Abstract
Efficient duplication of the genome requires the concerted action of helicase and DNA polymerases at replication forks1, to avoid stalling of the replication machinery and consequent genomic instability2-4. In eukaryotes, the physical coupling between helicase and DNA polymerases remains poorly understood. Here we define the molecular mechanism by which the yeast Ctf4 protein links the Cdc45-MCM-GINS (CMG) DNA helicase to DNA polymerase α (Pol α) within the replisome. We use X-ray crystallography and electron microscopy to show that Ctf4 self-associates in a constitutive disk-shaped trimer. Trimerization depends on a β-propeller domain in the carboxy-terminal half of the protein, which is fused to a helical extension that protrudes from one face of the trimeric disk. Critically, Pol α and the CMG helicase share a common mechanism of interaction with Ctf4. We show that the N-terminal tails of the catalytic subunit of Pol α and the Sld5 subunit of GINS contain a conserved Ctf4-binding motif that docks onto the exposed helical extension of a Ctf4 protomer within the trimer. Accordingly, we demonstrate that one Ctf4 trimer can support binding of up to three partner proteins, including the simultaneous association with both Pol α and GINS. Our findings indicate that Ctf4 can couple two molecules of Pol α to one CMG helicase within the replisome, providing a new paradigm for lagging-strand synthesis in eukaryotes that resembles the emerging model for the simpler replisome of E. coli5-8. The ability of Ctf4 to act as a platform for multivalent interactions illustrates a mechanism for the concurrent recruitment of factors that act together at the fork.
Recent evidence indicates that the leading- and lagging-strand polymerases are anchored to the helicase by replisome components that lack counterparts in bacteria9-13. The yeast Ctf4 protein is among the best characterized of these factors: its role is to link the CMG helicase with Pol α, the polymerase subunit of the Pol α/primase complex that initiaties Okazaki fragments during lagging-strand synthesis9,10. Ctf4 is part of a conserved family of replication factors that includes human And1 and fission yeast Mcl1, and is required for efficient DNA synthesis, normal cell cycle progression and genomic stability13-18. In addition to their role in DNA replication, Ctf4 and And1 perform an important yet poorly understood function in sister chromatid cohesion19-22.
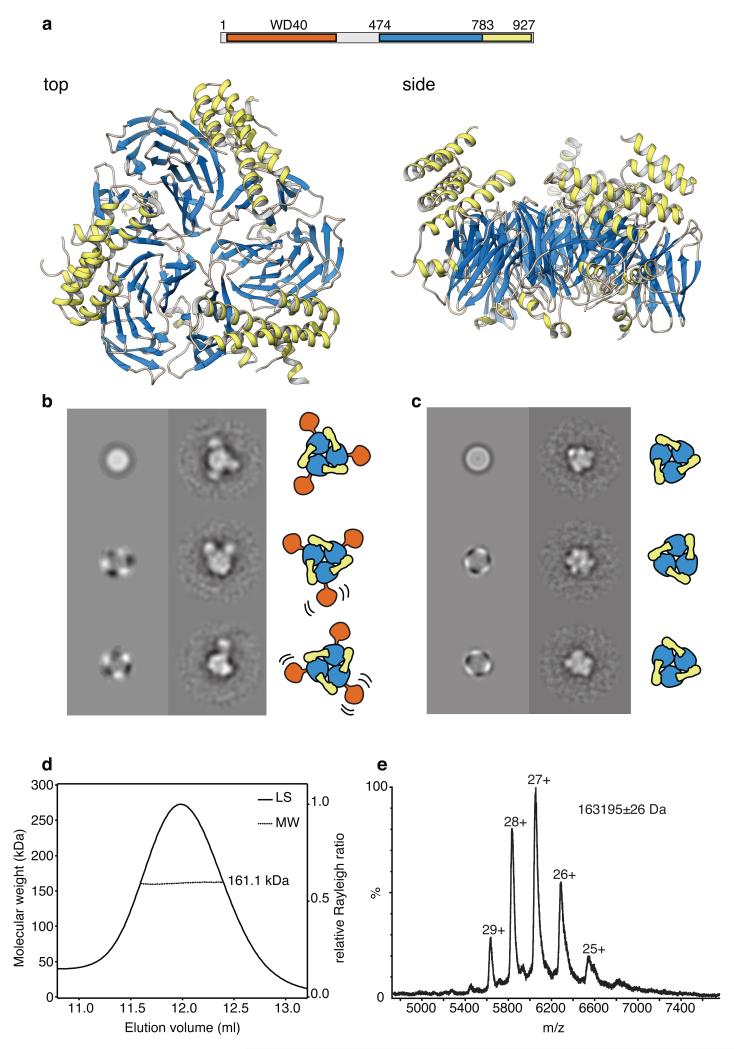
Earlier work had shown that Ctf4 binds directly to the GINS subunit of the CMG helicase and to the catalytic subunit of Pol α, via the carboxy-terminal half of the protein that does not include an annotated WD40 domain in the N-terminus of Ctf410. We identified by bioinformatic analysis a second WD40 domain in the C-terminal half of yeast Ctf4, juxtaposed to a predicted helical region. Crystallographic analysis of residues 471 to 927 (C-end; 457 amino acids) of yeast Ctf4 (Ctf4CTD) confirmed the presence of a six-bladed β-propeller domain fused to a helical bundle of six α-helices arranged in a stack of helical hairpins (Extended Data Table 1 and Extended Data Fig. 1). Importantly, the structural analysis revealed a trimeric assembly of Ctf4 molecules, resulting from side-on packing of β-propeller domains (Fig. 1a). The homotypic association of the β-propeller domains generates a discoidal shape with three-fold symmetry. The helical domains of each Ctf4 protomer extend upwards and away from the plane of the trimer, like legs of a three-legged stool. The trimeric assembly appears constitutive as it buries a total surface area of 8100 Å2, an average of 2700 Å2 per interface. The existence of Ctf4 as a constitutive trimer mediated by self-association of its C-terminal domain was confirmed by single-particle EM analysis, which showed the presence of three-fold symmetry in particles of full-length Ctf4 and Ctf4CTD (Fig. 1b, c and Extended Data Fig. 2). The EM analysis of full-length Ctf4 further revealed that the N-terminal WD40 domains depart radially from the Ctf4CTD trimer, to which they are loosely connected (Fig. 1b). The presence of Ctf4 as a stable trimer in solution was demonstrated by multi-angle laser scattering (MALS) of Ctf4CTD and full-length Ctf4 (Fig. 1d and Extended Data Fig. 2) and non-denaturing nano-electrospray ionization mass spectrometry (native mass spectrometry) of Ctf4CTD (Fig. 1e).
Figure 1. Architecture of yeast Ctf4.
a, Ctf4 self-associates in a trimer of novel design. The panel shows top and side views of the crystal structure of the C-terminal region of yeast Ctf4 (Ctf4CTD; amino acids 471 to 927). The protein is drawn as ribbon, coloured according to its domain structure: the β-propeller domain is in light blue and the helical domain in yellow. Above the drawing, a bar diagram shows the domain structure of full-length yeast Ctf4 and the extent of the region crystallized in our study. b, Analysis of full-length Ctf4 by single-particle electron microscopy. Multivariate statistical symmetry analysis detects a threefold symmetry component for the full-length Ctf4 particle. Reference-free class averages of full-length Ctf4 reveal a core structure flexibly linked to up to three satellite domains. c, Analysis of Ctf4CTD by single-particle electron microscopy. The C-terminal domain of Ctf4 maintains a trimeric structure, as shown by multivariate statistical symmetry analysis and reference-free class averages. d, Size exclusion chromatography - multi-angle laser scattering analysis of yeast Ctf4CTD. The light scattering is plotted alongside the fitted molecular weights. The protein eluted in a single peak, corresponding to a measured molecular weight of 161.1 kDa. The predicted molecular weight for the trimeric species is 163.1 kDa. e, Native mass-spectrometry analysis of yeast Ctf4CTD. The measured molecular weight of 163195 Da matches closely the predicted molecular weight of 163148 Da for a trimeric species.
We had previously shown that Ctf4 binds to the amino-terminal portion of Pol1, the yeast orthologue of Pol α10. By progressive truncations of this largely unstructured region of Pol1, we identified a short linear motif spanning residues 137 to 149 that is necessary and sufficient for the association with Ctf4 in vitro. The motif has a mixed acidic and hydrophobic nature and is conserved from yeast to humans (Fig. 2b). Alanine scanning mutagenesis of the motif revealed that conserved residues F140, D142, I143, L144 and F147 are essential for the interaction with Ctf4 (Fig. 2c). The results of the biochemical experiments with recombinant proteins were confirmed by immunoprecipitation of Pol1 from extracts of yeast cells that were synchronized in G1-phase before release into S-phase. Whilst wild-type Pol1 associated with Ctf4 and thus with the components of the Cdc45-MCM-GINS complex (Fig. 2d, control), the Pol1-A allele with alanine substitutions at D141, D142, L144 and F147 was unable to interact with either Ctf4 or the CMG (Fig. 2d, pol1-A).
Figure 2. Pol α contains a Ctf4-interacting motif that binds to the helical domain of Ctf4.
a, Identification of the Ctf4-binding motif of Pol1, the yeast orthologue of Pol α. GST-tagged constructs spanning progressively smaller N-terminal regions of Pol1 were tested for interaction with Ctf4CTD in pull-down experiments on glutathione sepharose beads. The top panel shows the boundaries of the GST-Pol1 constructs; the bottom panel shows the result of the pull-down experiments, analysed by SDS-PAGE. The last lane on the right-hand side of the gel contains only Ctf4CTD. The position of the Ctf4CTD band in the pull-down experiments is highlighted by a box. The asterisk marks the position of GST-Pol1121-348, which overlaps partially with Ctf4CTD. b, Multiple sequence alignment of the Ctf4-binding motif of yeast Pol1 (Sc; S. cerevisiae) with Pol α sequences from S. pombe (Sp), D. rerio (Dr), D. melanogaster (Dm) and H. sapiens (Hs). Invariant residues are highlighted in green, identical residues in yellow and similar residues in cyan. The asterisk marks amino acids that are essential for interaction with Ctf4CTD (see panel c). c, Alanine-scanning mutagenesis of the Ctf4-binding motif. Pol1 residues 137 to 149 were fused to GST and each amino acid between 140 and 149 (except G145) was mutated to alanine. The effect of each single-point mutation on the interaction with Ctf4CTD was tested by GST pull-down and analysed by SDS-PAGE. d, The budding yeast strains POL1-9MYC (Control) and pol1-A-9MYC (pol1-A, containing the D141A, D142A, L144A and F147A mutations in the endogenous POL1 locus) were grown at 24°C, arrested in G1-phase and released into S-phase for 30 minutes. The MYC-tagged proteins were isolated from cell extracts by immunoprecipitation on anti-MYC beads and the indicated proteins were detected by immunoblotting with the corresponding antibodies23. e, Co-crystal structure of Ctf4CTD bound to a peptide corresponding to the Ctf4-binding motif of Pol α. Ctf4 is drawn as in Fig. 1a, the Ctf4-binding motif of Pol α is drawn as green ribbon. f, Detailed view of the interaction between the Ctf4-binding motif of Pol α (green tube) and the helical domain of Ctf4 (yellow ribbon). The side chains of Pol1 residues F140, D142, I143, L144, F147 and Ctf4 residue R904 are shown as sticks.
To define the structural basis for the interaction between Ctf4 and Pol α, we soaked a 13 amino-acid peptide corresponding to Pol1 sequence 137-IDNFDDILGEFES-149 in the Ctf4CTD crystals. For the soaking experiments, we used a different crystal form of the Ctf4CTD trimer that is easier to grow; this form captures a topologically open conformation of the Ctf4CTD trimer resembling a cracked ring (Extended Data Fig. 3, Video S1). No differences are observed in protomer structure between closed and open forms of the Ctf4 trimer, with the exception of the helical domain located at the gap in the open form that becomes disordered in the electron density map. The potential functional significance of the open and closed forms of the Ctf4 trimer and their interconversion in solution is presently unclear.
The crystal structure of the Ctf4CTD - Pol α complex shows that the helical domain protruding from the discoidal trimer is responsible for binding the polymerase (Fig. 2e). In the structure, amino acids 140-FDDILGEFES-149 of Pol1 fold into a two-turn α-helix that packs in antiparallel fashion against helices α3 and α5, on the outward-facing side of the helical domain of one Ctf4 protomer. The Ctf4-binding motif of Pol1 occupies each of the two binding sites available in this crystal form of the Ctf4CTD trimer. The interaction with the Pol1 peptide does not induce an appreciable conformational change in the helical domain of Ctf4CTD nor does it alter its position in the trimeric structure. The side chains of Pol1 residues F140, I143, L144 and F147, which were critical for the interaction in the pull-down assay, become buried at the interface and pack against a continuous hydrophobic surface formed by Ctf4 residues L867, A871, A894, A897, I901 (Fig. 2f). The interaction is augmented by polar contacts on the perimeter of the hydrophobic interface, between acidic residues D141, D142, E146 and E148 of Pol1 and basic residues K864, R868, R893, K900 and R904 of Ctf4 (Extended Data Figs. 4a and 5). The salt bridge between the conserved residues D142 of Pol1 and R904 in helix α5 of Ctf4 appears to be particularly important for binding, as the D142A mutation abolishes the interaction (Fig. 2c).
We next set out to investigate the mode of Ctf4 interaction with the CMG helicase. The association between Ctf4 and GINS is sufficiently strong to be assayed by size-exclusion chromatography10 (Fig. 3a). We found that the interaction is dependent upon the unstructured tail at the N-terminus of the Sld5 subunit of GINS (Fig. 3a). Sequence comparison of fungal Sld5 orthologues revealed a conserved pattern of amino acids that is highly similar to the Ctf4-binding motif of Pol1 (Fig. 3b). Further dissection of the amino-terminal tail of Sld5 confirmed that the initial 18 residues, containing the Ctf4-binding motif, were essential for the interaction (Fig. 3c). Our identification of a Ctf4-binding site in Sld5 is in agreement with earlier reports9,10, which had identified Sld5 as a Ctf4-binding subunit of GINS. Whether a conserved Ctf4-binding motif is present in the Sld5 sequence of higher eukaryotes is presently unclear. Alanine scanning mutagenesis of the Ctf4-binding motif of Sld5 highlighted the importance of the same pattern of conserved hydrophobic residues that were essential for Pol1 binding to Ctf4. Alanine mutation of I5, I8 and L9, corresponding to residues F140, I143 and L144 of Pol1, abolished the association of Sld5 with Ctf4, whereas mutation of L12, equivalent to Pol1 F147, weakened the interaction (Fig. 3d).
Figure 3. The Sld5 subunit of yeast GINS shares a common mechanism of Ctf4 binding with Pol α.
a, Analysis of the Ctf4 - GINS interaction by gel filtration chromatography, using Ctf4CTD and versions of GINS that contain either full-length (top panel) or N-terminally truncated Sld5 (Sld5ΔN; bottom panel). b, Multiple sequence alignment of the N-terminus of fungal Sld5 sequences (Sc, Saccharomyces cerevisiae; Ag, Ashbya gossypii; An, Aspergillus niger; Gz, Gibberella zeae; Ca, Candida albicans). Invariant residues are highlighted in green, identical residues in yellow and similar residues in cyan. The Ctf4-binding motif of yeast Pol1 is reported below the alignment. c, Mapping of the Ctf4-binding sequence in the N-terminus of Sld5 by GST-pull down analysis. The top panel shows the boundaries of the GSTSld5 constructs tested for interaction with Ctf4CTD; the bottom panel shows the results of the pull-down experiments, analysed by SDS-PAGE. The band marked with an asterisk corresponds to free GST. d, Alanine-scanning mutagenesis of the Ctf4-binding motif. Residues 1 to 20 of yeast Sld5 were fused to GST and each position between 3 and 13 (except A10) was mutated to alanine. The effect of each single-point mutation on the interaction with Ctf4CTD was tested by GST pull-down and analysed by SDS-PAGE. e, Co-crystal structure of Ctf4CTD bound to a peptide corresponding to the Ctf4-binding motif of Sld5. Ctf4 is drawn as in Fig. 1a, the Ctf4-binding motif of Sld5 is drawn as red ribbon. f, Detailed view of the interaction between the Ctf4-binding motif of Sld5 (red tube) and the helical domain of Ctf4 (yellow ribbon). The side chains of Sld5 residues I3, I5, D7, I8, L9, L12 and Ctf4 residue R904 are shown as sticks.
To ascertain whether the similarity in the two Ctf4-binding motifs extended to their mechanism of interaction, we soaked the Ctf4CTD crystals with the peptide MDINIDDILAELDKETTAV, corresponding to amino acids 1 to 19 of yeast Sld5. The crystallographic analysis revealed a near-identical mode of interaction between Sld5 and Ctf4 as previously observed for the Ctf4-binding motif of Pol1 (Figs. 3e and 4a and Extended Data Figs. 4b and 5). The Ctf4-binding sequence of Sld5 includes an additional hydrophobic contact of I3 with Ctf4 residues A871, C874 and I901, which might account for its tighter (~5-fold) association with Ctf4 compared to Pol1 (Fig. 3f and Extended Data Fig. 6a). Conversely, the contribution of polar contacts appears diminished, as disruption of the salt bridge between D7 in Sld5 and R904 in Ctf4 does not impair appreciably the interaction (Fig. 3d, f). Collectively, these findings establish that Pol1 and GINS contain a Ctf4-binding motif that is conserved in sequence and function.
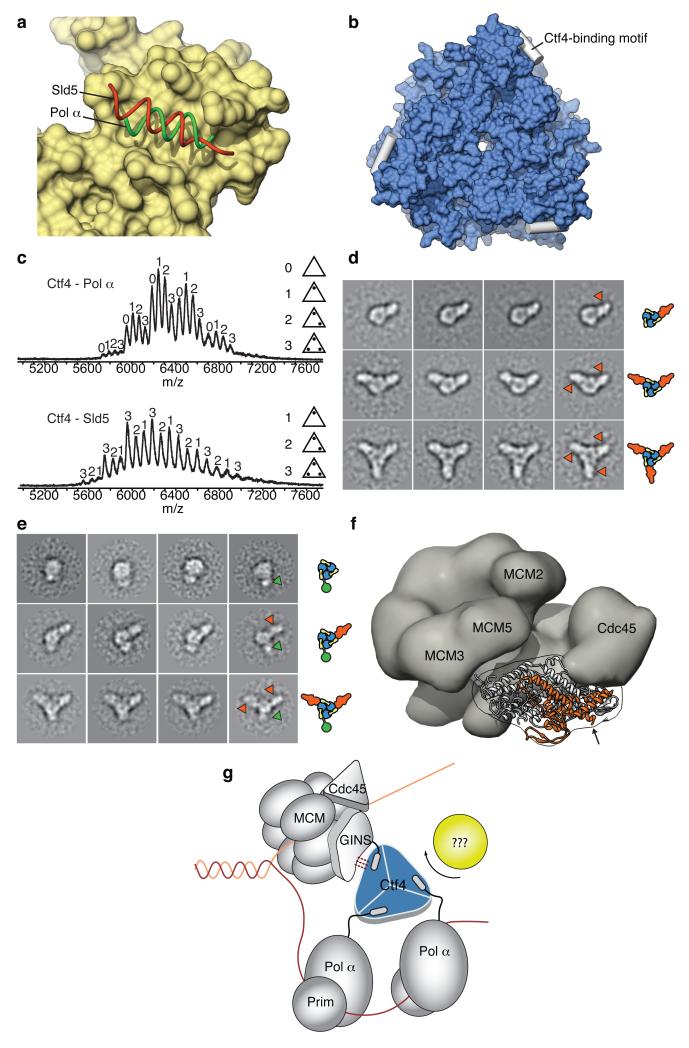
Figure 4. The Ctf4 trimer coordinates the recruitment of replication factors to the fork.
a, Superposition of the structures of Ctf4CTD bound to the Ctf4-binding motif of Pol α (green tube) and Sld5 (red tube). Ctf4CTD is displayed as molecular surface, in light brown. b, Ctf4CTD can associate in principle with up to three partner proteins. To illustrate this point, the Ctf4-binding motif of Pol α was modelled in each of the three binding sites of Ctf4CTD. The helical Ctf4-binding motif is shown as a white cylinder, and Ctf4CTD is drawn as a molecular surface, in light blue. c, Native mass spectrometry analysis of the Ctf4CTD trimer in the presence of peptides corresponding to the Ctf4-binding motifs of Pol α (top) and Sld5 (bottom). d, Single-particle electron microscopy analysis of the interaction of GINS with the Ctf4CTD trimer. Reference-free class averages of Ctf4CTD bound to one (top row), two (middle row) or three copies (bottom row) of GINS are shown. e, Reference-free class averages of the Ctf4CTD - Pol1NTD (top row), Ctf4CTD - Pol1NTD - GINS (middle row) and Ctf4CTD - Pol1NTD - (GINS)2 (bottom row) heteroassemblies. f, The panel shows the crystal structure of human GINS (ref. 25) docked into the electron microscopy reconstruction of the CMG helicase (ref. 24). The Sld5 subunit of GINS is coloured orange and the rest of GINS is shown in white. The density for the MCM and Cdc45 subunits of the CMG helicase is shown as a semi-transparent grey surface, whereas the density of the GINS tetramer is shown as an outline. The position of MCM2, MCM3, MCM5 and Cdc45, which surround GINS in the helicase complex, is indicated. An arrow marks the amino-terminal residue in the Sld5 structure. g, A model of Ctf4 function at the replication fork, as the physical bridge between the CMG helicase and the DNA polymerase α/primase complex. The additional contacts between Ctf4 and GINS suggested by the EM analysis (panel d) are indicated by dashed lines.
In yeast cells, Ctf4 appears to associate more tightly with the CMG helicase than with Pol α, since the association of Ctf4 with CMG resists buffers containing 700 mM salt, whereas the Ctf4-dependent association of Pol α with the replisome is lost at 300 mM salt23. Consistent with this, Ctf4 remains associated with CMG in cells containing mutations that disrupt the Ctf4-binding site of Sld5 (Extended Data Fig. 6b). It thus seems likely that Ctf4 has a more complex interaction with the CMG than with Pol α and that additional contacts between Ctf4 and CMG remain to be characterized.
Our findings predict that Ctf4 can support simultaneous interactions of varying stoichiometry and with multiple partners (Fig. 4b). Indeed, native mass-spectrometry analysis of Ctf4CTD in the presence of the Ctf4-binding sequences of Pol α and Sld5 showed reconstitution of complexes with 1:1, 1:2 and 1:3 Ctf4-to-peptide stoichiometries (Fig. 4c). To determine whether a Ctf4 trimer could support concomitant binding of three partner molecules, we analysed by EM reconstituted Ctf4CTD - GINS complexes (Extended Data Fig. 7). In agreement with the mass-spectrometry data, the EM analysis demonstrated the presence of 1, 2 or 3 copies of GINS bound to one Ctf4CTD trimer (Fig. 4d, Video S2), each arranged radially around the Ctf4CTD trimer. Interestingly, each GINS molecule occupies a fixed position relative to the Ctf4 trimer, indicating that the interface between Ctf4 and GINS extends beyond the contact provided by the flexible N-terminal tail of Sld5. The Ctf4 - GINS interface discernible in our EM averages is likely to be important to sustain the association between Ctf4 and the CMG helicase in the replisome.
The reported function of Ctf4 as a physical link between helicase and polymerase prompted us to determine whether GINS and Pol α can simultaneously associate with the Ctf4 trimer. We visualised by EM reconstituted hetero-assemblies of Ctf4CTD bound concurrently to GINS and the N-terminal region of Pol1 (residues 1 to 351) fused to Protein A (Pol1NTD) (Extended Data Figs. 8 and 9). As predicted by the trimeric nature of Ctf4, we could detect Ctf4CTD - GINS - Pol1NTD complexes of varied stoichiometries, with partial or full occupancy of the Ctf4 trimer (Fig. 4e). These data establish a structural basis for Ctf4 as the bridging factor between the CMG helicase and DNA polymerase α in eukaryotic replication.
Inside the cell, the appropriate stoichiometry will presumably be determined by the constraints imposed upon replisome assembly during replication initiation. Within the replisome, one binding site of the Ctf4 trimer is likely to engage in a constitutive interaction with GINS, to anchor Ctf4 to the CMG helicase at the fork. In the molecular model of the CMG24, the GINS structure25 has the Sld5 N-terminus favorably positioned for binding Ctf4, in agreement with our biochemical findings (Fig. 4f). As replisomes formed at a replication origin need not remain physically tethered for efficient replication26, it is unlikely that Ctf4 acts by coupling two CMG helicases. The other two binding sites of the CMG-bound Ctf4 trimer would remain available for interaction with Pol α, indicating that two copies of the Pol α/primase complex might work together during lagging-strand synthesis (Fig. 4g). Such coupling of helicase and polymerase in the eukaryotic replisome would be functionally analogous to the emerging model of the E. coli replisome, where two DNA polymerases cooperate in lagging-strand synthesis to increase processivity and efficiency of nucleotide polymerisation5-7.
In addition to its function as a helicase-polymerase bridge, Ctf4 appears ideally suited to fulfill a wider role in replication, as a platform for coordinating the activity of replication factors at the fork. In this model, one Ctf4 protomer would keep the trimer constitutively anchored to the CMG, whereas other replisome components, including Pol α, would engage with the helicase in a dynamic interaction mediated by the Ctf4-binding motif identified here. We note that this model of Ctf4 function is reminiscent of the way the Proliferating Cell Nuclear Antigen (PCNA) interacts with replication factors such as Fen1 and DNA Ligase I27. Thus, in addition to bridging CMG helicase and Pol α, Ctf4 might recruit to the fork other factors required for efficient replication under normal conditions or needed to deal with exceptional situations during replicative stress.
METHODS
DNA constructs for X-ray crystallography, MALS and MS of Ctf4CTD and biochemical analysis of the Ctf4CTD - Pol1 and Ctf4CTD - Sld5 interactions
Fold recognition analysis in Phyre228 predicted that the C-terminal half of yeast Ctf4, responsible for interactions with GINS and Pol α, contained a WD40 domain fused to an α-helical region. A region of yeast Ctf4 comprising amino acids (aa) 471-927 (natural C-end; Ctf4CTD) was PCR amplified from S. cerevisiae genomic DNA and cloned into a bacterial pRSFDuet-1 T7 expression plasmid (Novagen) via unique BamHI and AvrII sites. Using PCR primer extension, a TEV protease site was introduced at the start of the Ctf4CTD open reading frame sequence and after the N-terminal His6-affinity tag encoded by the pRSFDuet-1 vector.
The DNA Polymerase α (Pol α)- and Sld5-GST fusion constructs used in pull-down experiments were generated by insertion of the appropriate nucleotide sequence into the NcoI and XhoI sites of the pGAT2 T7 expression plasmid encoding a thrombin-cleavable N-terminal GST fusion protein29.
A construct for bacterial expression of yeast GINS was prepared starting from vector pKL65310, by subcloning one expression cassette comprising psf3 and psf1ΔC (aa 1-164) into the NcoI and NotI sites in the first MCS of a pRSFDuet-1 expression plasmid, and another expression cassette comprising psf2 with an N-terminal His6 affinity tag and sld5 into the second MCS of pRSFDuet-1, resulting in the polycistronic pGINS-Duet-1 expression plasmid. The GINSSld5ΔN construct used for analytical gel filtration experiments was derived from the pGINS-Duet-1 vector, by replacing the second expression cassette with a modified cassette that encodes, in addition to His6-psf2, a version of sld5 coding for a truncated protein lacking the first 48 aa at its amino-terminus.
DNA constructs for electron microscopy and MALS of full-length Ctf4
Full-length S. cerevisiae Ctf4 and Ctf4 N-terminal deletion (Ctf4CTD, aa 461-927) constructs were both cloned into the pET28c vector (Novagen) to express a N-terminal His6 affinity tag. The S. cerevisiae GINS Psf1 C-terminal deletion (ΔCT, aa 1-164) construct was subcloned from a previously described GINS operon-containing plasmid10 into the pET28c vector and carries a N-terminal Strep III tag in the GINS Psf3 subunit. The Pol1-protein A fusion was subcloned into the pET Strep II-TEV LIC vector (QB3 MacroLab) by ligation independent cloning30. This construct contains in the following order: a N-terminal Strep II tag, the N-terminal domain (aa 1-351) of S. cerevisiae Pol1, the protein A region of the TAP tag31 and a C-terminal His7 affinity tag.
Protein expression and purification for X-ray crystallography, MALS and MS of the Ctf4CTD and biochemical analysis of the Ctf4CTD - Pol1 and Ctf4CTD - Sld5 interactions
Ctf4CTD was over-expressed in E. coli strain BL21(DE3)Rosetta2 with IPTG induction and overnight expression at 20°C in LB medium. After over-expression, 4 liters of cells were harvested and resuspended in 50 mM Tris pH 7.0, 500 mM NaCl, 10% (w/v) glycerol, 1 mM DTT and protease inhibitors (Sigma). Cells were lysed via sonication, the crude extract was clarified by centrifugation and the supernatant was applied to a 4 ml-column of nickel agarose resin (Sigma) using gravity flow. The column with bound Ctf4CTD was washed in buffer supplemented with 20 mM imidazole and Ctf4CTD elution was performed with buffer supplemented with 200 mM imidazole. Eluted Ctf4CTD was further purified by gel filtration chromatography over a Superdex 200 16/60 HiLoad column (GE Healthcare) in 25 mM HEPES pH 7.0, 200 mM NaCl and 10% (w/v) glycerol and peak fractions were pooled, concentrated to 10 mg/ml, flash frozen in liquid nitrogen and stored in small aliquots at −80°C. Selenomethionine labelling of Ctf4CTD was achieved by metabolic inhibition of the methionine pathway32 and overnight expression as for the wild-type protein. The selenomethionine-labelled protein was purified in the same way as the native Ctf4CTD except that all buffers were supplemented with 10 mM DTT.
GINS constructs were over-expressed in E. coli strain BL21(DE3)Rosetta2 with IPTG induction and overnight expression at 25°C in LB medium. After over-expression, 8 liters of cells were harvested and resuspended in 50 mM Tris pH 7.0, 500 mM NaCl, 10% (w/v) glycerol, 1 mM DTT and protease inhibitor. Cells were lysed via sonication, the crude extract was clarified by centrifugation and the supernatant was applied to a 3 ml-column of nickel agarose resin (Sigma) using gravity flow. GINS bound to beads was eluted with buffer supplemented with 10 mM imidazole. The salt concentration of the eluted GINS sample was adjusted to below 160 mM NaCl and the protein was applied to an ion-exchange 6 ml-Resource Q column pre-equilibrated in 20 mM HEPES pH 8.0, 160 mM NaCl and eluted with a buffer gradient of 0.16 M to 0.5 M NaCl over 40 column volumes. Peak fractions containing GINS were pooled and further purified by gel filtration over a Superdex 200 16/60 column as described for Ctf4CTD. Purified GINS samples were flash frozen in liquid nitrogen and stored in small aliquots at −80°C.
Protein expression and purification for electron microscopy and MALS
Each expression construct was transformed into BL21 (DE3)-CodonPlus cells (Stratagene) and 2 to 4 liters of cells were grown to an optical density of 0.5 before induction with 1 mM IPTG, at 37°C for 2 hours. Each 2 L cell pellet was resuspended in 40 ml lysis buffer and cells were lysed via sonication. The resulting lysate was subject to the following purification steps.
Ctf4 purification
Cleared lysate containing His-tagged full-length Ctf4 or Ctf4CTD was incubated with 1 ml Ni-NTA resin (QIAGEN), washed with 20 ml Buffer A (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 20 mM imidazole) and eluted five times each with 1 ml Buffer A containing 250 mM imidazole. The resulting elution was dialysed in 2 L 100 mM NaCl, 20 mM Tris pH 8.0, 1 mM DTT for 2 hours with fresh buffer exchanged after the first hour. The dialysed elution was further purified by Mono Q (GE Healthcare) ion exchange through a 0.1-1 M NaCl gradient in 20 mM Tris pH 8.0, 1 mM DTT over 40 ml with 0.5 ml elutions. Peak fractions from the Mono Q were concentrated and polished via Superdex 200 16/600 HiLoad or 10/300 GL (GE Healthcare) size exclusion chromatography in Buffer B (150 mM NaCl, 20 mM Tris pH 8.0). Peak elutions were pooled and concentrated to 5 mg/ml and stored at −80°C in 2 nmol aliquots.
GINS purifications
Cleared lysate containing Strep III tagged GINSPsf1ΔC was incubated with 1 ml StrepTactin resin (IBA Life Sciences), washed with 20 ml Buffer C (150 mM NaCl, 100 mM Tris pH 8.0) and eluted five times each with 1 ml Buffer C supplemented with 2.5 mM desthiobiotin (IBA Life Sciences). Proteins were stored at −80°C with a concentration of 1 mg/ml in 2 nmol aliquots.
Pol1 purification
Cleared lysate containing N-terminal Strep II- and C-terminal His-tagged Pol1-NTD-ProteinA (hereafter referred to as Pol1NTD) were purified first via Ni-NTA, followed by StrepTactin affinity using the same method described above.
The typical yield from 2 L cells for full-length Ctf4, Ctf4CTD, GINSPsf1ΔC and Pol1NTD are around 0.15 mg, 0.65 mg, 2.5 mg and 1.5 mg respectively. The identity of all proteins was confirmed by trypsinization/mass spectrometry using a LTQ OrbitrapXL instrument (Protein Analysis and Proteomics, LRI).
In vitro reconstitution of recombinant protein complexes for electron microscopy
For the Ctf4 - GINS complex, 2 nmol of recombinant yeast Ctf4 (full-length or CTD) and GINSPsf1ΔC were co-incubated in 500 mM sodium acetate for 10 minutes on ice with a reaction volume of around 200 μl. To achieve high reproducibility the following procedure was followed. The reconstitution mix was initially dialysed in 500 mM sodium acetate, 25 mM Hepes pH 7.6, 0.5 mM DTT for 1 hour at 4°C in dialysis tubes with 6,000-8,000 Da MWCO (GeBAflex). The dialysis buffer was changed hourly to contain progressively 400 mM, 300 mM, 200 mM, 150 mM sodium acetate. 100-150 μl of the final reconstituted complex was separated via glycerol gradient sedimentation. For the Ctf4CTD - Pol1NTD complex and the Ctf4CTD - GINSPsf1ΔC - Pol1NTD complex 2 nmol of recombinant Ctf4CTD was used, 1:2 and 1:1:1 mole ratios were applied respectively and the dialysis was performed in buffers containing progressively 400 mM, 300 mM, 200 mM, 100 mM, 50 mM sodium acetate.
Glycerol gradient sedimentation with GraFix
Glycerol gradient sedimentation of full-length Ctf4, Ctf4CTD and complexes of Ctf4CTD with GINSPsf1ΔC and Pol1NTD was performed based on the GraFix method33. Briefly, 5ml 10% to 30% or 15% to 35% glycerol gradients were poured either with or without 0% to 0.1% glutaraldehyde gradient. The protein or reconstituted protein complex was loaded on top of the gradient and centrifuged at 50,000 rpm, 4°C in a SW 55 Ti ultracentrifuge rotor (Beckman Coulter) for 16 hours. Fractions were collected manually from the top of the gradient, resolved through a 4%-to-12% polyacrylamide-gradient gel (Biorad) in MOPS buffer at room temperature and silver stained for analysis.
Crystallisation and structure determination of Ctf4CTD
Ctf4CTD crystals were grown by vapour diffusion in hanging drop, mixing equal volumes of Ctf4CTD protein at 10 mg/ml and 0.2 M tri-sodium citrate pH 6.2, 7-9% PEG 8000 and 0.45-0.9 M NaCl at 19°C. Ctf4CTD crystals appeared within 2-3 days and grew to full size over the course of two weeks. For structure determination, selenomethionine-labelled Ctf4CTD crystals were grown against 0.2 M tri-sodium citrate pH 6.2 and 8-10% PEG 8000 at 19°C, using the same protein concentration and drop ratio as for the native protein.
X-ray diffraction data for selenomethionine-labelled Ctf4CTD crystals were collected at the peak wavelength of the selenium K-edge (λ=0.97938 nm) at beamline I03 of the Diamond Light Source, Oxford, UK. The data were integrated with XDS34, space group symmetry was assigned in POINTLESS and intensities scaled in AIMLESS35. The selenomethionine protein crystallised in the orthorhombic space group P22121 with unit cell dimensions of a=107.1 Å, b=118.1 Å, c=155.7 Å and one Ctf4CTD trimer per asymmetric unit. The position of the selenium atoms was determined using the single-wavelength anomalous dispersion (SAD) method in PHENIX Autosol, an interpretable electron density map was calculated to a resolution of 2.7 Å and an initial model was generated using the PHENIX AutoBuild function36. The crystallographic model was extended and completed by repeated cycles of manual building in Coot and crystallographic refinement with PHENIX Refine36,37. The final model was refined using data to 2.7 Å, to R-work and R-free values of 0.1895 and 0.2284 and a Molprobity score of 1.1538. The following amino acids were not included in the final model due to missing or poor electron density and are presumed to be disordered: 471 to 473, 644 to 647, 797 to 813 and 926 to 927 in chain A; 471 to 473, 644 to 647, 664 to 670, 794 to 813 and 924 to 927 in chain B; 471 to 473, 664 to 670, 794 to 813 and 926 to 927 in chain C. Statistics of data processing and crystallographic refinement are reported in Extended Data Table 1.
X-ray diffraction data for the native Ctf4CTD crystals were collected at beamline I04 of the Diamond Light Source, Oxford, UK and the data were processed as for the seleno-methionine dataset. The native protein crystallized in the same orthorhombic space group P22121 as the selenomethionine protein crystals, but with different unit cell dimensions a=88.9 Å, b=100.0 Å, c=219.3 Å, caused by an alternative set of crystal contacts made by the Ctf4CTD trimer in the asymmetric unit. The structure of native Ctf4CTD was solved by molecular replacement in PHASER39, using the structure of one protomer of the selenomethionine Ctf4CTD trimer as search model. The final model was refined using data to 3.0 Å resolution, to R-work and R-free values of 0.1674 and 0.2049 and a Molprobity score of 1.42. The following amino acids were not included in the final model due to missing or poor electron density and are presumed to be disordered: 471 to 473, 664 to 670, 792 to 813 in chain A; 471 to 473, 797 to 813 in chain B; 471 to 473, 664 to 670, 777 to 927 (helical domain) in chain C. In this crystal form, the Ctf4CTD structure adopts a more open conformation where one interface between Ctf4 protomers widens to become a narrow gap and the helical domain of one of the Ctf4CTD protomers at the interface becomes disordered (Extended Data Figure 4). Statistics of data processing and crystallographic refinement are reported in Extended Data Table 1.
Co-crystallisation of Ctf4CTD with Pol α and Sld5
For co-crystallisation experiments, the peptides IDNFDDILGEFES and MDINIDDILAELDKETTAV, corresponding to aa 137 to 149 of yeast Pol1 and aa 1 to 19 of yeast Sld5 respectively, were synthesized. The Pol1 peptide was solubilised in the same buffer as purified Ctf4CTD to a concentration of 340 μM; the Sld5 peptide was solubilised in water to a concentration of 2 mM. Soaking was performed by adding 1 μl of Pol1 peptide or 0.5 μl of Sld5 peptide to a 2 μl crystallization drop containing native Ctf4CTD crystals. The crystals were soaked with the peptide for 24 hours at 19 °C, back-soaked in crystallization buffer and flash-frozen in liquid nitrogen. X-ray diffraction data for Ctf4CTD crystals soaked with the Pol α and Sld5 peptides were collected on beamline I04 of the Diamond Light Source and processed as for the native crystals. The position of the Ctf4-binding motifs of Pol α and Sld5 in the crystals structure of Ctf4CTD was readily identified by inspection of Fo-Fc difference Fourier maps. Amino acids 140 to 149 of Pol1 and 3 to 15 of Sld5 were built in the electron density map and the structures of Ctf4CTD bound to Pol α and Sld5 were then further refined using Coot and PHENIX Refine to R-work/R-free values of 0.1718/0.2099 and 0.1787/0.2141, respectively. Molprobity scores for the Ctf4CTD - Pol α and Ctf4CTD - Sld5 structures were 1.33 and 1.32, respectively. Statistics of data processing and crystallographic refinement are reported in Extended Data Table 1.
Sample preparation for EM
Negative stain analysis was performed using 400 mesh carbon coated grids (Agar Scientific). Carbon was evaporated onto freshly cleaved mica with a Q150TE coater (Quorum Technologies) and incubated overnight prior to floating. Dried carbon grids were glow discharged for 30-60 seconds at 45 mA using a 100x glow discharger (Electron Microscopy Sciences). A 4-μl drop of the peak fraction from each GraFix-processed sample was applied onto the grid. Subsequently, grids were sequentially laid on top of five distinct 75 μl drops of 2% uranyl formate solution, and stirred for 10 seconds each time, before blotting to dryness.
EM data collection
Negative stain analyses of all complexes were performed using a Tecnai LaB6 G2 Spirit transmission electron microscope (FEI) operating at 120 keV (Electron Microscopy Unit, London Research Institute). Images were recorded using a 2k x 2k GATAN Ultrascan 100 camera at a nominal magnification of 30,000× (3.45 Å/pixel at the specimen level). Between 100 and 350 micrographs were collected for each dataset.
Single-particle analysis
CTF corrected image stacks were prepared in the EMAN2 environment40. Single-particle symmetry analysis was performed as described41. Reference free two-dimensional class averages were calculated using the one-step rotation & classification approach as described24, followed by routine MSA/MRA IMAGIC protocols42.
GST pull-downs
For each Pol α and Sld5 construct to be tested for interaction with Ctf4CTD, a 25-ml E. coli BL21(DE3) culture overexpressing the GST-fusion construct was pelleted, resuspended in buffer 50 mM Tris pH 7.0, 500 mM NaCl, 10% (w/v) glycerol, 1 mM DTT and protease inhibitors (Sigma) and lysed by sonication. Following centrifugation, the soluble extract was mixed with 50 μl of Glutathione Sepharose beads (GE Healthcare) preequilibrated in the same buffer and incubated under rotation at 4°C for 1 hour. Unbound protein was removed by 3 consecutive washes with 1 ml of buffer, followed by 3 1-ml washes with pull-down buffer (20 mM HEPES pH 7.2, 150 mM NaCl, 5% (w/v) glycerol, 0.1% Igepal CA-630, 1 mM TCEP and 1% BSA). Subsequently, 500 μl of purified Ctf4CTD protein at a concentration of 2 mg/ml was added to the Sepharose beads and binding was allowed to take place for an additional hour at 4°C. The binding reaction was stopped by two consecutive washes with 1 ml of pull-down buffer and a final 1 ml wash with pull-down buffer without BSA. The Sepharose beads were mixed with SDS loading dye and Ctf4CTD interactions with the respective bait proteins were detected via SDS-PAGE. As a control, Ctf4CTD was tested for unspecific interaction with the Glutathione Sepharose resin and with GST and in both cases no interaction was detected.
Yeast strains and growth
The yeast strains CC2619 (POL1-9MYC pep4Δ::URA3 ADE2) and CC10682 (pol1-A POL1-9MYC pep4Δ::URA3 ADE2) were grown at 24°C in rich medium (1% yeast extract, 2% peptone, 40 g/ml adenine) with 2% glucose as carbon source. Cells were synchronised in G1-phase by adding 7.5 μg/ml alpha factor mating pheromone for 70% of one generation time, followed by additional aliquots of 2.5 μg/ml every 20 minutes up to 1.5 generation times.
Immunoprecipitation of proteins from yeast cell extracts
MYC-tagged proteins were isolated from yeast cell extracts as described previously43.
Gel filtration
All proteins were purified as described above. Putative complexes were reconstituted prior to analytical gel filtration by mixing stoichiometric molar ratios of Ctf4CTD with either GINSPsf1ΔC or GINSPsf1ΔC,Sld5ΔN followed by centrifugation at 16,000g for 10 minutes at 4°C to remove potential aggregates. 100 μl samples of Ctf4CTD, GINSPsf1ΔC, GINSPsf1ΔC,Sld5ΔN, Ctf4CTD-GINSPsf1ΔC and Ctf4CTD-GINSPsf1ΔC,Sld5ΔN were subsequently fractionated over a Superdex S200 HR 10/300 column (GE Healthcare) preequilibrated in 20 mM HEPES pH 7.2, 140 mM KCl.
MALS analysis of Ctf4CTD
100 μl of Ctf4CTD protein at a concentration of 2 mg/ml was loaded onto a Superdex S200 HR 10/300 gel-filtration column (GE Healthcare) in 25 mM Hepes pH 7.0, 200 mM NaCl at a flow rate of 0.5 ml/min. The column was controlled using an Äkta Purifier System (GE Healthcare) and was linked to a DAWN 8+ 8-angle light scattering detector (Wyatt Technology) with a fused silica sample cell using a laser wavelength of 664 nm. The change in the refractive index was detected using an Optilab T-rEX refractometer with extended range (Wyatt Technology) using a wavelength of 658 nm. Data collection and analysis was carried out using the ASTRA6 software package (Wyatt Technology). Molecular weight determination across the sample peak was carried out using a Zimm-plot derived global fitting algorithm with a fit degree of 1 and a dn/dc value of 0.1850 ml/g.
MALS analysis of full-length Ctf4
Around 100 μg of Ctf4 FL protein was loaded onto a Wyatt MP-030S5 HPLC size-exclusion chromatography column (Wyatt) mounted on an AKTA Micro (GE Healthcare) chromatography. The column was equilibrated in a buffer containing 150 mM NaCl, 20 mM Tris pH 8.0, 1 mM DTT. The chromatography system was coupled to an 8-angle light scattering detector (DAWN 8+) and a refractive index detector (OptiLab TRex) (Wyatt Technology). Data were collected every 0.5 seconds. Data analysis was carried out using ASTRA VI.
Fluorescence polarization
Both the Pol1 peptide (137-IFDNDDILGEFES-149) and the yeast Sld5 peptide (1- MDINIDDILAELDKETTAV -19) were synthesized with an amino-terminal fluorescein label. The lowest concentration of peptide at which the binding studies could be performed was determined via peptide calibration curves. Fluorescence anisotropy measurements were recorded in a PHERAstar Plus multi-detection plate reader (BMG Labtech) equipped with fluorescence polarization optic module (λex=485 nm; λem=520 nm) at 25°C. Each data point is the mean of 200 flashes/well. The voltage gain was set by adjusting the target mP values of fluorescein-labeled peptides relative to that of fluorescein (35 mP). Serial dilutions of Ctf4CTD were made in 20 mM HEPES, pH 7.2, 140 mM KCl and 5% (w/v) glycerol in the presence of 40 nM (Sld5) or 50 nM (Pol1) fluorescein-labeled peptide. Each data point is the mean of three independent experiments. Curve fitting to the experimental data was performed in ProFit 6.2 (QuantumSoft) using a Robust fitting algorithm in combination with a Lorentzian error distribution analysis.
Native mass spectrometry
In preparation for non-denaturing nanoelectrospray ionization mass spectrometry (native mass spectrometry), protein samples were subjected to two successive rounds of buffer exchange into 500 mM ammonium acetate using illustra NAP-5 columns (GE Healthcare). For reconstitution of the Ctf4CTD - Pol α and Ctf4CTD - Sld5 complexes, Ctf4CTD was incubated with a 10-fold or 5-fold molar excess of Pol1 peptide 137-IDNFDDILGEFES-149 or Sld5 peptide 1-MDINIDDILAELDKETTAV-19, respectively, for 30 minutes prior to buffer exchange. After buffer exchange, samples were concentrated to at least 50 μM in preparation for mass spectrometric analysis. Native mass spectra were recorded on a Synapt HDMS instrument (Waters, Manchester, UK), and calibrated using caesium iodide (100 mg ml−1) as described previously44. Typical parameter values were: capillary voltage 1.8 kV, cone voltage 40-80 V, cone gas 40 L h−1, extractor 1.2-2.2 V, ion transfer stage pressure 3.61-3.44 mbar, trap collision energy 10-15 V, transfer collision energy 10-20.0 V, trap and transfer pressure 5.29-5.33 × 10−2 mbar, IMS pressure 5.01-5.02 × 10−1 mbar, TOF analyser pressure 1.17-1.18 × 10−6 mbar. Micromass MassLynx 4.1 was used for data acquisition and processing.
Artwork
All structural drawings were prepared with UCSF Chimera45.
Supplementary Material
The animation shows morphing between the open and closed forms observed in crystal structures of the Ctf4CTD trimer. The protein is drawn as ribbon, coloured according to its domain structure: the β-propeller domain is in light blue and the helical domain in yellow.
The video shows a sequence of reference-free class averages of Ctf4CTD - GINS complexes, arranged in order of increasing GINS occupancy around the Ctf4CTD trimer.
a The 6-bladed β-propeller of Ctf4CTD. The structure is drawn as a ribbon diagram, coloured blue to light green from the N- to the C-terminus. The six blades of the propeller are labelled WD1 to 6, from N- to C-end. The helical domain has been omitted for clarity. b Two views of the Ctf4CTD protomer, highlighting the helical domain of Ctf4CTD, coloured yellow to red from the amino- to the carboxy-terminus. The α-helices are labelled α1 to α6. The β-propeller domain is in light grey.
a Silver-stained SDS PAGE gel showing the purified full-length yeast Ctf4 and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. b Representative micrograph of the full-length yeast Ctf4. c Zoomed-in view of the micrograph in panel b. d Reference-free class averages of the full-length protein complex highlight the presence of a ring shaped core linked to mobile, satellite densities. Box size 448 Å. e Multi-angle light scattering reveals that Ctf4 forms stable homotrimers in solution (absolute molecular mass equates to 322 kDa, with a 0.6% error; expected molecular mass for a monomer: 106.9 kDa). f Silver-stained SDS-PAGE gel showing the purified Ctf4CTD and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. g Representative micrograph of the Ctf4CTD complex. h Zoomed-in view of the micrograph in panel g. i Reference-free class averages of the Ctf4CTD protein complex highlight the presence of an oligomerization core. Box size 448 Å.
Panels a and b show side-by-side comparisons of the symmetric, closed form of the Ctf4CTD trimer (left-hand side) with the pseudo-symmetric, open form (right-hand side), used for study of the interaction with the Ctf4-binding motifs of Pol α and Sld5. Panel a shows a side view of the two crystal forms, drawn as ribbons, with the β-propeller domain of the three protomers coloured blue, cyan and light blue, respectively and the helical domain coloured yellow. Panel b shows a top view of the two crystal forms, drawn as ribbons, with the symmetric, closed form of the Ctf4CTD trimer in light gray and the pseudo-symmetric, open form in dark gray. The helical domains have been removed for clarity. Panels c, d and e show superpositions of the Cα traces of the protomers of the symmetric, closed form of the Ctf4CTD trimer, the protomers of the pseudo-symmetric, open Ctf4CTD trimer and all protomers of the two crystal forms, respectively. In panels c and d, the protomers are coloured according to secondary structure, with β-strands in cyan and α-helices in yellow; the crystal form of the superimposed protomers is highlighted by a space-fill model of the structure in the top-left corner of the panel. In panel e, the Ctf4CTD protomers of the closed and open forms are coloured light and dark gray, respectively.
Ctf4CTD is drawn as a yellow ribbon, the Ctf4-binding motifs of Pol α and Sld5 as green and red tubes. The side chains of amino acids at the interface are shown as sticks, with carbon atoms coloured white (Ctf4) or light brown (Pol α and Sld5), oxygen atoms in red, nitrogen atoms in blue and sulphur atoms in yellow. The bidentate salt link between D142 (Pol1) or D7 (Sld5) and R904 of Ctf4 is shown as solid pink lines.
Only the region corresponding to the crystal structure described in the paper is reported. Observed secondary structure elements of Ctf4 and predicted secondary structure elements of Mcl1 and And1 are boxed and shaded in green and yellow for β-sheets and α-helices, respectively. The extent of the WD40 domains of the six-bladed β-propeller and the α-helices of the helical domain are illustrated above the alignment. Ctf4 residues that form the interface with Pol α and Sld5 are marked by an asterisk.
a Binding affinity of Ctf4CTD for the Ctf4-binding motifs of Pol α (top panel) and Sld5 (bottom panel). Affinity was measured by fluorescence anisotropy of fluorescein-labelled peptides in the presence of increasing amounts of Ctf4. See full Methods for experimental details. b CMG still associates with Ctf4 and Pol1 in yeast cells with mutations in the Ctf4-binding motif of Sld5. The budding yeast strains MCM4-5FLAG (Control) and MCM4-5FLAG sld5-Δ2-9 (the endogenous copy of SLD5 was modified to create sld5-Δ2-9, such that the encoded protein lacks amino acids 2-9) were grown at 24°C, arrested in G1-phase, and then released into S-phase for 30 minutes. Mcm4-5FLAG was then isolated from cell extracts by immunoprecipitation, and the indicated proteins were detected by immunoblotting with the corresponding antibodies23 (top panel). An analogous experiment was performed with MCM4-5FLAG (Control) and MCM4-5FLAG sld5-GA (the endogenous copy of SLD5 was modified to create sld5-GA, such that amino acids 5-9 were changed from Ile-Asp-Asp-Ile-Leu to Gly-Ala-Gly-Ala-Gly) (bottom panel).
Comparison between GINS - Ctf4CTD complexes prepared by glycerol gradient or GraFix. a SDS PAGE gel of non-crosslinked GINS - Ctf4CTD complex. A red box highlights the fraction imaged by EM. b Representative micrograph for the non-cross-linked preparation highlights small, globular particles. c SDS PAGE gel of cross-linked GINS - Ctf4CTD complex. A red box highlights the fraction imaged by EM. d Representative micrograph for the cross-linked preparation highlights elongated features compatible with one, two or three GINS docked onto a Ctf4 trimerization core. e Zoomed-in view of the same micrograph. f Representative class averages of the Ctf4CTD - GINS complex show a mixture of complexes with clearly discernible stoichiometry: Ctf43 - GINS, Ctf43 - (GINS)2, Ctf43 - (GINS)3.
a Silver-stained SDS PAGE gel showing the purified Ctf4CTD - Pol1NTD complex and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. (b) Representative micrograph of the Ctf4CTD - Pol1NTD complex. (c) Zoomed-in view of the same micrograph. (d) Reference-free class averages of the Ctf4CTD - Pol1NTD complex highlight the presence of a ring-shaped core linked to one mobile globular density. The box size is 448 Å.
a Silver-stained SDS-PAGE gel showing the purified GINS - Ctf4CTD - Pol1NTD complex and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. (b) Representative micrograph of the GINS - Ctf4CTD - Pol1NTD complex. (c) Zoomed-in view of the same micrograph. (d-e) Reference-free class averages of the complex highlight the presence of a ring-shape core linked to 2 or 3 peripheral features. The peripheral features fall into two categories: either elongated radially departing features similar to those seen for the Ctf4CTD - GINS complex, or smaller globular densities that were assigned to Pol1NTD. The box size is 448 Å.
Acknowledgments
We would like to thank Lucy Collinson and Raffaella Carzaniga (LRI) for EM access, David Firth and Bram Snijder (LRI) for mass spectrometry work, Joseph Maman for help with SEC-MALS and Philip Zegerman and Julian Gannon for comments on the manuscript. This work was supported by the Cambridge Gates PhD program (A.C.S.), CRUK (A.C. and K.L), MRC (K.L.) and a Wellcome Trust SRF award in basic biomedical science (L.P.).
Footnotes
The authors declare no competing financial interest.
Coordinates and structure factors for Ctf4CTD (selenomethionine-labeled protein), Ctf4CTD (native), Ctf4CTD - Pol α and Ctf4CTD - Sld5 complexes are available from the Protein Data Bank under accession codes 4C8H, 4C8S, 4C93 and 4C95, respectively.
References
- 1.DePamphilis ML, Bell SD. Genome duplication. Garland Science; 2011. pp. xiii–449. [Google Scholar]
- 2.Zegerman P, Diffley JFX. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst.) 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 3.Cortez D. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit. Rev. Biochem. Mol. Biol. 2012;47:222–235. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- 5.Lia G, Michel BND, Allemand J-FSS. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science. 2012;335:328–331. doi: 10.1126/science.1210400. [DOI] [PubMed] [Google Scholar]
- 6.Georgescu RE, Kurth I, O’Donnell M. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat. Struct. Mol. Biol. 2011;19:113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a Triple DNA Polymerase Replisome. Mol. Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and Architecture of Active DNA Replication Machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka H, et al. Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes to cells. 2009;14:807–820. doi: 10.1111/j.1365-2443.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- 10.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta S, et al. Dpb2 Integrates the Leading-Strand DNA Polymerase into the Eukaryotic Replisome. Curr. Biol. 2013;23:543–552. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Lou H, et al. Mrc1 and DNA Polymerase epsilon Function Together in Linking DNA Replication and the S Phase Checkpoint. Mol. Cell. 2008;32:106. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu W, et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouprina N, et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miles J, Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol. Cell. Biol. 1992;12:5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosnell JA, Christensen TW. Drosophila Ctf4 is essential for efficient DNA replication and normal cell cycle progression. BMC Molecular Biology. 2011;12:13. doi: 10.1186/1471-2199-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bermudez VP, Farina A, Tappin I, Hurwitz J. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 2010;285:9493–9505. doi: 10.1074/jbc.M109.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams D, McIntosh J. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryotic cell. 2002;1:758–773. doi: 10.1128/EC.1.5.758-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka H, et al. Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes to cells. 2009;14:949–963. doi: 10.1111/j.1365-2443.2009.01322.x. [DOI] [PubMed] [Google Scholar]
- 20.Petronczki M, et al. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell. Sci. 2004;117:3547–3559. doi: 10.1242/jcs.01231. [DOI] [PubMed] [Google Scholar]
- 21.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshizawa-Sugata N, Masai H. Roles of Human AND-1 in Chromosome Transactions in S Phase. J. Biol. Chem. 2009;284:20718–20728. doi: 10.1074/jbc.M806711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell. Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 24.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beattie TR, Bell SD. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 2012;31:1556–1567. doi: 10.1038/emboj.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 29.Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 30.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 32.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic Structures of the Human Immunophilin FKBP-12 Complexes with FK506 and Rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 33.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Costa A, et al. Structural studies of the archaeal MCM complex in different functional states. J. Struct. Biol. 2006;156:210–219. doi: 10.1016/j.jsb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 42.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 43.Kilkenny ML, De Piccoli G, Perera RL, Labib K, Pellegrini L. A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J. Biol. Chem. 2012;287:23740–23747. doi: 10.1074/jbc.M112.368951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The animation shows morphing between the open and closed forms observed in crystal structures of the Ctf4CTD trimer. The protein is drawn as ribbon, coloured according to its domain structure: the β-propeller domain is in light blue and the helical domain in yellow.
The video shows a sequence of reference-free class averages of Ctf4CTD - GINS complexes, arranged in order of increasing GINS occupancy around the Ctf4CTD trimer.
a The 6-bladed β-propeller of Ctf4CTD. The structure is drawn as a ribbon diagram, coloured blue to light green from the N- to the C-terminus. The six blades of the propeller are labelled WD1 to 6, from N- to C-end. The helical domain has been omitted for clarity. b Two views of the Ctf4CTD protomer, highlighting the helical domain of Ctf4CTD, coloured yellow to red from the amino- to the carboxy-terminus. The α-helices are labelled α1 to α6. The β-propeller domain is in light grey.
a Silver-stained SDS PAGE gel showing the purified full-length yeast Ctf4 and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. b Representative micrograph of the full-length yeast Ctf4. c Zoomed-in view of the micrograph in panel b. d Reference-free class averages of the full-length protein complex highlight the presence of a ring shaped core linked to mobile, satellite densities. Box size 448 Å. e Multi-angle light scattering reveals that Ctf4 forms stable homotrimers in solution (absolute molecular mass equates to 322 kDa, with a 0.6% error; expected molecular mass for a monomer: 106.9 kDa). f Silver-stained SDS-PAGE gel showing the purified Ctf4CTD and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. g Representative micrograph of the Ctf4CTD complex. h Zoomed-in view of the micrograph in panel g. i Reference-free class averages of the Ctf4CTD protein complex highlight the presence of an oligomerization core. Box size 448 Å.
Panels a and b show side-by-side comparisons of the symmetric, closed form of the Ctf4CTD trimer (left-hand side) with the pseudo-symmetric, open form (right-hand side), used for study of the interaction with the Ctf4-binding motifs of Pol α and Sld5. Panel a shows a side view of the two crystal forms, drawn as ribbons, with the β-propeller domain of the three protomers coloured blue, cyan and light blue, respectively and the helical domain coloured yellow. Panel b shows a top view of the two crystal forms, drawn as ribbons, with the symmetric, closed form of the Ctf4CTD trimer in light gray and the pseudo-symmetric, open form in dark gray. The helical domains have been removed for clarity. Panels c, d and e show superpositions of the Cα traces of the protomers of the symmetric, closed form of the Ctf4CTD trimer, the protomers of the pseudo-symmetric, open Ctf4CTD trimer and all protomers of the two crystal forms, respectively. In panels c and d, the protomers are coloured according to secondary structure, with β-strands in cyan and α-helices in yellow; the crystal form of the superimposed protomers is highlighted by a space-fill model of the structure in the top-left corner of the panel. In panel e, the Ctf4CTD protomers of the closed and open forms are coloured light and dark gray, respectively.
Ctf4CTD is drawn as a yellow ribbon, the Ctf4-binding motifs of Pol α and Sld5 as green and red tubes. The side chains of amino acids at the interface are shown as sticks, with carbon atoms coloured white (Ctf4) or light brown (Pol α and Sld5), oxygen atoms in red, nitrogen atoms in blue and sulphur atoms in yellow. The bidentate salt link between D142 (Pol1) or D7 (Sld5) and R904 of Ctf4 is shown as solid pink lines.
Only the region corresponding to the crystal structure described in the paper is reported. Observed secondary structure elements of Ctf4 and predicted secondary structure elements of Mcl1 and And1 are boxed and shaded in green and yellow for β-sheets and α-helices, respectively. The extent of the WD40 domains of the six-bladed β-propeller and the α-helices of the helical domain are illustrated above the alignment. Ctf4 residues that form the interface with Pol α and Sld5 are marked by an asterisk.
a Binding affinity of Ctf4CTD for the Ctf4-binding motifs of Pol α (top panel) and Sld5 (bottom panel). Affinity was measured by fluorescence anisotropy of fluorescein-labelled peptides in the presence of increasing amounts of Ctf4. See full Methods for experimental details. b CMG still associates with Ctf4 and Pol1 in yeast cells with mutations in the Ctf4-binding motif of Sld5. The budding yeast strains MCM4-5FLAG (Control) and MCM4-5FLAG sld5-Δ2-9 (the endogenous copy of SLD5 was modified to create sld5-Δ2-9, such that the encoded protein lacks amino acids 2-9) were grown at 24°C, arrested in G1-phase, and then released into S-phase for 30 minutes. Mcm4-5FLAG was then isolated from cell extracts by immunoprecipitation, and the indicated proteins were detected by immunoblotting with the corresponding antibodies23 (top panel). An analogous experiment was performed with MCM4-5FLAG (Control) and MCM4-5FLAG sld5-GA (the endogenous copy of SLD5 was modified to create sld5-GA, such that amino acids 5-9 were changed from Ile-Asp-Asp-Ile-Leu to Gly-Ala-Gly-Ala-Gly) (bottom panel).
Comparison between GINS - Ctf4CTD complexes prepared by glycerol gradient or GraFix. a SDS PAGE gel of non-crosslinked GINS - Ctf4CTD complex. A red box highlights the fraction imaged by EM. b Representative micrograph for the non-cross-linked preparation highlights small, globular particles. c SDS PAGE gel of cross-linked GINS - Ctf4CTD complex. A red box highlights the fraction imaged by EM. d Representative micrograph for the cross-linked preparation highlights elongated features compatible with one, two or three GINS docked onto a Ctf4 trimerization core. e Zoomed-in view of the same micrograph. f Representative class averages of the Ctf4CTD - GINS complex show a mixture of complexes with clearly discernible stoichiometry: Ctf43 - GINS, Ctf43 - (GINS)2, Ctf43 - (GINS)3.
a Silver-stained SDS PAGE gel showing the purified Ctf4CTD - Pol1NTD complex and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. (b) Representative micrograph of the Ctf4CTD - Pol1NTD complex. (c) Zoomed-in view of the same micrograph. (d) Reference-free class averages of the Ctf4CTD - Pol1NTD complex highlight the presence of a ring-shaped core linked to one mobile globular density. The box size is 448 Å.
a Silver-stained SDS-PAGE gel showing the purified GINS - Ctf4CTD - Pol1NTD complex and GraFix gel of the same preparation. A red box highlights the fraction imaged by EM. (b) Representative micrograph of the GINS - Ctf4CTD - Pol1NTD complex. (c) Zoomed-in view of the same micrograph. (d-e) Reference-free class averages of the complex highlight the presence of a ring-shape core linked to 2 or 3 peripheral features. The peripheral features fall into two categories: either elongated radially departing features similar to those seen for the Ctf4CTD - GINS complex, or smaller globular densities that were assigned to Pol1NTD. The box size is 448 Å.