Abstract
More than 400 serotypes of Shiga toxin-producing Escherichia coli (STEC) have been implicated in outbreaks and sporadic human diseases. In recent years STEC strains belonging to serogroup O178 have been commonly isolated from cattle and food of bovine origin in South America and Europe. In order to explore the significance of these STEC strains as potential human pathogens, 74 German and Argentinean E. coli O178 strains from animals, food and humans were characterized phenotypically and investigated for their serotypes, stx-genotypes and 43 virulence-associated markers by a real-time PCR-microarray. The majority (n = 66) of the O178 strains belonged to serotype O178:H19. The remaining strains divided into O178:H7 (n = 6), O178:H10 (n = 1), and O178:H16 (n = 1). STEC O178:H19 strains were mainly isolated from cattle and food of bovine origin, but one strain was from a patient with hemolytic uremic syndrome (HUS). Genotyping of the STEC O178:H19 strains by pulsed-field gel electrophoresis revealed two major clusters of genetically highly related strains which differ in their stx-genotypes and non-Stx putative virulence traits, including adhesins, toxins, and serine-proteases. Cluster A-strains including the HUS-strain (n = 35) carried genes associated with severe disease in humans (stx2a, stx2d, ehxA, saa, subAB1, lpfAO113, terE combined with stx1a, espP, iha). Cluster B-strains (n = 26) showed a limited repertoire of virulence genes (stx2c, pagC, lpfAO113, espP, iha). Among O178:H7 strains isolated from deer meat and patients with uncomplicated disease a new STEC variant was detected that is associated with the genotype stx1c/stx2b/ehxA/subAB2/espI/[terE]/espP/iha. None of the STEC O178 strains was positive for locus of enterocyte effacement (LEE)- and nle-genes. Results indicate that STEC O178:H19 strains belong to the growing group of LEE-negative STEC that should be considered with respect to their potential to cause diseases in humans.
Keywords: E. coli O178, STEC, Shiga toxins, virulence, genotyping, PFGE, real-time PCR micro array
Introduction
Shiga toxin-producing Escherichia coli (STEC) is a zoonotic pathogen of significant public health concern. Worldwide, STEC has been associated with both outbreaks and sporadic cases of human disease, ranging from mild diarrhea to hemorrhagic colitis (HC) and life-threatening hemolytic uremic syndrome (HUS) (Melton-Celsa et al., 2012). In Argentina, HUS is endemic with 500 cases per year and an incidence of 17/100,000 children less than 5 years of age (Rivas et al., 2010). In contrast, Germany notifies about 65 HUS cases per year and an incidence of 0.1/100,000 population (Robert Koch-Institut, 2011). STEC strains produce one or both of two major types of potent cytotoxins called Shiga toxins Stx1 and Stx2, which function as primary virulence factors in human disease. Stx1 and Stx2 are immunologically not cross-reactive and display 57 and 60% nucleotide sequence identity in their A and B subunits, respectively (Muthing et al., 2009). The more homogenous Stx1 family consists of Stx1a, Stx1c, and Stx1d. The more heterogeneous Stx2 family comprises Stx2a and the variants designated Stx2b, Stx2c, Stx2d, Stx2e, Stx2f, and Stx2g (Scheutz et al., 2012). Stx2d differs from all known Stx types because it is activated in its biological activity by elastase, a constituent of the intestinal mucus (Melton-Celsa et al., 1996).
The STEC group is serologically highly diverse. More than 400 O:H types of STEC associated with infections in humans were recorded in 2005 (Scheutz and Strockbine, 2005). In the last years, the E. coli serotyping scheme was extended due to the emergence of STEC strains belonging to the newly defined O-serogroups O174 to O181 (Scheutz et al., 2004). Cattle have been recognized as the most important animal reservoir of STEC causing disease in humans (Hussein, 2007). However, there are only few data available about animal reservoir, epidemiology and human pathogenicity of the new emerging STEC types. STEC O178 was first described in 2004; the prototype strain (O178:H7, stx1, and stx2) was isolated from raw meat (Scheutz et al., 2004). STEC O178:H19 strains were frequently isolated from beef in Germany (Beutin et al., 2007; Werber et al., 2008; Slanec et al., 2009) and Argentina (Sanz et al., 2007; Kruger et al., 2011). Cattle were identified as important reservoir for STEC O178:H19 in South America (Cergole-Novella et al., 2007; Oliveira et al., 2008; Fernandez et al., 2010; Kruger et al., 2011; Masana et al., 2011; Tanaro et al., 2012) and in Europe (Blanco et al., 2004; Aidar-Ugrinovich et al., 2007; European Food Safety Authority and European Centre for Disease Prevention and Control, 2012). Sporadic cases of human infections with STEC O178:H19 were reported from Germany (Bielaszewska et al., 2006; Werber et al., 2008) and Brazil (De Toni et al., 2009). STEC O178:H19 strains were also reported as the cause of HUS cases in Belgium (Buvens et al., 2010) and Argentina (Giugno et al., 2007). Besides O178:H19, STEC O178:H7 producing stx2f were associated with diarrhea in humans (Prager et al., 2009).
At present, relatively few cases of human illness were associated with STEC O178 infections. Such cases might be underreported because serotyping of non-O157 STEC is only performed by a small number of reference laboratories worldwide (Bettelheim, 2007). On the other hand, it is possible that STEC O178 strains are less virulent for humans compared with other frequently isolated STEC types. A number of factors were shown to play a role in the virulence of STEC for humans. Some variants of the Stx2 gene, namely Stx2a, Stx2c and the mucus activatable Stx2d were significantly associated with HC and HUS in human patients (Friedrich et al., 2002; Jelacic et al., 2003; Bielaszewska et al., 2006). Besides production of Shiga toxins, the attaching and effacing phenotype encoded by the genomic locus of enterocyte effacement (LEE), and the presence of some non-LEE genomic O-islands (OI) such as OI-122, OI-71 and OI-57, are significantly associated with STEC types that are frequently involved in outbreaks and cause HC and HUS in humans (Karmali et al., 2003; Coombes et al., 2008; Konczy et al., 2008; Bugarel et al., 2010a,b). The LEE encodes for the outer-membrane adhesin intimin (eae), the translocated intimin receptor Tir, a type III secretion system, and effector proteins translocated by the secretion system. OI-122, OI-71 and OI-57 harbor a number of non-LEE encoded effector (nle) genes which encode potential virulence traits (Coombes et al., 2008; Konczy et al., 2008). Additional adhesins and putative virulence factors encoded outside of the LEE in genomic islands or on large plasmids, such as pO157 in LEE-positive strains and pO113 in LEE-negative strains, may play a role in pathogenesis. Plasmids pO157 and pO113 share several virulence genes such as ehxA, which encodes an EHEC enterohemolysin (Schmidt et al., 1995), espP, which encodes an extracellular serine protease (Brunder et al., 1999), and iha which was also found within a genomic island, and encodes an adherence-conferring protein similar to Vibrio cholerae IrgA (Tarr et al., 2000; Schmidt et al., 2001; Newton et al., 2009). However, pO113 lacked the pO157-encoded type II secretion system (etpD), a periplasmic catalase peroxidase (katP) (Schmidt et al., 1997; Brunder et al., 1999) and a homolog of the adherence-promoting protein ToxB (toxB) (Tatsuno et al., 2001). On the other hand, pO113 encodes a number of unique virulence-associated determinants, including the autoagglutinating adhesin Saa (saa) and the subtilase-like serine protease toxin SubAB (subAB) (Paton et al., 2004). Saa encoded by LEE-negative O113:H21 (Paton et al., 2001) and O91:H21 (Paton and Paton, 2002) strains was characterized as putative colonization factor that enables LEE-negative STEC strains to adhere to the intestinal epithelium of humans. SubAB might also contribute to the virulence of LEE-negative STEC strains in humans by a synergistic action with Stx (Paton et al., 2004). Furthermore, long polar fimbriae (LPF) (lpfA) closely related to LPF of Salmonella enterica serovar Typhimurium, were proposed to be novel adhesion factors (Doughty et al., 2002; Torres et al., 2002).
Due to the recent emergence of STEC O178 strains in cattle and food of bovine origin on both the South American and the European continent, we became interested in studying the virulence potential of such strains. For this, we compared E. coli O178 isolates from Argentina and Germany originating from animals, food and humans in regard to their virulence properties, including toxins, adhesins, and serine proteases. Furthermore, we analyzed the genetic relationships among these strains using data obtained by pulsed-field gel electrophoresis. Our results indicate that STEC O178:H19 belong to the growing group of LEE-negative strains that are able to cause severe illness in humans (Newton et al., 2009).
Materials and methods
Bacterial strains
A total of 74 E. coli serogroup O178 strains were characterized in this study (Table 1). All except three were isolated between 2005 and 2012 and originated from Argentina (n = 49) or Germany (n = 25). Forty-two of the strains were isolated from feces or carcasses of cattle originating from different farms and 19 from food, such as beef (n = 4), ground beef (n = 12), ground beef/pork (n = 2), and raw cows milk (n = 1). Six strains were obtained from other food sources, such as ground lamb (n = 1), red deer meat (n = 4), and salad (n = 1). Of the four strains from humans examined, three were isolated from patients with uncomplicated disease and one from a HUS patient. Three strains were from rectal swabs of dogs.
Table 1.
Characteristic traits of the E. coli O178 isolates investigated in this study.
| Presence of (putative) virulence genesa | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | H-typeb | Source | Origin | Year | stx-genotypec | ehxA | espP | iha | saa | subABd | tia | lpfAO113 | terE | pagC | espIe |
| CB10181 | 19 | beef | Germany | 2005 | stx1a stx2a | + | + | + | + | − | nd | + | − | − | − |
| CB10542 | 19 | ground beef | Germany | 2006 | stx1a stx2a stx2c | + | + | + | + | − | nd | + | − | − | − |
| CB10545 | 19 | ground beef | Germany | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| CB10659 | 19 | ground lamb | Germany | 2006 | stx2a | + | + | + | + | subAB1 | − | + | − | + | − |
| CB10751 | 19 | meat, deer | Germany | 2007 | stx2a | + | + | + | + | subAB1 | − | + | − | + | − |
| CB10836 | 19 | ground beef/pork | Germany | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| CB11017 | 19 | ground beef | Germany | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| CB11536 | 19 | ground beef/pork | Germany | 2008 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| CB11726 | 19 | ground beef | Germany | 2008 | stx2a stx2d | + | + | + | + | subAB1 | − | + | − | + | − |
| CB12008 | 19 | raw milk | Germany | 2009 | stx1a stx2a stx2d | + | + | + | + | subAB1 | − | + | + | − | − |
| CB12145 | 19 | ground beef | Germany | 2009 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| CB12220 | 19 | beef | Germany | 2009 | stx1a stx2a | + | + | + | + | subAB1 | − | + | + | − | − |
| CB13785 | 19 | beef | Germany | 2011 | stx1a stx2d | + | + | + | + | − | nd | + | − | − | − |
| CB13787 | 19 | beef | Germany | 2011 | stx1a stx2d | + | + | + | + | − | nd | + | − | − | − |
| CB14014 | 19 | ground beef | Germany | 2012 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| CB14018 | 19 | cattle | Germany | 2012 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| CB14051 | 19 | cattle | Germany | 2012 | stx1a stx2d | + | + | + | + | − | nd | + | − | − | − |
| 188/06 30 | 19 | cattle | Argentina | 2006 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| 276 | 19 | dog | Argentina | 2005 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| 377ii | 19 | dog | Argentina | 2005 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| 489/06 16 | 19 | cattle | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| 489/06 24 | 19 | cattle | Argentina | 2006 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| 489/06 36 | 19 | cattle | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| 612/06 21 | 19 | cattle | Argentina | 2006 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| 612/06 33 | 19 | cattle | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| 612/06 35 | 19 | cattle | Argentina | 2006 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| 612/06 9 | 19 | cattle | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| 96/00 | 19 | human (HUS) | Argentina | 2000 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| C11 | 19 | ground beef | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| C18iii | 19 | ground beef | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| C22 | 19 | ground beef | Argentina | 2006 | stx2c stx2b | − | + | + | − | − | nd | + | − | + | − |
| C27 | 19 | ground beef | Argentina | 2006 | stx1a stx2d | + | + | + | + | subAB1 | − | + | − | − | − |
| C39 | 19 | ground beef | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| C42 | 19 | ground beef | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−001 | 19 | cattle | Argentina | 2006 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−023 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−024 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| FP−065 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| FP−067 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| FP−070 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| FP−073 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| FP−075 | [19] | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| FP−080 | 19 | cattle | Argentina | 2007 | stx2a stx2d | + | + | + | + | subAB1 | − | − | − | − | − |
| FP−082 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | + | + | − |
| FP−087 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| FP−089 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−090 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | + | + | − |
| FP−100 | 19 | cattle | Argentina | 2007 | stx2a | + | + | + | + | subAB1 | − | + | − | + | − |
| FP−110 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | − | nd | + | + | − | − |
| FP−112 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−113 | 19 | cattle | Argentina | 2009 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−116 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−134 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−163 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−194 | 19 | cattle | Argentina | 2008 | stx2c | − | + | + | − | − | nd | + | + | + | − |
| FP−203 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | − | nd | + | − | − | − |
| FP−229 | [19] | cattle | Argentina | 2008 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| FP−245 | 19 | cattle | Argentina | 2008 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| I−005 | 19 | cattle | Argentina | 2006 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| I−011 | [19] | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| I−023 | [19] | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| I−039 | 19 | cattle | Argentina | 2007 | stx1a stx2a stx2d | + | + | + | + | subAB1 | − | + | + | + | − |
| I−058 | 19 | cattle | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| I−073 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | − | nd | + | − | − | − |
| I−115 | 19 | cattle | Argentina | 2007 | stx1a stx2a | + | + | + | + | subAB1 | − | + | − | − | − |
| P5a | [19] | dog | Argentina | 2007 | stx2c | − | + | + | − | − | nd | + | − | + | − |
| CB06224 | [7] | human (D) | Germany | 1996 | stx1c stx2b | + | + | + | − | subAB2 | + | + | − | − | + |
| CB06571 | [7] | human (AS) | Germany | 1996 | stx1c | + | + | + | − | subAB2 | + | + | + | − | + |
| CB09017f | [7] | human (E) | Germany | 2001 | − | − | − | − | − | − | nd | + | − | − | − |
| CB10124 | [7] | meat, deer | Germany | 2005 | stx1c stx2b | + | + | + | − | subAB2 | + | + | − | − | + |
| CB11675 | [7] | meat, deer | Germany | 2008 | stx1c stx2b | + | + | + | − | subAB2 | + | + | + | − | + |
| CB12072 | 7 | cattle | Germany | 2009 | − | − | − | − | − | − | nd | + | − | − | − |
| CB11890 | 10 | meat, deer | Germany | 2009 | stx1a | − | + | + | − | subAB1 | − | + | − | − | − |
| CB12372 | 16 | salad | Germany | 2009 | − | − | − | − | − | − | nd | + | − | − | − |
HUS, hemolytic uremic syndrome; D, diarrhea; E, enteritis; AS, asymptomatic carrier; +, the gene is present; −, the gene is absent.
The genes encode the following proteins: ehxa, enterohemolysin; espP, serine protease (EspP); iha; iron-regulated gene A homolog adhesin (Iha); saa, STEC autoagglunating adhesin (Saa); subAB, subtilase-like serine protease toxin (SubAB); tia, invasion determinant described in enterotoxigenic E. coli; lpfAO113, major subunit of long polar fimbriae of STEC O113; OI-43/48-terE (marker of ter cluster), tellurite resistance; OI-122-pagC (Z4321), homolog of PagC protein of Salmonella enterica serovar Typhimurium; espI, serine protease (EspI).
Twenty eight of the LEE- or OI-encoded virulence-associated genes (eae, espG [LEE], ent, nleB, nleE, efa1, efa2 [OI-122], nleF, nleH1-2, nleA, nleG, espM1, ecs1822 [OI-71], ureD [OI-43/48] (marker für ure cluster), ecs1763, espN, espM2, espX2, espX7, espO1-1, espY4-2, espX6, espW, espY1, espY3, nleH1-1, nleD-2, and espJ) as well as the plasmid pO157-encoded genes katP, etpD, and toxB as well as the lpfAO157 gene which encodes the major subunit of long polar fimbriae of EHEC O157:H7 EDL933, and the EAEC marker gene aggr were not detected in any of the strains (except the EPEC O178:H7 strain CB09017, see below).
[] indicates nonmotile strains that were analyzed for their fliC genotypes by PCR/RFLP of HhaI-digested fliC PCR products.
The standardized Stx nomenclature published by Scheutz et al. (2012) was used.
Two variants of the subAB were detected; subAB1 indicates subAB98NK2, plasmid pO113-encoded. No tia gene was detected in these strains. subAB2 indicates subABED32, chromosomal encoded on a putative PAI. The tia gene was detected in these strains. nd, not done.
Encoded by the locus of protease and adherence (LPA) which is located in the chromosome (Schmidt et al., 2001).
The EPEC O178:H7 strain CB09017 revealed genes eae, espG, and espF (LEE-encoded) and genes ent/espL2, nleB, nleE, efa1, efa2, nleF, nleH1-2, ecs1763, espM2, espW, espX1, nleH1-1, espR1, and espJ (OI-encoded).
Serotyping and genotyping of flagellar (fliC) genes
Typing of O (lipopolysaccharide) and H (flagellar) antigens was performed as described (Beutin et al., 2004). Nonmotile (NM) E. coli strains were analyzed for their flagellar (fliC) genotypes by PCR and restriction fragment length polymorphisms (RFLP) of HhaI-digested fliC PCR products (Beutin et al., 2005). fliC-genotypes were confirmed by real-time PCR microarray testing as previously described (Bugarel et al., 2010a).
Phenotypes
Stx production was determined using the Vero cell cytotoxicity assay and a commercially obtainable Stx enzyme immunoassay (Ridascreen-EIA, R-Biopharm AG, Darmstadt, Germany) as described previously (Beutin et al., 2007). Detection of the enterohemolysin or α-hemolysin phenotype of bacteria was performed on washed sheep blood (enterohemolysin) agar (Beutin et al., 2004). Sorbitol fermentation was tested on Sorbitol MacConkey (SMAC) agar, and B-D-glucuronidase activity on Fluorocult® E. coli 0157:H7 agar. Tellurite resistance was evidenced by the ability of strains to grow on cefixime-tellurite (CT)–SMAC agar and CHROMagar STEC. Media were obtained from Merck KGaA, Darmstadt, Germany, and CHROMagar, Paris, France.
Detection of single and multiple copies of stx genes and subtyping
All isolates were screened by PCR for the presence of stx1 and stx2 using common primers (Beutin et al., 2007). stx genes were subtyped using PCR/RFLP analysis or subtype-specific primers as described (Beutin et al., 2007). The PCR/RFLP approaches allow to discriminate between stx1a (formerly stx1), stx1c, and stx1d genes as well as between stx2a (formerly stx2), stx2c/stx2d (variants stx2v-ha, stx2v-hb, stx2-NV206, stx2-EC1586, stx2d-O73), and stx2g genes according to differences in their B-subunits. The PCR/RFLP method has the advantage that strains carrying multiple variants of the stx2 gene can be identified. Toxin genotypes stx2b (formerly stx2-O118 and stx2-OX3:H21), stx2e, and stx2f were detected by PCR using subtype-specific primers as described (Beutin et al., 2007). Controls used were C600(H19) stx1a (GenBank accession no. M16625), DG131/3 stx1c (Z36901), and MHI813 stx1d (AY17085), C600(W34) stx2a (X07865), CB2851 stx2v-ha (AJ605767), CB7753 stx2v-hb (AF479829), NV206 stx2-NV206 (AF329817), RL481/06 stx2-EC1586 (AM498375), C165-02 stx2d-O73 (DQ059012), 12422 stx2g (AY286000), EH250 stx2b (AF 043627), 3615-99 stx2e (AJ313016), and T4/97 stx2f (AJ0110730).
Detection of the mucus-activatable stx2 variant gene stx2d
All STEC strains were tested by PCR for the presence of the mucus-activatable stx2d using primers stx2d-F1, stx2d-R1, stx2d-R2, and stx2d-O55-R and following the new subtyping protocol developed by (Scheutz et al., 2012). In order to confirm the presence of stx2d and to further characterize the strains, we sequenced the stx2AB genes of all stx2d-positive and selected stx2d-negative strains (CB11017, 96/00 [CB12249], FP023 [CB12251], FP-087 [CB12260], I-005 [CB12275], 276-X [CB12282], 377-11 [CB12283], and C11 [CB12285]). Briefly, these strains were used for further PCRs with primers SLT-II-vc and CKS2, which flank the PstI site in the A-subunit and the complete B-subunit of the Stx2 toxin (Jelacic et al., 2003) as well as primers stx2all-f (CGGGCCTTTTTTAATCTGCG) and stx2all-r (ACCCACATACCACGAATCAG), which flank the complete stx2AB genes (this work). The resulting 890 bp-amplicons and 1334 bp-amplicons, respectively, were purified and sequenced at a DNA sequencing service unit (Eurofins MWG Operon, Ebersberg, Germany). The sequences were analyzed using Lasergene software (DNASTAR, Madison, WI) and Accelrys Gene software (Accelrys Inc., San Diego, CA). Using the Blast algorithmus, the sequences were compared with sequences deposited at NCBI (http://www.ncbi.nlm.nih.gov/blast/). The following sequences were used as references: stx2a (accession no X07865), stx2b (AF043627), stx2c (M59432), and stx2d (AF479829).
Nucleotide sequence accession number
The nucleotide sequences obtained have been entered into the EMBL database under accession numbers FR846369.1 to FR846379.1, and FR850037.1.
Accessory virulence genes
The presence of accessory virulence genes was determined by a real-time PCR microarray as previously described (Bugarel et al., 2010a). Strains were screened for the presence of 42 genes associated with human virulent STEC (enterohemorrhagic E. coli [EHEC]) strains (Table 2). These genes were the LEE-encoded eae gene encoding intimin, six non-LEE-encoded effector (nle) genes issued from OI-122 (pagC, ent/espL2, nleB, nleE, efa1, efa2), six genes issued from OI-71 (nleF, nleH1-2, nleA, nleG, espM1, ecs1822) which mainly encode translocated substrates of the type III secretion system, as well as three genes issued from OI-43/OI-48 (iha, ureD [marker for the ure gene cluster], terE [marker for the ter gene cluster]) which encode the iron-regulated gene homolog adhesin Iha, urease activity and tellurite resistance, respectively. The array was also designed for the detection of seven genes derived from EHEC O157 virulence plasmid pO157 and STEC O113 viulence plasmid pO113, respectively: ehxA (enterohemolysin), espP (extracellular STEC-specific serine protease), katP (catalase peroxidase), etpD (the type II secretion system), toxB (putative EHEC adhesin), saa (autoagglutinating adhesin), and subA (subtilase cytotoxin). Other putative virulence factors, such as long polar fimbriae (lpfAO157, lpfAO113) and a transcriptional regulator of enteroaggregative Escherichia coli (aggr) were additionally screened.
Table 2.
E. coli gene targets for the real-time PCR micro array.
| Gene (ORF name if chromosomal) | Encoded protein or family effector | Genetic support (mobile elements) |
|---|---|---|
| pagC (Z4321) | PagC-like membrane protein | OI-122a |
| ent (Z4326) | Ankyrin repeats | OI-122a |
| nleB (Z4328) | Non-LEE-encoded type III effector | OI-122a |
| nleE (Z4329) | NleE | OI-122a |
| efa1 (Z4332) | EHEC factor for adherence | OI-122a |
| efa2 (Z4333) | EHEC factor for adherence | OI-122a |
| nleF (Z6020) | Non-LEE-encoded type III effector | OI-71a |
| nleH1-2 (Z6021) | Non-LEE-encoded type III effector | OI-71a |
| nleA (Z6024) | Non-LEE-encoded type III effector | OI-71a |
| nleG (Z6010) | NleG | OI-71a |
| espM1 (Z2565) | Non-LEE-encoded type III effector | OI-71a |
| ecs1822 (Z6011) | Hypothetical protein | OI-71a |
| ureD (Z1142) | Urease-associated protein UreD | OI-43a & OI-48a |
| terE (Z1176) | Tellurite resistance cluster | OI-43a & OI-48a |
| Iha (Z1148) | iron regulated gene A homolog adhesin | OI-43a & OI-48a |
| espX1 (Z0025) | Pentapetide repeats | OI-1a |
| espY1 (Z0065) | SopD N-terminal domain | |
| espY3 (Z0521) | Non-LEE-encoded type III effector | OI-26a |
| nleH1-1 (Z0989) | NleH | OI-36a |
| nleD-2 (Z0990) | NleD | OI-36a |
| espX2 (Z1019) | Non-LEE-encoded type III effector | OI-37a |
| espV (Z1387) | AvrA | OI-44a |
| espK (Z1829) | Leucine-rich repeats | OI-50 (prophage CP-933N)a |
| espX7 (Z1822) | non-LEE-encoded type III effector | OI-50a |
| espO1-1 | OspE family | OI-50a |
| espN (Z1823) | CNF | OI-50a |
| ecs1763 (Z2040) | Hypothetical protein | OI-57a |
| espR1 (Z2242) | Leucine-rich repeats | OI-62a |
| espJ2 (Z3071) | EspJ | OI-79a |
| espM2 (Z3918) | IpgB | OI-108a |
| espW (Z3920) | HopW | OI-108a |
| lpfA (Z4971) | Long polar Fimbrial protein LpfA | OI-141 & OI-154a |
| eae (Z5110) | intimin | LEEa |
| espG (Z5142) | EspG | LEEa |
| espF1 (Z5100) | EspF (secreted effector) | LEEa |
| katP | Catalase peroxidase | EHEC-plasmidb |
| toxB | Adhesin | EHEC-plasmidb |
| etpD | Type II effector | EHEC-plasmidb |
| ehxA | Enterohemolysin | EHEC-plasmidb,c |
| espP | Serine protease EspP | EHEC-plasmidb,c |
| saa | STEC autoagglunating adhesin Saa | aEHEC-plasmidc |
| subAB | Subtilase cytotoxin SubAB | aEHEC-plasmidc or putative PAI |
Furthermore, all strains were tested by PCR for the presence of the espI gene encoding a serine protease, termed EsPI, that is located in the chromosome on the locus of protease and adherence (LPA). Primers espI-I and espI-II and cycling conditions are used as previously described (Schmidt et al., 2001).
All subA-positive strains were tested by PCR for the presence of the tia gene (Genbank accession no. U20318) using primers tia_lo and tia_sense as described by Tozzoli et al. (2010). tia encodes for an invasion determinant described in enterotoxigenic E. coli.
PFGE
Pulsed-field gel electrophoresis was performed using the standardized PulseNet protocol published previously (Gerner-Smidt and Scheutz, 2006). Briefly, agarose-embedded DNA was digested with 50 U of XbaI (Roche Diagnostics GmbH, Mannheim, Germany) for 4 h at 37°C. The restriction fragments were separated by electrophoresis in 0.5 × Tris-borate-EDTA buffer containing 50 μM thiourea at 14°C for 19 h by the use of a CHEF DR-III system (Bio-Rad, Munich, Germany), with pulse times of 2.16 to 54.17 s. XbaI-digested DNA of Salmonella enterica serovar Braenderup strain H9812 (Centers for Disease Control and Prevention, Atlanta, GA) was used as a molecular size marker. Similarities between restriction patterns were calculated with BioNumerics software (version 6.6, Applied Maths, Ghent, Belgium) using the Dice coefficient with band matching parameters of 1.0% optimization and 1.5% position tolerance. Interstrain relationships were assessed by cluster analysis using the Unweighted Pair-Group with Mathematical Average (UPGMA) method.
Results
Two serotypes dominate among stec O178 strains
Sixty four O178 strains were motile and could thus be serotyped for their H antigens. Among them were 61 O178:H19 and each one O178:H7, O178:H10, and O178:H16 strains. Ten strains were nonmotile and the flagellar types were determined by fliC genotyping. Five of these were O178:[H19] and five O178:[H7]. In total, 66 of the strains were serotyped as O178:H19, six as O178:H7 and each one as O178:H10 and O178:H16 (Table 1).
Phenotypes
Cytotoxic activity on Vero cells and production of Stx tested by an enzyme immunoassay was found with all 66 O178:H19, four O178:H7 and the O178:H10 strain(s). Two O178:H7 strains and the O178:H16 strain were Stx-negative with both methods. All strains possessing the ehxA gene produced enterohemolysin. No strain produced α-hemolysin. All strains fermented sorbitol and show B-D-glucuronidase activity. All strains possessing the terE gene grew on CT–SMAC agar and CHROMagar STEC.
Association between stx-genotypes and E. coli O178 serotypes
All but two solely stx1 positive O178 STEC strains (2.8%) carried stx2 genes (97.2%) and 33 strains of which (46.5%) carried both stx1 and stx2 genes (Table 1). All 30 stx1 positive O178:H19 strains and the single O178:H10 strain carried the stx1a subtype. The stx1c subtype was present in all four STEC O178:H7 strains (Table 1).
Thirty-one (47.0%) of the 66 STEC O178:H19 strains carried the stx2a gene, 32 (48.5%) the stx2c gene and eight (12.1%) the stx2d gene. Except in one case, combined presence of stx2a and stx2c was not observed, but the stx2a and the stx2d genes were present together in four (6.1%) of the 66 STEC O178:H19 strains. The stx2b gene was found in one (1.5%) of the 66 STEC O178:H19, but in three (75.0%) of the four STEC O178:H7 strains (Table 1). Other stx2-subtypes were not detected in any of the strains.
Detection of mucus-activatable stx2d by PCR and nucleotide sequencing
By PCR eight of the STEC O178:H19 strains (12.1%) were identified as stx2d-positive. The STEC O178:H7 and O178:H10 strains were found negative for stx2d (Table 1). To confirm the results, we determined the nucleotide sequences of the stx2AB genes of all stx2d-positive and 8 selected stx2d-negative STEC O178:H19 strains. The sequences were analyzed and compared with the published stx2-reference sequences in GenBank. It is noteworthy, that chromatograms derived from STEC strains carrying both stx2a and stx2d genes showed overlapping peaks at nucleotide positions which differ between stx2a and stx2d (data not shown). The sequences were translated into putative amino acid sequences of the A and B subunits and analyzed for the activatable type of a Stx2 toxin. Scheutz et al. (2012) defined the activatable type of a Stx2 toxin as the combination of the “activatable tail” in the Stx2A subunit and the END motif in the Stx2B subunit The activatable tail consists of the last 10 amino acids in the C-terminal end of the A2 subunit and has been identified as KSQSLYTTGE from position 288 to 297 in the mature toxin sequence (with the relevant amino acids serine at position 291 and glutamic acid at the final position 297); the motif END is found at position 14 to 16 in the B subunit. Only the Stx sequences of the eight O178:H19 strains CB11726, CB12008, FP-080 [CB12258], I-039 [CB12278], C27 [CB12288], CB13785, CB13787 and CB14051 tested as stx2d-positive by PCR, fulfilled these criteria, and are therefore considered to be activatable. The other strains revealed neither the activatable tail in the A subunit nor the END motif in the B subunit or only one of these characteristic traits (Figure 1).
Figure 1.
Alignment of the amino acid sequences of the C-terminal ends of the A-subunits and B-subunits of 8 stx2d-positive and 8 stx2d-negative O178:H19 strains investigated in this study. Amino acid sequences in bold show the activatable tail in the A-subunit and the END motif in the B-subunit. Amino acid sequences which contain both motifs, thus correspond to the mucus-activatable stx2d variant, are shaded. The following sequences were used as references: stx2a (GenBank accession no. X07865), stx2b (AF043627), stx2c (M59432), and stx2d (AF479829). Amino acid sequences underlined indicate double peaks in the chromatogram which reveal multiple stx2-toxin types in these strains. aPosition in mature toxin sequence.
Presence of accessory virulence-associated genes
Twenty eight of the LEE- or OI-encoded virulence-associated genes (eae, ent/espL2, nleB, nleE, efa1, efa2, nleF, nleH1-2, nleA, espG, ecs1763, espM1, espN, espM2, espX2, espX7, espO1-1, espY4-2, espX6, espW, espY1, espY3, nleH1-1, nleD-2, nleG, espJ, ecs1822, and ureD) were not present in any of the O178 strains investigated. All O178 strains were also tested negative for the pO157-encoded katP, etpD and toxB genes, as well as the lpfAO157 and aggr genes. An exception was the Stx-negative O178:H7 strain CB09017 from a human patient which was positive for the eae and for several nle genes (ent/espL2, nleB, nleE, efa1, efa2, nleF, nleH1-2, espG, espF1, ecs1763, espM2, espW, espX1, nleH1-1, espR1, and espJ). According to its virulence profile and the absence of the bfp gene encoding bundle-forming pili (data not shown), CB09017 was classified as an atypical EPEC strain. The LPA-encoded espI was not found in any of the E. coli O178:H19, O178:H10, and O178:H16 strains, but was detectable in four O178:H7 strains. Genes iha and espP were common in all STEC O178 strains, but were missing in the three Stx-negative strains. Characteristic traits and (putative) virulence genes detected in the O178 strains are listed in Table 1. pagC was detected in 36 of the O178:H19 strains (54.5%) and 31 (47.0%) were associated with PFGE cluster B, C, D and E strains (see below). iha which encodes the iron-regulated gene homolog adhesin IrgA was found in all O178:H19 strains, whereas terE which encodes tellurite resistance was detected in 16 O178:H19 strains (24.2%). Except for FP-080, in all STEC O178 strains lpfAO113 which encodes the major subunit of LPF of STEC O113 was detectable. Among the seven virulence genes derived from virulence plasmids, espP was found in all O178:H19 strains; ehxA and saa were detected each in 35 O178:H19 strains (53.0%), whereas subAB was present in 18 O178:H19 strains (27.3%) which belonged all to PFGE cluster A (see below). All subAB-positive O178:H19 strains were tia-negative. The four O178:H7 STEC strains isolated from human patients and from deer meat harbored ehxA, espP, subA, iha, in addition two of them harbored terE. In contrast to O178:H19 strains, they all were also tia- and espI-positive. The stx-negative O178:H7 strain isolated from cattle and the stx-negative O178:H16 strain isolated from salad carried none of the virulence genes investigated (except lpfAO113). The human EPEC O178:H7 strain did not harbor genes other than the eae and nle genes mentioned above. The O178:H10 STEC strain from deer meat carried espP, subAB, and iha genes, but no tia gene. None of the O178:nonH19-strains possessed pagC and saa genes (Table 1).
PFGE genotyping of STEC O178:H19 strains
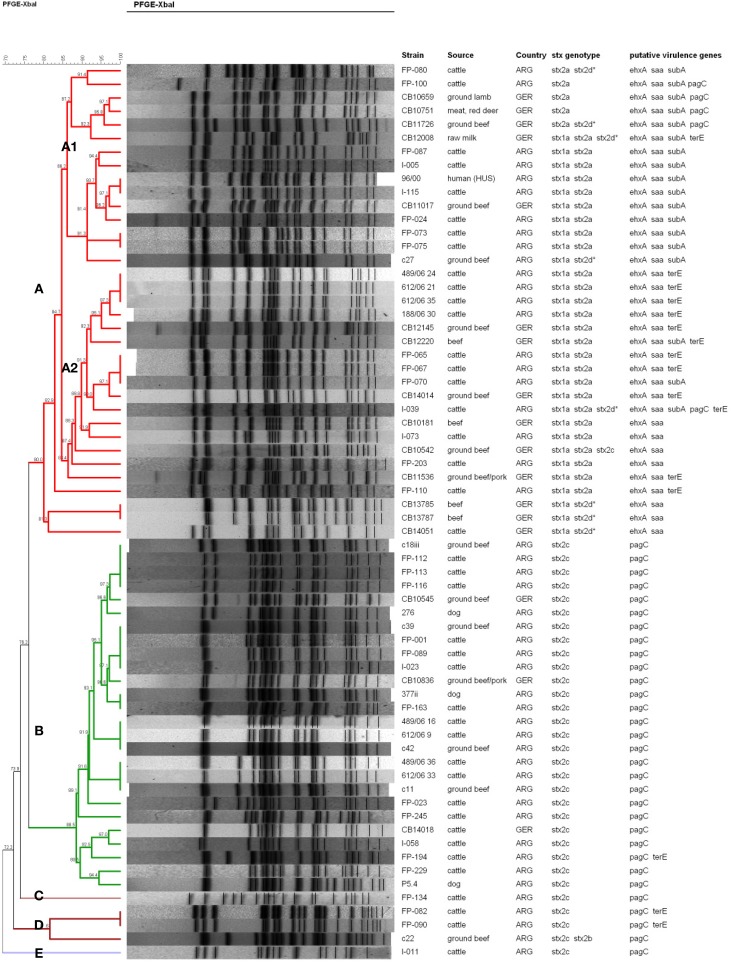
The genetic relatedness between the different E. coli O178 strains was investigated by XbaI-PFGE genotyping. E. coli O178 strains carrying other H-antigens/fliC types than H19 revealed individual PFGE patterns and proved to be genetically more distant (data not shown). The 66 O178:H19 strains revealed 47 different PFGE patterns. A dendrogram based on similarities of the PFGE patterns was created with the BioNumerics software v. 6.6 using the band-based Dice coefficient (Figure 2). Sixty-one of the O178:H19 strains were assigned to two major PFGE clusters designated A (35 strains) and B (26 strains) (Figure 2). The five remaining strains were assigned to clusters C (one strain), D (three strains), and E (one strain).
Figure 2.
PFGE-based clustering of 66 STEC O178:H19 strains investigated in this study. Clusters A–E were defined using a cut-off value of 80% similarity and are indicated by different colors. The degree of similarity (%) is shown on the scale at the top left of the figure. stx2d*, mucus-activatable stx2d. All STEC O178:H19 strains possessed espP and iha, and all except strain FP-080 lpfAO113. None of the STEC O178:H19 strains possessed katP, etpD, and toxB.
Clusters A to E were defined using a cut-off value of 80% similarity (Figure 2). Strains belonging to cluster A and B, respectively, show only 76.2% similarity in their PFGE patterns and also clearly differ in their virulence traits. Cluster A-O178:H19 STEC (>80.0% similarity) carry the pO113 virulence plasmid-genes ehxA, saa and espP together with the stx2a and/or stx2d, iha and lpfAO113 genes. Thirty cluster A-strains additionally carry the stx1a gene. Cluster A was further subdivided into subclusters A1 (15 strains, 86.2% similarity) and A2 (16 strains, 86.4% similarity) and, more distantly related, four additional strains. Subcluster A1-strains were additionally characterized by presence of the pO113-encoded subAB gene which was only found in three of the 15 subcluster A2-strains. Nearly 70% of the subcluster A2-strains carry an additional terE gene which was not detected in A1-strains (except in strain CB12008). The genetically highly related subclusters A1 and A2 encompassed both strains from Argentina and Germany. The STEC O178:H19 HUS-patient isolate from Argentina (96/00) grouped within subcluster A1 together with four isolates from Argentinean cattle and one isolate from German ground beef with a high degree of similarity (93.7%). The HUS strain 96/00 was found 100% similar to the strain I-115 from Argentine cattle and 97.1% similar to the German food strain CB11017.
The 26 cluster B-O178:H19 strains (88.5% similarity) were from cattle, beef, and from dogs, only three of these strains were from Germany. Cluster B-strains were characterized by the stx2c gene: the stx2v-ha variant was present in 22 strains (84.6%) and stx2v-hb in 4 strains (15.4%). Characteristically, all strains were also positive for pagC, lpfAO113, iha, and espP, and were, in contrast to cluster A–strains, negative for stx1a, ehxA, saa, and subAB. Cluster B contains strains of different sources and geographical origins with high similarities up to 100%. Among them are strains from three Argentine dogs which show high similarity (94.4 – 100%) to strains from cattle and food of bovine origin.
Discussion
Over the past years, STEC O178:H19 became one of the most prevalent serotypes isolated from dairy cows (Fernandez et al., 2010) and beef cattle (Tanaro et al., 2012) as well as in beef abattoirs (Masana et al., 2011) and in food of bovine origin in Argentina and Germany (Beutin et al., 2007; Sanz et al., 2007; Werber et al., 2008; Slanec et al., 2009; Kruger et al., 2011). Moreover, this serotype was prevalent in pets (Bentancor et al., 2012). In Argentina where the incidence of HUS is high, STEC O178:H19 has been already isolated from a human patient with HUS (Giugno et al., 2007). In Germany, some sporadic cases of human infections with STEC O178:H19 were reported (Bielaszewska et al., 2006; Werber et al., 2008). There are several reasons why STEC O178:H19 strains might represent an “emerging” pathogen. On one hand, the O178 serogroup was defined quite recently and was not part of the E. coli serotyping scheme before 2004 (Scheutz et al., 2004), and even today complete serotyping is rarely performed in routine investigations. On the other hand, modified cattle feeding management practices in Argentina is conceivable as another reason for the “emergence” of STEC O178 strains. The intensive feeding and high animal density in feedlot could give rise to a selective bacterial flora with E. coli serotypes that differ from those isolated previously in grazing-fed cattle (Parma et al., 2000; Padola et al., 2004).
The aim of this study was to characterize and to assess the potential risk of STEC O178 strains isolated from different sources for humans. Our findings indicate that STEC O178 strains lack typical properties that are closely associated with enterohemorrhagic E. coli (EHEC) strains that are frequently isolated from outbreaks and from human HC and HUS cases. STEC O178 strains were negative for the LEE-encoded eae gene as well as for non-LEE effector genes encoded by O-islands 122, 71 and 57 which have been identified as markers related to potentially highly virulent STEC (Karmali et al., 2003; Coombes et al., 2008; Konczy et al., 2008; Bugarel et al., 2010b). As an exception, a pagC-like gene encoding a putative PagC-like outer membrane adhesin is present in 36 (54.5%) of the O178:H19 isolates. This gene was shown to reside on the first of three modules composing OI-122 in the EDL933 genome (Konczy et al., 2008). However, in the LEE-negative O113:H21 strain CL3, pagC is not part of an OI-122-like structure, but rather a part of a unique hybrid genomic island containing segments of OIs 122 and 48 (Shen et al., 2004). Sequence analysis of the pagC-like gene in LEE-positive and LEE-negative STEC strains revealed significant differences in their composition (Konczy et al., 2008). LEE-positive pagC-strains always possessed module 2 and/or module 3 besides the pagC-module 1. LEE-negative pagC-strains always possessed exclusively the pagC-module 1 and differed in their pagC alleles from LEE-positive strains. LEE-positive pagC-strains were assigned to highly virulent human seropathotype A, B, or C strains, and LEE-negative pagC-strains were grouped either in seropathotype C (human strains) or seropathotypes D and E (bovine strains) with low or no virulence to humans (Karmali et al., 2003; Konczy et al., 2008). Accordingly, the O178:H19 pagC-strains from this study might be classified as seropathotype D or E strains at this point. The large majority of our pagC-strains are grouped in PFGE-cluster B with a high degree of similarity and carry a limited repertoire of virulence genes (stx2c, pagC, lpfAO113, iha, and espP) (Figure 2).
However, the presence of LEE- and nle-genes is not essential for STEC pathogenesis and sporadic cases and small outbreaks of STEC infections, including HC and HUS, have been caused by LEE-negative strains (Keskimaki et al., 1997; Bonnet et al., 1998; Paton et al., 1999). In the absence of LEE, mechanisms are emerging by which the LEE-negative STEC interact with the host intestinal mucosa and induce disease (Newton et al., 2009). Recently, a number of putative adhesins have been identified, such as the Long Polar Fimbrial Protein LPFAO113, the STEC autoagglutinating adhesin (Saa), and an iron-regulated gene homolog adhesin (Iha) (Paton et al., 1999; Tarr et al., 2000; Doughty et al., 2002).
The lpfAO113 gene, located within a novel fimbrial gene cluster, was initially reported in a LEE-negative EHEC O113:H21 strain isolated from a HUS patient. It was shown that deletion of the fimbrial subunit gene lpfAO113 resulted in decreased adherence of these bacteria to epithelial cells in vitro (Doughty et al., 2002). Remarkable, lpfAO113 was also found in other LEE-negative EHEC and in non-O157:H7 LEE-positive EHEC strains (Doughty et al., 2002).
Saa was first characterized from a LEE-negative O113:H21 STEC strain responsible for an outbreak of HUS (Paton et al., 1999). Over the past years the Saa-encoding gene has been detected in many LEE-negative STEC strains from animals, food and humans (Zweifel et al., 2004, 2005; Cergole-Novella et al., 2007; Oliveira et al., 2007, 2008; Vu-Khac and Cornick, 2008; Fernandez et al., 2010; Bandyopadhyay et al., 2011; Bosilevac and Koohmaraie, 2011) including samples from HUS patients (Paton et al., 2001; Paton and Paton, 2002; Jenkins et al., 2003). The saa-positive STEC O178:H19 isolates from our study belonged all to PFGE-cluster A which comprises O178:H19 strains with a high virulence potential. The saa-positive STEC isolates (47.3%) were all ehxA-positive, too, since both genes are encoded in the pO113 megaplasmid (Newton et al., 2009). By contrast, the saa and eae genes appear to be mutually exclusive within STEC strains, so that saa has been suggested to serve as an alternative adhesin of LEE-negative strains in human colonization. The saa gene from different STEC strains vary in size due to the number of direct repeated units at the 3′ end, and five saa-variants were identified until now (Paton et al., 2001; Lucchesi et al., 2006). However, binding properties of saa-carrying STEC to epithelial cells exhibit marked variation with no correlation to the number of C-terminal repeats and to the Saa-expression (Toma et al., 2008). Clarification of the role of saa in human infections needs further investigation, however, its high prevalence in bovine and ovine STEC suggest a role in the attachment to bovine and ovine gut (Jenkins et al., 2003; Zweifel et al., 2004; Fernandez et al., 2010; Tanaro et al., 2012).
Pathogenicity islands OI-43 and OI-48 are duplicate genomic islands present in EHEC O157:H7 strains. OI-43/48 carry three functional gene clusters, encoding urease (ure cluster), tellurite resistance (ter cluster), and putative adhesins Iha (iha) and AIDA-1 (aidA-1). Iha similar too Saa, was suggested to serve as adhesin in LEE-negative STEC strains. The role of Iha in human virulence is not clear yet, as it was found in similar frequencies in STEC seropathotypes highly associated with severe diseases as well as in less or apathogenic strains (Ju et al., 2013). It is noteworthy, that iha has different origins. Besides the OI-43/48 origin, it was also found in plasmid pO113 of LEE-negative O113:H21 (Newton et al., 2009) as well as on the newly described genomic island LPA in STEC O91:H- (Schmidt et al., 2001). In our study iha is present in all STEC O178 strains, but is missing in the three Stx-negative strains. In contrast to iha, OI-43/48 encoded ure and ter genes were closely associated with seropathotypes causing severe disease and outbreaks (Ju et al., 2013). Urease helps STEC to survive in the stomach and amends its ability to colonize the calf and human intestinal tracts. The role of tellurite resistance genes is not completely clear. However, it was postulated that ter genes might encode an adhesin-related gene product and/or might offer a selective advantage in the host environment and under stress conditions (Yin et al., 2009). No O178 strain in this study was ureD-positive, 24.3% of the strains were terE-positive. Tellurite resistance of these strains was evidenced by their ability to grow on CT-SMAC agar and CHROMagar STEC (data not shown).
Among other toxins a novel subtilase cytotoxin (SubAB) has been reported to contribute to the virulence and pathogenicity of LEE-negative STEC for humans. SubAB is the prototype of a new AB5 toxin family first detected in the LEE-negative O113:H21 STEC strain 98NK2 that caused an outbreak of HUS in Australia in 1998 (Paton et al., 2004). Since then the subAB operon has been detected in STEC isolates from different continents belonging to over 30 STEC serogroups (Irino et al., 2010; Paton and Paton, 2010). In Belgium SubAB was found in one O178:H19 HUS-associated strain (Buvens et al., 2010). SubAB is lethal for mice and induces pathological traits overlapping those seen in the HUS. It was suggested (Paton and Paton, 2010) that SubAB directly contributes to pathology in humans infected with STEC strains that produce both Stx and SubAB. The question, whether the production of SubAB correlates to the severity of STEC disease in humans needs further investigation. In our study, nearly 30% of the STEC O178:H19 strains carry subA, in all cases together with the saa, ehxA, and espP genes, suggesting their location on the pO113 plasmid as found with STEC O113:H21 (Newton et al., 2009) (Table 1). It is noteworthy, that all subAB-carrying STEC O178:H19 strains isolated in Germany and Argentina, among them the Argentinian HUS strain, group in the same PFGE-subcluster A1 indicating their genetic similarity (Figure 2). Argentina has a very high incidence of STEC infections and HUS cases, many of them caused by LEE-negative, but SubAB-positive STEC strains (Gerhardt et al., 2009). In future, SubAB-producing STEC strains should be payed more attention with regard to their role to cause or to augment STEC diseases. It has been suggested that different variants of subAB exist (Tozzoli et al., 2010; Orden et al., 2011). One called SubAB1 is encoded by the pO113 megaplasmid as found in the human pathogenic STEC O113:H21 98NK2 prototype strain. The other, called SubAB2, is chromosomally located close the tia gene (an invasion gene previously described in enterotoxigenic E. coli) on a subAB-tia putative pathogenicity island (Tozzoli et al., 2010; Orden et al., 2011; Michelacci et al., 2013). Virulence features of the SubAB-positive STEC O178:H19 strains in our study (saa-positive, tia-negative) suggest that they harbor the SubAB1 variant typical associated with bovine strains (Table 1). By contrast, the SubAB-positive STEC O178:H7 strains isolated from human patients and from deer meat revealed the stx genotype stx1c, stx2b and were tia-positive and saa-negative, suggesting the presence of subAB2 (Table 1). Further work is needed, to explore the role of SubAB toxins in severe disease.
The role of stx2a and stx2d genes as major virulence factors in STEC infections of humans was previously investigated (Friedrich et al., 2002; Persson et al., 2007; Scheutz et al., 2012). The mucus-activatable Stx2d was found in LEE-negative STEC causing severe disease including HC and HUS in humans (Melton-Celsa et al., 1996, 1998, 2002; Paton et al., 1999; Jelacic et al., 2003; Bielaszewska et al., 2006). Strains producing Stx2d proved to be highly virulent in experimental infections of streptomycin-treated mice (Melton-Celsa et al., 1998). Epidemiological and toxicity data point to a high pathogenic potential of Stx2d-producing STEC for humans. At present, limited data are available regarding the occurrence of strains harboring the stx2d gene in cattle, other livestock sources and foods (Gobius et al., 2003; Oliveira et al., 2008; Zheng et al., 2008; Slanec et al., 2009; Xia et al., 2010). In our study, we used a recently developed PCR (Scheutz et al., 2012) to identify the stx2d gene in 8 of the 74 STEC O178 strains (10.8%) investigated in this study. The PCR results were confirmed by nucleotide sequencing of Stx2 genes. Like Stx2a-strains, Stx2d-strains were exclusively found in PFGE-cluster A.
Generally, serogroup O178 belongs to those serogroups of E. coli that covers multiple lineages of STEC strains which express different H-types (Scheutz and Strockbine, 2005). Strains which express H10, H12, H16, H21, and H27 are very rare and often avirulent. O178:H7 strains are heterogeneous and include STEC, EPEC and avirulent strains. A particular H7-lineage has emerged that substantially differ from the O178:H19 lineage. Strains of this lineage analyzed in this study originate from deer and humans and are associated with the genotype stx1c/stx2b/ehxA/subAB2/espI/[terE]/espP/iha.
However, in this study we have focused on STEC O178:H19 strains which emerged in numerous countries worldwide. STEC O178:H19 strains are heterogeneous and split up into distinct PFGE-clusters of related strains. The two major PFGE-clusters A and B differ in their stx-genotypes and non-Stx putative virulence traits, including adhesins, toxins, and serine-proteases. PFGE-cluster B includes strains of low virulence attributed to the very limited spectrum of virulence genes (stx2c, pagC, lpfAO113, espP, iha). By contrast, PFGE cluster A comprises more virulent strains carrying virulence genes related to severe clinical outcomes (stx2a, stx2d, ehxA, saa, subAB1, lpfAO113, terE combined with stx1a, espP, iha). The toxic stx2a, the mucus-activatable stx2d, the enterohemolysin gene ehxA, and the novel adhesin/cytotoxin genes lpfAO113, saa, and subAB might enhance the pathogenicity of the strains, enabling them to adhere to the intestinal epithelium and to produce a potentially dangerous toxin. Such strains are an increasing risk for humans, because they, unlike other LEE-negative strains, may cause human diseases. Therefore, the risk represented by them to public health should be carefully monitored.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the technical staff in the different laboratories for the skillful assistance.
References
- Aidar-Ugrinovich L., Blanco J., Blanco M., Blanco J. E., Leomil L., Dahbi, et al. (2007). Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolated from calves in Sao Paulo, Brazil. Int. J. Food Microbiol. 115, 297–306 10.1016/j.ijfoodmicro.2006.10.046 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Mahanti A., Samanta I., Dutta T. K., Ghosh M. K., Bera A. K., et al. (2011). Virulence repertoire of Shiga toxin-producing Escherichia coli (STEC) and enterotoxigenic Escherichia coli (ETEC) from diarrhoeic lambs of Arunachal Pradesh, India. Trop. Anim. Health Prod. 43, 705–710 10.1007/s11250-010-9757-1 [DOI] [PubMed] [Google Scholar]
- Bentancor A., Rumi M. V., Carbonari C., Gerhardt E., Larzabal M., Vilte D. A., et al. (2012). Profile of Shiga toxin-producing Escherichia coli strains isolated from dogs and cats and genetic relationships with isolates from cattle, meat and humans. Vet. Microbiol. 156, 336–342 10.1016/j.vetmic.2011.10.030 [DOI] [PubMed] [Google Scholar]
- Bettelheim K. A. (2007). The non-O157 shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33, 67–87 10.1080/10408410601172172 [DOI] [PubMed] [Google Scholar]
- Beutin L., Krause G., Zimmermann S., Kaulfuss S., Gleier K. (2004). Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42, 1099–1108 10.1128/JCM.42.3.1099-1108.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Miko A., Krause G., Pries K., Haby S., Steege K., et al. (2007). Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73, 4769–4775 10.1128/AEM.00873-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Strauch E., Zimmermann S., Kaulfuss S., Schaudinn C., Mannel A., et al. (2005). Genetical and functional investigation of fliC genes encoding flagellar serotype H4 in wildtype strains of Escherichia coli and in a laboratory E. coli K-12 strain expressing flagellar antigen type H48. BMC Microbiol. 5:4 10.1186/1471-2180-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M., Friedrich A. W., Aldick T., Schurk-Bulgrin R., Karch H. (2006). Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43, 1160–1167 10.1086/508195 [DOI] [PubMed] [Google Scholar]
- Blanco M., Blanco J. E., Mora A., Dahbi G., Alonso M. P., Gonzalez E. A., et al. (2004). Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-^ı). J. Clin. Microbiol. 42, 645–651 10.1128/JCM.42.2.645-651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R., Souweine B., Gauthier G., Rich C., Livrelli V., Sirot J., et al. (1998). Non-O157:H7 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J. Clin. Microbiol. 36, 1777–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosilevac J. M., Koohmaraie M. (2011). Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 77, 2103–2112 10.1128/AEM.02833-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunder W., Schmidt H., Frosch M., Karch H. (1999). The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145 ( Pt 5), 1005–1014 10.1099/13500872-145-5-1005 [DOI] [PubMed] [Google Scholar]
- Bugarel M., Beutin L., Fach P. (2010a). Low-density macroarray targeting non-locus of enterocyte effacement effectors (nle genes) and major virulence factors of Shiga toxin-producing Escherichia coli (STEC): a new approach for molecular risk assessment of STEC isolates. Appl. Environ. Microbiol. 76, 203–211 10.1128/AEM.01921-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugarel M., Beutin L., Martin A., Gill A., Fach P. (2010b). Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with Hemorrhagic Colitis and Hemolytic Uremic Syndrome in humans. Int. J. Food Microbiol. 142, 318–329 10.1016/j.ijfoodmicro.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Buvens G., Lauwers S., Pierard D. (2010). Prevalence of subtilase cytotoxin in verocytotoxin-producing Escherichia coli isolated from humans and raw meats in Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1395–1399 10.1007/s10096-010-1014-z [DOI] [PubMed] [Google Scholar]
- Cergole-Novella M. C., Nishimura L. S., Dos Santos L. F., Irino K., Vaz T. M., Bergamini A. M., et al. (2007). Distribution of virulence profiles related to new toxins and putative adhesins in Shiga toxin-producing Escherichia coli isolated from diverse sources in Brazil. FEMS Microbiol. Lett. 274, 329–334 10.1111/j.1574-6968.2007.00856.x [DOI] [PubMed] [Google Scholar]
- Coombes B. K., Wickham M. E., Mascarenhas M., Gruenheid S., Finlay B. B., Karmali M. A. (2008). Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74, 2153–2160 10.1128/AEM.02566-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Toni F., de Souza E. M., Pedrosa F. O., Klassen G., Irino K., Un R. L., et al. (2009). A prospective study on Shiga toxin-producing Escherichia coli in children with diarrhea in Parana State, Brazil. Lett. Appl. Microbiol. 48, 645–647 10.1111/j.1472-765X.2009.02569.x [DOI] [PubMed] [Google Scholar]
- Doughty S., Sloan J., Bennett-Wood V., Robertson M., Robins-Browne R. M., Hartland E. L. (2002). Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70, 6761–6769 10.1128/IAI.70.12.6761-6769.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority and European Centre for Disease Prevention and Control. (2012). The european union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J. 10:2597 10.2903/j.efsa.2012.2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D., Irino K., Sanz M. E., Padola N. L., Parma A. E. (2010). Characterization of Shiga toxin-producing Escherichia coli isolated from dairy cows in Argentina. Lett. Appl. Microbiol. 51, 377–382 10.1111/j.1472-765X.2010.02904.x [DOI] [PubMed] [Google Scholar]
- Friedrich A. W., Bielaszewska M., Zhang W. L., Pulz M., Kuczius T., Ammon A., et al. (2002). Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185, 74–84 10.1086/338115 [DOI] [PubMed] [Google Scholar]
- Gerhardt E., Miliwebsky E., Rivas M., Ibarra C. (2009). Detection of subtilase cytotoxin (SubAB) in Shiga toxin-producing Escherichia coli strains associated with hemolytic uremic syndrome in Argentine children, in 7th International Symposium on Shiga toxin (Verotoxin)-Producing Escherichia coli Infections (VTEC2009) (Buenos Aires: ). Abstr. P04.4.3, 101 [Google Scholar]
- Gerner-Smidt P., Scheutz F. (2006). Standardized pulsed-field gel electrophoresis of Shiga toxin-producing Escherichia coli: the PulseNet Europe Feasibility Study. Foodborne Pathog. Dis. 3, 74–80 10.1089/fpd.2006.3.74 [DOI] [PubMed] [Google Scholar]
- Giugno S. M., Bibiloni N., Rahman R., Miliwebsky E., Chinen I., Rivas M. (2007). Association between uremic hemolytic syndrome and infection by Shiga toxin-producing Escherichia coli. Acta Bioquím. Clín. Latinoam. 41, 27–33 [Google Scholar]
- Gobius K. S., Higgs G. M., Desmarchelier P. M. (2003). Presence of activatable Shiga toxin genotype (stx2d) in Shiga toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41, 3777–3783 10.1128/JCM.41.8.3777-3783.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein H. S. (2007). Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85, E63–E72 10.2527/jas.2006-421 [DOI] [PubMed] [Google Scholar]
- Irino K., Vieira M. A., Gomes T. A., Guth B. E., Naves Z. V., Oliveira M. G., et al. (2010). Subtilase cytotoxin-encoding subAB operon found exclusively among Shiga toxin-producing Escherichia coli strains. J. Clin. Microbiol. 48, 988–990 10.1128/JCM.00010-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelacic J. K., Damrow T., Chen G. S., Jelacic S., Bielaszewska M., Ciol M., et al. (2003). Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188, 719–729 10.1086/376999 [DOI] [PubMed] [Google Scholar]
- Jenkins C., Perry N. T., Cheasty T., Shaw D. J., Frankel G., Dougan G., et al. (2003). Distribution of the saa gene in strains of Shiga toxin-producing Escherichia coli of human and bovine origins. J. Clin. Microbiol. 41, 1775–1778 10.1128/JCM.41.4.1775-1778.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W., Shen J., Toro M., Zhao S., Meng J. (2013). Distribution of pathogenicity islands OI-122, OI-43/48, and OI-57 and a high-pathogenicity island in Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 79, 3406–3412 10.1128/AEM.03661-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali M. A., Mascarenhas M., Shen S., Ziebell K., Johnson S., Reid-Smith R., et al. (2003). Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41, 4930–4940 10.1128/JCM.41.11.4930-4940.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskimaki M., Ikaheimo R., Karkkainen P., Scheutz F., Ratiner Y., Puohiniemi R., et al. (1997). Shiga toxin-producing Escherichia coli serotype OX3:H21 as a cause of hemolytic-uremic syndrome. Clin. Infect. Dis. 24, 1278–1279 10.1086/513668 [DOI] [PubMed] [Google Scholar]
- Konczy P., Ziebell K., Mascarenhas M., Choi A., Michaud C., Kropinski A. M., et al. (2008). Genomic O island 122, locus for enterocyte effacement, and the evolution of virulent verocytotoxin-producing Escherichia coli. J. Bacteriol. 190, 5832–5840 10.1128/JB.00480-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A., Lucchesi P. M., Parma A. E. (2011). Verotoxins in bovine and meat verotoxin-producing Escherichia coli isolates: type, number of variants, and relationship to cytotoxicity. Appl. Environ. Microbiol. 77, 73–79 10.1128/AEM.01445-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi P. M., Kruger A., Parma A. E. (2006). Distribution of saa gene variants in verocytotoxigenic Escherichia coli isolated from cattle and food. Res. Microbiol. 157, 263–266 10.1016/j.resmic.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Masana M. O., D'Astek B. A., Palladino P. M., Galli L., Del Castillo L. L., Carbonari C., et al. (2011). Genotypic characterization of non-O157 Shiga toxin-producing Escherichia coli in beef abattoirs of Argentina. J. Food. Prot. 74, 2008–2017 10.4315/0362-028X.JFP-11-189 [DOI] [PubMed] [Google Scholar]
- Melton-Celsa A., Mohawk K., Teel L., O'Brien A. (2012). Pathogenesis of Shiga-toxin producing Escherichia coli. Curr. Top. Microbiol. Immunol. 357, 67–103 10.1007/82_2011_176 [DOI] [PubMed] [Google Scholar]
- Melton-Celsa A. R., Darnell S. C., O'Brien A. D. (1996). Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton-Celsa A. R., Kokai-Kun J. F., O'Brien A. D. (2002). Activation of Shiga toxin type 2d (Stx2d) by elastase involves cleavage of the C-terminal two amino acids of the A2 peptide in the context of the appropriate B pentamer. Mol. Microbiol. 43, 207–215 10.1046/j.1365-2958.2002.02733.x [DOI] [PubMed] [Google Scholar]
- Melton-Celsa A. R., Rogers J. E., Schmitt C. K., Darnell S. C., O'Brien A. D. (1998). Virulence of Shiga toxin-producing Escherichia coli (STEC) in orally-infected mice correlates with the type of toxin produced by the infecting strain. Jpn. J. Med. Sci. Biol. 51(Suppl.), S108–S114 10.7883/yoken1952.51.Supplement1_S108 [DOI] [PubMed] [Google Scholar]
- Michelacci V., Tozzoli R., Caprioli A., Martinez R., Scheutz F., Grande L., et al. (2013). A new pathogenicity island carrying an allelic variant of the Subtilase cytotoxin is common among Shiga toxin producing Escherichia coli of human and ovine origin. Clin. Microbiol. Infect. 19, E149–E156 10.1111/1469-0691.12122 [DOI] [PubMed] [Google Scholar]
- Muthing J., Schweppe C. H., Karch H., Friedrich A. W. (2009). Shiga toxins, glycosphingolipid diversity, and endothelial cell injury. Thromb. Haemost. 101, 252–264 10.1160/TH08-05-0317 [DOI] [PubMed] [Google Scholar]
- Newton H. J., Sloan J., Bulach D. M., Seemann T., Allison C. C., Tauschek M., et al. (2009). Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15, 372–380 10.3201/eid1503.080631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. G., Brito J. R., Carvalho R. R., Guth B. E., Gomes T. A., Vieira M. A., et al. (2007). Water buffaloes (Bubalus bubalis) identified as an important reservoir of Shiga toxin-producing Escherichia coli in Brazil. Appl. Environ. Microbiol. 73, 5945–5948 10.1128/AEM.00929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. G., Brito J. R., Gomes T. A., Guth B. E., Vieira M. A., Naves Z. V., et al. (2008). Diversity of virulence profiles of Shiga toxin-producing Escherichia coli serotypes in food-producing animals in Brazil. Int. J. Food Microbiol. 127, 139–146 10.1016/j.ijfoodmicro.2008.06.023 [DOI] [PubMed] [Google Scholar]
- Orden J. A., Horcajo P., de la Fuente R., Ruiz-Santa-Quiteria J. A., Dominguez-Bernal G., Carrion J. (2011). Subtilase cytotoxin-coding genes in verotoxin-producing Escherichia coli strains from sheep and goats differ from those from cattle. Appl. Environ. Microbiol. 77, 8259–8264 10.1128/AEM.05604-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padola N. L., Sanz M. E., Blanco J. E., Blanco M., Blanco J., Etcheverria A. I., et al. (2004). Serotypes and virulence genes of bovine Shigatoxigenic Escherichia coli (STEC) isolated from a feedlot in Argentina. Vet. Microbiol. 100, 3–9 10.1016/S0378-1135(03)00127-5 [DOI] [PubMed] [Google Scholar]
- Parma A. E., Sanz M. E., Blanco J. E., Blanco J., Vinas M. R., Blanco M., et al. (2000). Virulence genotypes and serotypes of verotoxigenic Escherichia coli isolated from cattle and foods in Argentina. Importance in public health. Eur. J. Epidemiol. 16, 757–762 10.1023/A:1026746016896 [DOI] [PubMed] [Google Scholar]
- Paton A. W., Paton J. C. (2002). Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40, 271–274 10.1128/JCM.40.1.271-274.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A. W., Paton J. C. (2010). Escherichia coli subtilase cytotoxin. Toxins (Basel.) 2, 215–228 10.3390/toxins2020215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A. W., Srimanote P., Talbot U. M., Wang H., Paton J. C. (2004). A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200, 35–46 10.1084/jem.20040392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A. W., Srimanote P., Woodrow M. C., Paton J. C. (2001). Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69, 6999–7009 10.1128/IAI.69.11.6999-7009.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A. W., Woodrow M. C., Doyle R. M., Lanser J. A., Paton J. C. (1999). Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37, 3357–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Olsen K. E., Ethelberg S., Scheutz F. (2007). Subtyping method for Escherichia coli shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45, 2020–2024 10.1128/JCM.02591-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager R., Fruth A., Siewert U., Strutz U., Tschape H. (2009). Escherichia coli encoding Shiga toxin 2f as an emerging human pathogen. Int. J. Med. Microbiol. 299, 343–353 10.1016/j.ijmm.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Rivas M., Padola N. L., Lucchesi P. M., Masana M. (2010). Diarrheagenic Escherichia coli in Argentina, in Pathogenic Escherichia coli in Latin America, ed Torres A. G. (Oak Park, IL: Bentham Science Publishers Ltd; ), 142–161 [Google Scholar]
- Robert Koch-Institut. (2011). Infektionsepidemiologisches Jahrbuch Meldepflichtiger Krankheiten für 2010. Berlin: Robert Koch-Institut [Google Scholar]
- Sanz M. E., Villalobos C., Elichiribehety E., Arroyo G. H. (2007). Prevalencia de escherichia coli verocitotoxigénico en productos cárnicos de la ciudad de Tandil. La industria cárnica latinoamericana, 27, 56–58 [Google Scholar]
- Scheutz F., Cheasty T., Woodward D., Smith H. R. (2004). Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include Verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS 112, 569–584 10.1111/j.1600-0463.2004.apm1120903.x [DOI] [PubMed] [Google Scholar]
- Scheutz F., Strockbine N. A. (2005). Genus I. Escherichia. The proteobacteria Part B the gammaproteobacteria, in Bergey's Manual of Systematic Bacteriology, eds Garrity G. M., Brenner D. J., Krieg N. R., Staley J. T. (New York, NY: Springer; ), 607–624 [Google Scholar]
- Scheutz F., Teel L. D., Beutin L., Pierard D., Buvens G., Karch H., et al. (2012). Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50, 2951–2963 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Beutin L., Karch H. (1995). Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Henkel B., Karch H. (1997). A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol. Lett. 148, 265–272 10.1111/j.1574-6968.1997.tb10299.x [DOI] [PubMed] [Google Scholar]
- Schmidt H., Zhang W. L., Hemmrich U., Jelacic S., Brunder W., Tarr P. I., et al. (2001). Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69, 6863–6873 10.1128/IAI.69.11.6863-6873.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Mascarenhas M., Rahn K., Kaper J. B., Karmali M. A. (2004). Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect. Immun. 72, 1496–1503 10.1128/IAI.72.3.1496-1503.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanec T., Fruth A., Creuzburg K., Schmidt H. (2009). Molecular analysis of virulence profiles and Shiga toxin genes in food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 75, 6187–6197 10.1128/AEM.00874-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaro J. D., Galli L., Lound L. H., Leotta G. A., Piaggio M. C., Carbonari C. C., et al. (2012). Non-O157:H7 Shiga toxin-producing Escherichia coli in bovine rectums and surface water streams on a beef cattle farm in Argentina. Foodborne Pathog. Dis. 9, 878–884 10.1089/fpd.2012.1182 [DOI] [PubMed] [Google Scholar]
- Tarr P. I., Bilge S. S., Vary J. C., Jr., Jelacic S., Habeeb R. L., Ward T. R., et al. (2000). Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68, 1400–1407 10.1128/IAI.68.3.1400-1407.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuno I., Horie M., Abe H., Miki T., Makino K., Shinagawa H., et al. (2001). toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 69, 6660–6669 10.1128/IAI.69.11.6660-6669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C., Nakasone N., Miliwebsky E., Higa N., Rivas M., Suzuki T. (2008). Differential adherence of Shiga toxin-producing Escherichia coli harboring saa to epithelial cells. Int. J. Med. Microbiol. 298, 571–578 10.1016/j.ijmm.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Torres A. G., Giron J. A., Perna N. T., Burland V., Blattner F. R., Avelino-Flores F., et al. (2002). Identification and characterization of lpfABCC'DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70, 5416–5427 10.1128/IAI.70.10.5416-5427.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzoli R., Caprioli A., Cappannella S., Michelacci V., Marziano M. L., Morabito S. (2010). Production of the subtilase AB5 cytotoxin by Shiga toxin-negative Escherichia coli. J. Clin. Microbiol. 48, 178–183 10.1128/JCM.01648-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu-Khac H., Cornick N. A. (2008). Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet. Microbiol. 126, 356–363 10.1016/j.vetmic.2007.07.023 [DOI] [PubMed] [Google Scholar]
- Werber D., Beutin L., Pichner R., Stark K., Fruth A. (2008). Shiga toxin-producing Escherichia coli serogroups in food and patients, Germany. Emerg. Infect. Dis. 14, 1803–1806 10.3201/eid1411.080361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Meng J., McDermott P. F., Ayers S., Blickenstaff K., Tran T. T., et al. (2010). Presence and characterization of shiga toxin-producing Escherichia coli and other potentially diarrheagenic E. coli strains in retail meats. Appl. Environ. Microbiol. 76, 1709–1717 10.1128/AEM.01968-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Wheatcroft R., Chambers J. R., Liu B., Zhu J., Gyles C. L. (2009). Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 75, 5779–5786 10.1128/AEM.00507-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Cui S., Teel L. D., Zhao S., Singh R., O'Brien A. D., et al. (2008). Identification and characterization of Shiga toxin type 2 variants in Escherichia coli isolates from animals, food, and humans. Appl. Environ. Microbiol. 74, 5645–5652 10.1128/AEM.00503-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel C., Blanco J. E., Blanco M., Blanco J., Stephan R. (2004). Serotypes and virulence genes of ovine non-O157 Shiga toxin-producing Escherichia coli in Switzerland. Int. J. Food Microbiol. 95, 19–27 10.1016/j.ijfoodmicro.2004.01.015 [DOI] [PubMed] [Google Scholar]
- Zweifel C., Schumacher S., Blanco M., Blanco J. E., Tasara T., Blanco J., et al. (2005). Phenotypic and genotypic characteristics of non-O157 Shiga toxin-producing Escherichia coli (STEC) from Swiss cattle. Vet. Microbiol. 105, 37–45 10.1016/j.vetmic.2004.10.007 [DOI] [PubMed] [Google Scholar]