Abstract
Eliciting protective neutralizing antibodies (NAbs) against HIV-1 is daunting because of the extensive genetic and antigenic diversity of HIV-1. Moreover, broad and potent responses are uncommon even during persistent infection, with only a subset of adults developing broadly neutralizing antibodies (bNAbs) that recognize variants from different HIV-1 clades1–8. It is not known whether bNAbs can also arise in HIV-1-infected infants, who typically progress to disease faster than adults9, presumably in part due to an immature immune system10. Here, we show that bNAbs develop at least as commonly in infants as in adults. Cross-clade NAb responses were detected in 20/28 infected infants, in some cases, within 1 year of infection. Among infants with the top quartile of responses, neutralization of Tier 2–3 variants from multiple clades was detected at 20 months post-infection. These findings suggest that, even in early life, there is sufficient B-cell functionality to mount bNAbs against HIV-1. Additionally, the relatively early appearance of bNAbs in infants may provide a unique setting for understanding the pathways of B-cell maturation leading to bNAbs.
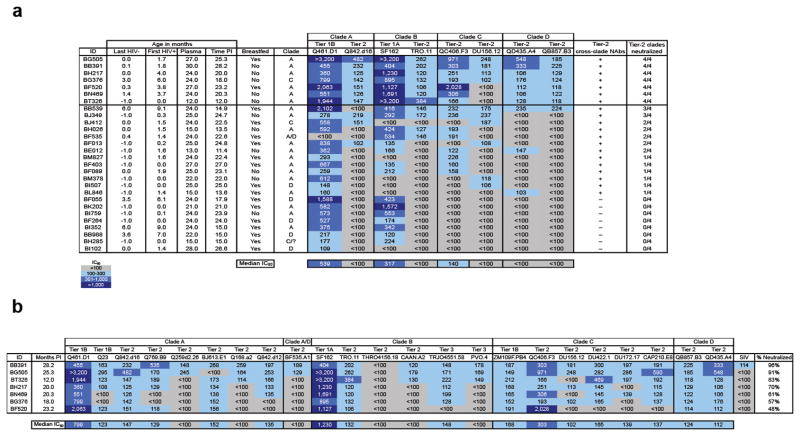
HIV-1-specific NAb breadth, which develops after several years of infection in some adults1–8,, has not been measured in infants. We assessed NAb breadth in 28 infants who acquired clades A, C, and D viruses in-utero, during delivery, or via breastfeeding (median time of first HIV-1 detection = 1.5 months, range = 0–9.1 months) in the Nairobi Breastfeeding Trial11, and who had plasma samples available ≥ 12 months of life. Samples at the last timepoint after birth (median = 24 months, range = 12–30 months) were first screened against 8 viruses representing clades A–D (Fig. 1a). Overall, 20/28 (71%) infants neutralized a Tier 2 virus12,13 from a different clade than the infecting virus (cross-clade responses) at a median time of 22.2 months post-infection (PI) (range = 11.4–28.2 months PI). Remarkably, 4 infants (BB539, BH026, BE012, and BL846) already had evidence of cross-clade responses by 11–15 months PI but we did not have samples to assess whether they ultimately developed more broad and potent NAbs at later timepoints.
Figure 1.
Neutralization of panel viruses. (a) Initial screen of 28 infant samples against 8 viruses. The first column shows infant ID, followed by the last HIV-1 negative timepoint, where ‘−1.0’ indicates testing of cord blood sample, and ‘0.0’ indicates delivery timepoint. The plasma timepoint tested is followed by time post-HIV-1 infection (PI), calculated as time from first HIV-1 detection. The column labeled ‘Clade’ indicates the infecting virus clade34. BH285 was infected with a clade C recombinant virus but portions of the envelope gene could not be readily assigned to any known subtype. Panel virus clades and tier designations12,13,17 are shown above virus names. In the second to last column, ‘+’ and ‘−’ indicate the presence or absence of cross-clade NAbs against a Tier 2 virus, respectively. The last column shows the number of Tier 2 virus clades neutralized. IC50 values, shown as reciprocal plasma dilutions from ≥ 2 independent experiments, are color coded as shown in the key. Gray shading indicates that 50% neutralization was not achieved at the lowest plasma dilution tested (1:100). The 7 samples that neutralized a Tier 2 virus from all 4 clades are shown above the horizontal line. (b) Neutralization profile of 7 samples with bNAbs against 23 viruses. Infant IDs are shown in the first column, followed by the months post-infection at which samples were tested. IC50 values are from ≥ 2 independent experiments and are color coded as in (a). The last column shows percentage of viruses neutralized. SIV was included as a negative control.
Based on this initial screen, 7/28 samples that neutralized Tier 2 viruses from all 4 clades were tested against 15 other viruses representing various neutralization sensitivities12,14,15 (Fig 1b). All 7 samples neutralized ≥ 2 viruses across 4 clades at a median time of 20.3 months PI (range = 12–28.2 months PI). BB391 and BG505 had the most impressive responses, neutralizing 91%–96% of viruses. Purified IgG from these 2 infants neutralized HIV-1 variants, but not SIV (Supplementary Table 1), verifying that IgG antibodies mediated HIV-1-specific NAb breadth. Notably, BT326 and BG376 had cross-clade breadth against Tier 2 and Tier 3 viruses as early as 12–18 months PI.
To confirm that bNAbs were due to de novo responses generated by infants in response to infection and to determine kinetics of NAb breadth, we tested longitudinal samples, beginning at the earliest timepoint after birth (Fig. 2 and Supplementary Fig. 1). For some infants tested at birth (BT326, BG376, BF520), we observed high NAb titers against a few viruses, which were likely due to passive NAbs as they waned by ~3 months, consistent with the kinetics of passive HIV-1 NAb decay16,17. These titers rebounded and peaked at the last timepoint, reflecting the development of de novo responses. For all infants, there was evidence of de novo responses at ~12 months PI that increased in potency over time (geometric mean IC50 = 201–570 at the last timepoint). Of note, by 12 months of life (~8–12 months PI), BT326 and BN469 already had broad de novo responses, defined by neutralization of ≥ 1 virus across 4 clades with IC50 > 1004, while BG376 and BF520 developed similar breadth by 18 and 15 months of life (12 and 11.2 months PI, respectively; Supplementary Fig. 1). These results confirm that de novo responses mediated NAb breadth, and suggest that bNAbs can develop within the first year of life and of HIV-1 infection.
Figure 2.
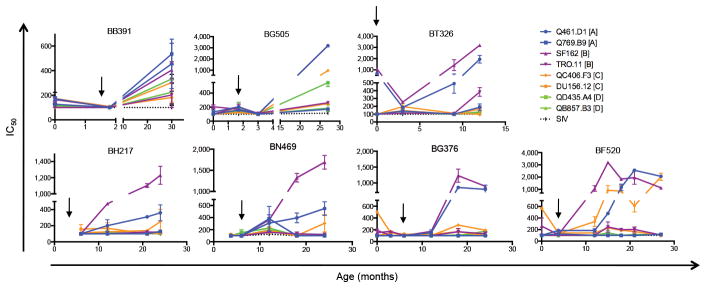
Kinetics of NAb breadth for 7 infants. Graphs show IC50 values against 8 viruses, 2 from each subtype A–D (shown in the key in upper right corner) over time. Viruses are color-coded by clade, as shown in the key. Black arrows denote when HIV-1 was first detected. Error bars indicate standard error of the mean based on 2 independent experiments.
We compared NAb breadth of the top 2 infants with bNAbs to that of adults by testing against 6 viruses (Supplementary Fig. 2) used to identify HIV-1-infected adults with the top 1% of bNAb responses (‘elite neutralizers’)4. BB391 and BG505 had scores of 2.1, which fall short of the rare subset of adults with elite bNAbs (score ≥ 2.5)4. Nevertheless, by < 2.5 years of HIV-1 infection, these infants had scores similar to those of the top 3 of 463 (0.7%) adult samples initially screened for bNAbs at ≥ 3 years PI in the prior study4, and to those of QB850 and QA255 (neutralization scores of 2.3 and 1.6, respectively), 2 adults with the broadest responses at ~5 years PI in previous screens of 48 and 70 women, respectively5,13. Thus, bNAbs in these 2 infants at ~2.5 years PI are approaching those found in the top 1% of adults identified from larger screens at later times PI.
To determine whether infant NAb responses were correlated to passive NAbs from the mother, we tested plasma from the first week of life. While there was no correlation between passive and de novo NAb titers overall (Pearson’s r = 0.29, P = 0.19), excluding BG505, an outlier in this analysis, resulted in a significant correlation (Pearson’s r = 0.52, P = 0.02, Fig. 3a). This correlation was generally observed among infants with (Pearson’s r = 0.7, P = 0.03, Fig. 3b; excluding BG505) or without (Pearson’s r = 0.53, p = 0.09, Fig. 3c) NAbs against Tier-2 viruses from ≥ 2 different clades (Fig. 1a). Including BG505 in the analysis for the former group resulted in a weaker correlation (Pearson’s r = 0.58, P = 0.06). These results suggest that antigenic features in the shared maternal/infant virus population may shape infant NAbs, but there may be unique factors contributing to de novo breadth in BG505. Indeed, there was no correlation between infant de novo responses for the subset of 7 infants with the greatest NAb breadth, either with passive NAbs or maternal NAbs (Supplementary Fig. 3a, b). These findings could suggest that viral determinants are not the only factor driving infant NAb breadth, although larger studies are needed to clarify this interesting possibility.
Figure 3.
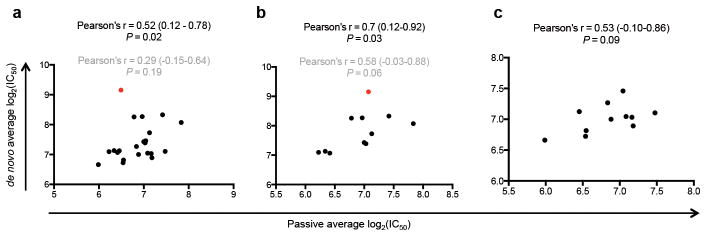
Association between passive and de novo NAbs. (a) Correlation between passive and de novo infant average log2(IC50) values. Samples were available for 22/28 infants. (b) Correlation between passive and de novo log2(IC50) for infants with NAbs against Tier-2 viruses from at least 2 different clades identified in Figure 1a and with plasma sample available within the first week of life (n = 11). (c) Correlation between passive and de novo log2(IC50) for infants without NAbs against Tier-2 viruses from at least 2 different clades identified in Figure 1a and with plasma sample available within the first week of life (n = 11). For all panels, infant de novo IC50 values were obtained from the last available timepoint (the plasma sample used in Fig. 1), while passive IC50 values were obtained within the first week of life. For panels (a) and (b), red dot represents data point for BG505. Pearson’s r, 95% confidence intervals, and P-values shown in black and gray indicate analysis excluding and including BG505, respectively.
Next, we analyzed factors associated with de novo NAb breadth, including set-point viral load (SVL)2,5, duration of HIV-1 infection1, total and envelope (Env)-specific IgG levels1 at the latest timepoint, and passive NAbs. A clade A Env, which was previously shown to have highly exposed NAb epitopes18 was used to assess binding. In univariate analyses, SVL and Env-specific IgG levels were significantly associated with NAb breadth (P = 0.002 and P = 0.0004, respectively, Supplementary Table 2), and this association persisted in multivariate analysis (P = 0.02 and P = 0.001, respectively, Supplementary Table 2). Moreover, SVL was significantly higher among infants with bNAbs identified in Figure 1 compared to those without bNAbs (P = 0.0004, Fig. 4a), and there was a trend for higher Env-specific IgG levels in the former compared to the latter group (P = 0.05, Fig. 4b).
Figure 4.
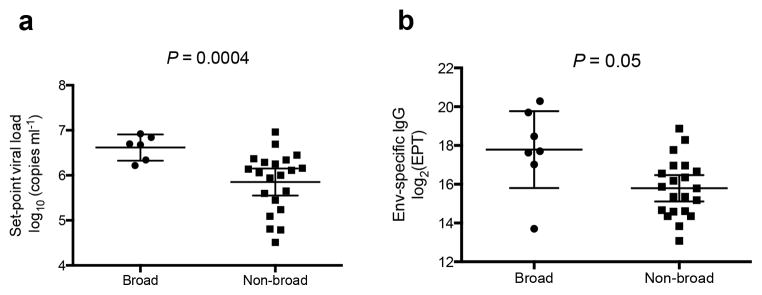
Comparison of set-point viral load and Env-specific IgG for infants with and without bNAbs. Unpaired t-test with Welch’s correction comparing (a) set-point viral load of infants with (n=6, excluding BB391, who had no SVL measurement) and without (n = 21) bNAbs, and (b) Env-specific (Q461.d1 gp120) log-transformed end-point titers (EPT) of plasma from infants with (n = 7) and without (n = 21) bNAbs identified in Figure 1. Horizontal lines in (a) and (b) represent mean and 95% confidence intervals.
Currently identified bNAbs target 4 main epitopes on Env: glycan-dependent sites in V1/V219 or V320, the CD4 binding site (CD4bs)21, and the membrane proximal external region (MPER)22. Using experimental methods that are currently standard for detecting these bNAbs2,3,19–22, we found no evidence for NAbs with these specificities in the 7 infant samples with NAb breadth, except for a modest anti-MPER response for BG505 (Supplementary Fig. 4). This was true using mutations in different viral envelopes (data not shown). Similarly, by using a computational algorithm to delineate polyclonal specificities23, we did not detect a consistent signal for a single dominant response against known epitopes (Goo, Georgiev, Kwong, Overbaugh, unpublished). We note, however, that the precision of the predictions may have been affected by the limited plasma availability, which necessitated the use of a reduced panel of only 13 isolates.23 Overall, our experimental and computational mapping analyses suggest that infant NAb breadth may be mediated by a polyclonal response and/or possibly a bNAb with novel specificity.
In summary, we have shown that HIV-1 bNAbs can develop early in life. Thus, despite having higher viral loads and faster disease progression compared to adults7, infants maintain sufficient B-cell function to mount bNAbs. Indeed, as seen in adults, higher SVL in infants was associated with bNAbs, supporting the hypothesis that early antigenic load drives NAb breadth2,5,6.
HIV-1-specific bNAbs isolated from adults after several years of infection display extensive somatic hypermutation19–22. Infants can achieve adult-like IgG diversity and concentration by 8–12 and 12–24 months, respectively10, raising the possibility that rapid somatic hypermutation early in life may contribute to the development of infant bNAbs. The predominance of IgG1 and IgG3 over IgG2 in infants10 may also enhance HIV-1 neutralizing activity24–26. Although HIV-1-specific passive IgG reduced viremia and accelerated NAb development in newborn macaques27, presumably by mitigating HIV-1-induced B-cell dysfunction28, we found a trend for a positive correlation between higher passive NAbs and infant SVL (data not shown), which in turn was associated with de novo breadth. Because only passive IgG matched to the infecting virus successfully reduced viremia in the prior study27, the failure of passive NAbs to control viremia in infants could be explained by the transmission of NAb escape variants16. Interestingly, an immunogen based on monomeric HIV-1 Env of the transmitted BG505 escape variant16 elicited NAbs against Tier 2 viruses in rabbits29 and the trimeric HIV-1 BG505 Env30, modified to increase stability, expressed epitopes for bNAbs31,32. It is interesting to speculate that the process of NAb escape from maternal and perhaps early de novo antibodies of vertically transmitted viruses could have indirectly exposed conserved epitopes that induce bNAbs, as shown in heterosexual infection33.
As infants are monitored frequently for infection in prevention of mother-to-child transmission efforts, pre-infection and longitudinal samples are available in many cohort studies. Given the growing interest in the field to delineate pathways to HIV-1-specific bNAbs, these samples may be especially valuable, as some infants appear to develop bNAbs relatively rapidly, and at their early age, may have been exposed to fewer non-HIV-specific antigens than their adult counterparts.
Online methods
Study population
Plasma samples were obtained from antiretroviral-naïve mother-infant pairs enrolled in the Nairobi Breastfeeding Trial11. Ethical review committees of the University of Nairobi, University of Washington, and Fred Hutchinson Cancer Research Center approved the study. In addition, the Kenyan Ministry of Health gave permission to conduct the Nairobi Breastfeeding Trial. Informed consent was obtained for all participants.
We screened infants for bNAbs in this study if they had plasma samples available at least after 12 months of birth in order to increase our chances of detecting de novo antibodies. Twenty-eight infants met this criterion, of which 17 were breast-fed and 11 formula-fed. Infants were defined as HIV-1 infected by PCR using PBMC or dried blood spot DNA, and by HIV-1 RNA testing using the Gen-Probe HIV-1 Viral Load assay11. Time post-infection is defined as the time from the infant’s first HIV-1 positive nucleic acid test.
For each of the 28 infants, we measured the presence of bNAbs at the last available plasma sample. To determine kinetics of NAb breadth in 7 infants with the greatest breadth at the latest time point, we tested earlier available samples, beginning at either the first week of life (5 infants), month 3.7 (1 infant), or month 6 (1 infant). Maternal plasma samples were obtained from the last HIV-1 negative timepoint for their corresponding infants, or the earliest timepoint if the infant was predicted to be infected in-utero. HIV-1 clade was determined by a combination of heteroduplex mobility assay of V1-C3 (21/28 subjects), and sequence analysis of V1-V5 (7/28 subjects) of maternal virus envelopes, as described previously34. Viral loads were quantified by the Gen-Probe HIV-1 viral load assay11. We also screened plasma samples from 2 HIV-1-infected adult women, QB850 and QA255, who were participants of a heterosexual transmission cohort in Mombasa, Kenya for HIV-1-specific NAbs using samples from ~5 years PI. QB850 is a top broad neutralizer identified in a screen of 12 superinfected and 36 singly-infected women13, while QA255 is a top broad neutralizer in a similar screen of 70 singly infected women5.
Pseudovirus panels and neutralization assay
The 8-virus panel used in the initial screen for bNAbs includes 2 Tier 1 and 6 Tier 2 viruses representing clades A (Q461.D1, Q842.d16), B (SF162, TRO.11), C (QC406.F3, DU156.12), and D (QD435.A4, QB857.B3). Tier designation was taken directly from Seaman, et al.12 when available; for other viruses, tier designation was defined by comparing IC50 values against pooled plasma13,17 to envelope variants of known tier designation based on Seaman, et al.12. Viruses were obtained from infected Kenyans and were representative of major circulating subtypes, which are relevant to the infant cohort examined here. These viruses have been used to screen prior Kenyan cohorts 13,15,17, and detailed virus characteristics have been described elsewhere15. Infant plasma samples that could neutralize a Tier 2 virus from all 4 clades at an IC50 > 100 were classified as having bNAbs, and were further screened against an additional 15 viruses, including 6 clade A (Q23.17, Q769.B9, Q259d2.26, BJ613.E1, Q168.a2, Q842.d12), 1 clade A/D (BF535.A1), 4 clade B (THRO4156.18, CAAN.A2, TRJO4156.18, PVO.4), and 4 clade C (ZM109F.PB4, DU422.1, DU172.17, CAP210.E8) viruses. We tested longitudinal infant samples and maternal pre-transmission samples against 2 viruses each from clades A, B, C, and D (Q461.D1, Q769.B9, SF162, TRO.11, QC406.F3, DU156.12, QD435.A4, QB857.B3). A negative control pseudovirus from the envelope of simian immunodeficiency virus (SIV), SIVMneCl8, was also tested against all plasma samples to confirm that neutralizing activity was HIV-1-specific. Plasma samples from BB391 and BG505, the top 2 broad neutralizers, were tested against a 6-virus panel that was previously used to identify elite neutralizers4. This panel consists of 1 clade A (94UG103), 2 clade B (JRCSF, 92BR020), 2 clade C (IAVI C22, 93IN905), and 1 clade CRF01_A3 (92TH021) virus.
To generate pseudoviruses, envelope plasmids were co-transfected with Q23Δenv an envelope-deficient proviral plasmid, as previously described16. Neutralization assays were performed using TZM-bl cells as previously described16. Heat-inactivated plasma samples (56°C, 1hr) were tested using six 2-fold serial dilutions, starting at 1:100. IC50 values represent the reciprocal plasma dilution resulting in a 50% reduction of virus infectivity16. For viruses not neutralized at the lowest dilution tested (1:100), IC50 values of 100 were assigned for analyses. For viruses that did not reach 50% neutralization at the highest dilution tested (1:3,200), IC50 values of 3,200 were assigned for analyses. IC50 values represent the average of at least 2 independent experiments.
Neutralization scores
For BB391 and BG505, the top 2 broad infant samples, and QB850 and QA255, the top 2 broad adult samples in a Kenyan heterosexual transmission cohort5,13, we calculated a ‘neutralization score’ using a scoring procedure that was deemed most suitable for identifying samples with bNAbs (‘Score 1’), as described in Simek et al.4. Briefly, an individual’s neutralization score is the average of log-transformed titers against 6 viruses in Supplementary Fig. 2. We obtained log-transformed titers by dividing IC50 values by 100 followed by a log-base 2 transformation and adding 1: (Y = log2(IC50/100)+1). All IC50 values below the limit of detection of 100 were assigned a log-transformed titer of 0 for calculating neutralization scores.
Immunoglobulin G (IgG) purification and quantification
We purified total IgG from 50 μl of plasma using the Melon Gel IgG purification kit (Thermo Scientific) in a final volume of 500 μl (1:10 dilution in purification buffer). Purified IgG was quantified and tested in neutralization assays at a starting concentration of 2 mg ml−1 for BG505 or 1 mg ml−1 for BB391, which had more limited sample, against Q461.D1, Q842.d16, TRO.11, QC406.F3, QD435.A4, and SIV.
Total and HIV-1-specific ELISA
Total IgG ELISA in plasma samples from mother-infant pairs were quantified using a human IgG ELISA kit (Immunology Consultants Laboratory) according to manufacturer’s instructions. We performed HIV-1 envelope-specific IgG ELISAs as previously described1, using 25 ng/well of purified recombinant gp120 (Immune Technology Corp, New York, NY) from Q461.D1, a Tier 1B clade A variant with optimal exposure of multiple epitopes18. Plasma samples were serially diluted 2-fold starting at a dilution of either 1:1,000 or 1:10,000 to obtain an end-point titer (EPT), defined as the reciprocal dilution at which the average optical density (OD) value was greater than or equal to twice the average OD value of background. Total IgG levels and EPT values used in analyses were the average of at least 2 independent experiments.
Expression and purification of RSC3 proteins
Proteins were expressed and purified as described by Wu, et al.21, with slight modifications. Briefly, proteins were produced by transient transfection of 293F cells (1.2 x 106 in 400 ml) using 293fectin (Invitrogen). Culture supernatants were harvested 5 days after transfection, filtered through a 0.45 μm filter, and concentrated by centrifugation in 100 kDa Centricon-Plus 70 filter tubes (Millipore) with buffer exchange into PBS. Proteins were first purified by DEAE sepharose (GE) ion exchange chromatography, followed by His-Select Nickel (Sigma) affinity chromatography.
Epitope mapping
To screen for PG9/16- and PGT128-like19,20 NAbs, we compared neutralization of a commonly used clade A wildtype virus, Q23.172,33 to that of N160K or N332A mutants, respectively in most infant plasma samples. Mutants were generated by overlap PCR and verified by sequencing. BG376 and BN469 could not neutralize Q23.17 at the lowest dilution tested (1:100) and were instead screened against JRCSF and corresponding mutants, which have been used to detect PG- and PGT-like antibodies in previous studies19,20. Results were confirmed using a Tier 1B clade A virus, Q461.D1 and its corresponding N160K mutant (data not shown). The effect of N332A could not be confirmed in Q461.D1 as this virus lacks N332. Thus, in the case of BG505 who had notably broad responses, we confirmed mapping results with the N332A mutant in the context of JRCSF (data not shown). In all experiments, PG9 and PGT128 were included as positive controls for N160K and N332A, respectively, while VRC0121, which targets the CD4 binding site, served as a control for non-specific effects of both mutants. We assessed for the presence of CD4bs (VRC01-like) antibodies by ELISA as described above for Q461.D1 gp120, except a resurfaced stabilized core (RSC3) gp120 protein and its mutant (RSC3Δ371I) were used21, and plasma samples were tested at a starting dilution of 1:100. EPT was calculated as described above. To map MPER specificities, we compared neutralization of a HIV-2 full-length construct (7312A) to that of a HIV-2/HIV-1 MPER chimera (7312-C1)22.
To assess NAb specificity using the computational method described by Georgiev, et al.23, IC50 values were obtained for the following 13 viruses: CAAN.A2, CAP210.E8, DU156.12, DU172.17, DU422.01, PVO.04, Q168.a2, Q23.17, Q842.d12, THRO4156.18, TRJO4551.58, TRO.11, ZM109.PB4. This panel represents the minimal virus collection validated by this method, although the antibody classification accuracy may be sensitive to the size of the virus panel23.
Statistical analyses
All analyses were performed in R 2.10.1 and GraphPad Prism 9.0. Infant de novo antibodies were assessed at the latest available timepoint for all analyses. Average log2(IC50) values used for analyses were based on neutralization of 6 Tier 2 and 2 Tier 1 viruses, including 2 clade A (Q461.D1, Q769.B9), 2 clade B (SF162, TRO.11), 2 clade C (QC406.F3, DU156.12), and 2 clade D (QD435.A4, QB857.B3) viruses.
For a subset of 22 infants who had plasma availability within the first week of life, we determined the association between passive and de novo antibodies. IC50 values for passive neutralization for 13/22 infants were available from a previous study investigating the breadth of passive NAb responses17 starting at a plasma dilution of 1:25, while neutralization data for the remaining 9/22 infants were generated in this study at a starting plasma dilution of 1:100 to preserve sample. For all analyses, viruses not neutralized at the lowest plasma dilution tested were assigned IC50 values of 25 or 100 (the lowest dilution tested in the prior or current study, respectively). We used Pearson’s correlation coefficient to compare the average log2(IC50) values of passive and de novo antibodies, and maternal and de novo antibodies.
Set-point viral load measurements, defined as the first available measurement within 4–12 months of infection9, were available for all infants except BB391. To compare set-point viral loads (log10 copies ml−1) and Env-specific IgG log2 end-point titers (EPT) between infants who developed bNAbs (n = 7) and those who did not (n = 21), as determined in Figure 1, we performed unpaired t-tests with Welch’s correction. Univariate linear regression models were used to identify factors associated with de novo NAb breadth, defined as the average log2(IC50) value against 8 viruses described above. In these models, logarithmic transformations were performed for passive NAb IC50 values against these viruses, set-point viral load, and Env-specific IgG EPT. Duration of HIV-1 infection was calculated by subtracting the timepoint of the first HIV-1 positive result from the plasma timepoint used for neutralization assays. For infants who were predicted to be infected in-utero based on an HIV-1-positive cord blood sample, or at delivery, the timepoint of first HIV-1 positive result was set to 0. Covariates significantly associated with de novo NAb breadth in univariate analyses (P < 0.05) were included in multivariate models. Analyses involving passive NAbs and set-point viral load were performed for 22/28 and 27/28 infants, respectively.
Supplementary Material
Supplementary Figure 1. Neutralization profile of maternal and longitudinal infant plasma against 8 viruses, including 2 Tier 1 and 6 Tier 2 viruses representing clades A–D.
Supplementary Figure 2. Comparison of neutralization profile of 2 infants with greatest NAb breadth (BB391 and BG505) and 2 adult samples (QB850, QA255).
Supplementary Figure 3. Association between maternal, passive, and de novo NAbs for 7 infants with bNAbs that neutralize Tier 2 virus from clades A–D.
Supplementary Figure 4. Epitope mapping of bNAbs in infants
Supplementary Table 1. Neutralization profile of purified IgG from BB391 and BG505
Supplementary Table 2. Univariate and multivariate linear regression analyses of factors associated with NAb breadth
Acknowledgments
We are grateful to the participants and staff of the Nairobi Breastfeeding Trial; P. Poignard (IAVI Neutralizing Antibody Center, The Scripps Research Institute) for providing envelope plasmids used for viruses in Supplementary Figure 2; P. Moore (Centre for HIV and STIs, National Institute for Communicable Diseases of the National Health Laboratory Services, Johannesburg, South Africa) and G. Shaw (Perelman School of Medicine, University of Pennsylvania) for providing 7312-A and 7312-C1 plasmids; X. Wu and J. Mascola (Vaccine Research Center, US National Institutes of Health) for providing RSC3 and RSC3d371I plasmids; N. Doria-Rose, S. O’Dell, and J. Mascola (Vaccine Research Center, US National Institutes of Health) for providing JRCSF N160K and N332A plasmids; S. Rainwater and B. Wang for assistance in obtaining viral clade information; V. Cortez for assistance in RSC3 protein purification; C. Milligan for sample database management; I. Georgiev, V. Cortez, D. Lehman, C. Milligan, and K. Ronen for helpful discussions; and B. Richardson for advice on statistical analyses.
This work was supported by NIH Grants AI076105 and AI103981.
Footnotes
Author contributions
L.G. designed and performed experiments, analyzed data, and wrote the manuscript. V.C. performed experiments. R.N. oversaw the cohort and clinical aspects. J.O. conceived and oversaw the study, and edited the manuscript.
References
- 1.Sather DN, et al. Factors Associated with the Development of Cross-Reactive Neutralizing Antibodies during Human Immunodeficiency Virus Type 1 Infection. J Virol. 2008;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray ES, et al. The Neutralization Breadth of HIV-1 Develops Incrementally over Four Years and Is Associated with CD4+ T Cell Decline and High Viral Load during Acute Infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikell I, et al. Characteristics of the Earliest Cross-Neutralizing Antibody Response to HIV-1. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simek MD, et al. Human Immunodeficiency Virus Type 1 Elite Neutralizers: Individuals with Broad and Potent Neutralizing Activity Identified by Using a High-Throughput Neutralization Assay together with an Analytical Selection Algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piantadosi A, et al. Breadth of Neutralizing Antibody Response to Human Immunodeficiency Virus Type 1 Is Affected by Factors Early in Infection but Does Not Influence Disease Progression. J Virol. 2009;83:10269–10274. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS. 2009;23:2405–2414. doi: 10.1097/QAD.0b013e32833243e7. [DOI] [PubMed] [Google Scholar]
- 7.Doria-Rose NA, et al. Breadth of Human Immunodeficiency Virus-Specific Neutralizing Activity in Sera: Clustering Analysis and Association with Clinical Variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hraber P, et al. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson BA, et al. Comparison of Human Immunodeficiency Virus Type 1 Viral Loads in Kenyan Women, Men, and Infants during Primary and Early Infection. J Virol. 2003;77:7120–7123. doi: 10.1128/JVI.77.12.7120-7123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegrist CA. Neonatal and early life vaccinology. Vaccine. 2001;19:3331–3346. doi: 10.1016/s0264-410x(01)00028-7. [DOI] [PubMed] [Google Scholar]
- 11.Nduati R, et al. Effect of breastfeeding and formula feeding on transmission of HIV-1: a randomized clinical trial. JAMA. 2000;283:1167–1174. doi: 10.1001/jama.283.9.1167. [DOI] [PubMed] [Google Scholar]
- 12.Seaman MS, et al. Tiered Categorization of a Diverse Panel of HIV-1 Env Pseudoviruses for Assessment of Neutralizing Antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortez V, Odem-Davis K, McClelland RS, Jaoko W, Overbaugh J. HIV-1 Superinfection in Women Broadens and Strengthens the Neutralizing Antibody Response. PLoS Pathog. 2012;8:e1002611. doi: 10.1371/journal.ppat.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS. 2007;21:693–702. doi: 10.1097/QAD.0b013e32805e8727. [DOI] [PubMed] [Google Scholar]
- 15.Blish CA, et al. Cross-Subtype Neutralization Sensitivity despite Monoclonal Antibody Resistance among Early Subtype A, C, and D Envelope Variants of Human Immunodeficiency Virus Type 1. J Virol. 2009;83:7783–7788. doi: 10.1128/JVI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, et al. Neutralization Escape Variants of Human Immunodeficiency Virus Type 1 Are Transmitted from Mother to Infant. J Virol. 2005;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch JB, et al. The Breadth and Potency of Passively Acquired Human Immunodeficiency Virus Type 1-Specific Neutralizing Antibodies Do Not Correlate with the Risk of Infant Infection. J Virol. 2011;85:5252–5261. doi: 10.1128/JVI.02216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blish CA, Nguyen MA, Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 2008;5:e9. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011:1–6. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2013;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiev IS, et al. Delineating Antibody Recognition in Polyclonal Sera from Patterns of HIV-1 Isolate Neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 24.Scharf O, et al. Immunoglobulin G3 from Polyclonal Human Immunodeficiency Virus (HIV) Immune Globulin Is More Potent than Other Subclasses in Neutralizing HIV Type 1. J Virol. 2001;75:6558–6565. doi: 10.1128/JVI.75.14.6558-6565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavacini LA, et al. Influence of heavy chain constant regions on antigen binding and HIV-1 neutralization by a human monoclonal antibody. J Immunol. 1995;155:3638–3644. [PubMed] [Google Scholar]
- 26.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nature Medicine. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 27.Ng CT, et al. brief communications. Nature Medicine. 2010;16:1117–1119. doi: 10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque MC, et al. Polyclonal B Cell Differentiation and Loss of Gastrointestinal Tract Germinal Centers in the Earliest Stages of HIV-1 Infection. PLoS Med. 2009;6:e1000107. doi: 10.1371/journal.pmed.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffenberg S, et al. Identification of an HIV-1 Clade A Envelope That Exhibits Broad Antigenicity and Neutralization Sensitivity and Elicits Antibodies Targeting Three Distinct Epitopes. J Virol. 2013;87:5372–5383. doi: 10.1128/JVI.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore PL, et al. Evolution of an HIV glycan–dependent broadly neutralizing antibody epitope through immune escape. Nature Medicine. 2012:1–6. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neilson JR, et al. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J Virol. 1999;73:4393–4403. doi: 10.1128/jvi.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Neutralization profile of maternal and longitudinal infant plasma against 8 viruses, including 2 Tier 1 and 6 Tier 2 viruses representing clades A–D.
Supplementary Figure 2. Comparison of neutralization profile of 2 infants with greatest NAb breadth (BB391 and BG505) and 2 adult samples (QB850, QA255).
Supplementary Figure 3. Association between maternal, passive, and de novo NAbs for 7 infants with bNAbs that neutralize Tier 2 virus from clades A–D.
Supplementary Figure 4. Epitope mapping of bNAbs in infants
Supplementary Table 1. Neutralization profile of purified IgG from BB391 and BG505
Supplementary Table 2. Univariate and multivariate linear regression analyses of factors associated with NAb breadth