Abstract
Our aim was to conduct a pilot case-control study of RNA expression profile using RNA sequencing of rectosigmoid mucosa of nine females with -diarrhea-predominant irritable bowel syndrome (IBS-D) with accelerated colonic transit and nine female healthy controls. Mucosal total RNA was isolated and purified, and next-generation pair-end sequencing was performed using Illumina TruSeq. Analysis was carried out using a targeted approach toward 12 genes previously associated with IBS and a hypothesis-generating approach. Of the 12 targeted genes tested, patients with IBS-D had decreased mRNA expression of TNFSF15 (fold change controls to IBS-D: 1.53, P = 0.01). Overall, up- and downregulated mRNA expressions of 21 genes (P = 10−5 to 10−8; P values with false detection rates are shown) were potentially relevant to IBS-D including the following: neurotransmitters [P2RY4 (P = 0.001), vasoactive intestinal peptide (VIP, P = 0.02)]; cytokines [CCL20 (P = 0.019)]; immune function [C4BPA complement cascade (P = 0.0187)]; interferon-related [IFIT3 (P = 0.016)]; mucosal repair and cell adhesion [trefoil protein (TFF1, P = 0.012)], retinol binding protein [RBP2 (P = 0.017)]; fibronectin (FN1, P = 0.009); and ion channel functions [guanylate cyclase (GUCA2B, P = 0.017), PDZ domain-containing protein 3 (PDZD3, P = 0.029)]. Ten genes associated with functions related to pathobiology of IBS-D were validated by RT-PCR. There was significant correlation in fold changes of the selected genes (Rs = 0.73, P = 0.013). Up- or downregulation of P2RY4, GUC2AB, RBP2, FNI, and C4BPA genes were confirmed on RT-PCR, which also revealed upregulation of farnesoid X receptor (FXR) and apical sodium-coupled bile acid transporter (IBAT/ASBT). RNA-Seq and RT-PCR analysis of rectosigmoid mucosa in IBS-D show transcriptome changes that provide the rationale for validation studies to explore the role of mucosal factors in the pathobiology of IBS-D.
Keywords: neurotransmitters, ion channels, cytokines, barrier, mucosal repair
the cause of loose consistency of bowel movements in patients with irritable bowel syndrome with diarrhea (IBS-D) is attributed in part to acceleration of colonic transit, which is documented in ∼45% of patients with IBS-D (11), providing less time for reabsorption of water in the colon (16). Circulating postprandial levels of serotonin are increased in patients with IBS-D (17; 21). Increased duodenal and rectosigmoid expression of secretory transmitters and fecal excretion of granins are also documented (18; 19; 29) although it is unclear whether these result in intestinal fluid and electrolyte secretion or accelerated transit with reduced colonic fluid absorption. There is one study that suggested that the ileal mucosa of patients with IBS, compared with that of healthy controls, responds to infusion of bile acids with greater fluid and electrolyte secretion (18). Mutations in the gene GUCY2C that controls the secretory guanylate cyclase C receptor have been reported in an extensive family who presented with chronic symptoms suggestive of IBS-D (20).
The mucosal functions of patients with IBS-D and controls appear to differ (7). IBS has been associated with changes in the rectosigmoid mucosal expression of immune and nonimmune protective factors, such as barrier function and mucus secretion (2). The literature to date (2; 4; 5; 8; 12; 22; 36; 39–41; 44; 45) is summarized in Table 1. However, there are only a few examples, typically derived from assessments of single genes in relatively small numbers of patients and controls, where the inherited genotype has been associated with altered mucosal gene expression. For example, differential expression of the TNFSF15 gene, which is associated with IBS, has been linked with functional alterations of mucosal immune and protective functions (36; 45). Although genotype can clearly influence the expression of related proteins in the colonic mucosa, function is more closely reflected in the expression of mRNA in the tissue. There is also evidence of alterations in jejunal mucosal abnormalities in the expression and distribution of apical junction complex proteins, specifically, increased protein expression of claudin-2, reduced occludin phosphorylation, enhanced redistribution from the membrane to the cytoplasm (24), increased myosin kinase expression, reduced myosin phosphatase, and, consequently, enhanced phosphorylation of myosin (24). In addition, the Barcelona group also reported that expression of zonula occludens 1 (ZO-1) was reduced in IBS-D at both gene and protein level, with protein redistribution from the tight junction to the cytoplasm (25). In a prior study, we have demonstrated a borderline significant difference in the ZO-1 intensity score in the small bowel mucosa (P = 0.06) of patients with IBS-D, compared with healthy controls with lower intensity in HLA-DQ2/8-positive relative to HLA-DQ2/8-negative patients with IBS-D (39).
Table 1.
Summary of changes in mucosal expression in IBS in the published literature
| Function or Phenotype | Mucosal Expression in Rectosigmoid Biopsies in IBS | Associated Genotype | Reference |
|---|---|---|---|
| Cellular defense functions, nonimmune-mediated | Upregulation of mucin gene (MUC20) and dual oxidase 2 (DUOX2); reduced caspase-1 (CASP1) and lysozyme (LYZ) | Not tested | Aerssens et al., 2008 |
| Mucosal cell immune activation: proinflammatory IL-8, IL-1 β | Upregulation of TLR2 and TLR4 mRNA in epithelial cells; | Not tested | Belmonte et al., 2012 |
| Upregulation of CCL-11, CCL-13 | No association with genotype | Swan et al., 2013 | |
| Serotonergic functions | Reduced SERT-P mRNA expression (normal in some studies) | 5-HTTLPR | Coates et al., 2004, Camilleri et al., 2007, Wang et al., 2012, Kerckhoffs et al., 2012 |
| Immune activation | Reduced TNFSF15 mRNA in PI-IBS | TNFSF15 (C allele rs6478108) | Swan et al., 2013, |
| Increased TNFSF15 mRNA in healthy | TNFSF15(G allele rs4263839) | Zucchelli et al., 2011 | |
| Increased IL13 mRNA | IL13 | Wouters et al., 2013 | |
| Immune recognition and TJ protein | Not tested | CDH1, TLR9, IL6 | Villani et al., 2010 |
| TJ protein mRNA expression | Reduced mRNA ZO-1 in IBS-D; downregulation of ZO-1 and occludin expression with no changes in mRNA expression | HLA DQ2/8 | Vazquez et al., 2012 |
| Bertiaux-Vandaele et al., 2011 |
IBS, irritable bowel syndrome; TLR, Toll-like receptor; PI-IBS, postinfectious IBS; TJ, tight junction; IBS-D, diarrhea-predominant IBS; ZO, zonnula occludens.
The development of next-generation RNA sequencing provides a novel opportunity to examine the entire transcriptome. Analysis of differential expression in well-matched patients and controls has the potential to assess differences in the transcriptome that may modify the function of the tissue (e.g., mucosa) in the disease of interest.
Our hypothesis was that RNA expression of mucosal biopsies from patients with IBS-D (with previously demonstrated accelerated colonic transit) differs from that of healthy controls. The aim of the study was to conduct RNA sequencing in stored mucosal samples obtained from the rectosigmoid region of nine female patients with IBS-D (with previously demonstrated accelerated colonic transit) and nine female healthy controls. We approached this aim using two strategies: first, we examined differential expression in the following 12 “targeted” genes associated with IBS in the literature: 5-HTTLPR, TNFSF15, FAAH, CNR1, KLB, FGFR4, GPBAR1, TLR9, C11orf30, ORMDL3, NPSR1, and HLA2/8; second, we sought associations with differential expression of any other gene mRNA, independent of any a priori hypothesis, that is, an “unbiased analysis” intended for hypothesis generation. The results obtained for 10 of the genes of interest were validated by a quantitative RT-PCR. An ancillary study used pathway analyses to explore how any identified alterations in mRNA expression might mediate the pathobiology of IBS.
MATERIALS AND METHODS
Design.
We conducted a case-control study of a convenient number of participants in this pilot study. We used stored samples from patients who had consented to the use of biospecimens (for future research) in prior studies (2; 38) conducted at Mayo Clinic in Rochester, MN. These samples were obtained in nine female patients with IBS-D and nine female healthy controls. RNA sequencing was conducted by the Mayo Medical Genomics Facility and bioinformatics by the Mayo Bioinformatics Core Facility to interpret differential expression in patients and controls. The study was approved by Mayo Clinic Institutional Review Board (IRB no. 13-001595, date of approval 3/13/2013).
Selection of participants.
We examined, using RNA sequencing, the RNA expression of rectosigmoid mucosal biopsies from nine female patients with IBS-D (age range, 25–63 yr; BMI range, 20–40.5 kg/m2) with rapid colonic transit (geometric center, GC24: 4.19–4.92) and from nine female healthy controls who had no gastrointestinal symptoms on screening with a gastrointestinal symptom questionnaire (age range, 19–54 yr; BMI range, 19.4–40.2 kg/m2) and with normal colonic transit (GC24: 1.15–2.63). All participants lived in the same geographic region within 120 miles of Rochester, MN. The samples of equal numbers of cases and controls were obtained from people of same sex and similar BMI range.
Sample storage.
Participants' rectosigmoid biopsies were preserved in a solution of RNALater, stored at −80°C. Total RNA was isolated using the RNeasy Plus Mini (Qiagen, Valencia, CA). Quality of the extracted RNA was assessed by the Mayo Gene Expression Core using Agilent Bioanalyzers. All samples had RNA Integrity Numbers (RIN) greater than 7.0.
In-depth RNA sequencing of colonic mucosa.
RNA sequencing was performed as paired-end 51 base reads on an Illumina HiSeq 2000 with three samples multiplexed per lane, using TruSeq SBS Sequencing Kit Version 3. Base calling was performed using Illumina's RTA version 1.12.4.2.
Bioinformatics analysis workflow.
Analysis of each sample (alignment statistics, in-depth quality control metrics, gene and exon expression levels, fusion transcripts, and single nucleotide variants) was done using Mayo Clinic's MAPRSeq v1.0, a computational workflow for analysis of RNA-Seq. Differential expression between samples from patients with IBS-D and healthy controls was computed using edgeR algorithm (32). EdgeR [or empirical analysis of digital gene expression in R (an open source programming environment)] is a bioconductor software package for examining differential expression of replicated count data; it involves determining whether counts for a transcript or exon are significantly different across experimental conditions. An important summary statistic is the number of reads in a class; for RNA-Seq, this read count has been found to be linearly related to the abundance of the target transcript (27). Further details of the analysis, including bioinformatics, are included in Supplemental Materials.
Comparison of expressions in rectosigmoid mucosa from IBS-D and healthy participants.
Gene expression variation was examined in the following genes, which have been associated with IBS in the literature based on genotype association studies (9; 39): 5-HTTLPR, TNFSF15, FAAH, CNR1, KLB, FGFR4, TGR5 (GPBAR1), PRDM1 (BLIMP1), TLR9, C11orf30, ORMDL3, NPSR1, and HLA2/8. The association of TNFSF15 with IBS symptom phenotype was significant with false detection rate (FDR) correction, and the strongest association was with constipation-predominant IBS (IBS-C) (45). Of these 13 genes, the association with colonic transit has been reported in 10, and the association of KLB was significant with FDR correction (43). In the published literature, no significant association with colonic transit was found with 5-HTTLPR, TNFSF15, or FGFR4 as individual genetic risk factors. However, FGFR4 significantly influenced the effect of KLB on colonic transit (43), and HLADQ2/8 significantly altered the effect of gluten on gut permeability (38) and was associated with accelerated small bowel transit in patients with IBS-D (37), in addition to the association with colonic transit (39). The second experimental approach assessed differential expression of any other gene mRNA, independent of any a priori hypothesis, that is an “unbiased” or hypothesis-generating analysis. An ancillary investigation used pathway analysis to assess the genes identified as differentially expressed by edgeR (adjusted P < 0.05).
mRNA expression using quantitative RT-PCR.
To confirm the results of the RNA-Seq study, we submitted the mRNA from these biopsies to quantitative RT-PCR using the Qiagen RT2 assays, which use a Sybr Green detection method. Thus we evaluated 20 genes of interest, including 10 (identified in Fig. 2) of those with significant P values on RNA-Seq that are associated with functions related to pathobiology of IBS-D, as well as other genes related to tight junction proteins (CLDN1, OCLN, ZO-1), bile acid homeostasis, receptor or absorption [FGFR4, GPBAR1, FXR, IBAT/ASBT (SLC10A2)], serotonin transporter protein (SLC6A4), and immune modulation (TNFSF15). The latter genes were selected based on recent data acquired in other studies of IBS-D in our laboratory or in the literature (1; 3; 37; 38; 45).
Fig. 2.
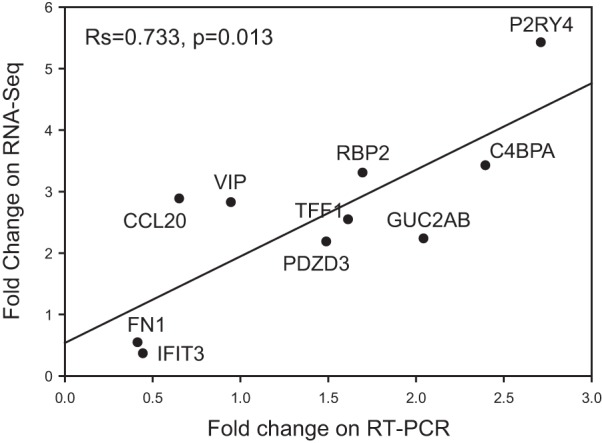
Correlation (Spearman) analysis comparing the mRNA fold change (IBS-D over healthy controls) for 10 genes selected from the RNA-Seq analysis.
Briefly, total RNA was purified from human rectosigmoid tissue using the Qiagen RNAeasy Plus kit (Qiagen), including on-column DNAse treatment to remove genomic DNA. RNA quality was assessed by Agilent Bioanalyzer, and 0.5 μg RNA (RIN>7) was reverse transcribed using the RT2 First Strand Kit (Qiagen). Resulting cDNA was amplified and gene expression quantified by Custom RT2 PCR Array (Qiagen) using a 7300 Real-time PCR Machine (Applied Biosystems, Foster City, CA). In this custom profile, we included 20 candidate genes, two housekeeping genes for normalization, and three control genes to assure reaction quality (see Supplemental Materials, gene expression method by RT-PCR). Each assay utilizes a proprietary and experimentally verified algorithm for designing gene-specific qPCR primers with uniform PCR efficiency and amplification conditions. The assay was conducted as recommended by the supplier (Qiagen). For data analysis, the RT2 Profiler PCR Array Data Analysis v3.5 software package was used. This package uses ΔΔCT-based fold change calculations and Student's t-test (two-tail, equal variance: http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php?target=upload) to assess the differences in mRNA fold changes between IBS-D and healthy control groups. In addition, we determined the Spearman correlation between by RNA-Seq and RT-PCR.
RESULTS
Sequence statistics based on sample analysis demonstrating quality control of the sequencing method.
The sequence statistics showed an average of 118,760,446 (min/max 99,967,896/150,273,460) total reads per sample was obtained, which is above the minimum of 50 million reads necessary for RNA sequencing analysis: 84.5% mapped to the genome and 12.9% to the splice-site junction reference sequence set; 2.6% did not map for various technical reasons. Thus extensive expression data were available from all cases and controls.
mRNA expression of genes previously associated with IBS-D: hypothesis-based analysis.
Gene expression variation was examined in the following genes, which have been associated with IBS in the literature: 5-HTTLPR, TNFSF15, FAAH, CNR1, KLB, FGFR4, TGR5 (GPBAR1), PRDM1(BLIMP1), TLR9, C11orf30, ORMDL3, NPSR1, and HLA2/8 (Table 2 shows gene expression variations in genes analyzed by DE Seq and edgeR).
Table 2.
Gene expression by edgeR analysis in healthy participants relative to IBS in the genes previously associated with IBS in the literature
| Chr | Gene ID | Start | Stop | Coding Length | Fold Change | Log Fold Change | P Value |
|---|---|---|---|---|---|---|---|
| Neurotransmitter-related genes | |||||||
| chr1 | FAAH | 46859939 | 46879520 | 2095 | 0.874993 | −0.1834 | 0.242634 |
| chr6 | CNR1 | 88849585 | 88875767 | 5955 | 0.837784 | −0.23277 | 0.507953 |
| chr17 | SLC6A4 | 28523378 | 28562954 | 4533 | 1.016064 | No data | No data |
| chr7 | NPSR1 | 34697897 | 34917944 | 1824 | 0.338832 | No data | No data |
| Genes related to immune function | |||||||
| chr3 | TLR9 | 52255096 | 52260179 | 3870 | 0.924332 | −0.10029 | 0.784644 |
| chr9 | TNFSF15 | 1.18E +08 | 117568408 | 6765 | 1.531795 | 0.691593 | 0.00985 |
| Genes related to bile acid synthesis and receptor | |||||||
| chr4 | KLB | 39408473 | 39453153 | 6079 | 1.05282 | 0.074556 | 0.769264 |
| chr5 | FGFR4 | 1.77E +08 | 176525126 | 3143 | 0.964932 | −0.08917 | 0.776776 |
| chr2 | GPBAR1 | 2.19E +08 | 219128582 | 2003 | 0.891095 | −0.16474 | 0.61718 |
| Genes related to membrane or barrier function | |||||||
| chr6 | PRDM1 | 1.07E +08 | 106557814 | 5398 | 1.040026 | 0.09534 | 0.725122 |
| chr11 | C11orf30 | 76156069 | 76262589 | 5511 | 1.133884 | 0.208539 | 0.289325 |
| chr17 | ORMDL3 | 38077296 | 38083884 | 2139 | 0.977262 | −0.03066 | 0.865844 |
| Immunogenotype (histocompatibility locus) | |||||||
| chr6 | HLA-DQA1 | 32605183 | 32611429 | 1542 | 0.910484 | −0.13848 | 0.54487 |
| chr6 | HLA-DQA2 | 32709163 | 32714664 | 1152 | 1.176086 | 0.243684 | 0.59239 |
| chr6 | HLA-DQB1 | 32627241 | 32634466 | 1664 | 1.011884 | 0.086342 | 0.911969 |
| chr6 | HLA-DQB2 | 32723875 | 32731330 | 1122 | 1.219724 | 0.321712 | 0.296543 |
The significant difference in TNFSF15 is shown in boldface text, indicating higher fold change expression in health than in IBS-D.
There was decreased mRNA expression of TNFSF15 in rectosigmoid mucosa in nine patients with IBS-D compared with biopsies from nine healthy controls; the relative decrease in TNFSF15 expression was about 70% (Table 2). No other changes in expression of genes previously associated with IBS were found.
Differential expression of mRNA: unbiased hypothesis-generating analysis.
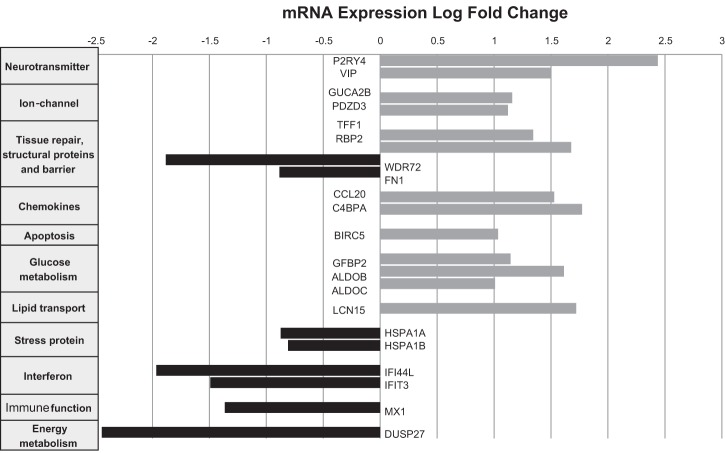
Differential expression analysis was run through edgeR algorithm, which identified 21 genes [13 upregulated, 8 downregulated in IBS-D relative to health (Fig. 1)] with an FDR-corrected P value of P < 0.05; the entire list of 21 genes with differentially expressed mRNA is shown in Table 3. The functions of these 21 genes are further described in Table 4. Many of the genes could be mapped to several biologically relevant pathways or mechanisms in IBS-D, as summarized here. Neurotransmitter functions were demonstrated when P2RY4 and VIP showed higher expression in IBS-D compared with healthy controls. Ion channel functions were upregulated in IBS-D relative to healthy controls; GUCA2B receptor associated with enterocyte chloride channel activation in response to uroguanylin and PDZD3 (also known as IKEPP, PDZK2, NHERF4); this PDZ protein associates with guanylate cyclase C and regulates cGMP production following receptor stimulation and mediates chloride secretion. Cytokines and complement were examined, and C4BPA complement cascade and CCL20 showed higher expression in IBS-D compared with healthy controls. Regarding immune function, TNFSF15 [P = 0.01 (Table 2)] and IFIT3 had lower expression in IBS-D relative to healthy controls. For mucosal repair and cell adhesion, TFF1 gastrointestinal trefoil protein and RBP2 retinol binding protein showed increased expression in IBS-D; conversely, FN1 fibronectin had lower expression in IBS-D relative to healthy controls.
Fig. 1.
Downregulated and upregulated mRNA expression in rectosigmoid mucosa in healthy controls compared with diarrhea-predominant irritable bowel syndrome (IBS-D). Gene expression fold change calculated by edgeR; negative numbers reflect lower mRNA expression of each gene of interest in patients with IBS-D compared with healthy controls.
Table 3.
Gene expression identified in hypothesis-free analysis by edgeR showing relative expression in IBS-D compared to health
| Chr | GeneID | Coding Length | Fold Change | logFC | P Value | P (FDR) |
|---|---|---|---|---|---|---|
| Neurotransmitter-related genes | ||||||
| chrX | P2RY4 | 1639 | 5.80 | 2.53 | 7.03E-08 | 0.001094 |
| chr6 | VIP | 1586 | 2.66 | 1.41 | 2.19E-05 | 0.02133 |
| Ion-channel-related genes | ||||||
| chr1 | GUCA2B | 597 | 2.15 | 1.10 | 1.15E-05 | 0.017229 |
| chr11 | PDZD3 | 2363 | 2.19 | 1.13 | 3.47E-05 | 0.029047 |
| Stress protein-related genes | ||||||
| chr6 | HSPA1A | 2429 | 0.54 | −0.89 | 7.97E-07 | 0.006201 |
| chr6 | HSPA1B | 2520 | 0.58 | −0.80 | 1.69E-05 | 0.018718 |
| Tissue repair, structural proteins, and barrier genes | ||||||
| chr21 | TFF1 | 492 | 2.66 | 1.41 | 4.56E-06 | 0.011828 |
| chr3 | RBP2 | 688 | 3.17 | 1.66 | 1.03E-05 | 0.017229 |
| chr15 | WDR72 | 7309 | 0.27 | −1.87 | 2.23E-06 | 0.008686 |
| chr2 | FN1 | 8975 | 0.54 | −0.89 | 2.82E-06 | 0.008766 |
| chr11 | MMP1 | 2055 | 0.35 | −1.50 | 0.000114 | 0.070962 |
| chr1 | ITLN1 | 1182 | 2.62 | 1.39 | 0.000183 | 0.098276 |
| Glucose metabolism-related genes | ||||||
| chr2 | IGFBP2 | 1429 | 2.40 | 1.26 | 1.46E-06 | 0.007553 |
| chr9 | ALDOB | 2426 | 3.18 | 1.67 | 1.22E-05 | 0.017229 |
| chr17 | ALDOC | 1651 | 2.09 | 1.06 | 1.41E-05 | 0.01827 |
| Lipid transport-related genes | ||||||
| chr9 | LCN15 | 762 | 3.35 | 1.74 | 2.61E-05 | 0.023844 |
| chr9 | ABCA1 | 10502 | 0.55 | −0.87 | 7.62E-05 | 0.053883 |
| Interferon-related genes | ||||||
| chr1 | IFI44L | 5874 | 0.23 | −2.14 | 7.13E-06 | 0.015853 |
| chr10 | IFIT3 | 2688 | 0.31 | −1.69 | 8.25E-06 | 0.016043 |
| chr10 | IFIT1 | 1860 | 0.36 | −1.48 | 0.000173 | 0.098276 |
| Chemokine-related genes | ||||||
| chr2 | CCL20 | 842 | 2.70 | 1.43 | 1.70E-05 | 0.018718 |
| chr1 | C4BPA | 2243 | 3.58 | 1.84 | 1.80E-05 | 0.018718 |
| chr17 | CCL3 | 796 | 0.31 | −1.67 | 0.000196 | 0.098334 |
| Immune function- and viral infection-related genes | ||||||
| chr21 | MX1 | 3605 | 0.36 | −1.48 | 4.67E-05 | 0.034633 |
| chr21 | MX2 | 2961 | 0.38 | −1.38 | 0.000164 | 0.098144 |
| chr12 | OAS2 | 6169 | 0.41 | −1.29 | 8.73E-05 | 0.059058 |
| Apoptosis-related gene | ||||||
| chr17 | BIRC5 | 2700 | 2.11 | 1.08 | 3.55E-05 | 0.029047 |
| Energy metabolism-related gene | ||||||
| chr1 | DUSP27 | 4034 | 0.19 | −2.38 | 3.80E-05 | 0.029561 |
Bold P values are significant with false detection rate (FDR).
Table 4.
Summary of functions of 21 genes with significant differential expression in IBS-D rectosigmoid mucosa
| Gene Symbol | Aliases and Descriptions | Gene ID | Summary of Gene Function or Product |
|---|---|---|---|
| Neurotransmitters | |||
| P2RY4 | purinergic receptor P2Y | 5030 | G protein-coupled receptor responsive to uridine nucleotides, partially responsive to ATP, and not responsive to ADP |
| VIP | vasoactive intestinal peptide | 7432 | Encodes G protein-coupled receptor, relaxes smooth muscle, stimulates cAMP to produce intestinal secretion |
| Ion Channels | |||
| GUCA2B | guanylate cyclase activator 2B (uroguanylin) | 2981 | Gene encodes uroguanylin expressed in the stomach and intestine, functions as endogenous ligand for guanylate cyclase-C receptor increasing cyclic GMP, chloride, and water secretion |
| PDZD3 | PDZ domain-containing protein 3; alias IKEPP; PDZK2; NHERF4 | 79849 | Encodes protein-protein interaction molecules of structural proteins and the tight junction protein, zonula occludens 1; functions in sodium-hydrogen exchange (NHE3) as well as guanylyl cyclase C (GCC, or GUCY2C)-induced chloride and water secretion (through cGMP signaling that leads to cystic fibrosis transmembrane conductance regulator phosphorylation) |
| Mucosal repair, apoptosis, and cell adhesion | |||
| FN1 | fibronectin 1 | 2335 | Encodes fibronectin, a glycoprotein at the cell surface and in extracellular matrix, is involved in cell adhesion and migration processes including wound healing and host defense |
| TFF1 | trefoil factor 1 | 7031 | Gene on chromosome 21 for stable secretory proteins expressed in GI mucosa and likely protect the mucosa from insults, stabilizes mucus layer, and affects healing of epithelium. |
| RBP2 | retinol-binding protein 2 | 5948 | RBP2 is present in the small intestinal epithelium and involved in the uptake and/or intracellular metabolism of vitamin A, necessary for growth, differentiation of epithelial tissues |
| BIRC5 | baculoviral IAP repeat-containing 5 | 332 | Inhibitor of apoptosis (IAP) gene family, expression is high during fetal development, low in adult tissues. |
| Glucose metabolism | |||
| IGFBP2 | insulin-like growth factor-binding protein 2 | 3485 | Metabolism of proteins; regulation of insulin-like growth factor (IGF) transport and uptake by insulin-like growth factor binding proteins (IGFBPs) |
| ALDOB | aldolase B and C, fructose-bisphosphatases | 229 | Catalyzes the conversion of hexose (fructose-1,6-bisphosphate) to 2 trioses (glyceraldehyde 3-phosphate and dihydroxyacetone phosphate). Vertebrates have 3 aldolase isozymes A, B, and C |
| Defects in ALDOB cause hereditary fructose intolerance. | |||
| ALDOC | 230 | Gene encodes aldolase expressed specifically in the hippocampus and Purkinje cells of the brain | |
| Stress protein-related genes | |||
| HSPA1A | heat shock 70-kDa protein 1A | 3303 | Intronless genes located in the major histocompatibility complex class III region; encodes a 70-kDa heat shock protein that stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and in organelles |
| HSPA1B | heat shock 70-kDa protein 1B | 3304 | |
| Chemokine-related genes | |||
| CCL20 | chemokine (C-C motif) ligand 20 | 6364 | Encodes gene for protein that provides key homing signals for cells of the adaptive immune system, both proinflammatory and suppressive T cell subsets |
| C4BPA | complement component 4-binding protein α | 722 | Gene on human chromosome 1 encodes C4b-binding protein that controls activation of the complement cascade through the classical pathway |
| Immune function, defense against infections | |||
| IFI44L | interferon-induced protein 44-like | 10964 | A type I interferon-stimulated gene that encodes a protein that has modest activity against hepatitis C virus |
| IFIT3 | interferon-induced protein with tetra-tricopeptide repeats 3 | 3437 | Genes involved in expression of INF-induced genes, hallmark of host antiviral immunity |
| MX1 | myxovirus resistance protein 1 | 4599 | Encodes interferon-inducible Mx protein responsible for protection against influenza virus infection and a member of both the dynamin family and the family of large GTPases |
| Absorptive mechanisms | |||
| LCN15 | lipocalin 15 | 389812 | Gene encodes proteins with high affinity to small lipophilic molecules |
| Other functions | |||
| DUSP27 | dual specificity phosphatase 27 | 92235 | Encodes protein with role in MAPK signaling cascades |
| WDR72 | WD repeat-containing protein 72 | 256764 | Encodes a protein with seven WD40 domains. WD or β-transducin repeats typically fold together to form a circular solenoid protein domain called the WD40 domain. Mutations in amelogenesis imperfecta type IIA3 |
Other genes appeared to be less specific to the pathobiology of IBS-D, and these included: 1) cellular intermediary carbohydrate metabolism [such as aldolase B and C genes ALDOB, ALDOC, and insulin growth factor binding protein (IGFBP2)] showed increased expression in IBS-D. 2) Cellular energy metabolism gene DUSP27 showed reduced expression in IBS-D compared with controls. 3) Lipid transport LCN15 is overexpressed in IBS-D relative to controls. 4) Protection against viral infection and stress-related proteins MX1, HSPA1A, and HSPA1B showed reduced expression in IBS-D compared with controls.
Comparison of RNA-Seq fold expression with results on RT-PCR.
Figure 2 demonstrates a comparison of fold changes between patients with IBS and healthy subjects in mRNA expression levels, as measured by RNA-Seq and RT Q-PCR. This includes 10 selected genes identified by RNA-Seq that are, at least theoretically, associated with functions related to pathobiology of IBS-D. In general, there was a significant Spearman correlation between fold change expression by the two techniques [Rs = 0.73, P = 0.013 (Fig. 2)].
The RT-PCR results mirror those of RNA-Seq in terms of the up- or downregulation of genes in IBS-D relative to healthy controls. In addition (Table 5), the RT-PCR identified significant upregulation (P < 0.05) of P2RY4 (also significant on RNA-Seq) and of C4BPA, borderline significant upregulation of GUCA2B and RBP2, and significant downregulation of FN1. Thus results of 5 of the 10 genes selected from RNA-Seq analysis were confirmed on RT-PCR.
Table 5.
Summary of mRNA expression of genes of interest using RT-PCR
| Chr | GeneID | Coding Length | Fold Change | 95% CI | P Value |
|---|---|---|---|---|---|
| GENES IDENTIFIED IN RNA-SEQ AS SIGNIFICANT OR BORDERLINE | |||||
| Neurotransmitter-related genes | |||||
| chrX | P2RY4 | 1639 | 2.714 | (0.94, 4.49) | 0.024 |
| chr6 | VIP | 1586 | 0.949 | (0.00001, 1.94) | 0.433 |
| Ion-channel function-related genes | |||||
| chr1 | GUCA2B | 597 | 2.046 | (0.60, 3.50) | 0.061 |
| chr11 | PDZD3 | 2363 | 1.492 | (0.72, 2.27) | 0.217 |
| Tissue repair, structural proteins, and barrier genes | |||||
| chr21 | TFF1 | 492 | 1.616 | (0.41, 2.82) | 0.514 |
| chr3 | RBP2 | 688 | 1.699 | (0.58, 2.82) | 0.056 |
| chr2 | FN1 | 8975 | 0.417 | (0.21, 0.62) | 0.012 |
| Interferon-related genes | |||||
| chr10 | IFIT3 | 2688 | 0.447 | (0.11, 0.78) | 0.209 |
| Chemokine-related genes | |||||
| chr2 | CCL20 | 842 | 0.654 | (0.00001, 1.45) | 0.637 |
| chr1 | C4BPA | 2243 | 2.398 | (0.19, 4.61) | 0.040 |
| OTHER GENES OF INTEREST MEASURED BY RT-PCR | |||||
| Tight junction protein-related genes | |||||
| 3 | CLDN1 | 3481 | 0.5123 | (0.24, 0.78) | 0.10 |
| 5 | OCLN | 6438 | 1.0341 | (0.48, 1.59) | 0.76 |
| 15 | ZO-1 | 7950 | 0.7658 | (0.45, 1.09) | 0.35 |
| Bile acid homeostasis-, receptor-, and absorption-related genes | |||||
| 5 | FGFR4 | 3122 | 0.8182 | (0.61, 1.03) | 0.17 |
| 2 | GPBAR1 | 2003 | 0.8386 | (0.56, 1.12) | 0.34 |
| 12 | FXR/NH1R4 | 2884 | 1.7404 | (0.94, 2.54) | 0.038 |
| 13 | IBAT/ASBT | 3777 | 1.7337 | (0.28, 3.19) | 0.09 |
| Serotonin transporter gene | |||||
| 17 | SLC6A4 | 6543 | 0.9596 | (0.20, 1.72) | 0.87 |
| Immune function-related gene | |||||
| 9 | TNFSF15 | 6687 | 0.4718 | (0.16, 0.79) | 0.057 |
P values in italics reflect borderline significance, and those in bold reflect significance.
Moreover (Table 5), the RT-PCR demonstrated borderline decreased expressions of the tight junction protein claudin 1 and an immune modulator (TNFSF15). Conversely, there was significantly increased expression of the nuclear receptor, FXR, and borderline increased expression of apical sodium-coupled bile acid transporter (IBAT/ASBT). Table 6 shows that the altered mRNA expression of these genes was not detected on RNA-Seq.
Table 6.
mRNA expression using RNA-Seq of additional genes of interest identified by the RT-PCR
| Chr | GeneID | Coding Length | logFC | P Value | P (FDR) | |
|---|---|---|---|---|---|---|
| Additional tight junction protein-related genes | ||||||
| Chr 3 | CLDN1 | 3452 | 0.60 | 0.081 | 1 | |
| Chr 5 | OCLN | 6576 | −0.029 | 0.827 | 1 | |
| Chr 15 | ZO-1 | 7164 | 0.068 | 0.793 | 1 | |
| Bile acid homeostasis-, receptor-, and absorption-related genes | ||||||
| Chr 5 | FGFR4 | 3143 | −0.089 | 0.777 | 1 | |
| Chr 2 | GPBAR1 | 2003 | −0.165 | 0.617 | 1 | |
| Chr 12 | NH1R4 | 2589 | 0.947 | 0.011 | 0.609 | |
| chr13 | IBAT/ASBT | No data | ||||
SLC6A4 and TNFSF15 were already documented in Table 2.
DISCUSSION
This is the first report of an RNA-Seq study of the expression of genes in patients with IBS-D compared with controls. This analysis builds on a prior analysis that we conducted several years ago, in which we examined the rectosigmoid mucosal expression of RNA from 30 patients with IBS conducted using Affimetrix Gene chips. The former study showed functional alterations of several components of the host mucosal immune response to microbial pathogens. The current study shows highly relevant alterations of mRNA expression that reflect alterations in functions that predispose to the pathophysiology of IBS-D.
Observations from mucosal RNA-Seq analysis.
In the section of the current study that was guided by an a priori hypothesis, patients with IBS-D had reduced expression of TNFSF15, which is involved in innate immune responses (15; 33). Our prior study had shown an association of TNFSF15 genotype, particularly with IBS-C (45). Thus, genotype TNFSF15 (G allele) was associated with IBS [P = 2.2 × 10−5; odds ratio (OR) 1.37; 95% CI 1.19 to 1.58] and was predominantly due to a significant association with IBS-C (P = 8.7 × 10−7; OR 1.79; 95% CI 1.41 to 2.26). We did observe that there was a correlation of rs4263839 genotype (GG genotype) with gene expression of TNFSF15 (mRNA levels), both in peripheral blood leucocytes and in rectal mucosal biopsies from healthy individuals (45). IBS-C (and G allele carriers) would have greater tissue expression of TNFSF15, and we hypothesize that IBS-D would be associated with reduced tissue expression of TNFSF15, consistent with our demonstration of reduced expression of TNFSF15 mRNA in patients with IBS-D with rapid colonic transit; further studies need to be conducted in the future to validate current findings.
In the unbiased analysis section of our study, we made several novel observations that led to testable hypotheses that are relevant to the pathobiology of IBS-D and that require confirmation in the future in validation cohorts. It is important to note that the increased or decreased expressions in tissues are typically >1.5 (Fig. 1). Peart et al. (30) and Raouf et al. (31) declared genes to be differentially expressed if they showed a fold change of at least 1.5 and also satisfied P < 0.05 after adjustment for multiple testing, as was used in the present analysis.
First, there was increased expression of two neurotransmitter ligand or receptor genes, VIP and P2RY4, that are involved in secretomotor and peristaltic reflexes, intrinsic sensory afferents and mucosal blood flow. Second, there was increased expression of two genes associated with ion channels or ion secretion, GUCA2B and PDZD3, and both control the enterocyte secretion of chloride ions through the effects of guanylate cyclase C receptor in response to endogenous ligands such as intestinal uroguanylin, which is secreted with mucins from goblet cells in normal physiological processes (13; 23). These two mechanisms may lead to increased fluid secretion and conceivably contribute to looser stool consistency in patients with IBS-D. The ileal epithelium has been shown to be more sensitive to the secretory effects of bile acids in patients with IBS-D (28). The relevance of genetic control of guanylate cyclase C receptor on the impact of clinical symptoms was clearly demonstrated in a report of an entire family diagnosed as having chronic functional diarrhea; in that family, there was a dominantly inherited, fully penetrant disease in 32 members that resulted from a missense mutation in GUCY2C (20).
A third group of overexpressed molecules, TFF1 and RBP2, is related to tissue repair or barrier function; such expression would be important to restore normal epithelial structure, for example, in the one-third of patients with IBS-D who have evidence of malabsorption of endogenous bile acids (34; 42) that have detergent effects resulting in increased mucosal permeability. On the other hand, in the same group of barrier-related genes, WDR72 and fibronectin 1 (FN1) are underexpressed in IBS-D relative to controls and may be responsible for the increased mucosal permeability and, possibly, the associated visceral hypersensitivity of IBS (10).
Fourth, the overexpression of a lipid transport-related gene, lipocalin 15 (LCN15), may also be a compensatory change to the slight increase in fecal fat observed in patients with IBS-D compared with controls and patients with IBS and constipation (34; 42). A fifth observation is that the spectrum of genes associated with interferons, responses to viruses (including myxoviruses), responses to stress (such as heat shock proteins), and innate immune responses (e.g., TNFSF15) was underexpressed in patients with IBS-D compared with controls. The underexpression of antimicrobial mechanisms, such as caspase 1 and lysozyme in mucosal biopsies previously demonstrated by Aerssens et al. (2), is consistent with reduced resistance to microbes. These characteristics may conceivably predispose to postinfectious IBS (35) or the pauci-inflammatory variant of IBS, which is most commonly observed in IBS-D relative to other groups (14). The current study did not replicate the prior observations in the study by Aerssens et al. (2), in which five genes were expressed at higher level in mucosa from patients with IBS, with fold changes in expression <1.53 for all genes. Of these five genes with increased expression in IBS, two genes are of unclear function, one was apparently unrelated to gut function (creatine kinase, brain), two reflect mucin production (MUC20), and one gene (VSIG2) is involved in myeloid cell functions. Hence, the current data have failed to confirm upregulation of mucin gene expression, and, possibly, one gene may be associated with immune activation. It is worth noting that, although all patients in the two studies were recruited and studied in our laboratory, the report by Aerssens et al. (2) included a heterogeneous group of 36 patients (15 with IBS-C and 21 with IBS-D), whereas the current study included nine patients selected by the most accelerated colonic transit in an attempt to have as homogeneous a group as possible for the RNA-Seq analysis. Therefore, the participants in the two studies had very different phenotypes, and, because there are diverse peripheral mechanisms leading to IBS (7), we hypothesize that these are associated with differential expression of genes or posttranslational modification in the mucosa in the different subgroups of IBS. Further prospective studies of the subgroup of IBS related to epidemiologically proven relation to enteric infection are indicated to ascertain whether the observed reductions in expression occur more frequently in postinfectious cases of IBS and to appraise the time course of the disordered expression.
Disorders in expression of factors protective against bacterial infection, such as cytokine and tissue repair overexpression in the current study, overexpression of mucin genes [MUC20 and V-set and immunoglobulin domain containing, 2 gene (VSIG2)] in the previous study by Aerssens et al. (2), as well as overexpression of Toll-like receptors demonstrated by Brint et al. (6), are all consistent with a role of microbiota and immune activation in the pathophysiology of IBS. Conversely, the underexpression of genes that protect the mucosa from luminal antigens or the inhibition of caspases that result in reduction in the apoptosis of immune cells may ultimately lead to the low-grade inflammation in IBS. Such reductions in gene expression identified in the Aerssens et al. study (2) were scavenger receptor cysteine-rich type 1 protein (M160), CD163 antigen-like 1, V-set and immunoglobulin domain-containing 4 (VSIG4), caspase 1, apoptosis-related cysteine peptidase (CASP1), IL-1β (convertase), neutrophil cytosolic factor 4 (NCF4), and lysozyme (LYZ). In the current study, downregulation of TNFSF15, IFIT3, MX1, HSPA1A, and HSPA1B is consistent with the same concept.
Observations from RT-PCR analysis of mRNA expression.
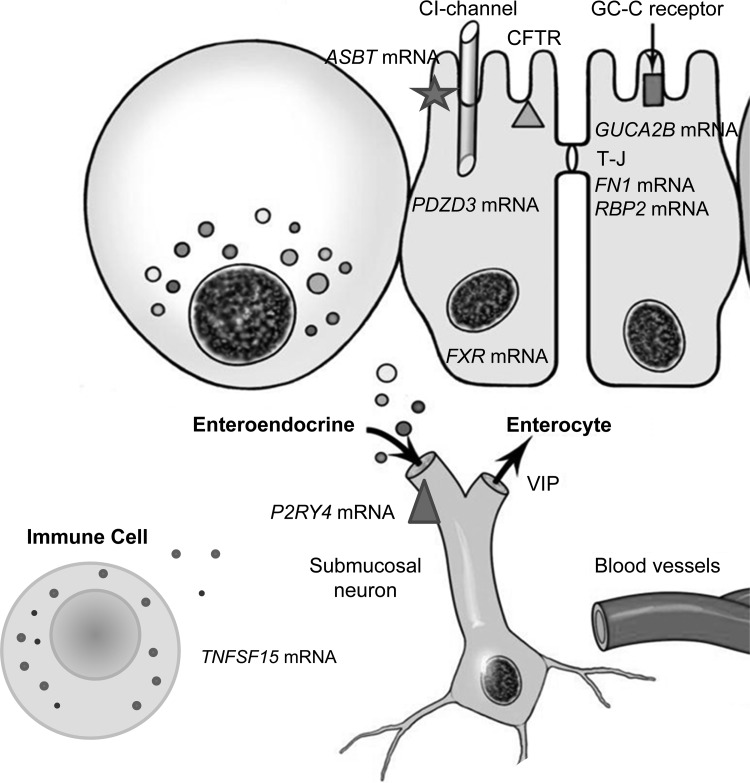
The RT-PCR analysis demonstrated numerical up- or downregulation of genes that were in the same direction as observed on the RNA-Seq analysis, and fold changes were confirmed by both methods for five genes (P2RY4, VIP, GUCA2B, C4BPA, and FN1); there was a significant correlation between the expressions by the two methods, suggesting that the RNA-Seq data are valid. In addition, the RT-PCR analysis identified significantly increased expression of FXR gene as well as interesting numerical fold changes (all P ≤ 0.10) in claudin 1, ASBT, and TNFSF15 genes. These findings are consistent with alterations in the mucosal expression of barrier, bile acid absorption, feedback regulation of synthesis, or immune activation. Importantly, ASBT expression is increased, and this is consistent with the literature that shows no evidence of mutation of the gene as a cause of bile acid malabsorption in patients with IBS (26). Conversely, the increased expression of the nuclear receptor, FXR, is consistent with the increased production of FGF-19 and feedback regulation of hepatic bile acid synthesis in patients with IBS-D. These interesting data provide hypotheses for replication in future studies. Figure 3 summarizes the overall findings in the current studies of mRNA expression. Pathway and ingenuity analyses provided confirmatory information on the relevance of these gastrointestinal mechanisms in IBS and identified common cellular and metabolic processes that may not be specifically related to the pathobiology of IBS.
Fig. 3.
Cartoon summarizing altered mRNA expression in cellular elements of the rectosigmoid mucosa in patients with IBS-D, based on the more stringent statistical analyses. TJ, tight junction; GC-C, guanylate cyclase; CFTR, cystic fibrosis transmembrane regulator; VIP, vasoactive intestinal peptide.
There are key limitations of this study. First, it is a pilot case-control study that requires replication. However, given the expense involved in such RNA-Seq studies, we perceive that this study provides the basis for planning future studies with regard to alterations in mucosal function, and it may allow greater focus on such mucosal mechanisms of secretion and barrier and immune functions in patients with IBS-D. Replication in a different cohort of patients with IBS-D and rapid colonic transit will inform researchers on the generalizability of the current findings. Regrettably, there are no other RNA-Seq studies in the current literature with which we can compare or contrast the results of the current study. However, the replication of several of the RNA-Seq fold changes by the RT-PCR data provides reassurance that the RNA-Seq data offer a rationale for future hypothesis-based studies that will be necessary to replicate these data. A second limitation is that we are unable to determine whether alterations of the transcriptome observed in the rectosigmoid mucosa of the patients with IBS-D are the cause or consequence of their condition. A third limitation is that the observations here pertain to a subgroup of patients with IBS-D and extremely fast colonic transit.
In conclusion, this pilot study demonstrates the potential utility of in-depth analysis, based on deep RNA-Seq of stored rectosigmoid mucosal biopsies, to develop greater understanding of the pathobiology of IBS-D and to help develop hypotheses for future studies. RNA-Seq and RT-PCR appear to be complementary in identifying changes in rectosigmoid mucosal mRNA expression of genes of interest. Rectosigmoid mucosa analysis by RNA-Seq and RT-PCR in IBS-D shows transcriptome changes affecting neurotransmitters, ion channels, cytokines, immune function, cell adhesion, or barrier function that provide the rationale for future validation studies to explore the role of mucosal factors in the pathobiology of IBS-D. Overall, these data continue to support the large body of evidence demonstrating the disturbances of peripheral functions in patients with IBS (7).
GRANTS
This study is supported by NIH RO1-DK92179.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: M.C. and G.F. conception and design of research; M.C., A.A.N., and E.W.K. analyzed data; M.C. and E.W.K. interpreted results of experiments; M.C. prepared figures; M.C. drafted manuscript; M.C., P.C., A.A., I.A.B., A.A.N., S.J.G., G.F., and E.W.K. edited and revised manuscript; M.C., P.C., A.A., and G.F. approved final version of manuscript; P.C., A.A., I.A.B., and S.J.G. performed experiments.
ACKNOWLEDGMENTS
The authors thank Cindy Stanislav for secretarial assistance.
REFERENCES
- 1.Acosta A, Carlson P, Busciglio I, Gibbons SJ, Farrugia G, Klee E, Camilleri M. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea. Gastroenterology 146, Suppl 1: S18, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aerssens J, Camilleri M, Talloen W, Thielemans L, Göhlmann HW, Van Den Wyngaert I, Thielemans T, De Hoogt R, Andrews CN, Bharucha AE, Carlson PJ, Busciglio I, Burton DD, Smyrk T, Urrutia R, Coulie B. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 194–205, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Areeshi MY, Haque S, Panda AK, Mandal RK. A serotonin transporter gene (SLC6A4) polymorphism is associated with reduced risk of irritable bowel syndrome in American and Asian population: a meta-analysis. PLoS One 8: e75567, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmonte L, Beutheu Youmba S, Bertiaux-Vandaële N, Antonietti M, Lecleire S, Zalar A, Gourcerol G, Leroi AM, Déchelotte P, Coëffier M, Ducrotté P. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One 7: e42777–e42786, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertiaux-Vandaële N, Youmba SB, Belmonte L, Lecleire S, Antonietti M, Gourcerol G, Leroi AM, Déchelotte P, Ménard JF, Ducrotté P, Coëffier M. The expression and the cellular distribution of the tight junction proteins are altered in irritable bowel syndrome patients with differences according to the disease subtype. Am J Gastroenterol 106: 2165–2173, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol 106: 329–336, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 367: 1626–1635, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Camilleri M, Andrews CN, Bharucha AE, Carlson PJ, Ferber I, Stephens D, Smyrk TC, Urrutia R, Aerssens J, Thielemans L, Göhlmann H, van den Wyngaert I, Coulie B. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology 132: 17–25, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camilleri M, Katzka DA. Invited Review. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Genetic epidemiology and pharmacogenetics in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 302: G1075–G1084, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilleri M, Lasch K, Zhou W. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. The confluence of increased permeability, inflammation, and pain in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 303: G775–G785, 2012 [DOI] [PubMed] [Google Scholar]
- 11.Camilleri M, McKinzie S, Busciglio I, Low PA, Sweetser S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Prospective study of motor, sensory, psychologic, and autonomic functions in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 772–781, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, Moses PL. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology 126: 1657–1664, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Cohen MB, Witte DP, Hawkins JA, Currie MG. Immunohistochemical localization of guanylin in the rat small intestine and colon. Biochem Biophys Res Commun 209: 803–808, 1995 [DOI] [PubMed] [Google Scholar]
- 14.Cremon C, Gargano L, Morselli-Labate AM, Santini D, Cogliandro RF, De Giorgio R, Stanghellini V, Corinaldesi R, Barbara G. Mucosal immune activation in irritable bowel syndrome: gender-dependence and association with digestive symptoms. Am J Gastroenterol 104: 392–400, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol 9: 271–285, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debongnie JC, Phillips SF. Capacity of the human colon to absorb fluid. Gastroenterology 74: 698–703, 1978 [PubMed] [Google Scholar]
- 17.Dunlop SP, Coleman NS, Blackshaw E, Perkins AC, Singh G, Marsden CA, Spiller RC. Abnormalities of 5 hydroxytryptamine metabolism in irritable bowel syndrome. Clin Gastroenterol Hepatol 3: 349–357, 2005 [DOI] [PubMed] [Google Scholar]
- 18.El-Salhy M, Gundersen D, Hatlebakk JG, Gilja OH, Hausken T. Abnormal rectal endocrine cells in patients with irritable bowel syndrome. Regul Pept 188C: 60–65, 2013 [DOI] [PubMed] [Google Scholar]
- 19.El-Salhy M, Wendelbo I, Gundersen D. Serotonin and serotonin transporter in the rectum of patients with irritable bowel disease. Mol Med Rep 8: 451–455, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Fiskerstrand T, Arshad N, Haukanes BI, Tronstad RR, Pham KD, Johansson S, Håvik B, Tønder SL, Levy SE, Brackman D, Boman H, Biswas KH, Apold J, Hovdenak N, Visweswariah SS, Knappskog PM. Familial diarrhea syndrome caused by an activating GUCY2C mutation. N Engl J Med 366: 1586–1595, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Houghton LA, Atkinson W, Whitaker RP, Whorwell PJ, Rimmer MJ. Increased platelet depleted plasma 5 hydroxytryptamine concentration following meal ingestion in symptomatic female subjects with diarrhoea predominant irritable bowel syndrome. Gut 52: 663–670, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerckhoffs AP, ter Linde JJ, Akkermans LM, Samsom M. SERT and TPH-1 mRNA expression are reduced in irritable bowel syndrome patients regardless of visceral sensitivity state in large intestine. Am J Physiol Gastrointest Liver Physiol 302: G1053–G1060, 2012 [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Taylor-Blake B, Light AR, Goy MF. Guanylin, an endogenous ligand for C-type guanylate cyclase, is produced by goblet cells in the rat intestine. Gastroenterology 109: 1863–1875, 1995 [DOI] [PubMed] [Google Scholar]
- 24.Martínez C, Lobo B, Pigrau M, Ramos L, González-Castro AM, Alonso C, Guilarte M, Guilá M, de Torres I, Azpiroz F, Santos J, Vicario M. Diarrhoea-predominant irritable bowel syndrome: an organic disorder with structural abnormalities in the jejunal epithelial barrier. Gut 62: 1160–1168, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Martínez C, Vicario M, Ramos L, Lobo B, Mosquera JL, Alonso C, Sánchez A, Guilarte M, Antolín M, de Torres I, González-Castro AM, Pigrau M, Saperas E, Azpiroz F, Santos J. The jejunum of diarrhea-predominant irritable bowel syndrome shows molecular alterations in the tight junction signaling pathway that are associated with mucosal pathobiology and clinical manifestations. Am J Gastroenterol 107: 736–746, 2012 [DOI] [PubMed] [Google Scholar]
- 26.Montagnani M, Love MW, Rössel P, Dawson PA, Qvist P. Absence of dysfunctional ileal sodium-bile acid cotransporter gene mutations in patients with adult-onset idiopathic bile acid malabsorption. Scand J Gastroenterol 36: 1077–1080, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Oddsson E, Rask-Madsen J, Krag E. A secretory epithelium of the small intestine with increased sensitivity to bile acids in irritable bowel syndrome associated with diarrhoea. Scand J Gastroenterol 13: 408–416, 1978 [PubMed] [Google Scholar]
- 29.Ohman L, Stridsberg M, Isaksson S, Jerlstad P, Simrén M. Altered levels of fecal chromogranins and secretogranins in IBS: relevance for pathophysiology and symptoms? Am J Gastroenterol 107: 440–447, 2012 [DOI] [PubMed] [Google Scholar]
- 30.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, Holloway AJ, Johnstone RW. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA 102: 3697–3702, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, Aparicio S, Marra M, Eaves C. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3: 109–118, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih DQ, Kwan LY, Chavez V, Cohavy O, Gonsky R, Chang EY, Chang C, Elson CO, Targan SR. Microbial induction of inflammatory bowel disease associated gene TL1A (TNFSF15) in antigen presenting cells. Eur J Immunol 39: 3239–3250, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin A, Camilleri M, Vijayvargiya P, Busciglio I, Burton D, Ryks M, Rhoten D, Lueke A, Saenger A, Girtman A, Zinsmeister AR. Bowel functions, fecal unconjugated primary and secondary bile acids, and colonic transit in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 11: 1270–1275, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology 136: 1979–1988, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, Neal KR, Whorwell PJ, Hall IP, Spiller RC. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut 62: 985–994, 2013 [DOI] [PubMed] [Google Scholar]
- 37.Vazquez-Roque MI, Camilleri M, Carlson P, McKinzie S, Murray JA, Brantner TL, Burton DD, Zinsmeister AR. HLA-DQ genotype is associated with accelerated small bowel transit in patients with diarrhea-predominant irritable bowel syndrome. Eur J Gastroenterol Hepatol 23: 481–487, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez-Roque MI, Camilleri M, Smyrk T, Murray JA, Marietta E, O'Neill J, Carlson P, Lamsam J, Janzow D, Eckert D, Burton D, Zinsmeister AR. A controlled trial of gluten-free diet in irritable bowel syndrome-diarrhea: effect on bowel frequency and intestinal functions. Gastroenterology 144: 903–911; e3, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez-Roque MI, Camilleri M, Smyrk T, Murray JA, O'Neill J, Carlson P, Lamsam J, Eckert D, Janzow D, Burton D, Ryks M, Rhoten D, Zinsmeister AR. Association of HLA-DQ gene with bowel transit, barrier function and inflammation in irritable bowel syndrome with diarrhea. Am J Physiol Gastrointest Liver Physiol 303: G1262–G1269, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villani AC, Lemire M, Thabane M, Belisle A, Geneau G, Garg AX, Clark WF, Moayyedi P, Collins SM, Franchimont D, Marshall JK. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology 138: 1502–1513, 2010 [DOI] [PubMed] [Google Scholar]
- 41.Wang YM, Chang Y, Chang YY, Cheng J, Li J, Wang T, Zhang QY, Liang DC, Sun B, Wang BM. Serotonin transporter gene promoter region polymorphisms and serotonin transporter expression in the colonic mucosa of irritable bowel syndrome patients. Neurogastroenterol Motil 24: 560–565, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Wong BS, Camilleri M, Carlson P, McKinzie S, Busciglio I, Bondar O, Dyer RB, Lamsam J, Zinsmeister AR. Increased bile acid biosynthesis is associated with irritable bowel syndrome with diarrhea. Clin Gastroenterol Hepatol 10: 1009–1015; e3, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong BS, Camilleri M, Carlson PJ, Guicciardi ME, Burton D, McKinzie S, Rao AS, Zinsmeister AR, Gores GJ. A klothoβ variant mediates protein stability and associates with colon transit in irritable bowel syndrome with diarrhea. Gastroenterology 140: 1934–1942, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wouters MM, Lambrechts D, Knapp M, Cleynen I, Whorwell P, Agréus L, Dlugosz A, Schmidt PT, Halfvarson J, Simrén M, Ohlsson B, Karling P, Van Wanrooy S, Mondelaers S, Vermeire S, Lindberg G, Spiller R, Dukes G, D'Amato M, Boeckxstaens G. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut. In press [DOI] [PubMed] [Google Scholar]
- 45.Zucchelli M, Camilleri M, Nixon Andreasson A, Bresso F, Dlugosz A, Halfvarson J, Törkvist L, Schmidt PT, Karling P, Ohlsson B, Duerr RH, Simren M, Lindberg G, Agreus L, Carlson P, Zinsmeister AR, D'Amato M. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 60: 1671–1677, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]