Abstract
Although multiple reports show that defective genetic networks underlie the aetiology of autism, few have translated into pharmacotherapeutic opportunities. Since drugs compete with endogenous small molecules for protein binding, many successful drugs target large gene families with multiple drug binding sites. Here we search for defective gene family interaction networks (GFINs) in 6,742 patients with the ASDs relative to 12,544 neurologically normal controls, to find potentially druggable genetic targets. We find significant enrichment of structural defects (P≤2.40E−09, 1.8-fold enrichment) in the metabotropic glutamate receptor (GRM) GFIN, previously observed to impact attention deficit hyperactivity disorder (ADHD) and schizophrenia. Also, the MXD-MYC-MAX network of genes, previously implicated in cancer, is significantly enriched (P≤3.83E−23, 2.5-fold enrichment), as is the calmodulin 1 (CALM1) gene interaction network (P≤4.16E−04, 14.4-fold enrichment), which regulates voltage-independent calcium-activated action potentials at the neuronal synapse. We find that multiple defective gene family interactions underlie autism, presenting new translational opportunities to explore for therapeutic interventions.
 The autism spectrum disorders are complex genetic traits characterized
by various neurodevelopmental deficits. Here, the authors analyse defective gene family
interaction networks in autism cases and healthy controls and identify potential gene
family interactions that may contribute to autism aetiology.
The autism spectrum disorders are complex genetic traits characterized
by various neurodevelopmental deficits. Here, the authors analyse defective gene family
interaction networks in autism cases and healthy controls and identify potential gene
family interactions that may contribute to autism aetiology.
The autism spectrum disorders (ASDs) represent a group of highly heritable childhood neuropsychiatric disorders characterized by a variable phenotypic spectrum of neurodevelopmental deficits of impaired socialization, reduced communication and restricted, repetitive, or stereotyped behaviour1. ASDs are four times more common in boys2,3, and the most recent prevalence estimates across the United States range from 1%4 to 2%5, although a recent study reported a prevalence as high as 2.6% in a general school-aged population in South Korea6. The ASDs have an estimated heritability as high as 90%7 based on data on monozygotic twin concordance studies8,9,10, whereas recent estimates of the sibling recurrence risk range from 19% to 22%11,12.
Despite being highly heritable, the vast majority of family studies suggest that the ASDs do not segregate as a simple Mendelian disorder, but rather display clinical and genetic heterogeneity consistent with a complex trait13. Indeed, recent studies estimate that the ASDs may comprise up to 400 distinct genetic and genomic disorders that phenotypically converge14,15. Common variants such as single-nucleotide polymorphisms seem to contribute to ASD susceptibility, but, taken individually, their effects appear to be small16. However, there is increasing evidence that the ASDs can arise from rare or ‘private’ highly penetrant mutations that segregate in families but are less generalizable to the general population17,18,19. Many genes implicated thus far, which are involved in chromatin remodelling, metabolism, mRNA translation and synaptic function, seem to converge in common pathways or genetic networks affecting neuronal and synaptic homeostasis16.
Such remarkable phenotypic and genotypic heterogeneity when coupled to the private nature of mutations in the ASDs has hindered identification of new genetic risk factors with therapeutic potential. However, it is noteworthy that many of the rare gene defects implicated in the ASDs belong to gene families. For instance, rare defects impacting multiple members of both the post-synaptic neuroligin (NLGN) gene family20 as well as their pre-synaptic neurexin molecular-interacting partners21,22 have long been reported in patients with ASDs. In addition, a number of other defective gene families with important functional roles have subsequently been well-characterized including ubiquitin conjugation23, gamma-aminobutyric acid receptor signalling24,25,26,27 and cadherin/protocadherin cell junction proteins28 in the brain. Furthermore, multiple defects in voltage-gated calcium channels have been found in schizophrenia29, and a defective network of metabotropic glutamate (GRM) receptor signalling was found in both ADHD30 and schizophrenia31,32,33,34,35,36, two neuropsychiatric disorders that are highly coincident with the ASDs. Also, the vast majority of significant defective genes identified from recent whole-exome sequences belong to gene families17,18,19.
Many studies have found defective genetic networks in the ASDs21,23,37,38,39,40 (see ref. 16 for review), and we complement these in this work by uncovering new networks and implicating specific defective gene families that may be enriched for novel potential therapeutic targets. Drug-binding sites on proteins usually exist out of functional necessity33, and gene families derive from gene duplication events that present additional binding sites for a given drug to exert its effects. Most successful drugs achieve their activity by competing for a binding site on a protein with an endogenous small molecule41; therefore, many successful pharmacologic gene targets are within large gene families. Indeed, nearly half of the pharmacologic gene targets fall into just six gene families: G-protein-coupled receptors (GPCRs), serine/threonine and tyrosine protein kinases, zinc metallopeptidases, serine proteases, nuclear hormone receptors and phosphodiesterases41. Moreover, many large gene families are localized to pre- and post synaptic neuronal terminals to coordinate the highly complex and evolutionarily conserved process of neurotransmission42, which is thought to be compromised to varying degrees in the autistic brain43. Therefore, we hypothesize that we may select more druggable targets for the ASDs by enriching for defective interaction networks defined by gene families.
Here we perform a large genome-wide association study (GWAS) of structural variants that disrupt gene family protein interaction networks in patients with autism. We find multiple defective networks in the ASDs, most notably rare copy-number variants (CNVs) in the metabotropic glutamate receptor (mGluR) signalling pathway in 5.8% of patients with the ASDs. Defective mGluR signalling was found in both ADHD30 and schizophrenia31,32,33,34,35,36, two common neuropsychiatric disorders that are highly coincident with the ASDs. Furthermore, we find other attractive candidates such as the MAX dimerization protein (MXD) network that is implicated in cancer, and a Calmodulin 1 (CALM1) gene interaction network that is active in neuronal tissues. The numerous defective gene family interactions we find to underlie autism present many novel translational opportunities to explore for therapeutic interventions.
Results
To identify and comprehensively characterize defective genetic networks underlying the ASDs, we performed a large-scale genome association study for copy-number variation (CNVs) enriched in patients with autism. By combining the affected cases from previously published large ASD studies21,23,28,44 with more recently recruited cases from the Children’s Hospital of Philadelphia, we executed one of the largest searches for rare pathogenic CNVs in ASDs to date. In sum, 6,742 genotyped samples from patients with the ASDs were compared with those from 12,544 neurologically normal controls recruited at The Children’s Hospital of Philadelphia (CHOP).
These cases were each screened by neurodevelopmental specialists to exclude patients with known syndromic causes for autism. Genotyping was performed at CHOP for the vast majority of the ASD cases as well as all the controls. After cleaning the data to remove sample duplicates and performing standard QC for CNVs, we first inferred the continental ancestry of 5,627 affected cases and 9,644 disease-free controls using a training set defined by populations from HapMap 3 (ref. 45) and the Human Genome Diversity Panel46 (Table 1). Using this QC criteria, we estimated that the sensitivity and specificity of calling CNVs is ~\n70% and 100%, respectively, across 121 different genomic regions assayed by PCR (Methods). Across all ethnicities, there was an increased burden of CNVs in cases versus controls, a statistically significantly difference (P≤0.001) in the larger European (63.3 versus 54.5 Kb, respectively) and African-derived (70.4 versus 48.0 Kb, respectively) populations.
Table 1. Distribtion of CNVs across samples and estimated ancestry.
| Continental ancestry | Case | Control | Total |
|---|---|---|---|
| Europe | |||
| Number of samples | 4,602 | 4,722 | 9,324 |
| *CNV burden (Kb) | 63.3 | 54.5 | |
| Africa | |||
| Number of samples | 312 | 4,169 | 4,481 |
| *CNV burden (Kb) | 70.4 | 48.0 | |
| America | |||
| Number of samples | 485 | 276 | 761 |
| CNV burden (Kb) | 59.1 | 58.4 | |
| Asia | |||
| Number of samples | 201 | 350 | 551 |
| CNV burden (Kb) | 56.1 | 54.1 | |
| Other | |||
| Number of samples | 27 | 127 | 154 |
| CNV burden (Kb) | 51.5 | 49.4 | |
| All Ethnicities | |||
| Number of samples | 5,627 | 9,644 | 15,271 |
| *CNV burden (Kb) | 63.0 | 51.7 | |
CNV=copy-number variation. The table shows the distribution of cases, controls and CNV coverage across estimated continental ancestry. For groups of cases and controls across estimated ancestries, the table lists the numbers of subjects that passed quality control and their group-wise CNV burden, defined as the average span of CNVs in Kb for each group.
*Statistically significant (P≤0.01 by PLINK permutation test) differences in CNV burden are marked with an asterix(*).
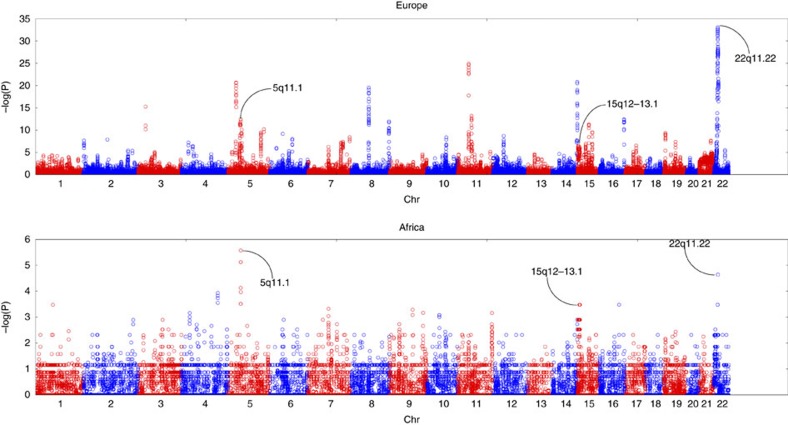
We then searched for pan-ethnic CNV regions (CNVRs) discovered in the European-derived data set (4,602 cases versus 4,722 controls; P≤0.0001 by Fisher’s exact test) and replicated in an independent ASD data set of African ancestry (312 cases versus 4,169 controls; P≤0.001 by Fisher’s exact test) with subsequent measurement of overall significance across the entire multi-ethnic discovery cohort (5,627 cases versus 9,644 controls) for maximal power (Fig. 1, Table 2). On the basis of these selection criteria, two large well-known ASD risk loci emerged that harboured multiple duplications in the Prader Willi/Angelman syndrome (15q11–13) critical region, and multiple deletions were detected in the DiGeorge syndrome (22q11) critical region, albeit notably smaller than the 22q11 deletion syndrome. A third locus harbouring deletions in poly ADP-ribose polymerase family 8 (PARP8) on chromosome 5q11 was also discovered. PARP8 was previously identified as associated with the ASDs in a Dutch population47, but it has not previously been described for its pan ethnic distribution across European-derived and African-derived populations.
Figure 1. Significance of CNVRs by GWAS of ASDs in European-derived or African-derived populations.
The Manhattan plots show the −log10 transformed P-value of association for each CNVR along the genome. Adjacent chromosomes are shown in alternating red and blue colours. The regions discovered in Europeans (P≤0.0001) that replicated in Africans (P≤0.001) are highlighted with black arrows labelled by chromosome band. GWAS of 4,634 cases versus 4,726 controls in Europeans is shown on top and GWAS of 312 cases versus 4,173 controls in Africans is shown below.
Table 2. Significant copy-number variable regions.
| CNVR | Genes | Bands | Size (Kb) | No. of SNP | No. of Case | No. of Control |
All
|
Europe
|
Africa
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | P -value | OR | P-value | OR | |||||||
| del | ZNF280B | 22q11.22 | 53.4 | 13 | 130 | 0 | 2.56E−57 | Inf | 1.94E−33 | Inf | 3.34E−04 | Inf |
| del | * PARP8 | 5q11.1 | 47.7 | 8 | 70 | 8 | 2.76E−22 | 15.1 | 3.84E−13 | 12.0 | 2.69E−06 | 40.9 |
| dup | * GABRB3 | 15q12 | 49.0 | 20 | 28 | 0 | 7.60E−13 | Inf | 1.50E−06 | Inf | 3.34E−04 | Inf |
| dup | * GABRG3 | 15q12 | 135.3 | 13 | 27 | 1 | 3.72E−11 | Inf | 1.60E−05 | 19.5 | 3.34E−04 | Inf |
| dup | * HERC2 | 15q13.1 | 84.4 | 2 | 24 | 0 | 4.12E−11 | Inf | 6.17E−06 | Inf | 3.34E−04 | Inf |
CNVR=copy-number variable region; OR=odds ratio. The table shows CNVRs distinguishing cases from controls significant across both European-derived populations (P≤0.0001 by Fisher’s exact test) and African-derived populations (P≤0.001). For each CNVR, the table lists the type (del or dup), the closest gene impacted, the chromosomal band, the approximate size of the defect (Kb), the number of contributing SNPs, the numbers of affected cases and controls, as well as P-value and odds ratio (OR) from Fisher’s exact test for across all populations, and subsets of European-derived and African-derived populations.
*Genes with an asterix (*) harbour CNVRs that disrupt their exons of directly, while those without the asterix are located in the genomic region around the intergenic CNVRs.
We examined the genetic interaction networks derived from gene families with members localized to the the Prader Willi/Angelman syndrome (15q11-13) critical region, the DiGeorge syndrome (22q11) critical region, and the novel PARP8 (5q11) region using a method previously applied to ADHD30; however, hardly any of the most significant genes harbouring significant CNVRs clustered within gene families. Consequently, we broadened our search for gene family interaction networks (GFINs) and searched the entire genome for GFINs with CNVs enriched in autism. For every gene family, we defined a GFIN as the genetic interaction network spawned by its multiple duplicated members. We used standard HUGO48 gene names to define 1,732 GFINs across which we searched for enrichment of network defects associated with the ASDs. However, because there is an a priori excess of CNV burden in ASD cases over disease-free controls (Table 1), larger GFINs are expected to display significant enrichment of case defects by virtue solely of their increased size and complexity. Therefore, for each GFIN, we used a network permutation test of case enrichment across 1,000 random sets of networked genes to control for the GFIN size and complexity. With this approach, we robustly identified network defects associated with the ASDs by minimizing statistical artefact derived from any a priori excessive CNV burden in cases over controls, as well as other unknown biases that may be inherent in the human interactome data49,50,51 that we mined.
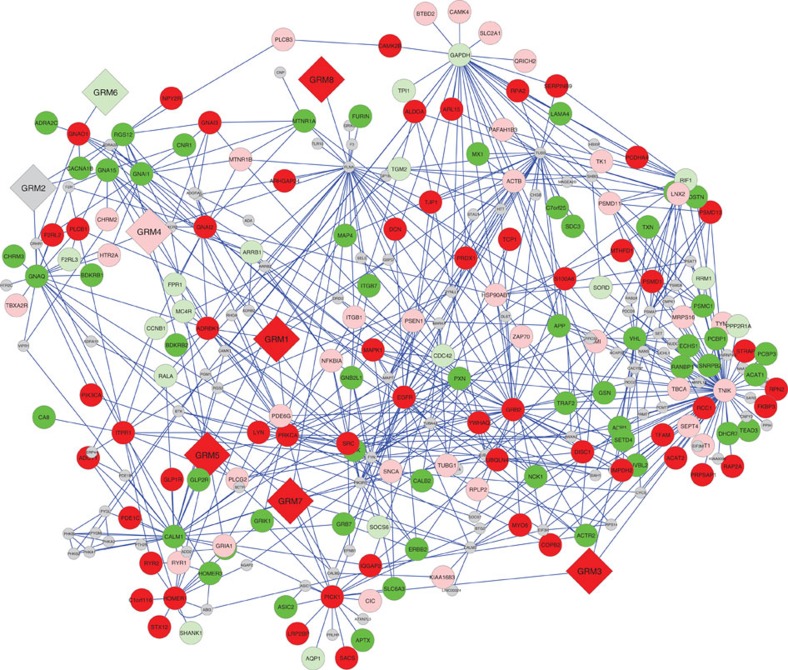
Out of 1,732 GFINs, we used the network permutation test to rank 1,557 GFINs with defined CNVs for enrichment of genetic defects in the ASDs. Among the top GFINs (Table 3) was the metabotropic glutamate receptor (mGluR) pathway defined by the GRM family of genes that impacts glutamatergic neurotransmission. The GRM family contains eight members, all of which were defined in the human interactome to cumulatively spawn a GFIN of 279 genes (Fig. 2). Across this GFIN for the GRM family of genes, we found CNV defects in 5.8% of European-derived ASD cases (265/4,602) versus only 3% of ethnically matched controls (153/4,722), a 1.8-fold enrichment of frequency (PFisher ≤2.40E−09). By 1,000 random network permutations, we found this excess of enrichment across cases in the mGluR pathway to also be statistically significant (Pperm ≤0.05). In addition, 69.2% (124/181) of the informative genes within our mGluR network showed an excess of CNVs among cases. However, the component genes that harbour the most significant CNVRs contributing to this overall network significance reveal that the duplicated mGluR genes themselves (GRM1, GRM3, GRM4, GRM5, GRM6, GRM7 and GRM8) fail to achieve significance individually, although there is a trend for an excess of CNV defects across a specific subset of mGluR receptors (GRM1, GRM3, GRM5, GRM7, GRM8) that is unique to cases (Supplementary Table 1).
Table 3. Top gene family interaction networks discovered.
|
Gene family
|
Enriched genes
|
Cases
|
Controls
|
Gene Network Association
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Size | No. | Frequency | No. | Frequency | No. | Frequency | P fisher | Enrichment | P perm |
| BRF | 2 | 242/326 | 0.742 | 567 | 0.123 | 370 | 0.078 | 3.30E−13 | 1.65 | 0.040 |
| CCL | 24 | 108/144 | 0.75 | 231 | 0.05 | 129 | 0.027 | 5.62E−09 | 1.88 | 0.008 |
| CCNT | 2 | 183/254 | 0.72 | 613 | 0.133 | 381 | 0.081 | 1.10E−16 | 1.75 | 0.007 |
| ELAVL | 4 | 108/156 | 0.692 | 327 | 0.071 | 152 | 0.032 | 6.87E−18 | 2.3 | 0.043 |
| ERCC | 7 | 263/369 | 0.713 | 836 | 0.182 | 560 | 0.119 | 7.67E−18 | 1.65 | 0.035 |
| GRM | 8 | 124/181 | 0.685 | 265 | 0.058 | 153 | 0.032 | 2.40E−09 | 1.82 | 0.043 |
| GTF2H | 5 | 152/223 | 0.682 | 391 | 0.085 | 233 | 0.049 | 3.21E−12 | 1.79 | 0.049 |
| KIAA | 106 | 268/373 | 0.718 | 988 | 0.215 | 647 | 0.137 | 3.12E−23 | 1.72 | 0.045 |
| KPNA | 7 | 256/367 | 0.698 | 560 | 0.122 | 369 | 0.078 | 1.26E−12 | 1.63 | 0.028 |
| MXD | 3 | 52/64 | 0.813 | 366 | 0.08 | 156 | 0.033 | 3.83E−23 | 2.53 | 0.042 |
| POU5F | 2 | 94/130 | 0.723 | 293 | 0.064 | 131 | 0.028 | 2.96E−17 | 2.38 | 0.041 |
| RAD | 7 | 218/309 | 0.706 | 535 | 0.116 | 339 | 0.072 | 9.68E−14 | 1.7 | 0.042 |
| SAP | 4 | 111/150 | 0.74 | 274 | 0.06 | 151 | 0.032 | 9.61E−11 | 1.92 | 0.040 |
| SMAD | 8 | 845/1,225 | 0.69 | 1,782 | 0.387 | 1,424 | 0.302 | 1.81E−18 | 1.46 | 0.039 |
| SMARCC | 2 | 106/147 | 0.721 | 239 | 0.052 | 131 | 0.028 | 1.22E−09 | 1.92 | 0.043 |
| SMC | 5 | 88/120 | 0.733 | 336 | 0.073 | 176 | 0.037 | 1.71E−14 | 2.03 | 0.034 |
The table shows significant gene family interaction networks (GFINs) by network permutation testing (Pperm≤0.05) enriched for CNV defects across at least 5% of cases. The table lists the name and size of gene family tested, the number and frequency of network genes enriched in the second degree gene interaction network, the number and frequency of cases harbouring defects across the network, the number and frequency of controls harbouring defects across the network, the significance of association by Fisher’s exact test, the enrichment of CNV defects in cases, and the significance of that enrichment by 1,000 random network permutations.
Figure 2. Enrichment of optimal CNVRs across mGluR network of genes.
Nodes of the network are labelled with their gene names, with red and green representing deletions and duplications, respectively, while grey nodes lack CNV data. Dark and light colours represent enrichment in cases and controls, respectively. The genes defining the network are shown as diamonds, while all other genes are shown as circles. Blue lines indicate evidence of interaction.
Many large studies of CNVs implicate genes within the glutamatergic signaling pathway in the aetiology of the ASDs21,23,37,38,39,40, and SNP52,53 and CNV duplications54 of GRM8 have been reported in association with the ASDs before in humans. Moreover, a recent functional study demonstrated that in mouse models of tuberous sclerosis and fragile X, two different forms of syndromic autism, the autistic phenotype was ameliorated by modulation of GRM5 in opposite directions for each syndrome, which suggests that GRM5 functional activity is central in defining the axis of synaptopathophysiology in syndromic autism55. Our GRM network findings implicate rare defects in mGluR signalling also contribute to the ASDs outside of fragile X and tuberous sclerosis, and we posit that functional mGluR synaptopathophysiology may be initiated from many dozens if not hundreds of defective genes within the mGluR pathway that may account for as much as 6% of the endophenotypes of the ASDs (Table 3).
In addition, we recently demonstrated the importance of mGluRs in ADHD30,56, a highly co-incident neuropsychiatric disorder within the autism spectrum. However, in contrast to ADHD where defects within the mGluR receptors themselves (GRMs) were among the most significant copy-number defects contributing to the overall network significance, we found that in the ASDs defects of component GRMs contributed only modestly to the overall significance of the mGluR pathway. Nonetheless, the defects within GRM1, GRM3, GRM5, GRM7 and GRM8 that we identified as unique to cases and thus enriched are the same GRMs we identified as being pathogenic in ADHD and may impact glutamatergic signalling.
Among the most highly ranked GFINs by permutation testing, the MAX dimerization protein (MXD) GFIN (PFisher ≤3.83E−23, enrichment=2.53, Pperm ≤0.042) was the most enriched. The MXD family of genes encode proteins that interact with MYC/MAX network of basic helix-loop-helix leucine zipper (bHLHZ) transcription factors that regulate cell proliferation, differentiation and apoptosis (MIM 600021)57; MXD genes are important candidate tumour suppressor genes as the MXD-MYC-MAX network is dysregulated in various types of cancer58. Interestingly an epidemiological link between autism and specific types of cancer has been reported59, and anticancer therapeutics were recently shown to modulate ASD phenotypes in the mouse through regulation of synaptic NLGN protein levels60. Within the component genes contributing to the MXD GFIN significance, duplications in PARP10 (P≤4.06E−11, OR=2.04) and UBE3A (1.50E−06, OR=inf) are the most significantly enriched (Supplementary Table 2). It is notable that we found PARP8 as significant across ethnicities as described earlier (Table 2), and we previously described the importance of structural defects in UBE3A in the ASDs23.
Other notable significant GFINs uncovered were POU class 5 homeobox (POU5F) GIFN (PFisher≤2.96E−17, enrichment=2.3, Pperm ≤0.008, and the SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily c (SMARCC) GFIN (PFisher ≤1.22E−09, enrichment=1.9, Pperm ≤0.035). The POU5F family of genes encodes for transcription factors containing a POU homeodomain, and their role has been demonstrated in embryonic development, especially during early embryogenesis, and it is necessary for embryonic stem cell pluripotency. Component genes of the SMARCC gene family are members of the SWI/SNF family of proteins, whose members display helicase and ATPase activities and which are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. Most interestingly, the KIAA family of genes ranked among the top GFINs (PFisher ≤3.12E−23, enrichment=1.6, Pperm ≤0.040). KIAA genes have been identified in the Kazusa cDNA sequencing project61 and are predicted from novel large human cDNAs; however, they have no known function.
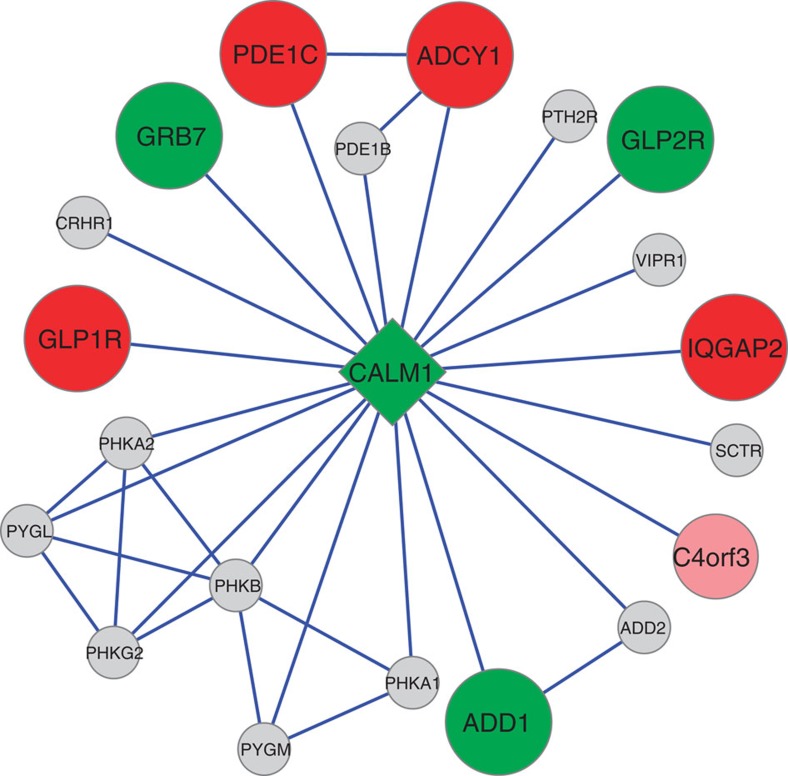
We also hypothesized that some component members of gene families may contribute disproportionately to the significance of a GFIN because they are highly connected to interacting gene partners that are enriched for CNV defects in ASD. Therefore, we decomposed the 1,732 gene families into their 15,352 component duplicated genes of which 1,218 had defined networks with data to test for significance by genome-wide network permutation. The calmodulin 1 (CALM1) gene interaction network ranked highest by network permutation testing of case enrichment for CNV defects across 1,000 random gene networks (Fig. 3, Table 4) and represents a novel and attractive candidate gene for the ASDs. Across the CALM1 network, we found CNV defects in 14/4,618 cases versus only 1/4726 controls (Pfisher ≤4.16E−04, enrichment=14.37, Pperm ≤0.002), and these defects were distributed such that 90% (9/10) of genes that harboured CNVs in the CALM1 interactome were enriched in cases. Closer inspection of the most significant CNVR contributing to the CALM1 network significance (Supplementary Table 3) revealed that no single gene was significant on its own; instead, with the exception of only one gene (PTH2R), each contributing CNVR tagged highly penetrant rare defects unique to cases. Calmodulin is the archetype of the family of calcium-modulated proteins of which nearly 20 members have been found. Calmodulin contains 149 amino acids that define four calcium-binding domains used for Ca2+-mediated coordination of a large number of enzymes, ion channels and other proteins including kinases and phosphatases; its functions include roles in growth and cell cycle regulation as well as in signal transduction and the synthesis and release of neurotransmitters [MIM 114180]57.
Figure 3. Enrichment of optimal CNVRs across CALM1 network.
The first degree-directed interaction network defined by CALM1 is shown.
Table 4. Most significant individual gene interaction networks ranked by permutation testing.
| Gene Family Member |
Enriched Genes
|
Cases
|
Controls
|
Gene Network Association
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Frequency | No. | Frequency | # | Frequency | P fisher | Enrichment | P perm | |
| AKAP13 | 7/7 | 1.00 | 16 | 0.0035 | 1 | 0.0002 | 1.14E−04 | 16.43 | 0.012 |
| BAG1 | 7/7 | 1.00 | 15 | 0.0032 | 1 | 0.0002 | 2.18E−04 | 15.40 | 0.014 |
| CALM1 | 9/10 | 0.90 | 14 | 0.0030 | 1 | 0.0002 | 4.16E−04 | 14.37 | 0.002 |
| CASP6 | 16/17 | 0.94 | 46 | 0.0100 | 6 | 0.0013 | 2.96E−09 | 7.91 | 0.012 |
| GTF2H3 | 23/26 | 0.88 | 42 | 0.0091 | 8 | 0.0017 | 3.66E−07 | 5.41 | 0.009 |
| MAP3K5 | 11/12 | 0.92 | 34 | 0.0074 | 4 | 0.0008 | 2.02E−07 | 8.76 | 0.012 |
| NCOR1 | 9/10 | 0.90 | 26 | 0.0056 | 2 | 0.0004 | 1.11E−06 | 13.37 | 0.004 |
| PARP1 | 5/5 | 1.00 | 5 | 0.0011 | 0 | 0.0000 | 2.95E−02 | inf | 0.012 |
| PTPN13 | 6/6 | 1.00 | 9 | 0.0019 | 0 | 0.0000 | 1.75E−03 | inf | 0.007 |
| TCEA1 | 22/26 | 0.85 | 39 | 0.0084 | 7 | 0.0015 | 5.94E−07 | 5.74 | 0.009 |
The table lists the name and gene family member tested, the number and frequency of network genes enriched, the number and frequency of cases harbouring defects, the number and frequency of controls harbouring defects, and the significance of association by Fisher’s exact test, the odds ratio of the effect size, and the significance of association by random permutation of network while controlling for number of genes tested.
Among other highly ranked first degree gene interaction networks were the nuclear receptor co-repressor 1 (NCOR1; Pfisher ≤1.11E−06, enrichment=13.37, Pperm ≤0.004) and BCL2-associated athanogene 1 (BAG1; Pfisher ≤2.18E−04, enrichment=15.40, Pperm ≤0.014) networks. NCOR1 is a transcriptional coregulatory protein that appears to assist nuclear receptors in the downregulation of DNA expression through recruitment of histone deacetylases to DNA promoter regions; it is a principal regulator in neural stem cells51. The oncogene BCL2 is a membrane protein that blocks the apoptosis pathway, and BAG1 forms a BCL2-associated athanogene and represents a link between growth factor receptors and antiapoptotic mechanisms. The BAG1 gene has been implicated in age-related neurodegenerative diseases, including Alzheimer’s disease62,63.
In summary, given the private nature of mutations in the ASDs, considering the cumulative contributions of rare highly penetrant genetic defects boosts our power to discover and prioritize significant pathway defects. As a result, our comprehensive, unbiased analytical approach has identified a diverse set of specific defective biological pathways that contribute to the underlying aetiology of the ASDs. Among GFINs robustly enriched for structural defects, the most enriched was that of the MXD family of genes that has been implicated in cancer pathogenesis58, thereby providing concrete genetic defects to explore the reported coincidence of specific cancers with the ASDs59. The most highly ranked component duplicated gene interaction network involves defects in CALM1 and its multiple interacting partners that are important in regulating voltage-independent calcium-activated action potentials at the neuronal synapse. Moreover, we found significant enrichment for defects within the GFIN for GRM that defines the mGluR pathway that has previously been shown to be defective in other neuropsychiatric diseases29,30. While specific mGluR gene family members have been shown to underlie syndromic ASDs55, our findings suggest that rare defects in mGluR signalling also contribute to idiopathic autism across the entire GFIN for GRM genes.
Consequently, in addition to specific neuronal pathways that are expected to be defective in the ASDs like those defined by GRM and CALM duplicate genes, we implicate completely novel biological pathways such as the MXD pathway specific forms of which may be associated with the ASDs59. Given the unmet need for better treatment for neurodevelopmental diseases64, the functionally diverse set of defective genetic interaction networks we report presents attractive genetic biomarkers to consider for targeted therapeutic intervention in ASDs and across the neuropsychiatric disease spectrum.
Methods
Ethics statement
The research presented here has been approved by the Children’s Hospital of Philadelphia IRB (CHOP IRB#: IRB 06-004886). Some patients and their families were recruited through CHOP outreach clinics. Written informed consent was obtained from the participants or their parents using IRB approved consent forms prior to enrollment in the project. There was no discrimination against individuals or families who chose not to participate in the study. All data were analysed anonymously and all clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Sample processing
The majority of cases (5,049 of 6,742) and all controls (12,544) were genotyped with genome-wide coverage using the Infinium II platform across various iterations of the HumanHap BeadChip with 550 K, 610 K, 660 K and 1 M markers by the Center for Applied Genomics at The Children’s Hospital of Philadelphia (CHOP). There were 1,693 cases genotyped by the AGP consortium. All cases and ~\n50% of controls were re-used from previously published large ASD studies21,23,28,44. All cases were diagnosed by ADI-R/ADOS and fulfilled standard criteria for ASDs. Duplicate samples were removed by selecting unique samples with the best quality (based on genotyping statistics used to QC samples) from clusters defined by single linkage clustering of all pairs of samples with high pairwise identity by state measures (IBS ≥0.9) across 140 K non-correlated SNPs. Ethnicity of samples was inferred by a supervised k-means classification (k=3) of the first 10 eigenvectors estimated by principal component analysis across the same subset of 140 K non-correlated SNPs. We used HapMap 3 (ref. 45) and the Human Genome Diversity Panel46 samples with known continental ancestry to train the k-means classifier implemented by the R Language for Statistical Computing65.
CNV inference and association
We called CNVs with the PennCNV algorithm66, which combines multiple values, including genotyping fluorescence intensity (Log R Ratio), population frequency of SNP minor alleles (B-allele frequency) and SNP spacing into a hidden Markov model. The term ‘CNV’ represents individual CNV calls, whereas ‘CNVR’ refers to population-level variation shared across subjects. Quality control thresholds for sample inclusion in CNV analysis included a high call rate (call rate ≥95%) across SNPs, low s.d. of normalized intensity (s.d. ≤0.3), low absolute genomic wave artefacts (|GCWF| ≤0.02) and low numbers of CNVs called (#CNVs ≤100). Genome-wide differences in CNV burden, defined as the average span of CNVs, between cases and controls and estimates of significance were computed using PLINK67. CNVRs were defined based on the genomic boundaries of individual CNVs, and the significance of the difference in CNVR frequency between cases and controls was evaluated at each CNVR using Fisher’s exact test.
Gene family interaction networks definition and association
We extended our previous work on ADHD30 here to rank all GFINs by a network permutation test. Specifically, using merged human interactome data from three different yeast two hybrid generated data sets49,50,51 accessed through the Human Interactome Database68, we defined the directed second-degree gene interaction network for all gene families here just as we did for the sole metabotropic glutamate receptor gene family network in ADHD. Specifically, here we use GFIN to refer to these gene family-derived interaction networks. In sum, we found 2,611 gene families with at least two members based on official HUGO48 gene nomenclature, and generated 1,732 GFINs using. For 1,557 GFINs with defined CNVs, we calculated an odds ratio of cumulative network enrichment over all genes harbouring CNVs within the network. Moreover, for each GFIN, we quantified its enrichment by a permutation test of 1,000 second-degree gene interaction networks derived from a random set of N genes, where N is the number of members of a given gene family. Because the CNVs we are focused on are so rare, we are relatively underpowered to achieve significance by permutation testing after correcting for multiple GFIN tests. However, we report all GFINs in the manuscript in order of their nominal/marginal significance.
Experimental validation of CNVs
Significant CNVRs that we identified were validated using commercially available qPCR Taqman probes run on the ABI GeneAmp 9700 system from Life Technology. Supplementary Data 1 lists 251 reactions that we tested using 121 different genomic probes across 85 different samples for which DNA was available. For deletions, our sensitivity=0.65, specificity=1.00, NPV=1.00 and PPV=0.88. For duplications, our sensitivity=0.68, specificity=0.99, NPV=0.94 and PPV=0.91.
Author contributions
D.H., Z.W., C.K., J.C., J.G. and H.H. conceived the study. D.H., A.K., K.T., F.M., and H.Q. performed computational analyses. A.M.H., L.V., R.P., and C.K. performed genotyping and experimental validation. H.H. and AGP consortium coordinated sample recruitment. D.H., C.K., Z.W., and H.H. interpreted the results. D.H. and H.H. wrote the manuscript. All authors read, edited and approved the final manuscript
Additional information
How to cite this article: Hadley, D. et al. The impact of the metabotropic glutamate receptor and other gene family interaction networks on autism. Nat. Commun. 5:4074 doi: 10.1038/ncomms5074 (2014).
Supplementary Material
Supplementary Tables 1-3
Experimental PCR validation of CNV predictions.
Acknowledgments
We thank all study participants and their families. We thank all the staff at the Center for Applied Genomics at CHOP for their invaluable contributions to recruitment of study subjects and genotyping of samples. We also gratefully acknowledge the resources provided by the AGRE Consortium and their participating families, and by the Autism Genome Project (AGP) Consortium and their participating families. The study was funded by an Institutional Development Fund from The Children’s Hospital of Philadelphia; The Margaret Q Landenberger Foundation; The Lurie Family Foundation; The Kubert Estate Fund and by U01HG005830. AGRE is a program of Autism Speaks and is at present supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to C.M. Lajonchere (PI) and formerly by grant MH64547 to D.H. Geschwind (PI). AGRE-approved academic researchers can acquire the data sets from AGRE at http://www.agre.org. There were 1,693 cases of the full AGP data sets that were genotyped by the AGP consortium. The full AGP data sets are made available from dbGaP at http://www.ncbi.nlm.nih.gov/gap. The remaining 5,049 cases and all 12,544 controls were all genotyped by the Center for Applied Genomics at the Children’s Hospital of Philadelphia.
Contributor Information
AGP Consortium:
Dalila Pinto, Alison Merikangas, Lambertus Klei, Jacob A.S. Vorstman, Ann Thompson, Regina Regan, Alistair T. Pagnamenta, Bárbara Oliveira, Tiago R. Magalhaes, John Gilbert, Eftichia Duketis, Maretha V. De Jonge, Michael Cuccaro, Catarina T. Correia, Judith Conroy, Inês C. Conceição, Andreas G. Chiocchetti, Jillian P. Casey, Nadia Bolshakova, Elena Bacchelli, Richard Anney, Lonnie Zwaigenbaum, Kerstin Wittemeyer, Simon Wallace, Herman van Engeland, Latha Soorya, Bernadette Rogé, Wendy Roberts, Fritz Poustka, Susana Mouga, Nancy Minshew, Susan G. McGrew, Catherine Lord, Marion Leboyer, Ann S. Le Couteur, Alexander Kolevzon, Suma Jacob, Stephen Guter, Jonathan Green, Andrew Green, Christopher Gillberg, Bridget A. Fernandez, Frederico Duque, Richard Delorme, Geraldine Dawson, Cátia Café, Sean Brennan, Thomas Bourgeron, Patrick F. Bolton, Sven Bölte, Raphael Bernier, Gillian Baird, Anthony J. Bailey, Evdokia Anagnostou, Joana Almeida, Ellen M. Wijsman, Veronica J. Vieland, Astrid M. Vicente, Gerard D. Schellenberg, Margaret Pericak-Vance, Andrew D. Paterson, Jeremy R. Parr, Guiomar Oliveira, Joana Almeida, Cátia Café, Susana Mouga, Catarina Correia, John I. Nurnberger, Anthony P. Monaco, Elena Maestrini, Sabine M. Klauck, Hakon Hakonarson, Jonathan L. Haines, Daniel H. Geschwind, Christine M. Freitag, Susan E. Folstein, Sean Ennis, Hilary Coon, Agatino Battaglia, Peter Szatmari, James S. Sutcliffe, Joachim Hallmayer, Michael Gill, Edwin H. Cook, Joseph D. Buxbaum, Bernie Devlin, Louise Gallagher, Catalina Betancur, and Stephen W. Scherer
References
- Muhle R., Trentacoste S. V. & Rapin I. The genetics of autism. Pediatrics 113, e472–e486 (2004). [DOI] [PubMed] [Google Scholar]
- Fombonne E. The prevalence of autism. JAMA 289, 87–89 (2003). [DOI] [PubMed] [Google Scholar]
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2002 Principal Investigators; Centers for Disease Control and Prevention. Prevalence of autism spectrum disorders--autism and developmental disabilities monitoring network, 14 sites, United States, 2002. MMWR Surveill Summ Morb Mortal Wkly report Surveill Summ/CDC 56, 12–28 (2007). [PubMed] [Google Scholar]
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2006 Principal Investigators; Centers for Disease Control and Prevention (CDC). Prevalence of autism spectrum disorders - Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ Morb Mortal Wkly report Surveill Summ/CDC 58, 1–20 (2009). [PubMed] [Google Scholar]
- Blumberg SJ, Ph D & Bramlett M. D. Changes in Prevalence of Parent-reported Autism Spectrum Disorder in School-aged U. S. Children: 2007 to 2011–2012 Hyattsville (2013) 20782. Available http://www.cdc.gov/nchs/data/nhsr/nhsr065.pdf. [PubMed] [Google Scholar]
- Kim Y. S. et al. Prevalence of autism spectrum disorders in a total population sample. Am. J. Psychiatry 168, 904–912 (2011). [DOI] [PubMed] [Google Scholar]
- Folstein S. E. & Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat. Rev. Genet. 2, 943–955 (2001). [DOI] [PubMed] [Google Scholar]
- Folstein S. & Rutter M. Infantile autism: a genetic study of 21 twin pairs. J. Child Psychol. Psychiatry 18, 297–321 (1977). [DOI] [PubMed] [Google Scholar]
- Steffenburg S. et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J. Child Psychol. Psychiatry 30, 405–416 (1989). [DOI] [PubMed] [Google Scholar]
- Bailey A. et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 25, 63–77 (1995). [DOI] [PubMed] [Google Scholar]
- Ozonoff S. et al. Recurrence risk for autism spectrum disorders: a baby siblings research consortium study. Pediatrics 128, e488–e495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino J. N., Zhang Y., Frazier T., Abbacchi A. M. & Law P. Sibling recurrence and the genetic epidemiology of autism. Am. J. Psychiatry 167, 1349–1356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolevzon A., Smith C. J., Schmeidler J., Buxbaum J. D. & Silverman J. M. Familial symptom domains in monozygotic siblings with autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 129B, 76–81 (2004). [DOI] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380, 42–77 (2011). [DOI] [PubMed] [Google Scholar]
- Iossifov I. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet G., Ey E. & Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu. Rev. Genomics Hum. Genet. 14, 191–213 (2013). [DOI] [PubMed] [Google Scholar]
- Sanders S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S. et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 34, 27–29 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P. et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 39, 319–328 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J. T. et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum J. D. et al. Association between a GABRB3 polymorphism and autism. Mol. Psychiatry 7, 311–316 (2002). [DOI] [PubMed] [Google Scholar]
- Collins A. L. et al. Investigation of autism and GABA receptor subunit genes in multiple ethnic groups. Neurogenetics 7, 167–174 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D. Q. et al. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am. J. Hum. Genet. 77, 377–388 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami N. et al. Identification of rare recurrent copy number variants in high-risk autism families and their prevalence in a large ASD population. PLoS ONE 8, e52239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459, 528–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J. T. et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc. Natl Acad. Sci. USA 107, 10584–10589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J. et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat. Genet. 44, 78–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y. et al. Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatr. Genet. 13, 71–76 (2003). [DOI] [PubMed] [Google Scholar]
- Li Z.-J. et al. The association between glutamate receptor gene SNP and schizophrenia. Fa Yi Xue Za Zhi 24, 369–374 377 (2008). [PubMed] [Google Scholar]
- Shibata H. et al. Association study of polymorphisms in the group III metabotropic glutamate receptor genes, GRM4 and GRM7, with schizophrenia. Psychiatr. Res. 167, 88–96 (2009). [DOI] [PubMed] [Google Scholar]
- Ohtsuki T. et al. A polymorphism of the metabotropic glutamate receptor mGluR7 (GRM7) gene is associated with schizophrenia. Schizophr. Res. 101, 9–16 (2008). [DOI] [PubMed] [Google Scholar]
- Bolonna A. A., Kerwin R. W., Munro J., Arranz M. J. & Makoff A. J. Polymorphisms in the genes for mGluR types 7 and 8: association studies with schizophrenia. Schizophr. Res. 47, 99–103 (2001). [DOI] [PubMed] [Google Scholar]
- Takaki H. et al. Positive associations of polymorphisms in the metabotropic glutamate receptor type 8 gene (GRM8) with schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 128B, 6–14 (2004). [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca D. et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol. Psychiatry 18, 1090–1095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman S. R. et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70, 898–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y. et al. Protein interactome reveals converging molecular pathways among autism disorders. Sci. Transl Med. 3, 86ra49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh H. J. et al. Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism. PLoS. Genet. 9, e1003523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L. & Groom C. R. The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 (2002). [DOI] [PubMed] [Google Scholar]
- Hadley D. et al. Patterns of sequence conservation in presynaptic neural genes. Genome Biol. 7, R105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi H. Y. Postnatal neurodevelopmental disorders: meeting at the synapse? Science 302, 826–830 (2003). [DOI] [PubMed] [Google Scholar]
- Anney R. et al. A genomewide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 19, 4072–4082 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann H. M. et al. A human genome diversity cell line panel. Science 296, 261–262 (2002). [DOI] [PubMed] [Google Scholar]
- Van der Zwaag B. et al. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE 4, e5324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGO Gene Nomenclature Committee Home Page|HUGO Gene Nomenclature Committee (n.d.). Available http://www.genenames.org/ Accessed 18 February (2013).
- Venkatesan K. et al. An empirical framework for binary interactome mapping. Nat. Methods 6, 83–90 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.-F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005). [DOI] [PubMed] [Google Scholar]
- Hermanson O., Jepsen K. & Rosenfeld M. G. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419, 934–939 (2002). [DOI] [PubMed] [Google Scholar]
- Li H. et al. The association analysis of RELN and GRM8 genes with autistic spectrum disorder in Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 194–200 (2008). [DOI] [PubMed] [Google Scholar]
- Serajee F. J., Zhong H., Nabi R. & Huq AHMM The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J. Med. Genet. 40, e42 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó I. et al. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum. Mol. Genet. 18, 1795–1804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach B. D., Osterweil E. K. & Bear M. F. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480, 63–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J. et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 15, 637–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMIM - Online Mendelian Inheritance in Man (n.d.). Available http://omim.org/ Accessed 18 February (2013).
- Nair S. K. & Burley S. K. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205 (2003). [DOI] [PubMed] [Google Scholar]
- Kao H.-T., Buka S. L., Kelsey K. T., Gruber D. F. & Porton B. The correlation between rates of cancer and autism: an exploratory ecological investigation. PLoS ONE 5, e9372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas C. G. et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno R. et al. HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucleic Acids Res. 32, D502–D504 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E., Tsvetkov P. & Ginzburg I. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of Tau protein. J. Biol. Chem. 282, 37276–37284 (2007). [DOI] [PubMed] [Google Scholar]
- Elliott E., Laufer O. & Ginzburg I. BAG-1M is up-regulated in hippocampus of Alzheimer’s disease patients and associates with tau and APP proteins. J. Neurochem. 109, 1168–1178 (2009). [DOI] [PubMed] [Google Scholar]
- McMahon F. J. & Insel T. R. Pharmacogenomics and personalized medicine in neuropsychiatry. Neuron 74, 773–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (n.d.) R: A language and environment for statistical computing. Available http://www.r-project.org/ Accessed 18 February (2013).
- Wang K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Interactome Database (n.d.). Available http://interactome.dfci.harvard.edu/H_sapiens/ Accessed 18 February (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1-3
Experimental PCR validation of CNV predictions.