remodeled tRNA geometry in the PTC, leading to very low elongation rates (Figure 7 E). Accordingly, SecM stalling is a dynamic phenomenon that involves precisely tuned interactions over the tunnel entrance and the constriction point formed by L4 and L22 at the minimum.
SecM stalls ribosomes in a heterogeneous population over different positions on the mRNA
The extensive period of reduced elongation rates leads to a distribution of stalling ribosomes over 3~4 codons (codons 19~21) after the terminal proline. That some ribosomes can elongate beyond Pro18 is corroborated by both bulk translation experiments, showing significant Lys22 incorporation, and labeled tRNA transit experiments, with ribosomes stably incorporating Phe21 and Lys22 tRNAs. Interestingly, most SecM homologues across different species of prokaryotes identified in (Yap and Bernstein, 2009) retain 3~5 codons past the terminal proline before a stop codon, perhaps to provide a buffer zone.
These results seem to contradict previous biochemical studies, which suggested that SecM stalled ribosomes uniformly contain Gly17 in the P site (Muto et al., 2006; Vazquez-Laslop et al., 2010). Mechanisms have been proposed, fitting cryo-EM densities of the nascent chain in stalled ribosomes with a poly-alanine backbone assuming that Gly is attached to the P-site tRNA (Gumbart et al., 2012). This discrepancy may stem from the long timescales required before stalled ribosomes can be probed, thereby enriching for very stably stalled ribosomes at the Gly codon.
Ribosomes that did not stall at Gly would move away from the stall region given the 1-hour time-scale. The full-length SecM sequence used in both studies has a stop codon 4 codons downstream from the final Pro. The full-length sequence was then translated in the presence of release factors and ribosome recycling factors, releasing ribosomes that are not stably stalled through the stop codon. This may explain why the toe-printing experiment in Vazquez-Laslop et al. shows one position for stalled ribosomes on the mRNA. On the other hand, a similar experiment by Muto et al. shows additional bands, possibly suggesting 2 additional stalling positions downstream from Gly.
Experiments in Muto et al., using probes targeting peptidyl-tRNAGly or tRNAPro, show signs of stalling over either Gly or Pro. They additionally observe that the fraction of ribosomes stopping with a tRNAPro increases markedly when the SecM construct was truncated to end at Pro or when a stop codon was inserted immediately after Pro (translated without release factors). These modifications eliminate all codons beyond Pro, consolidating all ribosomes that could have proceeded further onto the Pro codon. These results suggest that translation could proceed beyond the terminal proline and that ribosomes at the beginning of the stalling region may have the least chance of spontaneously resuming normal elongation.
SecM-induced stalling is stable over an hour
Our bulk translation experiments showed Leu incorporation that is 8 codons downstream of the stall region. In our intersubunit FRET experiments, 7% of the ribosomes elongated past the stall region of codons 18~22 during the short 5 minute observation window. Although these ribosomes slow down between codons 16~23, they return to normal rates of elongation once past codon 22 (Figure 1 C). Perhaps by moving enough amino acids into the exit tunnel, some ribosomes spontaneously break the interactions between the nascent peptide and the exit tunnel. This is similar to the proposed N-terminal “tugging” mechanism for alleviation of stalling, but from the opposite direction (Butkus et al., 2003). Assuming that the time to clear the stall is a simplified exponential distribution and that 7% of ribosomes clear the stall in 5 minutes, the exponential lifetime of stalling is on the order of an hour. This is consistent with prior single-molecule experiments that observed GFP anchored with SecM disappearing with in ~2 hours (Uemura et al., 2008). Additionally, one bulk study recovers only 20~30% of the ribosomes as stalled complexes after a 1 hour translation reaction (Evans et al., 2005), in agreement with 40% of the ribosomes remaining stalled based on our estimated lifetime. On the other hand, in vivo prokaryotic translation takes place over seconds to minutes; a stall lasting tens of minutes is effectively permanent, allowing for the downstream SecA to be expressed until the translocon relieves the stall. Mechanisms that are more severe may be too stable to relieve using biologically accessible processes.
The dimension of time in stalling and translation
Our work provides a dynamic framework to understand peptide-induced stalling in translation. Using single-molecule fluorescence techniques, we directly showed that SecM stalling unfolds within one minute and over 4 codons, slowing elongation dynamics significantly over a region of 5 codons and lasting on the order of an hour. Stalling involves precisely positioned interactions between the exit tunnel and the nascent peptide, with each interaction setting the stage for the next. Although these effects are too transient for biochemical and structural studies to capture, they are nonetheless key elements linking stalling to elongation dynamics.
While we have provided a broad canvas to explain translational stalling, the limitations of single-molecule fluorescence should be considered- limited access to shorter timescales (<100 ms), and the relatively narrow conformational and compositional resolution of individual signals. Therefore, some important questions that remain to be answered using other approaches include the contribution from other parts of the sequence and the mechanism for relieving stalled ribosomes. Full structural understanding of stalling will require a comprehensive collection of nascent chain-ribosome complexes that embrace the evolution of transient interactions required for SecM-induced stalling, which are not captured in the end state alone.
Our results highlight the utility of single-molecule methods that directly observe translation from multiple perspectives spanning the seconds to minutes timescale across multiple codons. By tracking elongation on natural mRNAs we can directly probe many processes in elongation, including co-factor dependent stall sequences such as ErmCL (Vazquez-Laslop et al., 2008), stronger stall sequences that are detrimental in vivo, and ribosomal frameshifting induced by specific mRNA sequences.
Experimental procedures
Reagents and buffers for translation experiments
30S subunits with an inserted hairpin loop where helix 44 of the 16S rRNA is surface-exposed on the 30S body and 50S subunits with an inserted hairpin loop where helix 101 of the 23S rRNA is surface exposed were prepared as described (Dorywalska et al., 2005). Dye-labeled DNA oligos complementary to the hairpin loops were hybridized to the mutant subunits (Dorywalska et al., 2005; Marshall et al., 2008). The 30S subunit is labeled with Cy3B and 50S subunit with Black Hole Quencher 2, which is a FRET quencher (Chen et al., 2012b).
Translation factors (IF2, EF-Tu, EF-G, EF-Ts), S1, fluorescently labeled Cy5-EF-G, elbow-labeled tRNA Phe (Phe-(Cy5)tRNAPhe) and tRNA Lys (Lys-(Cy2)tRNALys) were prepared as described (Blanchard et al., 2004; Chen et al., 2013b; Marshall et al., 2008). All experiments were conducted in a Tris polymix buffer consisting of 50 mM Tris-acetate (pH 7.5), 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA, 5 mM putrescine-HCl and 1 mM spermidine. All experiments had GTP as the energy source and were performed at 22°C
Total tRNAs from E. coli were purchased from Sigma-Aldrich (St. Louis, MO, USA), charged with a tRNA synthetase mixture purified from E. coli S30 extract, and then purified for single-molecule experiments (Blanchard et al., 2004; Lesley, 1995; Pavlov and Ehrenberg, 1996).
The SecM mRNA constructs used in this study contain: 1) 21 bases at the 5’ end that hybridize to a 3’-biotinylated DNA oligo to immobilize the mRNA via neutravidin, 2) a 5’ UTR and Shine-Dalgarno sequence based on T4 bacteriophage gene 32, 3) a version of the SecM sequence, and 4) a spacer sequence. The SecM mRNAs were transcribed in vitro from plasmids purchased from GenScript (Piscataway, NJ, USA) according to published protocols (McKenna et al., 2007). The gp32 mRNA is the gene 32 mRNA sequence from the T4 bacteriophage up to codon 225 with a hybridization sequence for the 3’ biotin DNA oligo added to the 5’ end. It is transcribed from a previously prepared plasmid (Blanchard et al., 2004). All mRNA sequences are listed in the Supplemental Data section in Supplemental Information.
Bulk translation experiments
Two mixtures are made in preparation for the bulk translation experiments: ribosome mix and factor mix. Both mixtures contain the Tris-based polymix buffer with 1 mM GTP, and 1 mM ATP along with 3 mM phosphoenolpyruvate (PEP), 0.05 mg/ml pyruvate kinase, and 0.007 mg/ml myokinase for energy regeneration. The ribosome mix contains 500 nM 30S subunits, 500 nM 50S subunits, 1.5 μM mRNA, 500 nM S1, 2 μM IF2, and 1 μM initiator tRNA (fMettRNAfMet). The ribosome mix is first incubated without 50S subunits for 5 minutes at 37 °C to form 30S pre-initiation complexes (PIC) and then with 50S subunits for 5 minutes at 37 °C to form 70S initiation complexes (IC). The factor mix contains 30 μM total E. coli tRNA, 20 μM of each amino acid (including the tritium labeled amino acid), 45 μM EF-Tu, 5 μM EF-Tu, 1 μM EF-G, and 15 μl of tRNA synthetase mixture for every 100 μl of total volume. The factor mix is incubated at 37 °C for 15 minutes to charge the tRNAs and form ternary complex (Pavlov and Ehrenberg, 1996).
Immediately before the experiment, the two mixes are warmed to room temperature in a water bath for 2 minutes. 10 μl of each mix is put into 40 μl of ice-cold 7.5% trichloroacetic acid (TCA) as a 0 time-point control. The reaction is started by mixing equal volumes of the two mixes and then incubated in the room temperature water bath. At 15 s, 45 s, 120 s, 300 s, and 600 s, 20 μl of the reaction is quenched in 40 μl of ice-cold 7.5% TCA. The precipitant in TCA is spun down in a table-top centrifuge at 13,200 rpm for 15 minutes at 4 °C. The pellets are dissolved in 50 μl of 0.5 M KOH and incubated at room temperature for 15 minutes to release the peptides from the tRNA. 500 μl of 5% TCA is added to each solution to reprecipitate the released peptide. The entire mixture is then trapped on glass fiber filters and washed with 5 ml of 5% TCA. The glass fiber filters are then placed into scintillation vials and counted in a Beckman scintillation counter with 5 ml of Cytoscint scintillation cocktail (MP Biomedicals, Solon, OH, USA) for tritium activity.
Intersubunit FRET experiments
30S pre-initiation complexes (PICs) were formed as described (Marshall et al., 2008; Tsai et al., 2013). 0.25 μM Cy3B-30S, pre-incubated with stoichiometric S1, 1 μM IF2, 1 μM fMettRNAfMet, 1 μM mRNA hybridized with stoichiometric 3’ biotin DNA oligos, and 4 mM GTP were incubated at 37 °C for 5 minutes to form 30S PICs in the Tris-based polymix buffer.
Before use, we pre-incubate a SMRT Cell V3 from Pacific Biosciences (Menlo Park, CA, USA), a zero-mode waveguide (ZMW) chip containing 150,000 ZMW wells (Levene et al., 2003), with a 1 mg/ml Neutravidin solution in 50mM Tris-acetate pH 7.5 and 50 mM KCl at room temperature for 5 minutes. The cell is then washed with Buffer 6 (50 mM Tris-acetate pH 7.5, 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, and 0.5 mM EDTA). 40 μl of Buffer 6 is left in the cell to keep the cell surface wet. We then dilute the 30S PICs with our Tris-based polymix buffer containing 1 μM IF2 and 4 mM GTP down to 25~75 nM PIC concentration, depending on the specific mRNA used. The diluted PICs are then loaded into the SMRT cell at room temperature for 3 minutes to immobilize the 30S PICs. We wash away unbound material with our Tris-based polymix buffer containing 1 μM IF2, 4 mM GTP, 1 mM Trolox (to stabilize Cy5 photophysics), and a PCA/PCD oxygen scavenging system (2.5 mM 3,4-dihydroxybenzoic acid and 250 nM protocatechuate deoxygenase (Aitken et al., 2008)). 20 μl of the washing mix is left in the cell to keep the surface wet and to remove oxygen in the liquid. We then form ternary complexes between total charged E. coli tRNAs and EF-Tu(GTP) as described (Marshall et al., 2008).
Before an experiment, the cell is loaded into a PacBio RS sequencer modified by Pacific Biosciences for single-molecule fluorescence experiments (Chen et al., 2013a). To begin the elongation experiment, the instrument illuminates the SMRT cell with a green laser and then immediately delivers 20 μl of a delivery mixture containing 200 nM BHQ2-50S, 1 μM IF2, 160~320 nM EF-G, 2.5~5 μM total tRNA ternary complexes, 4 mM GTP, 1 mM Trolox, and the PCA/PCD oxygen scavenging system in our Tris-based polymix buffer onto the cell surface.
Intersubunit FRET to EF-G binding cross-correlation experiments
Cross-correlation experiments between our intersubunit FRET signal and our EF-G binding signal are conducted as our intersubunit FRET experiments, but with a different delivery mix and laser illumination. The delivery mix contains: 200 nM BHQ2-50S, 1 μM IF2,160 nM Cy5-EF-G, 2.5 μM total tRNA ternary complexes, 4 mM GTP, 1 mM Trolox, and the PCA/PCD oxygen scavenging system in our Tris-based polymix buffer. The instrument illuminates the SMRT Cell with both a green and a red laser.
Static intersubunit FRET experiments with puromycin
The SMRT V3 Cells for the experiments are prepared the same as our intersubunit FRET experiments. We first pre-form stalled 70S complexes by mixing the following: 250 nM Cy3B-30S 500 nM Cy5-50S, 1 μM IF2, 1 μM fMet-tRNAfMet, 2.5 μM mRNA, 1 μM EF-G, 5 μM total tRNA ternary complexes, and 4 mM GTP in our Tris-based polymix buffer. The solution is incubated at 37 °C for 10 minutes. After diluting the 70S mix with 4 mM GTP, 1 mM Trolox, and the PCA/PCD oxygen scavenging system in our Tris-based polymix buffer, the ribosomes are immobilized onto the SMRT cell. Unbound materials are washed away with the same dilution buffer, and the cell is then observed in the dilution buffer with only green laser illumination.
Translation experiments with labeled tRNAPhe and/or tRNALys
Translation experiments using labeled tRNAPhe are performed as our intersubunit FRET experiments with the following differences. We prepared a total tRNA mixture charged with all amino acids except Phe (total tRNA ΔPhe), and formed ternary complexes with it. Ternary complexes with Phe-(Cy5)tRNAPhe are separately formed. The delivery mix contains: 200 nM 50S, 1 μM IF2, 160~320 nM EF-G, 2.5~5 μM total tRNA ΔPhe ternary complexes, 200 nM Phe-(Cy5)tRNAPhe ternary complexes, 4 mM GTP, 1 mM Trolox, and the PCA/PCD oxygen scavenging system in the Tris-based polymix buffer. During the experiment, the SMRT Cell is illuminated with both a green and a red laser.
Experiments using both Cy5 labeled tRNAPhe and Cy2 labeled tRNALys are setup the same way with the following difference in the delivery mix: 200 nM 50S, 1 μM IF2, 320 nM EFG, 5 μM total tRNA ΔPhe ΔLys, 200 nM Phe-(Cy5)tRNAPhe, 200 nM Lys-(Cy2)tRNALys, 4 mM GTP, 1 mM Trolox, and the PCA/PCD oxygen scavenging system in the Tris-based polymix buffer. During the experiment, the ZMW chip is illuminated with a red, a green, and a blue laser.
Instrumentation for observing fluorescence from ZMW wells and data analysis
All single-molecule fluorescence experiments except for those involving Cy2 labeled tRNALys were conducted using a commercial PacBio RS sequencer that Pacific Biosciences modified to collect single-molecule fluorescence intensities from individual ZMW wells about 130 nm in diameter in 4 different dye channels corresponding to Cy3, Cy3.5, Cy5, and Cy5.5 (Chen et al., 2013a). The RS sequencer has two lasers for excitation at 532 nm and 632 nm. ZMW technology has been used to conduct single-molecule experiments (Tsai et al., 2012; Uemura et al., 2010) to observe single-molecule fluorescence at high labeled ligand concentrations (>50 nM) and to observe numerous molecule simultaneously (Levene et al., 2003). Experiments using both Cy5 labeled tRNAPhe and Cy2 labeled tRNALys were performed on a research ZMW microscope with an additional 488 nm blue laser, as described previously (Uemura et al., 2010). In all experiments, data was collected at 10 frames per second (100 ms exposure time) for 5 minutes (300 s, or 3000 frames). On the RS instrument, the power of the green laser is 0.48 μW/μm2 and the red laser is at 0.22 μW/μm2 when on. The laser power on the research instrument are 0.5 μW/μm2 for all three lasers.
Data analysis for all experiments are conducted with MATLAB (MathWorks, Torrance, CA, USA) scripts as used in previous experiments (Chen et al., 2013b; Tsai et al., 2013). Traces from the ZMW wells are filtered based on fluorescence intensity, fluorescence lifetime, and the change in intensity when the laser is turned on. Filtered traces exhibiting intersubunit FRET and/or single-molecule binding signals are then selected for data analysis. The FRET states are assigned as described based on a hidden Markov model based approach and visually corrected (Aitken and Puglisi, 2010; Tsai et al., 2013). EF-G and tRNA binding signals are manually assigned. All lifetimes were fitted to a single-exponential distribution using maximum-likelihood parameter estimation.
Supplementary Material
Highlights.
SecM stalling unfolds within minutes of translating the stall sequence
3 specific nascent chain-ribosome exit tunnel interactions are required for stalling
Stalling slows elongation rates up to 4 codons past the end of the stall sequence
Acknowledgements
Supported by NIH grants GM51266 and GM099687 (JDP), and the Wenner-Gren Foundation (MJ). We thank Alexey Petrov (Stanford) and Seán O’Leary (Stanford) for valuable discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
AT and GK conducted the experiments and data analysis; MJ and JC prepared and provided experimental materials; JC provided technical expertise with instrumentation on the RS sequencer; JDP designed the experiments; and all authors discussed the results and wrote the manuscript.
Competing financial interests
JDP is a consultant of Pacific Biosciences, a company commercializing sequencing technologies.
References
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophysical journal. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Petrov A, Puglisi JD. Single ribosome dynamics and the mechanism of translation. Annual review of biophysics. 2010;39:491–513. doi: 10.1146/annurev.biophys.093008.131427. [DOI] [PubMed] [Google Scholar]
- Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nature structural & molecular biology. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS biology. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus ME, Prundeanu LB, Oliver DB. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. Journal of bacteriology. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dalal RV, Petrov AN, Tsai A, O'Leary SE, Chapin K, Cheng J, Ewan M, Hsiung P, Lundquist P, et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proceedings of the National Academy of Sciences of the United States of America. 2013a doi: 10.1073/pnas.1315735111. in-press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nature structural & molecular biology. 2013b;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tsai A, O'Leary SE, Petrov A, Puglisi JD. Unraveling the dynamics of ribosome translocation. Current opinion in structural biology. 2012a;22:804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. Journal of the American Chemical Society. 2012b;134:5734–5737. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic acids research. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MS, Ugrinov KG, Frese MA, Clark PL. Homogeneous stalled ribosome nascent chain complexes produced in vivo or in vitro. Nature methods. 2005;2:757–762. doi: 10.1038/nmeth790. [DOI] [PubMed] [Google Scholar]
- Gumbart J, Schreiner E, Wilson DN, Beckmann R, Schulten K. Mechanisms of SecM-mediated stalling in the ribosome. Biophysical journal. 2012;103:331–341. doi: 10.1016/j.bpj.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chiba S. Arrest peptides: cis-acting modulators of translation. Annual review of biochemistry. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- Lesley SA. Preparation and use of E. coli S-30 extracts. Methods in molecular biology. 1995;37:265–278. doi: 10.1385/0-89603-288-4:265. [DOI] [PubMed] [Google Scholar]
- Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna SA, Kim I, Puglisi EV, Lindhout DA, Aitken CE, Marshall RA, Puglisi JD. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nature protocols. 2007;2:3270–3277. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- McNicholas P, Salavati R, Oliver D. Dual regulation of Escherichia coli secA translation by distinct upstream elements. Journal of molecular biology. 1997;265:128–141. doi: 10.1006/jmbi.1996.0723. [DOI] [PubMed] [Google Scholar]
- Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Molecular cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Molecular cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Oliver D, Norman J, Sarker S. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. Journal of bacteriology. 1998;180:5240–5242. doi: 10.1128/jb.180.19.5240-5242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Ehrenberg M. Rate of translation of natural mRNAs in an optimized in vitro system. Archives of biochemistry and biophysics. 1996;328:9–16. doi: 10.1006/abbi.1996.0136. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Chen J, O'Leary S, Tsai A, Puglisi JD. Single-molecule analysis of translational dynamics. Cold Spring Harbor perspectives in biology. 2012;4:a011551. doi: 10.1101/cshperspect.a011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MG, Rollo EE, Grodberg J, Oliver DB. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. Journal of bacteriology. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. Journal of molecular biology. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487:390–393. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Uemura S, Johansson M, Puglisi EV, Marshall RA, Aitken CE, Korlach J, Ehrenberg M, Puglisi JD. The impact of aminoglycosides on the dynamics of translation elongation. Cell reports. 2013;3:497–508. doi: 10.1016/j.celrep.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Iizuka R, Ueno T, Shimizu Y, Taguchi H, Ueda T, Puglisi JD, Funatsu T. Single-molecule imaging of full protein synthesis by immobilized ribosomes. Nucleic acids research. 2008;36:e70. doi: 10.1093/nar/gkn338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood KA, Swartz JR, Puglisi JD. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnology and bioengineering. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. The EMBO journal. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Molecular cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Current opinion in structural biology. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Molecular cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Molecular cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MN, Bernstein HD. The translational regulatory function of SecM requires the precise timing of membrane targeting. Molecular microbiology. 2011;81:540–553. doi: 10.1111/j.1365-2958.2011.07713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.