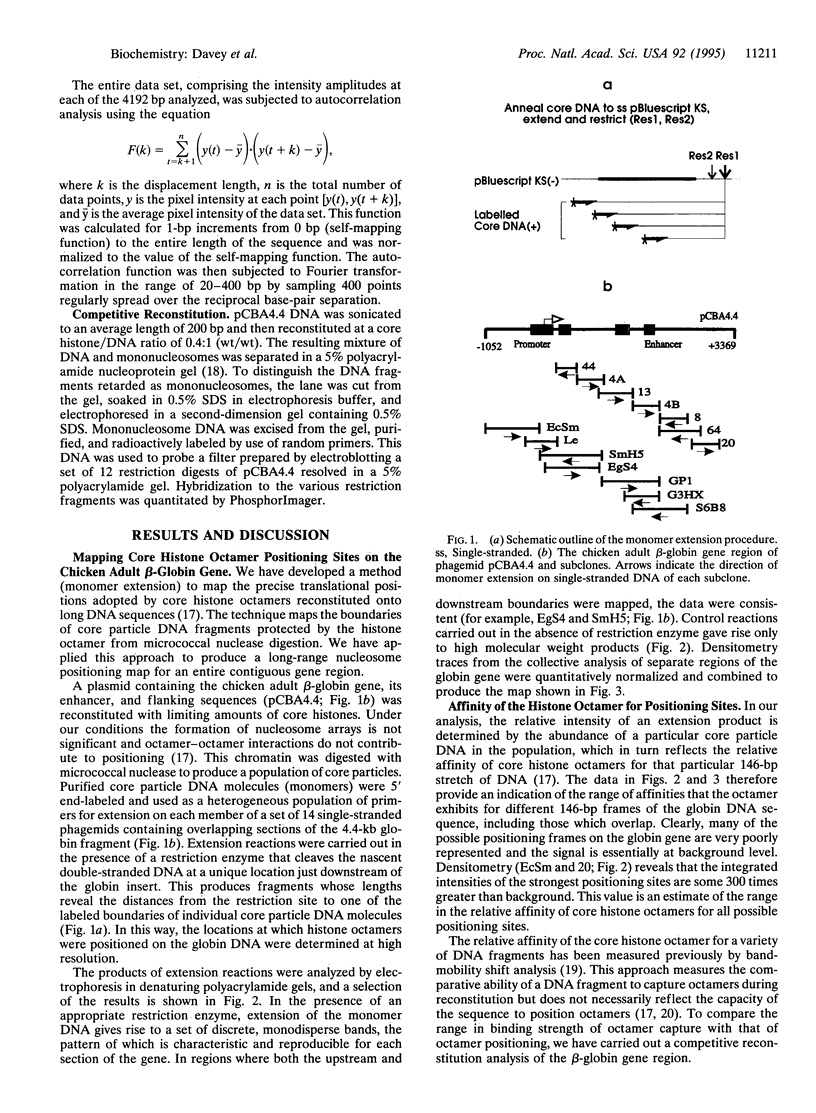
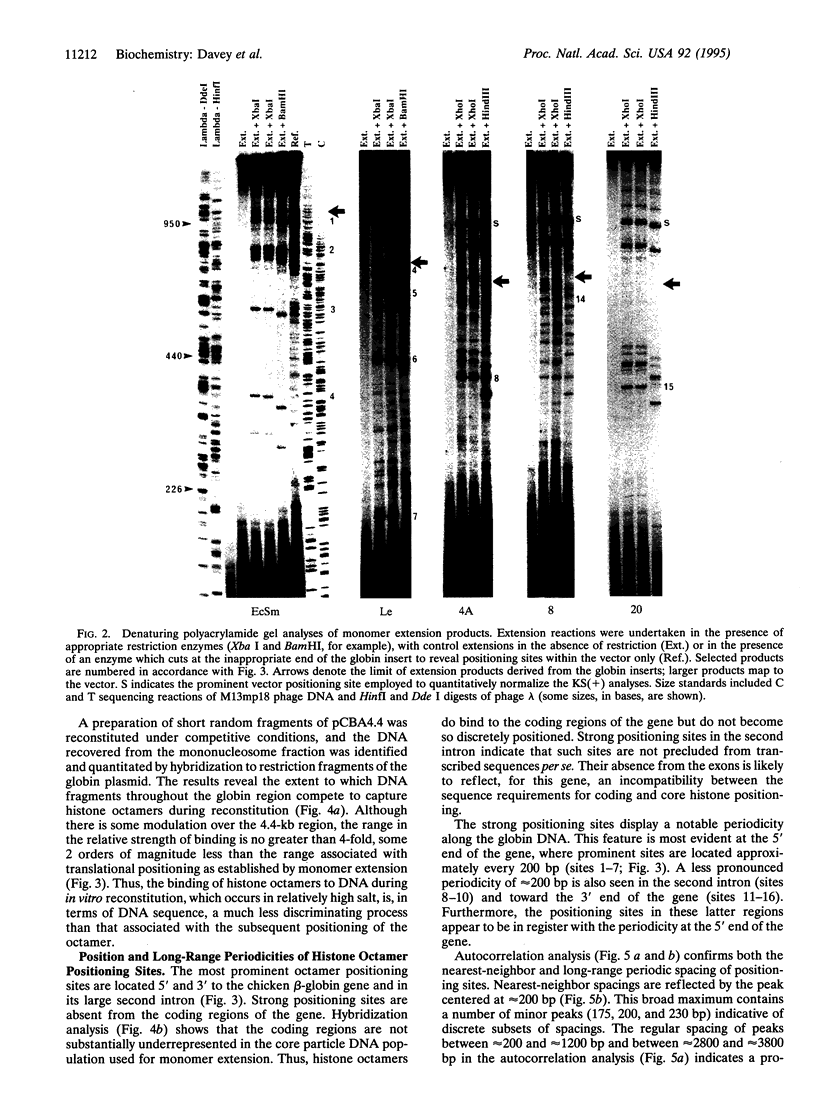
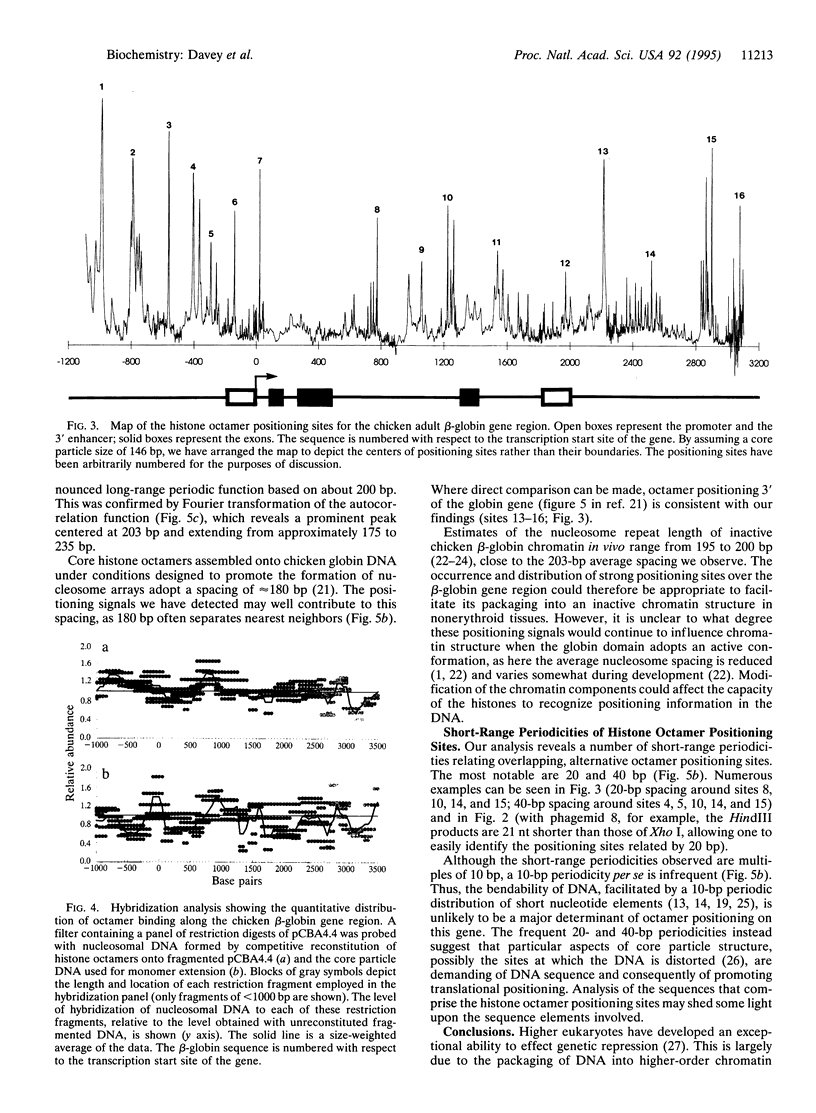
Abstract
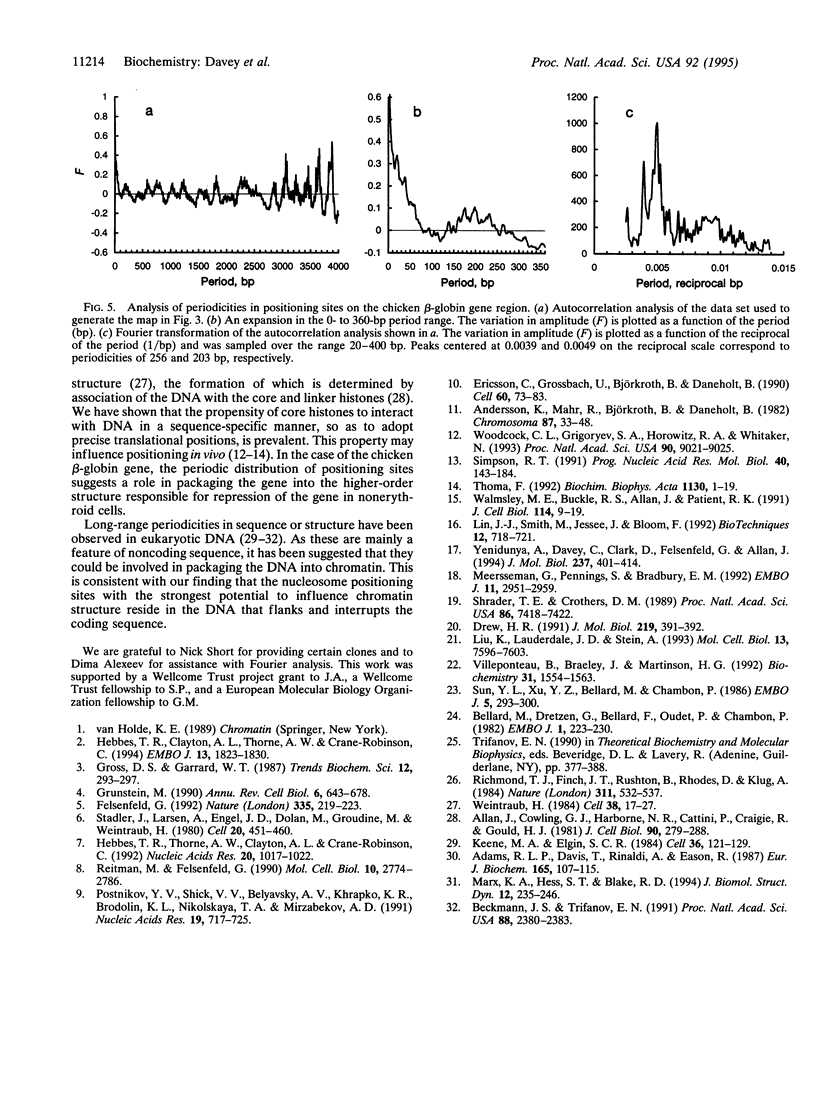
Positioned nucleosomes contribute to both the structure and the function of the chromatin fiber and can play a decisive role in controlling gene expression. We have mapped, at high resolution, the translational positions adopted by limiting amounts of core histone octamers reconstituted onto 4.4 kb of DNA comprising the entire chicken adult beta-globin gene, its enhancer, and flanking sequences. The octamer displays extensive variation in its affinity for different positioning sites, the range exhibited being about 2 orders of magnitude greater than that of the initial binding of the octamer. Strong positioning sites are located 5' and 3' of the globin gene and in the second intron but are absent from the coding regions. These sites exhibit a periodicity (approximately 200 bp) similar to the average spacing of nucleosomes on the inactive beta-globin gene in vivo, which could indicate their involvement in packaging the gene into higher-order chromatin structure. Overlapping, alternative octamer positioning sites commonly exhibit spacings of 20 and 40 bp, but not of 10 bp. These short-range periodicities could reflect features of the core particle structure contributing to the pronounced sequence-dependent manner in which the core histone octamer interacts with DNA.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Davis T., Rinaldi A., Eason R. CpG deficiency, dinucleotide distributions and nucleosome positioning. Eur J Biochem. 1987 May 15;165(1):107–115. doi: 10.1111/j.1432-1033.1987.tb11200.x. [DOI] [PubMed] [Google Scholar]
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson K., Mähr R., Björkroth B., Daneholt B. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma. 1982;87(1):33–48. doi: 10.1007/BF00333508. [DOI] [PubMed] [Google Scholar]
- Beckmann J. S., Trifonov E. N. Splice junctions follow a 205-base ladder. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2380–2383. doi: 10.1073/pnas.88.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Can one measure the free energy of binding of the histone octamer to different DNA sequences by salt-dependent reconstitution? J Mol Biol. 1991 Jun 5;219(3):391–392. doi: 10.1016/0022-2836(91)90179-a. [DOI] [PubMed] [Google Scholar]
- Ericsson C., Grossbach U., Björkroth B., Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990 Jan 12;60(1):73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994 Apr 15;13(8):1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Clayton A. L., Crane-Robinson C. Histone acetylation and globin gene switching. Nucleic Acids Res. 1992 Mar 11;20(5):1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Patterns of DNA structural polymorphism and their evolutionary implications. Cell. 1984 Jan;36(1):121–129. doi: 10.1016/0092-8674(84)90080-1. [DOI] [PubMed] [Google Scholar]
- Lin J. J., Smith M., Jessee J., Bloom F. DH11S: an Escherichia coli strain for preparation of single-stranded DNA from phagemid vectors. Biotechniques. 1992 May;12(5):718–721. [PubMed] [Google Scholar]
- Liu K., Lauderdale J. D., Stein A. Signals in chicken beta-globin DNA influence chromatin assembly in vitro. Mol Cell Biol. 1993 Dec;13(12):7596–7603. doi: 10.1128/mcb.13.12.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx K. A., Hess S. T., Blake R. D. Alignment of (dA).(dT) homopolymer tracts in gene flanking sequences suggests nucleosomal periodicity in D. discoideum DNA. J Biomol Struct Dyn. 1994 Aug;12(1):235–246. doi: 10.1080/07391102.1994.10508099. [DOI] [PubMed] [Google Scholar]
- Meersseman G., Pennings S., Bradbury E. M. Mobile nucleosomes--a general behavior. EMBO J. 1992 Aug;11(8):2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikov Y. V., Shick V. V., Belyavsky A. V., Khrapko K. R., Brodolin K. L., Nikolskaya T. A., Mirzabekov A. D. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 1991 Feb 25;19(4):717–725. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M., Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol Cell Biol. 1990 Jun;10(6):2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Sun Y. L., Xu Y. Z., Bellard M., Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986 Feb;5(2):293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. Nucleosome positioning. Biochim Biophys Acta. 1992 Feb 28;1130(1):1–19. doi: 10.1016/0167-4781(92)90455-9. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Brawley J., Martinson H. G. Nucleosome spacing is compressed in active chromatin domains of chick erythroid cells. Biochemistry. 1992 Feb 11;31(5):1554–1563. doi: 10.1021/bi00120a037. [DOI] [PubMed] [Google Scholar]
- Walmsley M. E., Buckle R. S., Allan J., Patient R. K. A chicken red cell inhibitor of transcription associated with the terminally differentiated state. J Cell Biol. 1991 Jul;114(1):9–19. doi: 10.1083/jcb.114.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984 Aug;38(1):17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenidunya A., Davey C., Clark D., Felsenfeld G., Allan J. Nucleosome positioning on chicken and human globin gene promoters in vitro. Novel mapping techniques. J Mol Biol. 1994 Apr 8;237(4):401–414. doi: 10.1006/jmbi.1994.1243. [DOI] [PubMed] [Google Scholar]