Summary
Knowledge of oncogenic mutations can inspire therapeutic strategies that are synthetically lethal, affecting cancer cells while sparing normal cells. Lenalidomide is an active agent in the activated B-cell-like (ABC) subtype of diffuse large B cell lymphoma (DLBCL), but its mechanism of action is unknown. Lenalidomide kills ABC DLBCL cells by augmenting interferon β (IFNβ) production, owing to the oncogenic MYD88 mutations in these lymphomas. In a cereblon-dependent fashion, lenalidomide downregulates IRF4 and SPIB, transcription factors that together prevent IFNβ production by repressing IRF7 and also amplify pro-survival NF-κB signaling by transactivating CARD11. Blockade of B cell receptor (BCR) signaling using the BTK inhibitor ibrutinib also downregulates IRF4 and consequently synergizes with lenalidomide in killing ABC DLBCLs, suggesting attractive therapeutic strategies.
Introduction
The activated B cell-like (ABC) subtype of diffuse large B cell lymphoma (DLBCL) is much less curable than the other common DLBCL subtypes – germinal center B cell-like (GCB DLBCL) and primary mediastinal B cell lymphoma (PMBL) – necessitating new therapeutic strategies (Alizadeh et al., 2000; Lenz et al., 2008b; Rosenwald et al., 2002; Rosenwald et al., 2003). ABC DLBCL tumors have constitutive NF-κB activity, which maintains their viability (Davis et al., 2001). Additionally, NF-κB induces expression of IRF4 (Davis et al., 2001; Saito et al., 2007), a key transcription factor in B cell differentiation and activation (Shaffer et al., 2009). IRF4 binds to a 10-base pair motif, termed the ETS/IRF composite element (EICE) (Kanno et al., 2005; Marecki and Fenton, 2000), in conjunction with one of two highly homologous ETS-family transcription factors, PU.1 and SPIB (Brass et al., 1996; Eisenbeis et al., 1995; Shaffer et al., 1997). SPIB is required for the survival of ABC DLBCL lines and is recurrently amplified and occasionally translocated in ABC DLBCL, suggesting an oncogenic function (Lenz et al., 2007; Lenz et al., 2008c). IRF4 is required for the survival of multiple myeloma cells but its role in ABC DLBCL has not been addressed (Shaffer et al., 2008).
The molecular basis for constitutive NF-κB activation in ABC DLBCL was elucidated using functional and structural genomics. Following BCR engagement, the signaling adapter CARD11 coordinates the activation of IκB kinase (IKK), a key regulator of NF-κB signaling (Thome et al., 2010). CARD11 is required for NF-κB activity and viability of ABC DLBCL lines (Ngo et al., 2006), and in ~10% of ABC DLBCLs, CARD11 acquires oncogenic mutations leading to spontaneous IKK and NF-κB activity (Lenz et al., 2008a). In other DLBCLs, BCR signaling engages wild-type CARD11 to activate NF-κB, a phenomenon termed chronic active BCR signaling (Davis et al., 2010). More than 20% of ABC DLBCL tumors have mutant forms of the CD79B and CD79A subunits of the BCR that augment receptor signaling, establishing the pathogenetic importance of the BCR pathway in ABC DLBCL (Davis et al., 2010).
The survival of ABC DLBCL lines also depends upon MYD88, a key adapter in Toll-like receptor signaling (Ngo et al., 2011). Oncogenic, gain-of-function mutations in MYD88 are among the most recurrent genetic aberrations in ABC DLBCL (Ngo et al., 2011). MYD88 promotes NF-κB and JAK/STAT3 signaling, thereby sustaining ABC DLBCL viability. Additionally, MYD88 mutants induce interferon β (IFNβ) production and autocrine type I interferon signaling, which paradoxically promotes cell cycle arrest and apoptosis (Stark et al., 1998).
New therapeutic strategies are being devised to exploit the separate oncogenic mechanisms in the DLBCL subtypes. A recent phase 2 clinical trial revealed that lenalidomide is an active agent in relapsed/refractory DLBCL (Hernandez-Ilizaliturri et al., 2011). Retrospective analysis showed a 55% response rate in non-GCB DLBCL (including ABC DLBCL cases) compared with a 9% response rate in GCB DLBCL. More than half of the responses in non-GCB DLBCL were complete, extending the progression-free survival of this cohort, although overall survival remained unchanged. In the present study, we investigated the molecular mechanisms underlying the toxicity of lenalidomide for ABC DLBCL cells in order to design rational strategies to optimize its therapeutic effect.
Results
Lenalidomide induces a lethal type I interferon response in ABC DLBCL
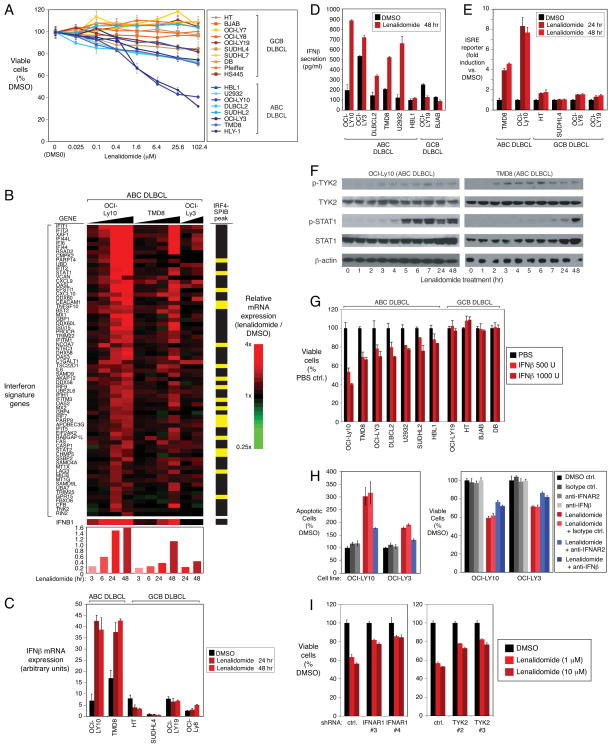
To understand the molecular basis for the efficacy and specificity of lenalidomide in treating lymphoma, we assessed its effect on the viability of cell line models of DLBCL. Lenalidomide treatment was toxic to most ABC DLBCL cell lines, whereas most GCB DLBCL lines were unaffected (Figure 1A). To investigate the mechanisms of this toxicity, we profiled gene expression changes in ABC DLBCL lines upon exposure to lenalidomide (Figure 1B). Lenalidomide increased the expression of 476 genes and reduced the expression of 272 genes (Table S1F,G). To gain biological insight into these lenalidomide-responsive genes, we used a database of gene expression signatures that reflect signaling and regulatory processes in normal and malignant cells (Shaffer et al., 2006). The most consistent signatures upregulated by lenalidomide were those associated with the type I interferon response (Table S1A, Figure 1B). Conversely, signatures of NF-κB, JAK and MYD88 signaling were downregulated by lenalidomide (Table S1B), suggesting that blockade of these pro-survival pathways contributes to lenalidomide toxicity (see below).
Figure 1. Lenalidomide induces a toxic type I interferon response in ABC DLBCL.
(A) Viability (MTS assay) of ABC and GCB DLBCL cell lines treated with lenalidomide for 4 days. Error bars show the standard error of the mean (SEM) of triplicates. (B) Relative expression of interferon signature genes over a time course of lenalidomide (10μM) treatment. Gene expression changes induced by lenalidomide are depicted according to the color scale shown. Average relative expression of interferon signature genes is at the bottom. Yellow bars: genes with overlapping IRF4/SPIB ChIP-seq peaks. (C, D) IFNβ mRNA expression and secretion in lenalidomide-treated (10μM) cells. Error bars show the SEM of triplicates. (E) Activity of an ISRE-driven luciferase reporter in cells treated with lenalidomide (10μM) or vehicle control (DMSO) at the indicated times. Error bars show the SEM of triplicates. (F) Western blot analysis of the indicated proteins in lenalidomide-treated (10μM) ABC DLBCL cells. p-: phospho-. (G) Viability (MTS assay) of DLBCL cells treated with the indicated amount of human recombinant IFNβ for 4 days. Error bars show the SEM of triplicates. (H) Measurement of viability (MTS assay; right) and apoptosis (PARP cleavage and caspase-3 activation by FACS; left) in ABC DLBCL cells treated with control compounds (DMSO or isotype-matched antibody), lenalidomide (1μM), or lenalidomide plus the indicated blocking antibodies (2.5μg/ml) for 4 days. Error bars show the SEM of triplicates. (I) Viability (MTS assay) of OCI-Ly10 ABC DLBCL cells in which the indicated shRNAs were induced for 2 days before treatment with DMSO or lenalidomide, as indicated, for 4 days. Error bars show the SEM of triplicates. See also Figure S1 and Table S1.
Lenalidomide increased interferon β (IFNβ) mRNA expression and protein secretion in the majority of ABC DLBCL lines, but not in most other DLBCL lines (Figure 1B,C,D). In ABC DLBCL lines, lenalidomide activated a reporter gene driven by an interferon-stimulated response element (ISRE), which did not occur in GCB DLBCL lines, even though they respond to exogenously added interferon (Figures 1E, S1B). Moreover, the drug induced phosphorylation of TYK2, a JAK family kinase associated with the type I interferon receptor, and STAT1, a transcription factor that is phosphorylated by TYK2 (Figure 1F).
Addition of IFNβ to cultures of ABC DLBCL lines induced cell death, with a potency that paralleled the effect of lenalidomide, suggesting that IFNβ might contribute to lenalidomide toxicity (Figures 1G, S1A). Indeed, antibodies against the interferon α/β receptor chain 2 (anti-IFNAR2) or IFNβ inhibited lenalidomide-induced death (Figure 1H). Likewise, silencing of the interferon α/β receptor chain 1 (IFNAR1) or TYK2 by RNA interference reduced lenalidomide toxicity (Figures 1I, S1C,I). Moreover, lenalidomide-induced STAT1 phosphorylation was blunted by anti-IFNAR2 antibodies or by IFNAR1 knockdown (Figure S1D).
Apoptosis induced by interferon is associated with induction of TRAIL (Oshima et al., 2001; Ucur et al., 2003). TRAIL (TNFSF10) mRNA and protein levels were increased by lenalidomide in ABC DLBCL cells and anti-IFNAR2 antibodies blocked this induction (Figures 1B, S1E,F,G). Anti-TRAIL antibodies partially rescued ABC DLBCL cells from lenalidomide-induced death (Figure S1H), suggesting that TRAIL induction contributes to lenalidomide toxicity but is not the only cell death mechanism involved (see below).
The IRF4 and SPIB regulatory network in ABC DLBCL
In a separate initiative, we defined the gene network controlled by the transcription factor IRF4, allowing us to appreciate an unexpected regulatory connection between IRF4 and lenalidomide. IRF4 expression is a hallmark of ABC DLBCL, secondary to the constitutive NF-κB activation and plasmacytic differentiation that characterizes this subtype (Alizadeh et al., 2000; Lam et al., 2005; Saito et al., 2007; Wright et al., 2003). Previously, we demonstrated that all multiple myeloma cell lines depend on IRF4 for survival (Shaffer et al., 2008). In a focused RNA interference screen, we observed that IRF4 knockdown was toxic to both ABC DLBCL and multiple myeloma lines, but not to a variety of other lymphoma and leukemia lines (Figure 2A; Table S2A). However, all cell lines were killed comparably when ribosomal or proteasomal proteins were knocked down. In confirmatory experiments, induction of an IRF4 shRNA killed ABC DLBCL and multiple myeloma cells in a time-dependent fashion, but GCB DLBCL lines were not affected (Figure 2B, Figure S2H). The toxicity of the IRF4 shRNA was reversed by ectopic expression of an IRF4 cDNA, confirming its specificity (Figure S2A). IRF4 mRNA and protein levels were reduced by ~40–60% by this shRNA, indicating that the ABC DLBCL lines are sensitive to partial IRF4 knockdown (Figure S2B). The cell cycle was not affected by IRF4 knockdown, but an increase in cells with sub-G1 DNA content was evident, indicating cell death (Figure S2C). IRF4 knockdown activated caspase-3, suggesting that apoptosis was initiated (Figure S2D,E), but a caspase inhibitor did not alter the kinetics of ABC DLBCL cell death (Figure S2F,G), suggesting that non-apoptotic cell death mechanisms are also invoked (Shaffer et al., 2008).
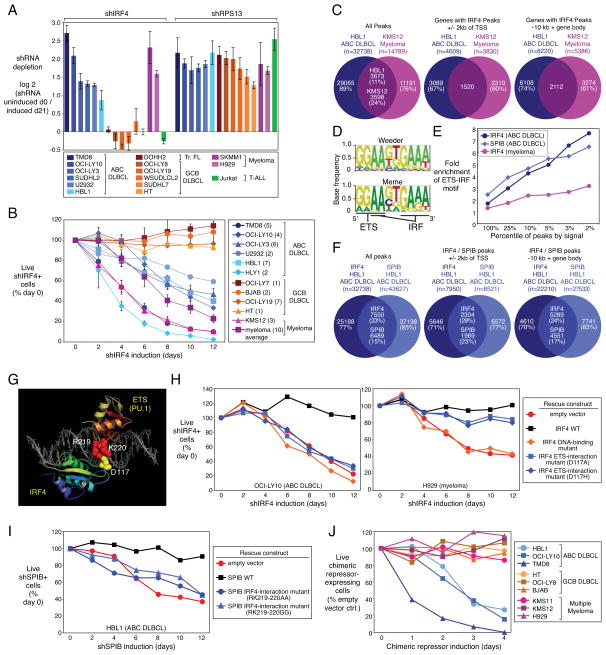
Figure 2. IRF4 and SPIB are required for ABC DLBCL viability.
(A) Toxicity of an IRF4 shRNA in a loss-of-function RNA interference screen of the indicated cell lines. Shown are the log2 ratios of shRNA abundance before induction (“uninduced d0”) versus abundance after 21 days in culture (“induced d21”). shRPS13 targets ribosomal protein S13, an essential gene in all cell types. Error bars show the SEM for quadriplicates. (B) Viability of shIRF4+ (GFP+) cells over time after induction as a percentage of live shIRF4+ cells following shIRF4 induction relative to day 0. The number of replicate infections is shown in parentheses. Error bars represent the SEM of replicates. (C) Overlap of IRF4 ChIP-Seq peaks in ABC DLBCL and multiple myeloma, based on all peaks (left), genes with an IRF4 peak within +/− 2kb of the TSS (middle), and genes with an IRF4 peak in a region encompassing the gene body and 10 kb upstream of the TSS (right). (D) Motif discovery using the Weeder and MEME algorithms based on the top 1000 ABC DLBCL IRF4 ChIP-Seq peaks by sequence tag abundance. The highest scoring motif is shown with core recognition motifs indicated. (E) Enrichment for the EICE motif in promoter-proximal peaks (+/− 2 kb from the TSS) based on IRF4 and SPIB ChIP-Seq data in ABC DLBCL or multiple myeloma, as a function of peak percentile, ranked by sequencing tag abundance. (F) Overlap of IRF4 and SPIB ChIP-seq peaks in ABC DLBCL, as in (C). (G) Crystal structure of the mouse IRF4 and PU.1 DNA binding domains interacting with an EICE, showing the interacting charged residues conserved in human IRF4 and SPIB. (H) Rescue experiment showing the viability of the indicated cell lines bearing empty vector, wild-type IRF4, or mutant IRF4 expression vectors, plotted as the fraction of shIRF4+ (GFP+) cells at times following shIRF4 induction relative to day 0. (I) Rescue experiment as in (H) with cells bearing the indicated SPIB expression vectors and an inducible SPIB-3′UTR-targeted shRNA. (J) Viability (FACS for live cells) of the indicated cell lines expressing an inducible IRF4-SPIB chimeric repressor, plotted as a percentage of live chimeric repressor+ cells relative to empty vector-bearing control cells, at various times following chimeric repressor induction. See also Figure S2 and Table S2.
To identify genes directly regulated by IRF4, we performed chromatin immunoprecipitation (ChIP) followed by high throughput DNA sequencing (ChIP-seq), in cell line models of ABC DLBCL (HBL1) and multiple myeloma (KMS12), as well as in a GCB DLBCL line that does not express IRF4 (OCI-Ly19). We identified significant binding events (“peaks”; see Supplemental Methods) in HBL1 and KMS12 that were not present in OCI-Ly19 (Figure 2C), and observed that IRF4 peaks were enriched near transcription start sites (TSSs) of protein-coding genes (Figure S2I). We confirmed 10 IRF4 binding sites in ABC DLBCL by conventional ChIP (Figure S2J).
Among IRF4 peaks in the ABC DLBCL line, 3,673 (11%) coincided with IRF4 peaks in the multiple myeloma line (Figure 2C). We defined a “whole gene” window from −10kb relative to the TSS and extending through the body of the gene, and observed that 2,112 genes had IRF4 peaks within this window in both ABC DLBCL and multiple myeloma (Figure 2C, Tables S2B,C). However, a substantial fraction (>60%) of the IRF4 target genes in these ABC DLBCL and multiple myeloma cell lines were unique to each tumor. For example, we previously identified a positive feedback loop between IRF4 and MYC in multiple myeloma cells whereby each factor binds the other’s promoter and drives expression (Shaffer et al., 2008). The IRF4 ChIP-Seq data confirmed IRF4 binding to the MYC locus in multiple myeloma but not ABC DLBCL (Figure S2K), despite high MYC expression in ABC DLBCL (Shaffer et al., 2006). IRF4 was itself an IRF4 target gene in multiple myeloma, suggesting positive autoregulation, but not in ABC DLBCL (Figure S2K). Conversely, many genes were bound by IRF4 in ABC DLBCL but not multiple myeloma, such as CD44 and CD40 (Figure S2K). These data suggest distinct IRF4 regulatory networks in these two malignancies, but analysis of more cell lines will be needed to fully elucidate these differences.
Using de novo DNA motif discovery algorithms, the most common sequence in ABC DLBCL IRF4 peaks was an exact match to the EICE motif (Figure 2D, Table S2D). IRF4 peaks in multiple myeloma were not enriched for this motif but rather for a direct repeat of an IRF binding site (GAAT(G/C)GAAT; Table S2D). Among promoter-proximal peaks, EICE enrichment steadily increased as a function of IRF4 peak intensity in ABC DLBCL (p=1.81E-24) and was located near the point of highest ChIP-seq intensity within each peak (Figures 2E, S2L). These data imply that IRF4 binds with an ETS family member to the EICE motif in ABC DLBCL but relies on other mechanisms to interact with its target genes in multiple myeloma.
IRF4 binds to the EICE motif with either PU.1 or SPIB, neither of which is expressed in myeloma cells. SPIB is characteristically expressed in ABC DLBCL and can be further upregulated by amplification or translocation of its genomic locus (Lenz et al., 2007; Lenz et al., 2008c). Given that ABC DLBCL lines require SPIB for survival (Lenz et al., 2008c), we suspected that SPIB was the relevant IRF4 binding partner in these cells. To perform ChIP-seq for SPIB, we engineered the HBL1 ABC DLBCL line to express biotinylated SPIB (SPIB biotag; see Methods). SPIB peaks were found preferentially near TSSs in ABC DLBCL (Figure S2M), and were enriched for the ETS-family DNA-binding motif (GGAA; Table S2D). SPIB and IRF4 peaks in ABC DLBCL overlapped 4.3-fold more often than expected by chance (p<10E-300) (Figure 2F, Table S2E,F). EICE motifs within IRF4 and SPIB binding peaks were enriched near TSSs (p = 1.91E-135) and their frequency increased as a function of peak intensity (Figures 2E, S2L,M). Overlapping IRF4-SPIB peaks were present at 3610 genes in ABC DLBCL, with 33% of those peaks containing at least one EICE (Table S2F).
The dependence of ABC DLBCLs on IRF4 and SPIB predicted that disruption of the IRF4-SPIB interaction would be deleterious to ABC DLBCL viability. A crystal structure of the mouse IRF4 and PU.1 DNA binding domains bound to an EICE motif allowed us to model the human IRF4-SPIB interaction (Escalante et al., 2002a; Escalante et al., 2002b). IRF4 contacts PU.1 across the DNA minor groove via charged residues that are conserved in human IRF4 (aspartic acid 117) and human SPIB (arginine 219 and lysine 220) (Figure 2G). We generated IRF4 and SPIB mutants to test whether this protein-protein interface is essential for ABC DLBCL survival. As expected, ectopic expression of the wild-type IRF4 coding region rescued ABC DLBCL and multiple myeloma lines from the toxicity of a 3′UTR-directed IRF4 shRNA whereas an IRF4 DNA-binding mutant was inactive (Figure 2H). IRF4 interaction mutants with histidine or alanine at position 117 were expressed as efficiently as wild-type IRF4 and were not toxic (data not shown), but did not sustain ABC DLBCL viability (Figure 2H). By contrast, these mutants did rescue multiple myeloma cells from IRF4 shRNA toxicity, demonstrating that they are functional in this context. Wild-type SPIB was able to rescue ABC DLBCL cells from SPIB knockdown, but SPIB interaction mutants with alanine or glycine substitutions at positions 219 and 220 were ineffective, while being equivalently expressed and non-toxic (Figure 2I, data not shown).
This mutational analysis indicated that the IRF4-SPIB interaction is critical for ABC DLBCL viability. To test this further, we created a fusion protein between the DNA binding domains of IRF4 and SPIB, based on previous work showing that a IRF4-PU.1 fusion functions as a sequence-specific transcriptional repressor (Brass et al., 1999). The IRF4-SPIB chimeric protein was acutely toxic to ABC DLBCL but not GCB DLBCL lines (Figure 2J, and see below). This chimeric repressor was not toxic to IRF4-dependent multiple myeloma lines, suggesting that it specifically represses genes that require both IRF4 and SPIB for expression.
Pathways regulated by IRF4-SPIB in ABC DLBCL
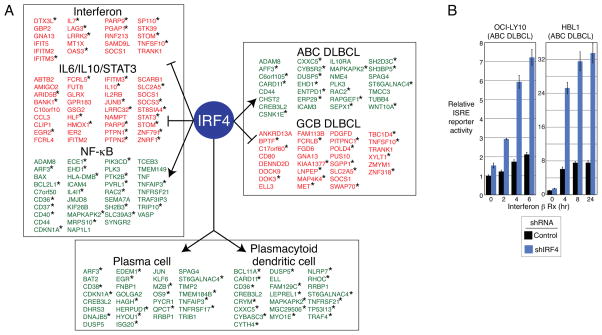
To determine the nature of the genes and pathways controlled by IRF4 and SPIB in ABC DLBCL, we profiled gene expression changes upon IRF4 knockdown, allowing us to define a set of genes that were consistently downregulated (n=435) or upregulated (n=410) (Table S3G,H, Figure S3A). Many of these genes were similarly regulated by SPIB and the chimeric IRF4-SPIB repressor (Table S3I,J,K). For example, among the downregulated genes, 46% and 42% also decreased in expression following SPIB knockdown and IRF4-SPIB chimeric repressor induction, respectively. Among IRF4-regulated genes, we defined “direct targets” as those that had an IRF4 binding peak within the “whole gene” window specified above (Figure 2C). Many IRF4 direct targets had overlapping IRF4-SPIB peaks (Figure 3A).
Figure 3. IRF4 controls essential gene expression programs in ABC DLBCL.
(A) IRF4 direct target genes grouped according to gene expression signatures (Shaffer et al., 2006). Signatures with significant enrichment for IRF4 targets were grouped by function (Table S3A). Genes that are induced or repressed by IRF4 are indicated in green and red, respectively. Asterisks indicate genes with an overlapping IRF4-SPIB ChIP-seq peak. (B) ISRE-driven luciferase reporter activity in ABC DLBCL lines with control or IRF4 shRNAs after 2 days of induction and subsequent addition of IFNβ (1000U). Error bars show the SEM of triplicates. See also Figure S3 and Table S3.
Gene expression signatures that were enriched among IRF4 targets were those that distinguish the DLBCL subtypes, characterize hematopoietic differentiation states, or are regulated by signaling pathways active in ABC DLBCL (Figure 3A, Table S3). Among IRF4 upregulated genes, a signature of genes more highly expressed in ABC DLBCL than GCB DLBCL was the most enriched (ABCDLBCL-4, p=1.18E-18). Represented are genes that specify the cell surface phenotype of ABC DLBCL (CD44, ENTPD1, IL10RA) as well as genes encoding important regulatory proteins, notably CARD11 (see below). Conversely, among IRF4 downregulated genes, a signature of genes more highly expressed in GCB DLBCL than ABC DLBCL was enriched (GCBDLBCL-3, p=3.84E-5). IRF4 upregulated genes also included ones more highly expressed in plasma cells than in mature B cells (PC-2, p=2.33E-08), in keeping with the essential role of IRF4 in plasmacytic differentiation (Klein et al., 2006; Sciammas et al., 2006). A signature of plasmacytoid dendritic cells was also enriched among IRF4 upregulated genes (DC-4, p=1.53E-08), consonant with the role of IRF4 in the differentiation of this lineage (Lehtonen et al., 2005; Schotte et al., 2003; Tamura et al., 2005).
Prominent among IRF4 direct targets were genes regulated by key ABC DLBCL signaling pathways. A signature of NF-κB activation was enriched among genes that were upregulated by IRF4 (NFKB-10, p=1.38E-17; Figure 3A). As discussed above, this NF-κB signature was downregulated by the treatment of ABC DLBCLs with lenalidomide (Table S1). Signatures that reflect autocrine IL-10 and/or IL-6 signaling in ABC DLBCL cells were enriched among genes that were repressed by IRF4, suggesting that IRF4 dampens JAK/STAT3 signaling in ABC DLBCL (IL10Up-1, p=1.83E-15; IL6Up-4, p=2.23E-11; Figure 3A, Table S3) (Lam et al., 2008). Finally, a signature of type I interferon signaling was significantly represented among IRF4-repressed target genes (IFN-3, p=5.80E-06; Figure 3A, Table S3). These interferon signature genes were induced by lenalidomide treatment of ABC DLBCL cells, and many of these induced genes had IRF4-SPIB intersection peaks (Figure 1B). Accordingly, IRF4 knockdown in ABC DLBCL lines increased their response to exogenous IFNβ, as measured by an ISRE reporter (Figure 3B, S3B).
Lenalidomide toxicity in ABC DLBCL is opposed by IRF4 and SPIB
This signature analysis suggested that IRF4 and SPIB cooperate to modulate type I interferon and NF-κB signaling in ABC DLBCL in a manner opposite to their regulation by lenalidomide. We therefore wondered if lenalidomide might have a direct effect on IRF4 or SPIB expression in ABC DLBCL. Indeed, IRF4 and SPIB mRNA and protein levels dropped rapidly upon lenalidomide treatment of ABC DLBCL cells (Figure 4A,B,C,D), suggesting that lenalidomide affects the expression of IRF4 and SPIB target genes by decreasing the levels of both factors.
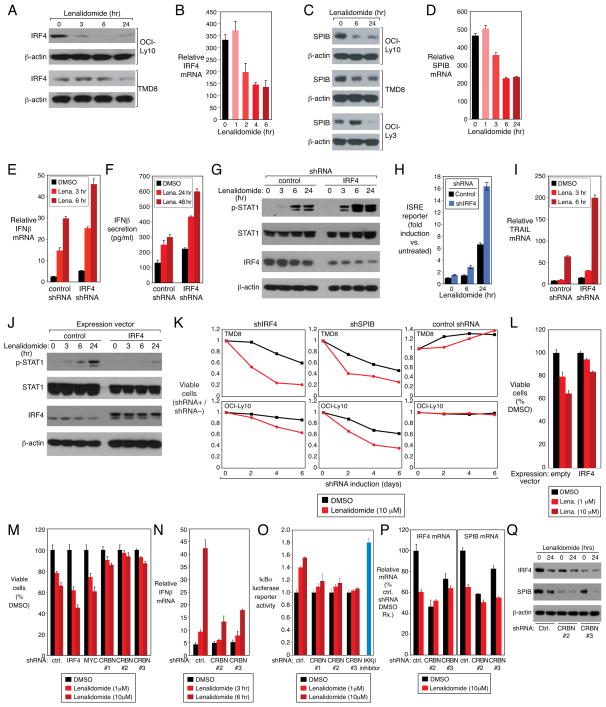
Figure 4. Lenalidomide toxicity in ABC DLBCL is opposed by IRF4 and SPIB.
(A,C) Western blot of IRF4, SPIB and β-actin proteins in ABC DLBCL cell lines treated with lenalidomide (10μM) over time. (B,D) IRF4 and SPIB mRNA expression quantified by Q-PCR, normalized to β2-microglobulin(B2M) expression, in the ABC DLBCL line OCI-Ly10 treated with lenalidomide (10μM). Error bars show the SEM of triplicates. (E,F) IFNβ mRNA expression and protein secretion in the OCI-Ly10 ABC DLBCL line induced for IRF4 or control shRNA expression for 2 days and treated with lenalidomide (10μM). Error bars represent the SEM of triplicates. (G)Western blot of the indicated proteins in OCI-Ly10, following induction of IRF4 or control shRNAs for 2 days and treatment with lenalidomide (10μM)for the indicated times. p-: phospho-. (H)ISRE -driven luciferase reporter activity in OCI-Ly10 with control or IRF4 shRNAs after lenalidomide (10μM) treatment. Error bars represent the SEM of triplicates. (I)TRAIL mRNA quantified by Q -PCR, normalized to B2M, in OCI-Ly10 cells with shRNA induction for 2 days and lenalidomide (10μM)treatment for the indicated times. Error bars show the SEM of triplicates. (J) Western blot analysis of the indicated proteins in OCI-Ly10 cells transduced with a flag epitope-tagged IRF4 expression vector or an empty vector, induced for 48 h, then treated with lenalidomide (10μM)for the indicated times. The lower IRF4 band is endogenous IRF4; the upper band is FLAG-tagged exogenous IRF4. p-: phosphorylation. (K) Viability of ABC DLBCL lines induced to express control, IRF4 or SPIB shRNAs and the treated with DMSO or lenalidomide (10μM) over a time course of induction. See text for details. (L) Viability of OCI-Ly10 cells transduced with an IRF4 expression or empty vector, induced for 24h and then treated with DMSO or lenalidomide for 4 days. Error bars represent the SEM of triplicates. (M) Viability (MTS assay) of OCI-Ly10 cells induced to express the indicated shRNAs for 2 days and treated with lenalidomide at the indicated concentrations for 4 days. ctrl.: control. Error bars show the SEM of triplicates. (N) IFNβ mRNA expression, measured by Q-PCR, in OCI-Ly10 cells induced to express CRBN shRNAs for 2 days and treated with lenalidomide (10μM) for the indicated times. Error bars show the SEM of triplicates. (O) TMD8 ABC DLBCL cells expressing an IκBα-luciferase fusion protein were induced to express control or CRBN shRNAs for 2 days, then treated with lenalidomide at the indicated concentrations or DMSO for 2 days. Luciferase activity was normalized to the DMSO control. As a positive control, cells were treated with the IKKβ inhibitor MLN120B (10μM) for 2 days. Error bars show the SEM of triplicates. (P) IRF4 and SPIB mRNA expression, quantified by Q-PCR, in TMD8 cells transduced with the indicated shRNAs. shRNA expression was induced for 2 days followed by lenalidomide treatment (10μM) for 24 hours. Error bars show the SEM of triplicates. (Q) Western blot for the indicated proteins in TMD8 cells induced to express CRBN or control shRNAs for 2 days, followed by treatment with lenalidomide (10μM) for 24 hours. See also Figure S4.
Given that lenalidomide only reduced IRF4 expression partially, we tested whether further silencing of IRF4 by RNA interference would enhance the interferon response in ABC DLBCL cells. Induction of IFNβ mRNA expression and secretion by lenalidomide was augmented by IRF4 knockdown (Figure 4E,F). IRF4 knockdown also increased lenalidomide-induced STAT1 phosphorylation, ISRE promoter activity, and TRAIL upregulation (Figure 4G,H,I). Conversely, ectopic expression of IRF4 suppressed lenalidomide-induced STAT1 phosphorylation (Figure 4J).
We next tested whether expression of IRF4 and SPIB in ABC DLBCL interferes with the toxicity of lenalidomide. We infected cells with vectors that expressed IRF4 or SPIB shRNAs along with green fluorescent protein (GFP), allowing us to visualize the subpopulation of cells that had been transduced with the shRNA. By comparing the viability of shRNA-transduced (GFP+) and shRNA-nontransduced (GFP−) cells, which were equally exposed to lenalidomide, we could discern how IRF4 or SPIB knockdown influenced lenalidomide toxicity. In the absence of lenalidomide, knockdown of IRF4 or SPIB alone was toxic for ABC DLBCLs, as expected (Figure 4K). The addition of lenalidomide accelerated the loss of cells bearing IRF4 and SPIB shRNAs relative to those bearing a control shRNA (Figure 4K). Conversely, ectopic expression of IRF4 counteracted lenalidomide toxicity in ABC DLBCL (Figure 4L). As well, the IRF4-SPIB chimeric repressor potentiated lenalidomide-induced IFNβ expression and increased lenalidomide toxicity (Figure S5F,G). Hence, IRF4 and SPIB regulate lenalidomide-induced interferon responses and toxicity in ABC DLBCL.
Cereblon mediates the toxic effect of lenalidomide in ABC DLBCL
Recent studies have demonstrated that the activity of thalidomide and lenalidomide is mediated by cereblon (CRBN), a component of an ubiquitin-ligase complex (Ito et al., 2010; Zhu et al., 2011). To address whether CRBN is required for the toxic effects of lenalidomide in ABC DLBCL, we identified three shRNAs that reduced CRBN mRNA expression by ~50% (Figure S4A). CRBN knockdown was moderately toxic for ABC DLBCL cells (Figure S4C), an effect that was reversed by ectopic expression of CRBN. CRBN depletion substantially reduced the toxicity of lenalidomide for ABC DLBCL cells (Figures 4M, S4B,C) and interfered with the ability of lenalidomide to induce an interferon response and block NF-κB signaling (Figures 4N,O, S4D,E). CRBN depletion lowered IRF4 mRNA and protein levels in ABC DLBCL cells and reduced the effect of lenalidomide on IRF4 levels (Figures 4P,Q, S4F). Similarly, SPIB mRNA and protein levels were also diminished upon CRBN knockdown (Figure 4P, Q). Thus, CRBN is required to maintain IRF4 and SPIB levels in ABC DLBCL, accounting for the toxicity of CRBN depletion. Moreover, these findings suggest that CRBN mediates most of the effects of lenalidomide in ABC DLBCL.
IRF4 represses the interferon pathway by inhibiting IRF7 expression
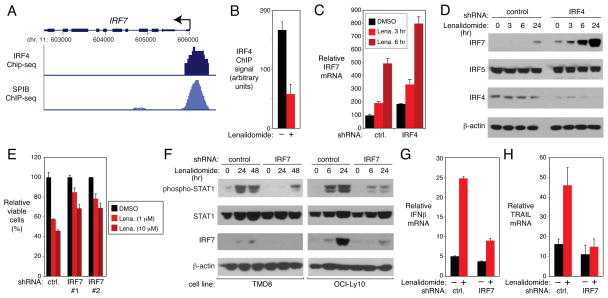
We noted that three IRF family members that regulate the type I interferon responses – IRF2, IRF7, and IRF9 – were bound by IRF4 and SPIB (Table S3F). We focused on a strong IRF4-SPIB peak in the promoter of the IRF7 gene (Figure 5A) since it is a master regulator of interferon responses that is strongly induced by IFNβ as part of a positive-feedback loop (Honda et al., 2005; Marie et al., 1998; Sato et al., 1998). Binding of IRF4 to the IRF7 promoter was confirmed by an independent ChIP assay, and lenalidomide reduced this binding (Figure 5B). IRF7 mRNA and protein were specifically induced in ABC DLBCL by lenalidomide and were further induced by knocking down IRF4 (Figure 5C,D). Together, these results suggest that IRF7 is negatively regulated by IRF4 and SPIB, and that lenalidomide can blunt this negative regulation.
Figure 5. IRF4-SPIB block interferon signaling by repressing IRF7.
(A) UCSC browser depiction of ChIP-seq data from HBL1 ABC DLBCL cells showing IRF4 and SPIB-biotag binding at the IRF7 promoter. TSS (arrow). (B) IRF4 binding at the IRF7 locus by ChIP in OCI-Ly10 ABC DLBCL cells treated with DMSO (“−”) or lenalidomide (10μM) for 24 hours. Error bars show SEM of triplicates. (C) Q-PCR quantification of IRF7 mRNA levels, normalized to B2M, in OCI-Ly10 cells with control (ctrl.) or IRF4 shRNAs, treated with lenalidomide(10μM) or DMSO. Error bars show SEM of triplicates. (D) Western blot of the indicated proteins in cells from (C).(E) Viability (MTS assay) of OCI-Ly10 cells induced to express the indicated shRNAs for 2 days and then treated with DMSO or lenalidomide(10μM) for 4 days. Error bars show SEM of triplicates. (F) Western blot of the indicated proteins in ABC DLBCL lines induced to express control or IRF7 shRNAs for 2 days and then treated with DMSO or lenalidomide(10μM)for the indicated times. (G,H) Q -PCR analysis, normalized to B2M, of IFNβ (G) or TRAIL (H) mRNA levels in OCI-Ly10 cells induced to express control or IRF7 shRNAs for 2 days and treated with DMSO (“−”) or lenalidomide (10μM; “+”) for 24 hours. Error bars show SEM of triplicates.
We next examined whether IRF7 expression is important for the toxicity of lenalidomide in ABC DLBCL cells. IRF7 knockdown attenuated the toxicity of lenalidomide in ABC DLBCL cells (Figure 5E). IRF7 depletion additionally impaired lenalidomide-induced STAT1 phosphorylation, IFNβ expression, and TRAIL mRNA induction (Figure 5F,G,H). Hence, IRF7 promotes lenalidomide toxicity by facillitating lenalidomide-induced IFNβ secretion and signaling.
IRF4-SPIB and CARD11 form an essential oncogenic loop in ABC DLBCL
While treatment with lenalidomide induces a toxic interferon response in ABC DLBCLs, blocking interferon signaling did not fully rescue the cells. We therefore wondered if there might be other mechanisms by which lenalidomide kills these lymphoma cells. Given that ABC DLBCL cells depend upon the NF-κB pathway for survival (Davis et al., 2001; Lam et al., 2005) and that lenalidomide suppressed the NF-κB gene expression signature (Table S1), we hypothesized that decreased NF-κB signaling contributes to the toxicity of lenalidomide in ABC DLBCL cells.
Since lenalidomide downregulates IRF4 and SPIB (Figure 4A,B), we were intrigued that CARD11 was a direct IRF4 and SPIB target (Figure 3A, Table S2B,E,F). CARD11 plays an essential role in the constitutive NF-κB activity that maintains ABC DLBCL viability (Davis et al., 2001; Lam et al., 2005; Ngo et al., 2006). The CARD11 locus had prominent, overlapping IRF4 and SPIB binding peaks located +705 bp relative to the TSS, coinciding with an evolutionarily conserved EICE motif (Figure 6A). IRF4 and SPIB binding was confirmed by independent ChIP assays, and IRF4 knockdown diminished this binding (Figure 6B). Gene expression profiling and quantitative PCR analysis showed that CARD11 mRNA levels are diminished by knockdown of IRF4 or SPIB, as well as by expression of the IRF4-SPIB chimeric repressor (Figures 6C, S5A). CARD11 protein levels were also reduced after IRF4 knockdown or chimeric repressor induction (Figure S5B,C).
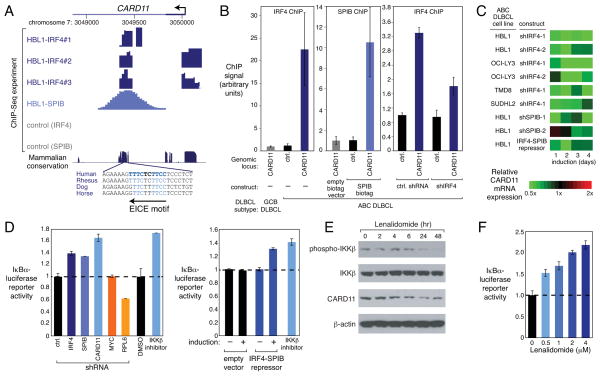
Figure 6. IRF4-SPIB and CARD11 form an essential oncogenic loop in ABC DLBCL.
(A) UCSC browser depiction of ChIP-seq data from the HBL1 ABC DLBCL line showing IRF4 (in triplicate) and SPIB-biotag binding at the CARD11 locus, with an evolutionarily conserved EICE binding motif indicated. Control (IRF4): OCI-Ly19 (IRF4-). Control (SPIB): HBL1 with empty biotag vector. TSS (arrow). (B) ChIP analysis in HBL1 cells for IRF4 and SPIB binding at the CARD11 peak identified in (A) or at a negative control (ctrl.) locus. IRF4 or control shRNAs were induced for 4 days. GCB DLBCL line: OCI-Ly19 (IRF4-) is IRF4-negative. Error bars show SEM of triplicates. (C) Relative CARD11 mRNA expression, depicted according to the color scale shown, from gene expression profiling of ABC DLBCL lines after induction of shIRF4 or the IRF4-SPIB chimeric repressor for 4 days. (D) IKK activity measured by an IκBα-luciferase reporter in TMD8 (ABC DLBCL) after induction of various shRNAs for 3 days (left) or the IRF4-SPIB chimeric repressor for 1 day (right). Also shown is the effect of 1 day exposure to an IKKβ inhibitor (MLN120B) or DMSO. (E) Western blot analysis of the indicated proteins following treatment of OCI-Ly10 cells with lenalidomide (10μM) for the indicated times. (F) IKK activity measured by an IκBα-luciferase reporter after treatment of TMD8 cells with lenalidomide at the indicated concentrations for 48 h. Error bars show the SEM of triplicates. See also Figure S5.
Since CARD11 coordinates the activation of IKK (Thome, 2004), the key regulatory kinase in the classical NF-κB pathway, we assessed IKK function using an ABC DLBCL reporter line engineered to express a fusion protein between luciferase and the IKK substrate IκBα (Lam et al., 2005). Phosphorylation of this fusion protein by IKK promotes its degradation, and thus IKK inhibition increases luciferase activity. Knockdown of either IRF4 or SPIB reduced IKK activity in the ABC DLBCL line TMD8, as did CARD11 knockdown or treatment with a small molecule IKK inhibitor, but two other toxic shRNAs targeting MYC and RPL6 did not (Lam et al., 2005) (Figure 6D). Moreover, induction of the chimeric IRF4-SPIB repressor also inhibited IKK activity (Figure 6D). Similar results were observed in an ABC DLBCL line, OCI-Ly3, that relies on an oncogenically active CARD11 mutant for survival (Lenz et al., 2008a), in keeping with an effect of IRF4 and SPIB on CARD11 transcription (Figure S5H). In accord with these functional experiments, knockdown of IRF4 decreased IKKβ phosphorylation, a modification associated with IKK activation downstream of CARD11 (Figure S5E). The effect of IRF4 on the NF-κB pathway was confirmed using an independent ABC DLBCL reporter system in which luciferase is driven by an NF-κB-dependent promoter (Figure S5D).
Lenalidomide treatment of an ABC DLBCL line reduced CARD11 expression and IKKβ phosphorylation, and inhibited IKK activity (Figure 6E,F). Together, these data suggest that IRF4 and SPIB act together to amplify NF-κB signaling in ABC DLBCL by transactivating CARD11 and that lenalidomide inhibits NF-κB by downregulating IRF4 and SPIB, breaking this positive feed-forward loop.
Synergism between lenalidomide and NF-κB pathway inhibitors
Since lenalidomide only partially inhibits IRF4 and SPIB expression (Figure 4A–D), we speculated that we could achieve greater toxicity by additionally blocking IKK, thereby further reducing NF-κB-dependent IRF4 expression. The IKKβ inhibitor MLN120B selectively kills ABC DLBCL cells, as does ibrutinib, a BTK kinase inhibitor that blocks signaling from the BCR to IKK (Davis et al., 2010; Lam et al., 2005). Treatment of ABC DLBCL cells with either MLN120B or ibrutinib alone decreased IRF4 protein levels, but when these agents were combined with lenalidomide, IRF4 became undetectable (Figure 7A). The combination of ibrutinib and lenalidomide induced a stronger interferon response than lenalidomide alone, as measured by ISRE reporter activity and STAT1 phosphorylation (Figure 7A,B). These two drugs also cooperated in blocking IKK activity (Figure 7C).
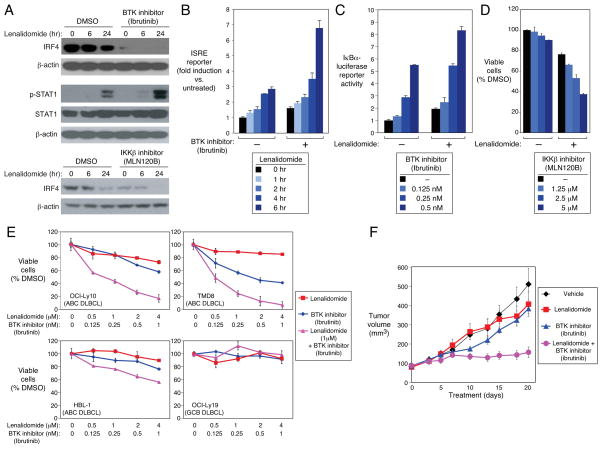
Figure 7. Synergy between lenalidomide and NF-κB pathway inhibitors in ABC DLBCL.
(A) Western blot of the indicated proteins in OCI-Ly10 cells treated with lenalidomide (5μM) alone or with MLN120B (10μM), ibrutinib(5nM) or DMSO for the indicated times. p-: phospho-. (B) ISRE-driven luciferase activity in OCI-Ly10 cells treated with lenalidomide (5μM) +/− ibrutinib (5nM) for the indicated times. Error bars show the SEM of triplicates. (C) IKK activity measured by an IκBα-luciferase reporter in TMD8 cells treated with ibrutinib at the indicated concentrations +/− lenalidomide (1μM) for 48 hours. Error bars show the SEM of triplicates. (D) Viability (MTS assay) of OCI-Ly10 treated with MLN120B at the indicated concentrations +/− lenalidomide (1μM) for 4 Days relative to DMSO-treated cells. Error bars show the SEM of triplicates. (E) Viability (MTS assay) of DLBCL lines treated with ibrutinib, lenalidomide, or both for 4 days at the concentrations indicated. Error bars show the SEM of triplicates. (F) OCI-Ly10 ABC DLBCL cells were established as a subcutaneous tumor (average 80 mm3) in immunodeficient mice, and then treated daily for 20 days with DMSO, lenalidomide (10 mg/kg), ibrutinib (3mg/kg), or lenalidomide plus ibrutinib by intraperitoneal injection. Tumor progression was monitored as a function of tumor volume. Error bars show the SEM of 5 mice per group. See also Figure S6.
To test for synergistic toxicity, an ABC DLBCL line was treated with the MLN120B at a range of doses that were only modestly toxic, in the presence or absence of low dose lenalidomide. The ABC DLBCL cells were killed more efficiently with the combination of these drugs than with either drug alone (Figure 7D). Similarly, synergistic toxicity was observed in ABC DLBCL lines when lenalidomide and ibrutinib were combined, but no toxicity was observed in GCB DLBCL lines that lack oncogenic activation of the BCR and MYD88 pathways (Figures 7E, S6A). A formal mathematical algorithm (Greco et al., 1990) confirmed the strong synergism between ibrutinib and lenalidomide in killing three ABC DLBCL lines (Figure S6B). Finally, we tested this drug combination in a xenograft mouse model created using the OCI-Ly10 ABC DLBCL cell line (Figures 7F, S6C). At the concentrations of lenalidomide and ibrutinib chosen, both drugs had little effect on the growth of the xenografts as single agents (Figures 7F, S6C), but were quite effective in combination in arresting the growth of established tumors.
Discussion
New treatments for ABC DLBCL should ideally exploit emerging insights into oncogenic pathways, which create opportunities for synthetic lethal interactions with drugs that target these pathways (Figure 8). The BCR and MYD88 signaling pathways promote ABC DLBCL viability by inducing NF-κB, and both pathways are affected by recurrent oncogenic mutations in ABC DLBCL. However, the penalty that ABC DLBCLs pay by acquiring oncogenic MYD88 mutations is the production of IFNβ (Ngo et al., 2011), which is toxic to these tumors. The present study revealed that IRF4 places a brake on IFNβ expression by repressing IRF7, allowing ABC DLBCLs with MYD88 mutations to survive and proliferate. Additionally, IRF4 sustains ABC DLBCL survival by transactivating CARD11 and potentiating NF-κB signaling. IRF4 emerges from these studies as a central regulatory hub in ABC DLBCL, making it an attractive therapeutic target. IRF4 and its regulatory partner SPIB were downregulated by treatment of ABC DLBCL lines with lenalidomide, a drug that has shown preferential activity against this lymphoma subtype in early phase clinical trials (Hernandez-Ilizaliturri et al., 2011). Lenalidomide toxicity for ABC DLBCL was associated with heightened IFNβ production and diminished NF-κB activity. Hence, lenalidomide toxicity in ABC DLBCL relies upon its modulation of oncogenically-activated signaling pathways, and therefore is an instance of “synthetic lethality” (Kaelin, 2005).
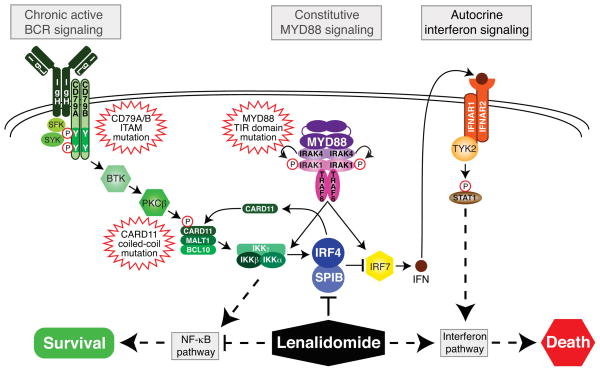
Figure 8. Exploiting synthetic lethality for the therapy of ABC DLBCL.
Recurrent oncogenic mutations in ABC DLBCL activate both the BCR and MYD88 pathways to drive pro-survival NF-κB signaling. However, MYD88 signaling also induces IFNβ, which is detrimental to ABC DLBCL survival. IRF4 and SPIB lie at the nexus of both pathways, promoting ABC DLBCL survival by repressing IRF7, thereby blocking IFNβ, and transactivating CARD11, thereby increasing NF-κB signaling. NF-κB factors transactivate IRF4, creating a positive feedback oncogenic loop. Lenalidomide targets this circuitry by down modulating IRF4 and SPIB, thereby increasing toxic IFNβ secretion and decreasing NF-κB activity.
This study highlights the central role of IRF4-SPIB heterodimers in ABC DLBCL biology, particularly in amplifying NF-κB signaling while blocking type I interferon signaling. In addition, IRF4 directly upregulates a large number of genes that distinguish ABC DLBCL from other lymphoma subtypes, many of which may contribute to viability or other attributes of these lymphoma cells. Remarkably, survival of ABC DLBCL cells depended on a single amino acid in IRF4 that mediates its interaction with SPIB on composite EICE motifs. IRF4 is clearly central to the action of lenalidomide in ABC DLBCL since enforced overexpression of IRF4 blocked the toxic effect of this drug, presumably by driving IRF4-SPIB interactions by mass action. IRF4 is similarly downregulated by lenalidomide in multiple myeloma (Li et al., 2011; Lopez-Girona et al., 2011; Zhu et al., 2011). In both ABC DLBCL and multiple myeloma, IRF4 levels are maintained by CRBN, a subunit of an ubiquitin ligase complex. We discovered that CRBN also controls SPIB levels in ABC DLBCL, which is not relevant to multiple myeloma as these cells do not express SPIB. Thalidomide, a chemically related cousin of lenalidomide, physically interacts with CRBN and blocks the ability of its ubiquitin ligase complex to autoubiquitinate (Ito et al., 2010). Further investigations are needed to discern how this ubiquitin ligase might control IRF4 and SPIB expression, apparently at the level of transcription.
Our findings provide a sound mechanistic basis for clinical trials in ABC DLBCL that rationally combine lenalidomide with other drugs that modulate NF-κB signaling. Drugs targeting NF-κB hold promise in cancer therapy despite some concerns about long-term suppression of this pathway (Baud and Karin, 2009; Gupta et al., 2010). In ABC DLBCL, NF-κB activity relies upon chronic active BCR signaling, which can be blocked by several drugs that are currently in clinical trials, including ibrutinib (targeting BTK) (Davis et al., 2010), fostamatinib (targeting SYK) (Friedberg et al., 2009), and CAL-101 (targeting PI(3) kinase δ) (Herman et al., 2010; Hoellenriegel et al., 2011). Additionally, NF-κB signaling can be inhibited by interfering with IκBα degradation, which can be achieved with the proteasome inhibitor bortezomib (Dunleavy et al., 2009) or the neddylation inhibitor MLN4924 (Milhollen et al., 2010). As a single agent, the BTK inhibitor ibrutinib is highly active against ABC DLBCL cells in vitro (Davis et al., 2010), and is showing clinical activity in a subset of patients with relapsed/refractory ABC DLBCL (L. Staudt, unpublished observations). We observed striking synergy between ibrutinib and lenalidomide in blocking IRF4 expression, increasing IFNβ production, and killing ABC DLBCL cells in vitro and in vivo, supporting clinical evaluation of this treatment regimen.
The effectiveness of this drug combination in ABC DLBCL capitalizes on recurrent genetic alterations in ABC DLBCL in two ways. First, the MYD88 L265P mutant promotes the abnormal synthesis and secretion of IFNβ. Second, mutations in the BCR subunits CD79A and CD79B promote chronic active BCR signaling, which activates NF-κB and induces IRF4, thereby dampening the toxic type I interferon response while augmenting the pro-survival NF-κB response. Hence, these recurrent oncogenic mutations in ABC DLBCL and the constitutive signaling pathways that they engage place IRF4 in a central regulatory position (Figure 8). Indeed, one reason that CD79B mutations often coexist with the MYD88 L265P mutation in ABC DLBCL tumors (Ngo et al., 2011) may be that the production of IRF4 in response to chronic active BCR signaling is necessary for the tumor to dampen the interferon response caused by the MYD88 L265P mutation. The rational therapeutic combinations proposed herein act in a synthetic lethal fashion to exploit this IRF4 addiction.
Experimental Procedures
(See Supplemental Experimental Procedures for details)
Cell culture and constructs
Methods for cell culture, plasmid transfection, retroviral transduction, and plasmid constructs were described previously (Lenz et al., 2008c; Ngo et al., 2006; Shaffer et al., 2008).
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitations were performed as described (Shaffer et al., 2008). Such ChIP-enriched DNA was either used in region-specific assessment of antibody binding by real-time PCR, or made into libraries for ChIP sequencing on an Genome Analyzer II (GAII, Illumina, Inc) according to manufacturer’s recommendations. See Supplemental Experimental Procedures for details.
Gene expression (Q-RTPCR and profiling)
Unless otherwise described, Q-RTPCR was performed on cDNA as previous described in (Sciammas et al., 2006; Shaffer et al., 2008; Shaffer et al., 2004), using pre-tested Assay-on-demand probe/primer sets from Applied Biosystems or primers designed for use with SYBR green using an ABI 7700 Taqman machine for 40 cycles with an annealing temperature of 60°C. Gene expression was normalized to the expression of beta-2-microglobulin for all samples. Gene expression profiling was performed using two-color human Agilent 4×44K gene expression arrays, exactly as described by the manufacturer, comparing signal from control cells (Cy3) and experimentally manipulated cells (Cy5). Array elements were filtered for those meeting confidence thresholds for spot size, architecture, and level above local background. These criteria are a feature of the Agilent gene expression software package for Agilent 4×44k arrays.
Cell viability (MTS) assay
Cells were plated in triplicate at a density of 15,000 cells per well in 96-well plates. Cell viability after indicated treatments was assayed by adding 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H tetrazolium and an electron coupling reagent (phenazine methosulphate; Promega), incubated for 3h and measured by the amount of 490nm absorbance using a 96-well plate reader. The background was subtracted using a media only control.
NF-κB reporter assays
The assay for IκB kinase activity using the IκBalpha-photinus luciferase reporter has been described (Lenz et al., 2008a), along with use of the IκB kinase inhibitor (Lam et al., 2005). In addition, cell lines were created with an NF-κB transcriptional reporter by transduction with lentiviral particles containing an inducible NF-κB -responsive luciferase reporter construct (SA Biosciences) and selected with puromycin. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) on a Microtiter Plate Luminometer (Dyn-Ex Technologies).
ISRE reporter assay
Cell lines were transduced with lentiviral particles containing an inducible ISRE-responsive luciferase reporter construct (SA Biosciences) and selected with puromycin. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) on a Microtiter Plate Luminometer (Dyn-Ex Technologies).
IFNβ Elisa
Human IFNβ was measured using ELISA kits from PBL InterferonSource. The results were normalized to live cell numbers.
Tumor model and therapy study
The xenograft tumor model of human ABC DLBCL lymphoma was established by subcutaneous (s.c.) injection of cells into nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (NCI-Frederick, Frederick, MD). The tumor growth was monitored by measuring tumor size in two orthogonal dimensions. See Supplemental Experimental Procedures for details. All animal experiments were approved by the National Cancer Institute Animal Care and Use Committee (NCI ACUC) and were performed in accordance with NCI ACUC guidelines.
Supplementary Material
Significance.
New therapies are needed for the activated B-cell-like (ABC) subtype of diffuse large B cell lymphoma (DLBCL), the most refractory subtype of this lymphoma. Oncogenic mutations activate the BCR and MYD88 pathways in ABC DLBCL, engaging NF-κB and IFNβ signaling. Lenalidomide, a drug showing clinical activity against DLBCL, kills ABC DLBCLs by inducing IFNβ and blocking NF-κB. Lenalidomide antagonizes a central regulatory hub in ABC DLBCL governed by the transcription factors IRF4 and SPIB, which together suppress IFNβ while augmenting NF-κB. Oncogenic BCR signaling to NF-κB induces IRF4 expression in ABC DLBCL. Inhibition of BCR signaling with the drug ibrutinib synergizes with lenalidomide to block IRF4 and kill ABC DLBCL cells, supporting clinical trials of this synthetically lethal drug combination.
Highlights.
Lenalidomide kills ABC DLBCLs by decreasing expression of IRF4 and SPIB
IRF4 and SPIB maintain ABC DLBCL viability
IRF4 and SPIB block toxic IFNβ production while enhancing pro-survival NF-κB activity
Lenalidomide and BCR pathway drugs synergize to inhibit IRF4 and kill ABC DLBCLs
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We also thank Carla Heise and Celgene for support. J.P. was supported by the UMD-NCI Partnership for Cancer Technology. We thank M. Celeste Simon for the anti-SPIB antibody. We wish to thank Kathleen Meyer for her assistance with GEO submissions, and the members of the Staudt lab for their assistance and helpful discussions. J.J.B. and S.B. are employees and shareholders of Pharmacyclics, Inc.
Footnotes
The other authors declare no competing financial interests.
Accession number: Gene expression data has been deposited under accessions GSE32456 and GSE33012. All ChIP-Seq data can be found under accession SRA025850.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappa B activity is required for survival of activated B Cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy K, Pittaluga S, Czuczman MS, Dave SS, Wright G, Grant N, Shovlin M, Jaffe ES, Janik JE, Staudt LM, Wilson WH. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113:6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell. 2002a;10:1097–1105. doi: 10.1016/s1097-2765(02)00703-7. [DOI] [PubMed] [Google Scholar]
- Escalante CR, Shen L, Escalante MC, Brass AL, Edwards TA, Singh H, Aggarwal AK. Crystallization and characterization of PU.1/IRF-4/DNA ternary complex. J Struct Biol. 2002b;139:55–59. doi: 10.1016/s1047-8477(02)00514-2. [DOI] [PubMed] [Google Scholar]
- Friedberg JW, Sharman J, Sweetenham J, Johnston PB, Vose JM, Lacasce A, Schaefer-Cutillo J, De Vos S, Sinha R, Leonard JP, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non Hodgkin’s lymphoma and chronic lymphocytic leukemia. Blood. 2009 doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco WR, Park HS, Rustum YM. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1990;50:5318–5327. [PubMed] [Google Scholar]
- Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799:775–787. doi: 10.1016/j.bbagrm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman SE, Gordon AL, Wagner AJ, Heerema NA, Zhao W, Flynn JM, Jones J, Andritsos L, Puri KD, Lannutti BJ, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116:2078–2088. doi: 10.1182/blood-2010-02-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Ilizaliturri FJ, Deeb G, Zinzani PL, Pileri SA, Malik F, Macon WR, Goy A, Witzig TE, Czuczman MS. Higher response to lenalidomide in relapsed/refractory diffuse large b-cell lymphoma in nongerminal center b-cell-like than in germinal center b-cell-like phenotype. Cancer. 2011 doi: 10.1002/cncr.26135. [DOI] [PubMed] [Google Scholar]
- Hoellenriegel J, Meadows SA, Sivina M, Wierda WG, Kantarjian H, Keating MJ, Giese N, O’Brien S, Yu A, Miller LL, et al. The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood. 2011 doi: 10.1182/blood-2011-05-352492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, Nong Y, Wen D, Adams J, Dang L, Staudt LM. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, Chan JW, Rosenwald A, Gascoyne RD, Staudt LM. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen A, Veckman V, Nikula T, Lahesmaa R, Kinnunen L, Matikainen S, Julkunen I. Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages. J Immunol. 2005;175:6570–6579. doi: 10.4049/jimmunol.175.10.6570. [DOI] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008a;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Lenz G, Nagel I, Siebert R, Roschke AV, Sanger W, Wright GW, Dave SS, Tan B, Zhao H, Rosenwald A, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204:633–643. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008b;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008c;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gill N, Lentzsch S. Recent advances of IMiDs in cancer therapy. Curr Opin Oncol. 2010;22:579–585. doi: 10.1097/CCO.0b013e32833d752c. [DOI] [PubMed] [Google Scholar]
- Li S, Pal R, Monaghan SA, Schafer P, Ouyang H, Mapara M, Galson DL, Lentzsch S. IMiD immunomodulatory compounds block C/EBP{beta} translation through eIF4E down-regulation resulting in inhibition of MM. Blood. 2011;117:5157–5165. doi: 10.1182/blood-2010-10-314278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A, Heintel D, Zhang LH, Mendy D, Gaidarova S, Brady H, Bartlett JB, Schafer PH, Schreder M, Bolomsky A, et al. Lenalidomide downregulates the cell survival factor, interferon regulatory factor-4, providing a potential mechanistic link for predicting response. Br J Haematol. 2011 doi: 10.1111/j.1365-2141.2011.08689.x. [DOI] [PubMed] [Google Scholar]
- Marecki S, Fenton MJ. PU.1/Interferon Regulatory Factor interactions: mechanisms of transcriptional regulation. Cell Biochem Biophys. 2000;33:127–148. doi: 10.1385/CBB:33:2:127. [DOI] [PubMed] [Google Scholar]
- Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Yanase N, Ibukiyama C, Yamashina A, Kayagaki N, Yagita H, Mizuguchi J. Involvement of TRAIL/TRAIL-R interaction in IFN-alpha-induced apoptosis of Daudi B lymphoma cells. Cytokine. 2001;14:193–201. doi: 10.1006/cyto.2001.0873. [DOI] [PubMed] [Google Scholar]
- Pal R, Monaghan SA, Hassett AC, Mapara MY, Schafer P, Roodman GD, Ragni MV, Moscinski L, List A, Lentzsch S. Immunomodulatory derivatives induce PU.1 down-regulation, myeloid maturation arrest, and neutropenia. Blood. 2010;115:605–614. doi: 10.1182/blood-2009-05-221077. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, Dalla-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- Schotte R, Rissoan MC, Bendriss-Vermare N, Bridon JM, Duhen T, Weijer K, Briere F, Spits H. The transcription factor Spi-B is expressed in plasmacytoid DC precursors and inhibits T-, B-, and NK-cell development. Blood. 2003;101:1015–1023. doi: 10.1182/blood-2002-02-0438. [DOI] [PubMed] [Google Scholar]
- Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Romesser PB, Staudt LM. IRF4: Immunity. Malignancy! Therapy? Clin Cancer Res. 2009;15:2954–2961. doi: 10.1158/1078-0432.CCR-08-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Peng A, Schlissel MS. In vivo occupancy of the kappa light chain enhancers in primary pro- and pre-B cells: a model for kappa locus activation. Immunity. 1997;6:131–143. doi: 10.1016/s1074-7613(00)80420-3. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, Lam LT, Davis RE, Staudt LM. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunological reviews. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O’Shea JJ, Singh H, Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucur E, Mattern J, Wenger T, Okouoyo S, Schroth A, Debatin KM, Herr I. Induction of apoptosis in experimental human B cell lymphomas by conditional TRAIL-expressing T cells. British journal of cancer. 2003;89:2155–2162. doi: 10.1038/sj.bjc.6601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Braggio E, Shi CX, Bruins LA, Schmidt JE, Van Wier S, Chang XB, Bjorklund CC, Fonseca R, Bergsagel PL, et al. Cereblon expression is required for the anti-myeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.