Summary
We performed a loss-of-function, RNA interference screen to define therapeutic targets in multiple myeloma, a genetically diverse plasma cell malignancy. Unexpectedly, we discovered that all myeloma lines require caspase-10 for survival, irrespective of their genetic abnormalities. The transcription factor IRF4 induces both caspase-10 and its associated protein cFLIPL in myeloma, generating a protease that does not induce apoptosis but rather blocks an autophagy-dependent cell death pathway. Caspase-10 inhibits autophagy by cleaving the BCL2-interacting protein BCLAF1, itself a strong inducer of autophagy that acts by displacing beclin-1 from BCL2. While myeloma cells require a basal level of autophagy for survival, caspase-10 tempers this response to avoid cell death. Drugs that disrupt this vital balance may have therapeutic potential in myeloma.
Introduction
Multiple myeloma is a malignant proliferation of plasma cells in the bone marrow. Autologous stem-cell transplantation and drugs such as lenalidomide and bortezomib have improved survival, yet myeloma remains largely incurable. One of the challenges in treating myeloma is its genomic and phenotypic heterogeneity (Kuehl and Bergsagel, 2012). Hence, an optimal therapy in myeloma would be one that would target aregulatory pathway that is essential in all subtypes of this cancer.
RNA interference screening can systematically identify genes and pathways that are essential for cancer cell proliferation and survival (Ngo et al., 2006). In some instances, these screens identify pathways that are affected by somatic mutations, thereby revealing “oncogene addiction”. In other cases, these screens identify essential genes that are not affected by structural abnormalities, a phenomenon dubbed “non-oncogene addiction” (Luo et al., 2009). In myeloma, one such non-oncogene addiction target is the transcription factor IRF4, which is required for the survival of all genetic subtypes of this cancer (Shaffer et al., 2008). Although the oncogene MYC is an important IRF4 target in myeloma, other targets must contribute to IRF4 addiction in myeloma.
Following ligand engagement of certain members of the tumor necrosis factor receptor superfamily, caspase-10 and its paralogue caspase-8 initiate a form of programmed cell death known as apoptosis (Wang et al., 2001; Wilson et al., 2009)). The recruitment of these caspases to membrane-associated “DISC” complexes induces their autoproteolysis, releasing p10/p18 dimers that initiate apoptosis by activating effector caspases. However, caspase-10 and caspase-8 have functions in addition to their pro-apoptotic role. Both proteins can activate the NF-κB pathway when overexpressed (Chaudhary et al., 2000), and caspase-8 deficiency is associated with defective T cell activation and immunodeficiency in humans and mice (Chun et al., 2002; Salmena et al., 2003). During T cell activation, macroautophagy (hereafter referred to as autophagy) is activated to meet the increased bioenergetic requirements of cell proliferation, but this autophagy is kept in check by caspase-8 (Bell et al., 2008; Hubbard et al., 2010; Yu et al., 2004).
Autophagy is a cellular process in which portions of the cytosol or entire organelles are sequestered into double membrane vesicles termed autophagosomes and subsequently fused with the lysosome, where the content is digested and recycled (reviewed in refs. (Levine and Kroemer, 2008; Rabinowitz and White, 2010). Autophagy is essential to maintain cell homeostasis, recycle damaged organelles, and overcome nutrient deprivation and metabolic stress. However, autophagy can be associated with a non-apoptotic form of cell death (Galluzzi et al., 2012), and hence this cellular reaction to stress must be tightly regulated (Bell et al., 2008; Shimizu et al., 2004; Yu et al., 2004). One level of regulation is dictated by the abundance of anti-apoptotic Bcl-2 family proteins, which sequester beclin-1, a key inducer of autophagy (Pattingre et al., 2005).
In addition to its regulation of autophagy, caspase-8 blocks a form of regulated necrosis by preventing the activation of RIP kinase 3 (RIPK3) (reviewed in (Green et al., 2011)). This regulatory pathway appears to require two caspase-8-interacting proteins, FADD and the caspase-like protein cFLIPL, forming a protease with limited activity that can block necrosis but cannot initiate apoptosis. cFLIPL pairs with caspase-8 and prevents its autocleavage while itself being a substrate of caspase-8, resulting in a caspase-8 complex containing a p43 cFLIP isoform (reviewed in (Budd et al., 2006)).
We report here an RNA interference-based genetic screen to discover therapeutic targets in genetically heterogeneous multiple myeloma cells, but not in lymphoma cells.
Results
Caspase-10 is essential for myeloma viability
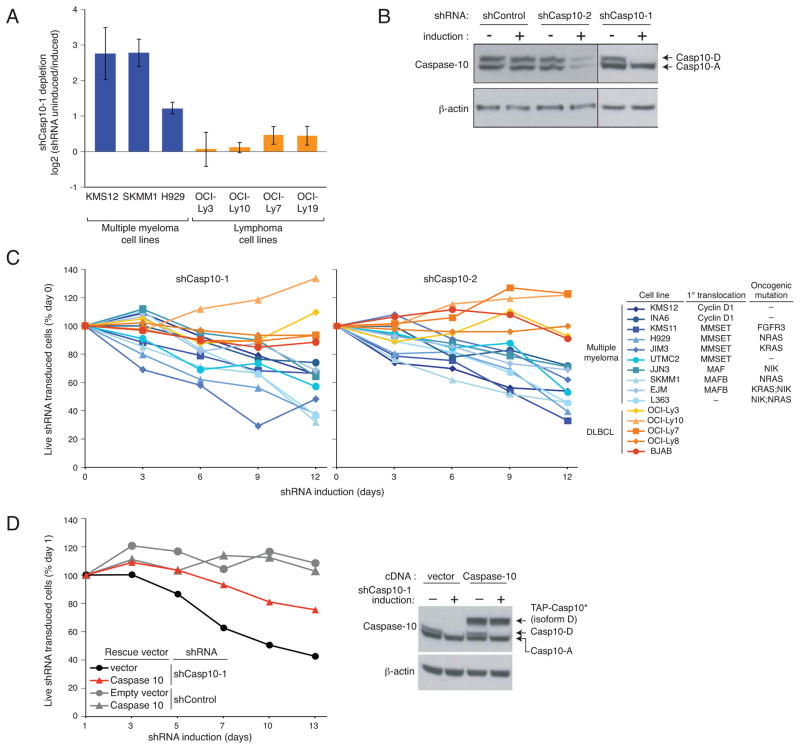
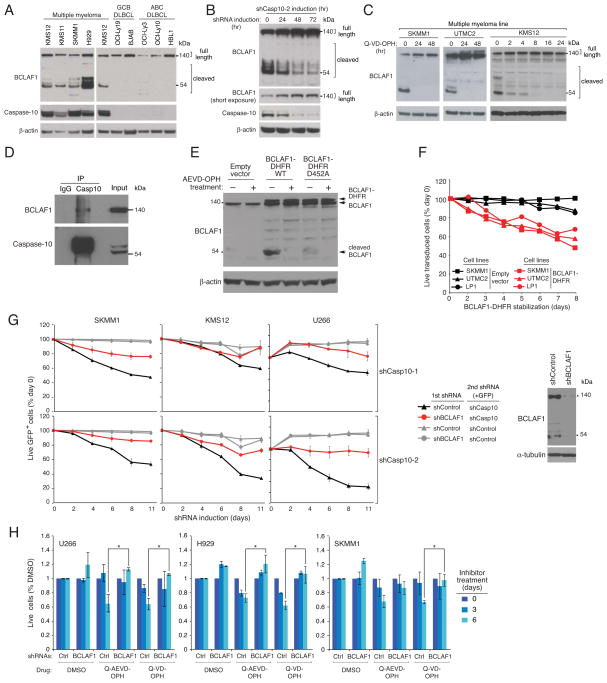
To uncover essential pathways required for myeloma proliferation and survival, we performed a loss-of-function RNA interference screen using a retroviral library to inducibly express short hairpin RNAs (shRNAs) (Ngo et al., 2006). An shRNA targeting caspase-10 was depleted during a 3-week culture of 3 myeloma cell lines, indicating its toxicity, but had no effect in 4 lymphoma lines (Figure 1A). To extend this finding, we cloned this shRNA and 5 additional caspase-10 shRNAs (Figures 1B, S1A, S1B) into a retroviral vector that allows co-expression of green fluorescent protein (GFP). In myeloma lines transduced with these vectors, the fraction of GFP+/shRNA+ cells declined over time, but no toxicity was observed in lymphoma lines (Figures 1C, S1C, S1D). One caspase-10 shRNA (#1) targets a sequence unique to the D splice isoform (Figure 1B), indicating its essential role in myeloma. The toxicity of this shRNA could be rescued by ectopic provision of a TAP-tagged caspase-10 isoform D that is resistant to this shRNA (TAP-Casp10*), showing that the toxicity of this shRNA for myeloma cells was not due to off-target effects (Figure 1D). Of note, caspase-10 shRNAs were toxic to all myeloma cell lines tested, regardless of their various oncogenic aberrations (Figure 1C).
Figure 1. Caspase 10 knockdown is toxic for all myeloma cell lines.
(A) Caspase-10 shRNA toxicity in an RNA interference screen was quantified as the ratio of shRNA abundance in shRNA-uninduced cells at time 0 vs. shRNA-induced cells after 21 days. Data represent the mean (± SEM) of 4 independent experiments. (B) Caspase-10 protein level in the SKMM1 myeloma line 4 days after the expression of 2 caspase-10 shRNAs (shCasp10-1, shCasp10-2). Caspase-10 A and D isoforms are indicated. (C) The indicated cell lines were infected with retroviruses co-expressing shCasp10-1 or shCasp10-2 and GFP. The percentage of GFP+ cells was measured over time by flow cytometry and normalized to the percentage of GFP+ cells before retroviral transduction. (D) SKMM1 cells were infected with a retrovirus expressing a TAP-tagged caspase-10 isoform D carrying mutations in the shCasp10-1 target sequence (TAP-Casp10*) or with an empty vector. The toxicity of shCasp10-1 and a control shRNA was measured as in (C). The endogenous caspase-10 and the TAP-Casp10* protein levels are shown in the right panel. See also Figure S1.
Myeloma viability depends on caspase-10 catalytic activity
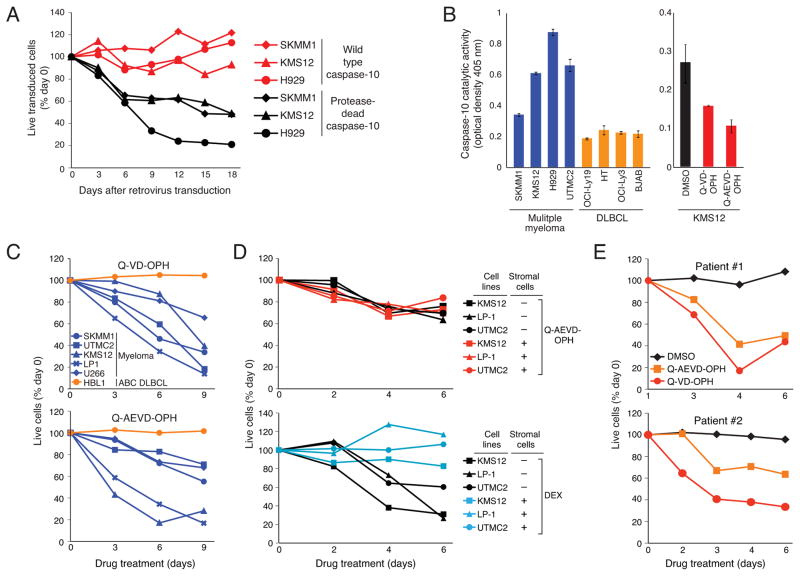
To determine if the pro-survival effect of caspase-10 in myeloma requires its protease activity, we generated a protease-dead version of the caspase-10 D isoform in which the catalytic site cysteine was replaced by serine. Wild type and protease-dead caspase-10 were cloned into a doxycyline-inducible vector that co-expresses GFP, allowing the abundance of caspase-10-transduced cells to be monitored over time (Figure 2A). After caspase-10 induction, myeloma cells expressing the protease-dead mutant were killed whereas those overexpressing wild type caspase-10 were not, suggesting that protease-dead caspase-10 functions in a dominant negative fashion to induce cell death.
Figure 2. Caspase 10 catalytic activity is essential to myeloma survival.
(A) The indicated myeloma lines were infected with a retrovirus co-expressing GFP with caspase-10 isoform D, either as the wild type or protease-dead mutant. The percentage of GFP+ cells was normalized to the percentage before retroviral transduction. (B) Caspase-10 activity in the indicated lines (Left panel) and in KMS12 myeloma cells treated for 3 hr as indicated (Right panel) was determined using an AEVD-pNA colorimetric assay. Shown are mean ± SEM from triplicates. (C) The indicated cell lines were treated with the caspase inhibitors Q-VD-OPH or Q-AEVD-OPH for the indicated times. Live cells (calcein+, propidium iodide (PI)−) were quantified by FACS using flow count fluorospheres (representative of 3 experiments). (D) The indicated myeloma lines were cultured alone or together with GFP-expressing HS-5 stromal cells for 16 hr. Cells were then exposed to Q-AEVD-OPH (25 μM) or dexamethasone (DEX, 0.5 μM) and the relative number of live myeloma cells (GFP−, PI−) was monitored by FACS. (E) CD138+ primary myeloma cells were cocultured with HS-5 stromal cells for 16 hr before adding 25 μM Q-AEVD-OPH or Q-VD-OPH, or DMSO. The relative number of viable myeloma cells was quantified by FACS. See also Figure S2.
Consistent with this hypothesis, myeloma lines had readily detectable proteolytic activity for the caspase-10 substrate AEVD-pNA, but this was not a feature of lymphoma cell lines (Figure 2B). This proteolytic activity was inhibited by the broad-spectrum caspase inhibitor Q-VD-OPH and a more selective caspase-10 inhibitor Q-AEVD-OPH (Figure 2B). Since the selectivity of synthetic caspase inhibitors is not absolute (McStay et al., 2008; Walsh et al., 2011), we further assess whether caspase-10 was responsible for the observed proteolytic activity. First, immunoprecipitated caspase-10 from different myeloma cell lines had proteolytic activity for AEVD-pNA (Figure S2A). Further, this proteolytic activity was decreased when caspase-10 was knocked down by RNA interference in myeloma cells (Figure S2B). Together, these data indicate that the elevated AEVD-pNA cleavage observed in myeloma cells is largely due to caspase-10 activity.
The caspase inhibitors Q-VD-OPH and Q-AEVD-OPH killed myeloma lines in a time-dependent manner but did not kill the lymphoma line HBL1 (Figure 2C). Since stromal cells are known to protect myeloma cells from the lethal effect of certain cytotoxic agents (Hideshima and Anderson, 2002), we investigated whether the stromal line HS-5 would mitigate the effect of caspase inhibitors on myeloma viability (Figure 2D). Whereas the toxic effect of dexamethasome was blocked by co-culture with HS-5 cells, the caspase-10 inhibitor Q-AEVD-OPH was still able to kill the myeloma cells while not affecting the viability of the stromal cells (data not shown). Finally, we isolated CD138+ neoplastic cells from newly diagnosed patients with myeloma and co-cultured them with HS-5 cells for 16 hr before adding Q-AEVD-OPH, Q-VD-OPH or DMSO. The caspase inhibitors decreased the number of viable myeloma cells in a time-dependent fashion (Figure 2E).
As caspase-8 and caspase-10 share structural and functional similarities, we examined whether caspase-8 might also regulate myeloma viability. Two shRNAs targeting caspase-8 reduced its expression by ~50% but were not toxic in the RNA interference screen (Figure S2C and S2D). To test if caspase-10 might function redundantly with caspase-8, we knocked down caspase-10 expression in myeloma lines and then treated them with a caspase-8 inhibitor (IETD-OPH) that does not inhibit caspase-10. As expected, caspase-10 knockdown was toxic, but inhibition of caspase-8 activity did not increase this toxicity, even though this inhibitor did reduce Fas-mediated apoptosis (Figure S2E). We conclude that caspase-10, but not caspase-8, maintains the viability of myeloma cells.
Caspase-10 inhibition of autophagic cell death in myeloma
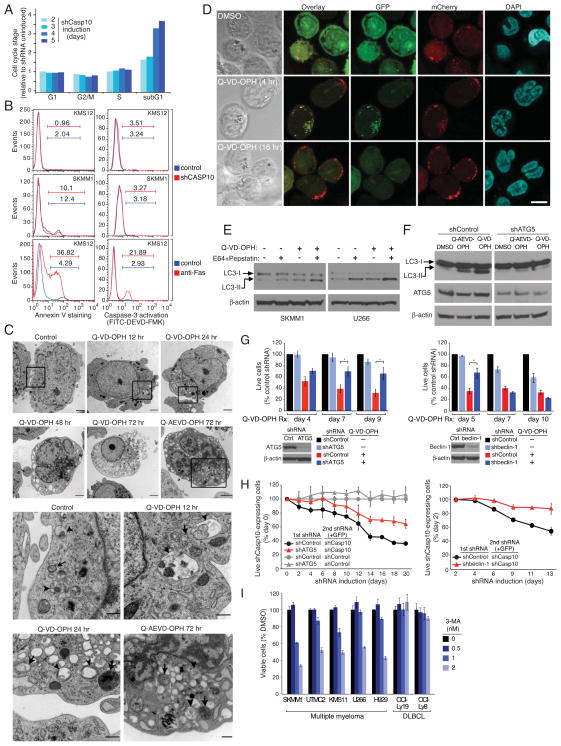
While the proportions of myeloma cells in the G1, S, and G2/M phases of the cell cycle were not affected by caspase-10 knockdown, the number of dead cells, as identified by sub-G1 DNA content, increased 3.5-fold (Figure 3A). Two hallmarks of apoptosis, phosphatidylserine exposure and caspase-3 activation, were not induced upon caspase-10 knockdown, but were induced following Fas crosslinking, as expected (Figure 3B). Moreover, treatment of myeloma cells with the caspase inhibitor Q-VD-OPH did not induce other apoptotic features, including cleavage of PARP, p23 (PTGES3) and caspase-7, but the apoptotis inducer etoposide did (Figure S3A). These experiments indicate that caspase-10 inhibits a non-apoptotic form of cell death in myeloma cells.
Figure 3. Caspase 10 inhibition in myeloma induces autophagy.
(A) Cell cycle distribution of KMS12 myeloma cells following shCasp10-2 induction determined by PI labeling and FACS analysis. Results were normalized to day 0 values. (B) shCasp10-2 was induced in the indicated myeloma lines for 5 days or left untreated. Phosphatidylserine (PS) exposure was monitored by Annexin V-PE binding (left). Caspase-3 activation was evaluated by incubation with FITC-DEVD-FMK (right). Apoptosis was induced by treatment with anti-Fas antibody CH11 (0.5 mg/ml) for 4 hr. (C) Representative electron microscopic images of UTMC2 myeloma cells treated with Q-VD-OPH (25 μM), Q-AEVD-OPH (25 μM) or vehicle for the indicated times. Lower panels show expanded view of the boxed areas in upper panel. Arrowheads indicate autophagosomes with a double membrane structure. Arrows indicate autophagolysosomes with degraded organelles. Scale bars: 1 μm. (D) Representative confocal images of SKMM1 myeloma cells expressing GFP-mCherry-tagged LC3 showing autophagosomes (green+red) and autolysosomes (red). Scale bar: 10 μm. (E) LC3-I and LC3-II levels in the indicated myeloma lines treated with QVD-OPH (25 μM) or with vehicle alone for 30 hr, and exposed to pepstatin A (10 μg/ml) plus E64-d (10 μg/ml) or vehicle in the last 12 hr. (F) ATG5 or control shRNAs were induced in SKMM1 myeloma cells for 3 days before treating with Q-VD-OPH (25 μM), Q-AEVD-OPH (25 μM) or DMSO for 2 more days, followed by immunoblot analysis of LC3, ATG5, and β-actin. (G) Left: Four myeloma lines (KMS11, KMS12, UTMC2, SKMM1) were infected with retroviruses expressing GFP with ATG5 or control shRNAs and induced for shRNA expression for 3 days before Q-AEVD-OPH (25 μM) exposure. Live GFP+, shRNA+ cells were quantified by FACS. Data from all myeloma lines were averaged. Error bars show mean ± SEM. *p<0.05. ATG5 protein level in KMS12 cells 4 days after shATG5 induction was determined by immunoblot. Right panel: Beclin-1 or control shRNAs were induced in KMS12 myeloma cells for 3 days, followed by treatment with Q-VD-OPH (25 μM) for the indicated times. Live GFP+, shRNA+ cells were quantified by FACS. Shown are means from 3 experiments ± SEM. *p<0.05. Beclin-1 protein level 4 days after beclin-1 shRNA induction was determined by immunoblot. (H) Myeloma lines were infected with retroviruses expressing indicated shRNAs, selected for integration for 4 days, then infected with retroviruses expressing GFP together with caspase-10 or control shRNAs. The relative abundance of GFP+ cells was measured by FACS. Data from two myeloma lines (U266 and KMS11) were averaged. Error bars show mean ± SEM. Data are representative of 3 independent experiments. (I) Myeloma and lymphoma lines were treated with 3-MA at the indicated concentrations for 72 hr, and viability was determined using MTS assay. See also Figure S3.
Ultrastructural analysis of UTMC2 myeloma cells treated with Q-VD-OPH or Q-AEVD-OPH revealed typical morphological features of autophagy (Figure 3C). By 12 hr after Q-VD-OPH treatment, early autophagosomes with double membranes accumulated as did a limited number of autophagic vacuoles. After 24 to 72 hr, large autophagic vacuoles containing disintegrated cellular structures appeared in the cytosol. The accumulation of autophagic vacuoles was accompanied by fewer intracellular organelles over time. By 48 hr, the endoplasmic reticulum network was barely visible and the mitochondria that remained were highly condensed. The characteristic hallmarks of apoptosis were absent (e.g. membrane blebbing, nuclear condensation), consistent with a non-apoptotic form of cell death. Similar results were obtained when caspase-10 was depleted using two different shRNAs (Figure S3B). Of note, autophagic vesicles were present at a low level in untreated myeloma cells (Figure 3C and Figure S3B), suggesting that autophagy is active at a basal rate in myeloma (see below).
To quantify autophagy and evaluate further the effect of caspase-10 inhibition on the formation of autophagosomes, myeloma cells were infected with a retrovirus expressing the autophagosome-associated protein LC3 fused to GFP. Upon initiation of autophagy, LC3 relocates from the cytosol to autophagosome membranes, where it plays a role in autophagosome enlargement (Reggiori and Klionsky, 2002), and cytosolic LC3 (LC3-I, 18 kDa) undergoes C-terminal proteolytic processing to a 16 kDa isoform, LC3-II. Autophagy is thus characterized by the accumulation of GFP-LC3 in punctate structures and by an increase in LC3-II levels (Kabeya et al., 2000). Treatment of 2 myeloma lines with Q-VD-OPH induced a time-dependent redistribution of GFP-LC3 into the punctate vesicles, in association with cell death (Figure S3C). By day 5, GFP-LC3 punctate structures were present in over 60% of caspase inhibitor-treated cells but in fewer than 5% of DMSO-treated cells.
We next wished to track autophagic flux induced by caspase-10 inhibition by distinguishing between early autophagosomes and later steps when these structures fuse with lysosomes, using a tandem GFP-mCherry-tagged LC3 (Kimura et al., 2007; Pankiv et al., 2007). Within the acidic lysosome, GFP fluorescence is quenched whereas mCherry fluorescence is stable. Hence, early autophagosomes produce yellow signals (green plus red) whereas autolysosomes produce red signals. Following exposure to the caspase inhibitor Q-VD-OPH, many myeloma cells developed overlapping green and red punctate structures by 4 hr and thus were engaged in an early phase of autophagy, but by 16 hr, most cells had only red puncta, indicating autolysosome formation (Figure 3D). Similarly, knockdown of caspase-10 expression induced both yellow and red puncta, as did the autophagy inducer rapamycin (Figure S3D). By contrast, DMSO-treated myeloma cells had diffuse green and red fluorescence as well as a few yellow and red punctate structures, consistent with a low degree of basal autophagy. Q-VD-OPH had no effect on control OCI-Ly19 lymphoma cells. Of note, the cell death induced by Q-VD-OPH did not cause chromatin condensation, as was evident in etoposide-treated cells undergoing apoptosis (Figure S3D). To quantify autophagic responses in myeloma, we used flow cytometry to monitor GFP and mCherry fluorescence in live cells following treatment with the caspase inhibitor Q-VD-OPH or DMSO (Figure S3E) (Hundeshagen et al., 2011). After 36 hr of Q-VD-OPH treatment, GFP fluorescence decreased while the mCherry signal was stable, indicating that autophagy had started and that some autophagosomes had already fused with the lysosomes. By 48 hr, both green and red signals faded, consistent with full degradation of the GFP-mCherry-LC3 fusion protein in the lysosome over time. Another way to assess autophagic flux through the lysosome is to monitor GFP-LC3 by immunoblotting, since initial lysosomal processing of GFP-LC3 releases an isolated GFP domain. Treatment of myeloma cells with Q-VD-OPH increased GFP cleavage, consistent with increased autophagic flux and lysosomal fusion (Figure S3F). To further evaluate LC3-II degradation in the lysosome, we used bafilomycin A1, which inhibits lysosomal acidification, as well as the lysosomal protease inhibitors pepstatin A and E64d. The LC3-II levels in myeloma cells were increased by Q-VD-OPH treatment, and these levels were further elevated by concurrent treatment with bafilomycin A1 or with lysosomal protease inhibitors (Figures 3E, S3G). Of note, both bafilomycin A1 and the lysosomal protease inhibitors increased LC3-II levels in myeloma cells even without caspase inhibition, consistent with a low level of basal autophagy. These data show that caspase-10 inhibition stimulates autophagic flux in myeloma cells by increasing the formation of autophagosomes that, over time, form autolysosomes.
We next tested whether the autophagic pathway is required for the death of myeloma cells following caspase-10 inhibition. Knockdown of the autophagy-related protein ATG5 decreased the induction of autophagy, as judged by LC3 processing (Figure 3F), and cell death induced by caspase inhibitors (Figure 3G). Similarly, knockdown of beclin-1 reduced Q-VD-OPH-induced death at an early time point (day 5), but depletion of beclin-1 was itself toxic at later time points (Figure 3G). Moreover, knockdown of caspase-10 was less toxic in myeloma cells induced to express shRNAs targeting the autophagy regulators beclin-1 or ATG5 than in cells expressing a control shRNA (Figure 3H). Hence, ATG5 and beclin-1 are important for the induction of cell death following caspase-10 inhibition, demonstrating that caspase-10 limits an autophagy-dependent death pathway in myeloma.
The fact that beclin-1 depletion was toxic for myeloma lines over time suggested that a basal level of autophagy is needed to maintain myeloma survival, as previously suggested (Hoang et al., 2009). Indeed, inhibition of autophagy using 3-methyladenine decreased the viability of all myeloma lines tested, but had no effect on lymphoma lines (Figure 3I).
IRF4 drives caspase-10 and cFLIPL expression in myeloma
We next wondered why myeloma cells are uniquely reliant upon caspase-10 activity. Caspase-10 protein is highly expressed in myeloma lines, with little if any expression in lymphoma lines (Figure 4A). Moreover, CASP10 mRNA levels are higher in normal bone marrow-derived plasma cells than in resting peripheral blood B cells (Figure 4B). Given that IRF4 is a master transcription factor specifying the myeloma and plasma cell phenotype (Shaffer et al., 2008), we hypothesized that IRF4 might transactivate CASP10. Indeed, knockdown of IRF4 in a myeloma cell line substantially reduced CASP10 mRNA and protein levels (Figures 4C and Figure S4A). Chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-Seq) revealed that IRF4 binds to three regions of the CASP10 locus in a myeloma line but not in a control lymphoma line that lacks IRF4 expression, which was confirmed by conventional ChIP (Figure S4B, S4C). The IRF4 peaks were centered over repeats of an IRF4 DNA binding motif (GAAA or TTTC). We conclude IRF4 upregulates CASP10 expression in normal and malignant plasma cells.
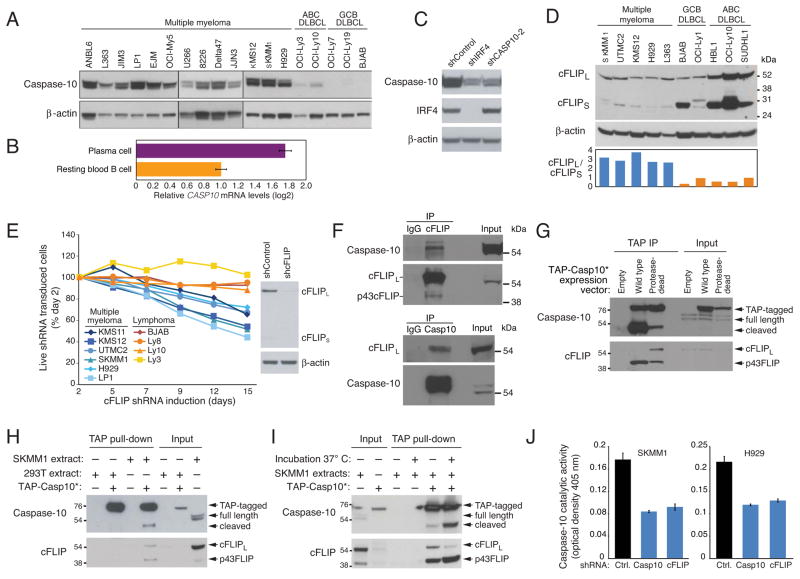
Figure 4. Myeloma cell viability requires cFLIP.
(A) Immunoblot analysis of caspase-10 and β-actin in the indicated lines. (B) Relative CASP10 mRNA expression levels in human resting blood B cells and bone marrow plasma cells quantified by Affymetrix microarray feature 210955_at using data in (Gutierrez et al., 2007). Mean normalized log2 signal values ± SEM are shown. (C) Immunoblot analysis of caspase-10, IRF4 and β-actin in UTMC2 myeloma cells induced for 4 days to express IRF4, caspase-10 or control shRNAs. (D) Immunoblot analysis of cFLIP expression in the indicated lines. Ratio of cFLIPL to cFLIPS was quantified by densitometry (bottom) (E) The indicated lines were infected with retroviruses co-expressing GFP and a cFLIP shRNA. Relative numbers of live GFP+/shRNA+ cells were monitored by FACS Right: immunoblot analysis of cFLIP in SKMM1 cells after induction of CFLAR or control shRNAs for 4 days. (F) Caspase 10 or cFLIP immunoprecipitates prepared from KMS12 myeloma cells were analyzed by immunoblotting for caspase 10 and cFLIP. Mouse IgG was used as an immunoprecipitation control and a light chain specific secondary antibody was used to improve reading around 55 kDa. Input proteins were also analyzed. (G) Wild type or protease-dead TAP-Casp10* were affinity purified on streptavidin beads from transduced SKMM1 cells and analyzed by immunoblotting for caspase-10 and cFLIP. Input extracts were also analyzed. (H) TAP-Casp10* purified from 293T cells on streptavidin beads was incubated overnight at 4°C with extracts from SKMM1 or 293T cells, and bound caspase-10 and cFLIP were analyzed by immunoblotting. An apparent cleavage product of caspase-10 is indicated. Input extracts were also analyzed. (I) TAP-Casp10* on streptavidin beads was incubated overnight at 4°C with extracts from the myeloma line SKMM1, then warmed to 37°C or kept at 4°C for 10 min followed by immunoblot analysis of caspase-10 and cFLIP. Input extracts were also analyzed. (J) Caspase-10 activity was measured using an AEVD-pNA colorimetric assay in whole cell extracts of SKMM1 and H929 myeloma lines after 4 days of caspase-10 or cFLIP shRNA induction (mean ± SEM, n = 3). See also Figure S4.
Another gene that is induced by IRF4 in myeloma cells is CFLAR (Shaffer et al., 2008), which encodes the caspase-like protein cFLIP (Shaffer et al., 2008). ChIP analyses confirmed that IRF4 binds to a promoter-proximal region in the CFLAR first intron (Figures S4D and S4E), coinciding with evolutionarily conserved tandem IRF4 binding motifs (TTTC). Two CFLAR splice variants exist, encoding a 55-kDa isoform (cFLIPL) and a 28-kDa isoform (cFLIPS). Both variants can block death receptor-induced apoptosis by heterodimerizing with caspase-8 and caspase-10, but cFLIPL has additional signaling functions, including a positive role in antigen-induced proliferation of T cells. cFLIPL was the predominant isoform in myeloma lines whereas cFLIPS was more abundant in lymphoma lines (Figure 4D).
Function of cFLIPL in myeloma
Since IRF4 upregulates both caspase-10 and cFLIPL in myeloma, we investigated whether their heterodimerization might play a role in caspase-10 function in these cells. An shRNA targeting both cFLIPL and cFLIPS was toxic for all myeloma lines tested but not for lymphoma lines (Figure 4E, S4F). This effect on myeloma viability was due to modulation of cFLIP since ectopic expression of cFLIPL rescued the SKMM1 myeloma line from the toxicity of the cFLIP shRNA (Figure S4G). Immunoprecipitation of endogenous caspase-10 in KMS12 myeloma cells enriched for cFLIPL and immunoprecipitation of cFLIP enriched for caspase-10 (Figure 4F), confirming that these two proteins heterodimerize in myeloma.
To investigate whether binding of cFLIPL to caspase-10 promotes caspase-10 activity, we used TAP-Casp10*, enabling streptavidin precipitation. In the myeloma line SKMM1, we expressed wild type TAP-Casp10* or a mutant version with an amino acid substitution that abrogates protease activity. Precipitation of wild type TAP-Casp10* pulled down a 43 kDa cFLIP isoform (p43FLIP), which was barely visible in the input lysate but could be detected in anti-cFLIP immunoprecipitation (Figure 4F upper panel), suggesting that the association of cFLIPL with TAP-Casp10* may promote its processing into p43FLIP, either within the myeloma cells or during the immunoprecipitation (Figure 4G, lower panel). By contrast, protease-dead TAP-Casp10* associated with full-length cFLIPL as well as with p43FLIP. The presence of p43FLIP under these conditions suggested that protease-dead TAP-Casp10* may associate with endogenous caspase-10, allowing some cFLIPL cleavage to proceed. This hypothesis is compatible with the analysis of TAP-Casp10* in these same immunoprecipitates (Figure 4G, upper panel). Wild type TAP-Casp10* was predominantly present as a ~47 kD fragment, presumably the product of autocatalytic cleavage occurring either in the myeloma cell or during the immunopreciptiation. Protease-dead TAP-Casp10* was mostly present as the full length isoform, but some cleaved TAP-Casp10* was present, potentially due to the presence of endogenous caspase-10 in the immunoprecipitates.
To test whether cFLIPL might induce the partial autocatalytic cleavage of caspase-10, we purified TAP-Casp10* using streptavidin beads from transfected HEK293T cells, which do not have detectable cFLIPL. Incubation of these TAP-Casp10*-coated beads with an extract from the SKMM1 myeloma line pulled down full-length cFLIPL as well as p43FLIP (Figure 4H). Incubation of TAP-Casp10* with the SKMM1 extract at 4° C yielded primarily the full length TAP-Casp10*, but also a small amount of a cleaved ~47 kDa TAP-Casp10* fragment, which was not observed following incubation with a HEK293T extract (Figure 4H). We next tested whether the processing of cFLIPL and the cleavage of caspase-10 are temperature-dependent, as would be expected if these events are due to the enzymatic activities of caspase-10. Following overnight incubation of TAP-Casp10*-coated beads with the SKMM1 extract at 4°C, warming to 37°C for 10 minutes significantly increased the cleavage of caspase-10 and the processing of cFLIPL into p43FLIP (Figure 4I). Finally, knockdown of cFLIP decreased caspase-10 proteolytic activity in myeloma lines, as did caspase-10 knockdown (Figure 4J). Together, these data suggest that the interaction between cFLIPL and caspase-10 in myeloma cells promotes partial activation of caspase-10.
Caspase-10 protects myeloma cells from BCLAF1-induced autophagy
To gain further insight into pathways regulated by caspase-10 in myeloma, we profiled gene expression following caspase-10 knockdown. BCLAF1, encoding BCL2-associated transcription factor 1, was the most consistently upregulated gene following caspase-10 inhibition in myeloma lines but not in lymphoma lines (Figure S5A), a finding confirmed by real-time PCR (Figure S5B). Although caspase-10 is unlikely to directly affect BCLAF1 transcription, these data suggested that BCLAF1 and caspase-10 might participate in a common regulatory pathway in myeloma cells. We noted that the degree of BCLAF1 protein induction following caspase-10 knockdown was quantitatively greater than the degree of BCLAF1 mRNA induction (Figures S5B, S5C), suggesting that caspase-10 might control BCLAF1 post-transcriptionally. Moreover, full length BCLAF1 protein co-existed with various smaller BCLAF1 species in myeloma lines, whereas full length BCLAF1 was predominant in lymphoma lines, raising the possibility that BCLAF1 might be targeted by a protease in myeloma cells (Figure 5A).
Figure 5. BCLAF1 is a substrate for caspase-10.
(A) Immunoblot analysis of BCLAF1 in the indicated cell lines. (B) A caspase-10 shRNA was induced in SKMM1 myeloma cells for the indicated times, and then caspase10, BCLAF1, and β-actin were analyzed by immunoblotting. (C) The indicated myeloma lines were treated with Q-VD-OPH (25 μM) for the indicated times, followed by immunoblotting for caspase-10, BCLAF1, and β-actin. (D) Caspase 10 immunoprecipitates prepared from SKMM1 myeloma cells were analyzed by immunoblotting using the indicated antibodies. Mouse IgG was used as an immunoprecipitation control. Input proteins were also analyzed. (E) SKMM1 myeloma cells were transduced with retroviruses expressing the wild type or the D452A mutant BCLAF1 fused to DHFR, BCLAF1 and β-actin were analyzed by immunblotting following 3 day induction of BCLAF1 expression. Representative of 3 experiments. (F) The indicated myeloma lines were infected with a retrovirus co-expressing mouse CD8α and BCLAF1-DHFR, or with empty vector. At time 0, the DHFR ligand trimethoprim was added to stabilize the BCLAF1-DHFR fusion protein. The percentage of LyT2+ cells was measured by FACS over time following induction of BCLAF1-DHFR, and normalized to day 0 values. (G) The indicated myeloma lines were infected with retroviruses expressing a mix of two BCLAF1 shRNAs or a control shRNA. Following shRNA induction for 2 days, cells were infected with a retrovirus co-expressing GFP and a caspase-10 shRNA or a control shRNA. Following shRNA induction, the percentage of GFP+ shCasp10+ cells was measured by FACS. Shown are means ± SEM. BCLAF1 and α-tubulin were analyzed by immunoblotting in SKMM1 cells after 4 days of shRNA induction. (H) The indicated myeloma lines were infected with retroviruses expressing a mix of two BCLAF1 shRNAs or a control shRNA, induced to express the shRNAs for 3 days, and then treated with 25 μM Q-VD-OPH or Q-AEVD-OPH, or with DMSO for the indicated times. Live calcein+, PI− cells were quantified by FACS, and normalized to DMSO values. Shown are means ± SEM *p<0.05. See also Figure S5.
Knockdown of caspase-10 in the SKMM1 myeloma line increased the abundance of full length BCLAF1 while decreasing the abundance of several smaller species, suggesting that caspase-10 is responsible for BCLAF1 cleavage in myeloma (Figure 5B). Similarly, treatment of myeloma lines with the caspase inhibitor Q-VD-OPH eliminated the smaller BCLAF1 isoforms, coinciding with an increase in the level of full length BCLAF1 (Figure 5C). By contrast, knockdown of caspase-8 had no effect on BCLAF1 cleavage in myeloma cells (Figure S5D). Immunoprecipitation of endogenous caspase-10 in SKMM1 myeloma cells enriched for BCLAF1, confirming their interaction (Figure 5D). However, the amount of endogenous BCLAF1 that co-precipitated with caspase-10 was low, suggesting that the cleavage of BCLAF1 by caspase-10 may cause the proteins to disassociate. We therefore expressed the proteolytically dead form of TAP-Casp10* or, as a control, an unrelated TAP-tagged protein (MyD88) in myeloma cells (Figure S5E). Full length BCLAF1 readily co-immunoprecipitated with this mutant TAP-Casp10* isoform but not with TAP-MyD88, supporting the notion that BCLAF1 is a substrate for caspase-10 in myeloma (Figure S5E). Of note, the abundance of BCLAF1 cleavage products decreased in cells expressing proteolytically dead TAP-Casp10*, suggesting that it acts in a dominant negative fashion, consistent with other data (Figure 2A).
In the MEROPS database of protease cleavage sites (Rawlings et al., 2004), the aspartic acid at position 452 of BCLAF1 is predicted to be a caspase-10 cleavage site. To test this, we sought to compare wild type BCLAF1 with a mutant isoform in which this aspartate is replaced by alanine (D452A). However, initial attempts to produce retroviruses expressing full length BCLAF1 were unsuccessful, potentially due to toxicity of BCLAF1 overexpression in the HEK293T retroviral producer cells (data not shown). To circumvent this problem, we engineered an inducible form of BCLAF1 by fusing it to an 18 kDa dihydrofolate reductase (DHFR)-derived “destruction” domain that targets fusion proteins for proteasomal degradation (Iwamoto et al., 2010). Proteins bearing this engineered destruction domain can be stabilized by the addition of the membrane-permeable ligand trimethoprim to the culture medium. Using this system, wild type and D452A BCLAF1-DHFR were induced in the SKMM1 myeloma line and the levels of full-length and cleaved products were assessed by immunoblotting (Figure 5E). Expression of wild type BCLAF1-DHFR yielded a 52 kDa fragment that was not present when the cells were treated with the caspase-10 inhibitor AEVD-OPH. This 52 kDa fragment was not detected in cells expressing D452A BCLAF1-DHFR, suggesting that D452 is a direct proteolytic target of caspase-10.
BCLAF1 was originally identified as a protein that binds to the adenoviral Bcl-2 homologue E1B19K as well as to several anti-apoptotic members of the BCL2 protein family and is toxic when overexpressed (Kasof et al., 1999; Renert et al., 2009). Indeed, induction of the BCLAF1-DHFR fusion protein was lethal to 3 myeloma lines (Figure 5F). We therefore investigated whether BCLAF1 is involved in the same cell death pathway in myeloma as caspase-10. We generated two shRNAs targeting BCLAF1 that lowered its mRNA and protein levels in myeloma cells without toxicity (Figures, S5F, S5G, S5H). We stably expressed these BCLAF1 shRNAs or a control shRNA in 3 myeloma lines. In the control lines, expression of a caspase-10 shRNA was toxic, as expected, but in BCLAF1 knockdown lines, the toxicity of the caspase-10 shRNA was mitigated (Figure 5G). BCLAF1 knockdown also blocked the myeloma cell death caused by Q-VD-OPH or Q-AEVD-OPH (Figure 5H) or cFLIP depletion (Figure S5I). We conclude that BCLAF1 is an important cleavage target of caspase-10/c-FLIPL that accumulates following caspase-10 inhibition, contributing to myeloma cell death.
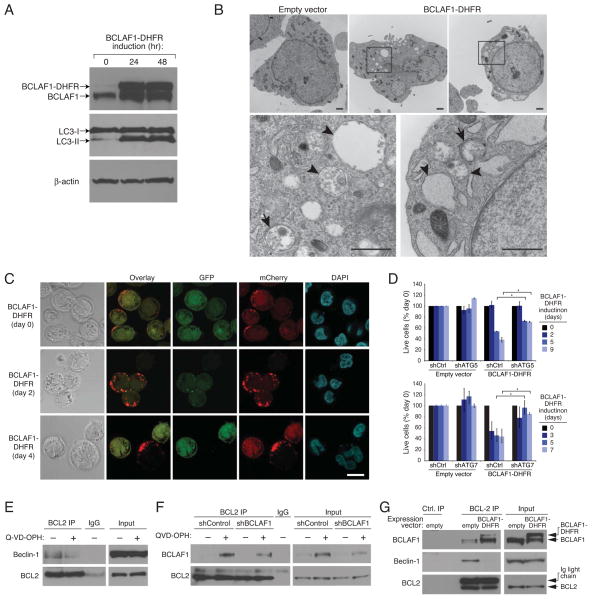
We next investigated whether the toxicity of BCLAF1 was associated with induction of autophagy. Induction of BCLAF1-DHFR expression in myeloma cells did not induce apoptosis (Figure S6A) but instead increased LC3-II conversion, consistent with autophagy (Figure 6A). Moreover, in myeloma cells expressing GFP-LC3, expression of BCLAF1-DHFR increased the percentage of cells with GFP-LC3 puncta compared with uninduced cells or cells bearing a control vector (Figure S6B). Electron micrographs of BCLAF1-DHFR-expressing cells revealed many vacuolated cells, with autophagosomes and late autophagic vacuoles visible at higher magnification (Figure 6B). As observed following caspase-10 inhibition, BCLAF1-DHFR-expressing cells had mitochondrial condensation and less endoplasmic reticulum, but the nucleus remained intact without evident chromatin changes (Figure 6B). We measured autophagic flux following BCLAF1-DHFR expression using GFP-mCherry-LC3 (Figure 6C). Control cells had a small number of LC3+ autophagosomes (yellow) and autolysosomes (red). Following BCLAF1-DHFR induction, the number of LC3+ autophagosomes increased after 2 days and by 4 days, LC3 was localized predominantly in autolysosomes. Hence, BCLAF1 overexpression in myeloma cells induces autophagy. Knockdown of the autophagy regulators ATG5 and ATG7 rescued myeloma cells from BCLAF1-induced death, demonstrating that BCLAF1 induces autophagic cell death (Figure 6D).
Figure 6. BCLAF1 promotes autophagy.
(A) Immunoblot analysis of LC3, BCLAF1, and β-actin in SKMM1 myeloma cells induced to express BCLAF1-DHFR for the indicated times. The autophagy-associated LC3-II isoform is indicated. (B) Electron micrographs of UTMC2 myeloma cells induced to express BCLAF1-DHFR for 3 days, or transduced with empty vector. Higher-power images (bottom) corresponding to boxed areas show autophagosomes with double membrane structure (arrows) and autophagolysosomes with degraded organelles (arrowheads). Scale bars: 1 μm. (C) Representative confocal images of SKMM1 myeloma cells expressing mCherry-GFP-LC3B and induced for BCLAF1-DHFR expression for the indicated times. Scale bar: 10 μm. (D) Two myeloma lines (SKMM1, KMS12) were first infected with retroviruses expressing BCLAF1-DHFR or with an empty vector, superinfected with retroviruses expressing an ATG5, ATG7 or control shRNAs and induced for shRNA expression for 2 days prior to BCLAF1-DHFR induction for the indicated periods of time. Live calcein+, PI− cells were quantified by FACS and data were normalized to DMSO values. Shown are means ± SEM. *p<0.05. Data are representative of 3 independent experiments. (E) KMS12 myeloma cells were treated with Q-VD-OPH (25 μM) or with vehicle for 48 hr, and then BCL2 immunoprecipitates were analyzed by immunoblotting for beclin-1 and BCL2. Mouse IgG was used as an immunoprecipitation control. Input proteins were also analyzed. (F) SKMM1 myeloma cells were induced to express shBCLAF1-1 for 3 days prior to Q-VD-OPH (25 μM) or vehicle treatment for 36 hr, and then BCL2 immunoprecipitates were analyzed by immunoblotting for BCLAF1 and BCL2. (G) SKMM1 myeloma cells were transduced with BCLAF1-DHFR or with an empty vector and induced to express BCLAF1-DHFR for 48 hr. Immunoprecipitates prepared using anti-BCL2 antibody-conjugated agarose beads were analysed by immunoblotting for BCLAF1, Beclin-1 or BCL2. A control precipitate prepared from SKMM1 cells transduced with an empty vector was also analyzed as were lysates. See also Figure S6.
The previous experiments suggested that the prevention of autophagic cell death by caspase-10 relies on its ability to cleave BCLAF1. Earlier work suggested that BCLAF1 binds to anti-apoptotic BCL2 family members (Kasof et al., 1999), as does the autophagy inducer beclin-1 (Pattingre et al., 2005). We therefore investigated whether caspase-10 activity might control the association of these two proteins with BCL-2. Beclin-1 could be co-immunoprecipitated with BCL-2 from myeloma cell extracts, but treatment of these cells with the caspase inhibitor Q-VD-OPH reduced this interaction (Figure 6E). By contrast, trace amounts of BCLAF1 could be detected in BCL-2 immunoprecipitates from myeloma cells, but caspase inhibition strongly increased the interaction of these two proteins (Figure 6F). Next, we expressed BCLAF1-DHFR in myeloma cells and observed that the amount of beclin-1 that could be co-immunoprecipitated with BCL-2 decreased (Figure 6G). These data are consistent with a model in which the increased interaction of BCLAF1 with BCL2 upon caspase-10 inhibition results in the dissociation of beclin-1 from BCL-2, thereby initiating autophagy.
Discussion
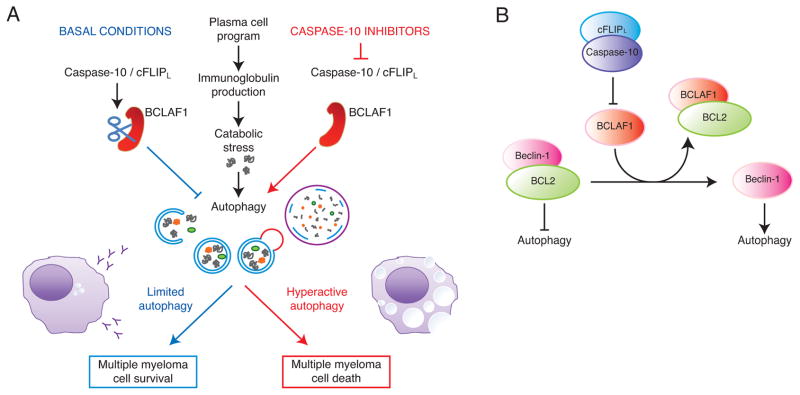
The identification of caspase-10 as essential for myeloma cell viability has unveiled a molecular pathway that regulates autophagy. Autophagy is required physiologically in normal plasma cells to moderate the expansion of the endoplasmic reticulum and immunoglobulin secretion, thereby maintaining cellular energy balance (Pengo et al., 2013) (Figure 7A). In addition, various types of endoplasmic reticulum stress, including misfolded proteins, activate the unfolded protein response, which in turn initiates autophagy (Hetz and Glimcher, 2009; Ogata et al., 2006). Myeloma cells may inherit the autophagy dependence of normal plasma cells and may also induce autophagy through the unfolded protein response. Indeed, myeloma cells had ultrastructural features consistent with low-level autophagy under basal conditions. Moreover, pharmacologic or genetic inhibition of autophagy caused myeloma cells to die, as previously described (Hoang et al., 2009). On the flip side, uncontrolled autophagy can reduce cellular viability under some circumstances, presumably when this catabolic process reduces the abundance of cellular organelles beyond a critical point (Galluzzi et al., 2012). Our studies demonstrate that a heterodimeric protease composed of caspase-10 and cFLIPL is used by myeloma cells to balance the pro-survival and pro-death effects of autophagy.
Figure 7.
Model of caspase-10 control of autophagic cell death in multiple myeloma. (A) Caspase-10 maintains the proper balance between pro-survival and pro-death autophagic responses. (B) BCLAF1 upregulation following caspase-10 inhibition leads to the dissociation of beclin-1 from BCL2, thereby augmenting autophagy.
All myeloma lines tested were caspase-10 dependent, irrespective of their underlying genetic abnormality, qualifying this phenomenon as “non-oncogene addiction” (Luo et al., 2009). The high expression of caspase-10 and cFLIPL in myeloma is due to the fact that their genes are transactivated by IRF4, a master regulator of the plasma cell phenotype (Shaffer et al., 2008). IRF4 levels in myeloma are higher than in normal plasma cells due to a positive autoregulatory loop with c-Myc (Shaffer et al., 2008), driving IRF4 targets genes to high levels in myeloma. By an unknown mechanism, myeloma cells preferentially express the cFLIPL isoform, which promotes caspase-10 activity, and not the cFLIPS isoform, which is inhibitory. These transcriptional and post-transcriptional influences promote high expression of caspase-10 and cFLIPL in myeloma, which may drive their heterodimerization and proteolytic activity. The present study implies that loss of this caspase-10 proteolytic activity is one of the many reasons that myeloma cells die upon IRF4 knockdown (Shaffer et al., 2008).
Caspase-10 inhibition in myeloma cells triggers an autophagic cell death that is reminiscent in some respects to the death invoked by caspase-8 inhibition in other cell types (Bell et al., 2008; Yu et al., 2004). In both cases, cell death can be blocked by inhibiting components of the autophagic machinery (e.g. beclin-1 and ATG5). Caspase-8 also regulates a necrotic death response, primarily by reducing RIPK3 activity. Caspase-10 does not seem to share this mechanistic feature since RIPK3 knockdown did not prevent death upon caspase-10 inhibition (data not shown). Caspase-8 itself was not required for myeloma survival, leading us to conclude that the caspase-10/cFLIPL protease performs an essential, non-redundant role in plasmacytic cells that limits the autophagic response.
BCLAF1 emerged from our studies as potent autophagy inducer that must be cleaved and inactivated by caspase-10 to protect myeloma from uncontrolled autophagy. By an unknown mechanism, BCLAF1 was also transcriptionally upregulated following caspase-10 inhibition, suggesting an orchestrated transcriptional and post-transcription regulatory module that promotes the expression of full length BCLAF1 under these circumstances. BCLAF1 function has been enigmatic. While BCLAF1 binds to anti-apoptotic BCL2 family proteins and can induce apoptosis when overexpressed in some contexts (Kasof et al., 1999), cells from BCLAF1-deficient mice do not have obvious defects in apoptosis (McPherson et al., 2009). In myeloma cells, BCLAF1 interacted with BCL2, and their association increased following caspase-10 inhibition. BCL2 residing on the endoplasmic reticulum regulates autophagy by sequestering the BH3-only protein beclin-1 (Pattingre et al., 2005). Release of beclin-1 from BCL2, and the subsequent initiation of autophagy, can be achieved by expression of the BH3-only proteins that compete for binding of beclin-1 to BCL2 (Maiuri et al., 2007). Hence, it is conceivable that BCLAF1 may induce autophagy by antagonizing the interaction of beclin-1 with BCL2 (Figure 7B). Consistent with this hypothesis, caspase-10 inhibition decreased the association of BCL2 with beclin-1 while increasing its association with BCLAF1. Although plausible, a variety of other functions have been ascribed to BCLAF1, including participation in mRNA splicing and transcriptional repression (reviewed in (Sarras et al., 2010)), which could also contribute to its function in myeloma.
The essential role of caspase-10/cFLIPL in myeloma raises the possibility that this enzyme might serve as a therapeutic target in this disease. Functional studies of caspase-10 have been limited by the fact that this caspase is not encoded in rodent genomes. Humans with inherited caspase-10 deficiency develop an autoimmune lymphoproliferative disorder, apparently due to the absence of pro-apoptotic caspase-10 signaling (Wang et al., 1999). Full auto-proteolytic processing of caspase-10 involves two internal cleavage sites and releases the active dimer that initiates apoptosis (Wachmann et al., 2010). The association of cFLIPL with caspase-10 in myeloma cells appears to prevent one of these two cleavage events, yielding a heterodimer composed of a partially cleaved caspase-10 subunit and a p43FLIP subunit. Conceivably, small molecules could be identified that would inhibit this caspase-10/cFLIPL heterodimer but not the fully cleaved caspase-10 homodimer. Such inhibitors might induce autophagic cell death of myeloma cells while not blocking caspase-10-dependent, physiological apoptosis.
Experimental Procedures
Molecular biology
The retroviral constructs for shRNA expression and the design of shRNA library sequences have been described (Ngo et al, 2006). shRNA target sequences as well as plasmid constructions are described in the Extended Experimental Procedures.
Study subjects
Bone marrow (BM) aspirates were collected from patients with newly diagnosed MM. After Ficoll–Hypaque gradient centrifugation, plasma cells were isolated from the mononuclear-cell fraction by immunomagnetic bead selection with the use of a monoclonal mouse antihuman CD138 antibody (Miltenyi-Biotec). Plasma cell purity (> 90%) was confirmed by 2-color flow cytometry using CD138+/CD45− and CD38+/CD45− antibodies (Becton Dickinson, San Jose, CA). All subjects provided written informed consent approving use of their samples for research purposes in accordance with the Declaration of Helsinki. All protocols were approved by the Institutional Review Board of the National Cancer Institute.
Cell Culture
Myeloma cell lines, lymphoma cell lines and the human stromal cell line HS-5 (kindly provided by Dr. Selina Chen-Kiang, Weill Cornell Medical College, New York, NY) were maintained as described in the Extended Experimental Procedures. For efficient retroviral infection and transduction, cell lines were engineered to express the murine ecotropic retroviral receptor (Lam et al., 2005) and the bacterial tetracycline repressor (TETR) (Ngo et al., 2006).
Caspase 10 activity
100 μg aliquots of cell lysate were prepared and used following the manufacturer’s recommendations to assay caspase 10 activity using the AEVD-pNA substrat in a colorimetric assay (Biovision, Mountain View, CA).
Cell Viability and Cell Death Measurement
To assess toxicity of a shRNA, retroviruses that coexpressed GFP were used as described (Ngo et al, 2006). In brief, flow cytometry was performed 2 days after retroviral infection to determine the initial GFP-positive proportion of live cells for each shRNA, then cells were subsequently cultured with doxycycline to induce shRNA and sampled over time. The GFP+ proportion at each time was monitored using a FACScan (Becton-Dickinson) and normalized to the initial value. For enumeration of live cells after vLyt-2 retroviral transductions, measured aliquots of cultures were stained on ice for 15 min with PE-conjugated anti–mouse CD8a (BD PharMingen). The LyT2+ proportion was monitored over by flow cytometry and normalized to the initial value. To assess Q-VD-OPH and Q-AEVD-OPH toxicity, cells were incubated with green calcein and propidium iodide (PI) at 37°C for 30 min. Flow count fluorospheres (Beckman Coulter, Fullerton, CA) were used to quantify the number of living cells (calcein positive, PI negative) by flow cytometry. Under each condition, 20.000 microbeads were added just prior to the analysis, 2000 microbeads were collected, and the number of living cells was determined using a FACScan (Becton-Dickinson).
Phosphatidylserine exposure and in-situ labelling of active caspase 3 were measured by flow cytometry using a FACScan (BD Biosciences) and data were processed with the CellQuest program (BD Biosciences). See Extended Experimental Procedures for details.
Immunoprecipitations, Pulldown Assays and Immunblot Analysis
TAP-tag Affinity Purifications, Immunoprecipitations and Pulldown Assays were performed using standard procedures from nondenaturing cell extracts. See Extended Experimental Procedures for details.
Electron microscopy
Cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (Electron Microscopy Sciences, Hatfield, PA). The cells were washed with 0.1 M sodium cacodylate buffer and post fixed with 1% osmium tetroxide/0.8% potassium ferricyanide in 0.1 M sodium cacodylate, followed by 1% tannic acid in distilled water, and stained en bloc with 1% aqueous uranyl acetate. The pellets were then dehydrated in graded ethanol series, infiltrated and embedded in Spurr’s resin. Thin sections were cut on a UC6 ultramicrotome (Leica Microsystems, Vienna, Austria) and stained with 4% aqueous uranyl acetate and Reynold’s lead citrate prior to viewing on a Tecnai BioTwin Spirit TEM (FEI, Hillsboro, OR). Digital images were acquired with a Hamamatsu XR-100 digital camera system (AMT, Danvers, MA.).
Fluorescence microscopy and confocal microscopy
Multiple myeloma cells were infected with a retrovirus expressing LC3-GFP fusion protein. After induction of autophagy, samples were examined using an epifluorescent microscope (Olympus BX61). For confocal microscopy, 1–2×105 cells were seeded on bottom glass 35 mm dishes coated with poly-lysine (MatTek Corp.) for 30 min at room temperature. The cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed two times in PBS. Nuclear counterstaining was performed with 0.1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in PBS for 10 min. Cells were washed 2 times with PBS and were examined using a Zeiss LSM510 laser scanning confocal microscope.
Supplementary Material
Significance.
A variety of translocations, mutations and copy number alterations drive the malignant phenotype of multiple myeloma. Faced with this genetic diversity, it is imperative that we design therapeutics to attack vulnerabilities that are shared by all myeloma subtypes. Using an unbiased genetic screen we discovered that all multiple myeloma subtypes tested require caspase-10 for survival. This instance of “non-oncogene addiction” appears to stem from a basal level of autophagy that is required to maintain myeloma cell viability. Caspase-10 modulates this autophagic response, preventing it from inducing cell death. Therapies targeting caspase-10 would exploit this regulatory pathway and could have broad efficacy across myeloma subtypes.
Highlights.
Caspase-10 activity maintains myeloma cells survival.
cFLIP drives caspase-10 activation in multiple myeloma.
Caspase-10 blocks autophagic cell death in myeloma.
Caspase-10 promotes survival by cleaving BCLAF1, an inducer of autophagic death.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors thank Mike Lenardo for helpful discussions and Kathleen Meyer for her assistance with GEO submissions.
Footnotes
Accession Numbers
IRF4 ChIP-seq data are at NCBI under accession SRA025850 and gene expression profiling data have been deposited in GEO under accession GSE43878.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA, Morrissette NS, Walsh CM. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008;105:16677–16682. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd RC, Yeh WC, Tschopp J. cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol. 2006;6:196–204. doi: 10.1038/nri1787. [DOI] [PubMed] [Google Scholar]
- Chaudhary PM, Eby MT, Jasmin A, Kumar A, Liu L, Hood L. Activation of the NF-kappaB pathway by caspase 8 and its homologs. Oncogene. 2000;19:4451–4460. doi: 10.1038/sj.onc.1203812. [DOI] [PubMed] [Google Scholar]
- Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell death and differentiation. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Oberst A, Dillon CP, Weinlich R, Salvesen GS. RIPK-dependent necrosis and its regulation by caspases: a mystery in five acts. Mol Cell. 2011;44:9–16. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez NC, Ocio EM, de Las Rivas J, Maiso P, Delgado M, Ferminan E, Arcos MJ, Sanchez ML, Hernandez JM, San Miguel JF. Gene expression profiling of B lymphocytes and plasma cells from Waldenstrom’s macroglobulinemia: comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma and normal individuals. Leukemia. 2007;21:541–549. doi: 10.1038/sj.leu.2404520. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Anderson KC. Molecular mechanisms of novel therapeutic approaches for multiple myeloma. Nat Rev Cancer. 2002;2:927–937. doi: 10.1038/nrc952. [DOI] [PubMed] [Google Scholar]
- Hoang B, Benavides A, Shi Y, Frost P, Lichtenstein A. Effect of autophagy on multiple myeloma cell viability. Mol Cancer Ther. 2009;8:1974–1984. doi: 10.1158/1535-7163.MCT-08-1177. [DOI] [PubMed] [Google Scholar]
- Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasof GM, Goyal L, White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol. 1999;19:4390–4404. doi: 10.1128/mcb.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. The Journal of clinical investigation. 2012;122:3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- McPherson JP, Sarras H, Lemmers B, Tamblyn L, Migon E, Matysiak-Zablocki E, Hakem A, Azami SA, Cardoso R, Fish J, et al. Essential role for Bclaf1 in lung development and immune system function. Cell death and differentiation. 2009;16:331–339. doi: 10.1038/cdd.2008.167. [DOI] [PubMed] [Google Scholar]
- McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell death and differentiation. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013 doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renert AF, Leprince P, Dieu M, Renaut J, Raes M, Bours V, Chapelle JP, Piette J, Merville MP, Fillet M. The proapoptotic C16-ceramide-dependent pathway requires the death-promoting factor Btf in colon adenocarcinoma cells. J Proteome Res. 2009;8:4810–4822. doi: 10.1021/pr9005316. [DOI] [PubMed] [Google Scholar]
- Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev. 2003;17:883–895. doi: 10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarras H, Alizadeh Azami S, McPherson JP. In search of a function for BCLAF1. Scientific World Journal. 2010;10:1450–1461. doi: 10.1100/tsw.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Wachmann K, Pop C, van Raam BJ, Drag M, Mace PD, Snipas SJ, Zmasek C, Schwarzenbacher R, Salvesen GS, Riedl SJ. Activation and specificity of human caspase-10. Biochemistry. 2010;49:8307–8315. doi: 10.1021/bi100968m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JG, Logue SE, Luthi AU, Martin SJ. Caspase-1 promiscuity is counterbalanced by rapid inactivation of processed enzyme. J Biol Chem. 2011;286:32513–32524. doi: 10.1074/jbc.M111.225862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ. Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci U S A. 2001;98:13884–13888. doi: 10.1073/pnas.241358198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zheng L, Lobito A, Chan FK, Dale J, Sneller M, Yao X, Puck JM, Straus SE, Lenardo MJ. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.