Abstract
MUTYH-associated polyposis (MAP) is an autosomal recessive adenomatous polyposis caused by biallelic germline mutations of the base-excision-repair gene MUTYH. In MAP patients of European origin, the combined allele frequency of the mutations p.Tyr179Cys and p.Gly396Asp ranges between 50 and 82%, while these mutations have not been identified in Far Eastern Asian populations, supporting the hypothesis that a founder effect has occurred at some point in European history. To investigate the natural history of the two common European MUTYH alleles, we genotyped six gene-flanking microsatellite markers in 80 unrelated Italian and German MAP patients segregating one or both mutations and calculated their age in generations (g) by using DMLE+2.2 software. Three distinct common haplotypes, one for p.Tyr179Cys and two for p.Gly396Asp, were identified. Estimated mutation ages were 305 g (95% CS: 271–418) for p.Tyr179Cys and 350 g (95% CS: 313–435) for p.Gly396Asp. These results provide evidence for strong founder effects and suggest that the p.Tyr179Cys and p.Gly396Asp mutations derive from ancestors who lived between 5–8 thousand years and 6–9 thousand years B.C., respectively.
Keywords: MUTYH-associated polyposis, MAP, mutation age, founder effect, colorectal cancer, colorectal adenoma
INTRODUCTION
MUTYH-associated polyposis (MAP) (OMIM no. 608456; http://www.ncbi.nlm.nih.gov/omim/) is an autosomal recessive colorectal polyposis syndrome caused by mutations of the base excision repair gene MUTYH.1, 2
MUTYH is located on chromosome 1p34.3–p32.1 and consists of 16 exons encompassing 1.65 kb (OMIM no. 604933, coding sequence: NM_001128425.1). Over the past years, its mutation spectrum has been described in series of MAP patients from different countries and with varying phenotypes (www.lovd.nl/MUTYH). In populations of European origin, the missense mutations c.536A>G;p.Tyr179Cys and c.1187G>A;p.Gly396Asp are by far the most common disease-causing variants, accounting for 50–82% of MUTYH mutations identified in MAP patients.3, 4 p.Tyr179Cys and p.Gly396Asp have been identified in Sephardi Jews, but not in Ashkenazi, in Finns or in Far Eastern Asians. Moreover, different variants have a larger role in other populations, pointing to ethnic and geographic differences in the pattern of MUTYH mutations.5 Screening of large series has shown that the combined p.Tyr179Cys and p.Gly396Asp allele frequency in healthy controls of European origin ranges from 0.2 to 1.14% (Table 1). These mutations are detected with similar frequencies in unselected series of colorectal cancer (CRC) patients (Table 1).
Table 1. p.Tyr179Cys and p.Gly396Asp allele frequencies in MAP and CRC patients and in controls from different populations.
| Study group | Country or population | N. subjects tested | p.Tyr179Cys allele frequency (%) | p.Gly396Asp allele frequency (%) | References |
|---|---|---|---|---|---|
| Classical or attenuated polyposis cases | Germany | 544 | 5.5 | 4.5 | 6, 7 |
| Italy | 110 | 9.2 | 4.1 | 8, 9, 10 | |
| Sweden | 15 | 16.8 | 3.2 | 3 | |
| Netherlands | 170 | 12 | 4.7 | 11 | |
| Czech Republic | 86 | 1.7 | 1.7 | 12, 13 | |
| France | 61 | 9 | 7.3 | 14, 15 | |
| Switzerland | 79 | 3.8 | 2.6 | 16 | |
| Portugal | 160 | 10.3 | 8.4 | 17, 18 | |
| UK-Denmark-Finland | 258 | 5.4 | 3.3 | 4, 19 | |
| Spain | 254 | 2.2 | 3.7 | 20, 21 | |
| Canada (European origin) | 20 | 9.9 | 5.1 | 22 | |
| North America | 188 | 10.9 | 5.8 | 23, 24 | |
| Brazil | 30 | 5 | 6.6 | 25 | |
| Australia | 120 | 7.9 | 4 | 26 | |
| Taiwan | 66 | 0 | 0 | 27 | |
| South Korea | 62 | 0 | 0 | 28 | |
| Japan | 56 | 0 | 0 | 29, 30 | |
| Indians/Pakistani | 6 | 0 | 0 | 19, 31 | |
| Singapore | 3 | 0 | 0 | 32 | |
| Morocco | 13 | 7.7 | 15.4 | 33 | |
| Tunisia | 10 | 0 | 5 | 34 | |
| Unselected CRC cases | UK (white) | 9268 | 0.5 | 0.9 | 35 |
| Spain | 1116 | 0.2 | 0.3 | 36 | |
| Canada-Australia-USA | 3811 | 0.4 | 1.2 | 37 | |
| North America | 444 | 0.9 | 0.7 | 24 | |
| Morocco | 49 | 0 | 0 | 33 | |
| Tunisia | 83 | 0.6 | 1.1 | 34 | |
| Arabs | 327 | 0 | 0.2 | 38 | |
| Moroccan Jews | 232 | 0.4 | 3.4 | 38 | |
| Ashkenazi Jews | 189 | 0 | 0 | 38 | |
| Controls | Germany | 116 | 0.4 | 0.4 | 6 |
| Italy | 247 | 0.2 | 0 | 39 | |
| Sweden | 466 | 0.2 | 0.1 | 40 | |
| Czech Republic | 100 | 0 | 0 | 12 | |
| France | 1121 | 0.2 | 0.7 | 41 | |
| UK (white) | 5064 | 0.3 | 0.7 | 35, 42 | |
| UK | 461 | 0.2 | 0.1 | 4, 43 | |
| Scotland | 2649 | 0.3 | 0.6 | 44, 45 | |
| Finland | 424 | 0 | 0 | 46 | |
| Spain | 1231 | 0.04 | 1.1 | 36, 47 | |
| Canada-Australia-USA | 2802 | 0.2 | 0.6 | 37 | |
| North America | 313 | 0.3 | 0.3 | 24 | |
| Canada | 2067 | 0.2 | 0.6 | 22, 48 | |
| Caucasian/Hispanic (US) | 32 | 0 | 1.6 | 49 | |
| Caucasian (US) | 360 | 0.3 | 0.6 | 49 | |
| Australia | 478 | 0.4 | 0.1 | 50 | |
| Chinaa | 101 | 0 | 0 | 51 | |
| Arabs | 304 | 0 | 0 | 38 | |
| Moroccans | 400 | 0.1 | 0.4 | 52 | |
| Sephardi Moroccan Jews | 541 | 0.4 | 1.8 | 53 | |
| Jews (US) | 450 | 0 | 0 | 49 |
Abbreviation: CRC, colorectal cancer.
The study population was represented by patients with gastric carcinoma.
These findings raise questions concerning the origin and spread of the p.Tyr179Cys and p.Gly396Asp mutations among European populations. In the present study, we have addressed this by performing haplotype analysis in 80 MAP families from Italy and Germany segregating one or both common MUTYH mutations. The estimated ages of the two mutations are consistent with the existence of ancestral haplotypes on which p.Tyr179Cys and p.Gly396Asp arose several thousand years ago, suggesting that separate founder effects account for the current genetic epidemiology of the two most common MUTYH mutations in individuals of European ancestry.
MATERIALS AND METHODS
A total of 144 genomic leukocyte DNA samples, derived from 31 families with p.Tyr179Cys, 29 with p.Gly396Asp and 20 with both mutations, were genotyped. One to 11 relatives were genotyped in 11 families segregating p.Tyr179Cys, in five families with p.Gly396Asp and in all families segregating both mutations. In the remaining families only the index case was available for genotyping. All families were of European ancestry and originated from different regions of Italy (n=42) or Germany (n=38).
Two hundred and eighty-eight control chromosomes, 164 from Italy and 124 from Germany, not carrying the two MUTYH recurrent mutations, were examined to estimate allele and haplotype frequencies in the general population. Controls included 62 unrelated individuals and 64 parents from 32 father-mother-child trios without history of CRC, as well as 18 members of MAP families who did not carry either MUTYH recurrent mutation.
Six short tandem repeat (STR) markers (D1S421, D1S451, D1S2677, D1S3175, D1S2874 and D1S2824), spanning a region of ∼3.5 Mb including the MUTYH locus on chromosome 1, were used for haplotype analysis. Primer sequences and conditions are available on request.
Allele frequency distributions in normal and mutated chromosomes were compared by Fisher's exact tests using PASW Statistic 18.03 software (IBM, NY, USA). Bonferroni correction was applied to correct for multiple comparisons, multiplying the nominal significance level (P) by the number of alleles tested for each locus. P<0.05 was considered as a cutoff for statistical significance. Haplotypes including the six microsatellites were manually constructed to minimize the number of recombinations.
Mutation age estimates were performed by DMLE+2.2 software (www.dmle.org)54 using the following parameters: haplotypes of microsatellite loci in mutation-bearing (MB) and control chromosomes; map distances between markers and mutation site inferred on the basis of physical distances (in Mb) given in the UCSC Genome Browser (GRCh37/hg19 assembly) considering 1 Mb=1 cM. The European population growth rate (genr) was estimated with the following formula: genr=log(Pt/P0)/g, in which genr is the population growth rate per generation, Pt is the estimated present population size, P0 is the estimated size of the population at reference time and g is the number of generations between these two time points. The total European population currently comprises 502 489 100 people (http://europa.eu/index_it.htm). Historical and demographic data indicate that about 19 000 000 people lived in this area in year 400 BC.55 Accordingly, the average genr of this population was estimated to be 0.034 from 400 BC to the present time, assuming 25 years/generation.56 The calculations were repeated using the genr value estimated for the last 20 000 years, corresponding to 0.0075.57 The proportion of MB chromosomes sampled (fc) was first calculated separately for Italy and Germany considering the following parameters: current Italian (60 742 397 inhabitants, http://demo.istat.it/) and German population size (82 438 000 inhabitants, www.destatis.de/); p.Tyr179Cys and p.Gly396Asp allele frequencies in Europeans: 0.3 and 0.7, respectively;35, 42 and the number of chromosomes investigated in the study. Mutation age was calculated both separately for the two countries and on combined Italian and German data using the average fc value estimated for the two populations (0.00014 for p.Tyr179Cys and 0.000043 for p.Gly396Asp).
RESULTS
Allele and haplotype analysis of polymorphic markers flanking the MUTYH locus was performed on the 288 control chromosomes, as well as on MAP probands and, when possible, on their relatives. For both mutations, not all chromosomes from the proband group were considered, as their phase could not be determined. Overall, 29 families (17 from Italy and 12 from Germany) with probands homozygous for p.Tyr179Cys or p.Gly396Asp, 19 (9 from Italy and 10 from Germany) with probands heterozygous for either mutation, and 13 (4 from Italy and 9 from Germany) with compound heterozygotes for both mutations were used to estimate allele frequencies and for haplotype analysis in chromosomes carrying either MUTYH variant.
Statistically significant differences in allele frequencies between wild-type and p.Tyr179Cys and p.Gly396Asp chromosomes were observed for markers located closer to MUTYH. In particular, D1S421, D1S451, D1S2677 and D1S3175 showed highly significant differences for p.Tyr179Cys, and D1S2677 and D1S3175 for p.Gly396Asp chromosomes (Table 2).
Table 2. Frequencies of the most common microsatellite alleles of the MUTYH region in p.Tyr179Cys and p.Gly396Asp cases and in controls.
|
Markers (allele size range in bp)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Study group | D1S421 (142-152) | D1S451b (240-270) | D1S2677b (159-181) | D1S3175 (153-169) | D1S2874c (226-244) | D1S2824 (234-256) | |||
| p.Tyr179Cys | |||||||||
| Most frequent alleles in cases | 148 | 252 | 167 | 157 | 228 | 230 | 240 | ||
| Cases (n=60 chromosomes) | |||||||||
| Allele frequencies | 0.95 | 0.90 | 0.97 | 0.87 | 0.37 | 0.40 | 0.35 | ||
| Controls (n=288 chromosomes) | |||||||||
| Allele frequencies | 0.57 | 0.23 | 0.38 | 0.60 | 0.19 | 0.32 | 0.31 | ||
| P-value | <0.01 | <0.001 | <0.001 | <0.001 | NS | NS | NS | ||
| p.Gly396Asp | |||||||||
| Most frequent alleles in cases | 148 | 250 | 252 | 175 | 177 | 157 | 230 | 238 | |
| Cases (n=49 chromosomes) | |||||||||
| Allele frequencies | 0.88 | 0.35 | 0.41 | 0.41 | 0.39 | 0.90 | 0.59 | 0.29 | |
| Controls (n=288 chromosomes) | |||||||||
| Allele frequencies | 0.57 | 0.38 | 0.23 | 0.007 | 0.05 | 0.60 | 0.32 | 0.24 | |
| P-value | NS | NS | NS | <0.001 | <0.001 | <0.001 | 0.21 | NS | |
Abbreviation: NS, not significant.
Alleles showing statistically significant association (corrected P-value <0.05) are underlined.
Two common alleles with similar frequencies were observed in p.Gly396Asp chromosomes at this locus.
Two common alleles with similar frequencies were observed in p.Tyr179Cys chromosomes at this locus.
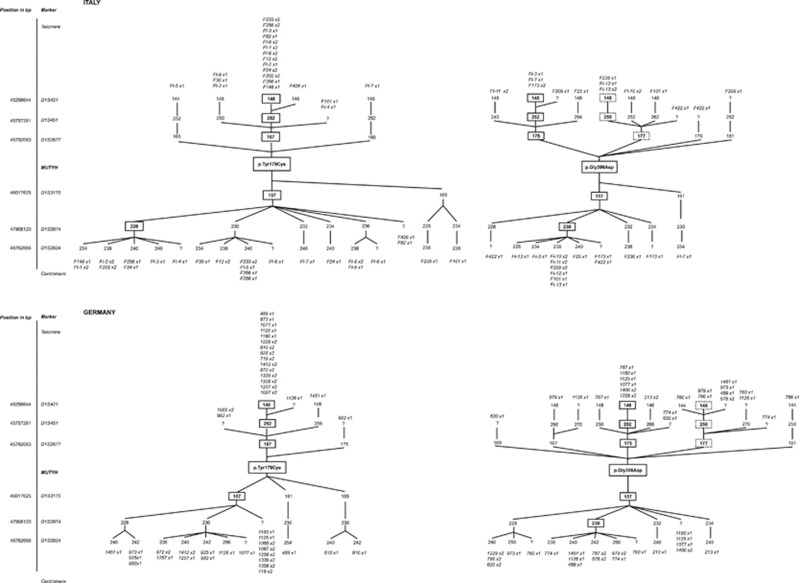
A common haplotype (148-252-167-157) spanning 0.76 Mb was found in 46 (76%) p.Tyr179Cys chromosomes (Figure 1). In addition, the genotypes observed in three of the six remaining p.Tyr179Cys heterozygotes whose phase at D1S451 was undefined were compatible with this common shared haplotype. Seven additional haplotypes (148-252-169-157, 144-252-165-157, 148-250-167-157, 146-252-167-157, 148-256-167-157, 148-252-167-161 and 148-252-167-165; the latter one was observed in four chromosomes) segregated with p.Tyr179Cys in the remaining chromosomes; these differed by at most two markers from the common haplotype.
Figure 1.
Haplotype branching trees of p.Tyr179Cys (right) and p.Gly396Asp (left) chromosomes in Italian (top panels) and German (bottom panels) families. The markers used are shown according to their position on the physical map (UCSC Genome Browser Feb. 2009 GRCh37/h19). Boxed numbers indicate STR alleles comprised in the ancestral haplotypes; for p.Gly396Asp, the two most common haplotypes (A and B) are indicated by solid and dashed lines, respectively; non boxed numbers correspond to additional alleles detected in the study families. Family codes are indicated above or below alleles in which they have been identified; for each family, the number of chromosomes with the corresponding allele is indicated as × 1 (1 copy) or × 2 (2 copies).
Two distinct haplotypes (referred to as haplotypes A and B) covering a region of 0.22 Mb were more common on p.Gly396Asp chromosomes (Figure 1). Haplotype A (175-157) was shared by 21 (42%) and haplotype B (177-157) by 19 (38%) p.Gly396Asp chromosomes, respectively. Five other haplotypes (179-157, 181-157, 175-161, 167-157 and 169-157), all sharing the 157 allele, were observed on the remaining p.Gly396Asp chromosomes.
Neither haplotype associated with p.Gly396Asp was observed in controls, and the p.Tyr179Cys-associated haplotype was present in only 2 out of 288 control chromosomes.
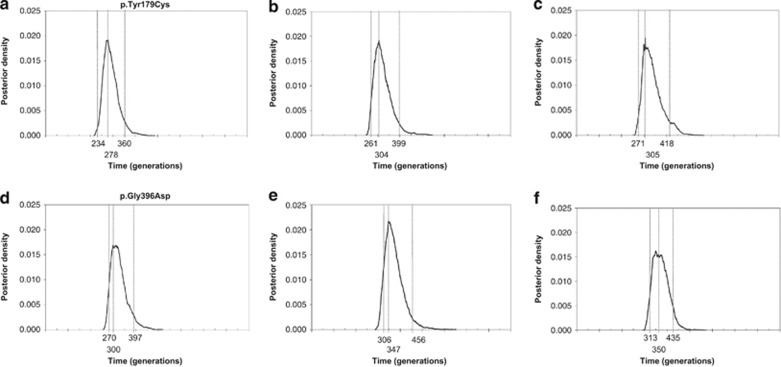
p.Tyr179Cys age was estimated as 278 generations (g) (95% credible set (CS): 234–360), corresponding to 6950 years (95% CS: 5850–9000), in the Italian population, and 304 g (95% CS: 261–399), corresponding to 7600 years (95% CS: 6525–9975), in the German population with genr 0.034. Combined data from both populations gave an age estimate of 305 g (95% CS: 271–418), corresponding to 7625 years (95% CS: 6775–10 450) (Figure 2a–c).
Figure 2.
Posterior probability distribution plots of p.Tyr179Cys and p.Gly396Asp ages (in generations), as estimated by DMLE+2.2 software using a population growth rate (genr) of 0.034. (a) Age estimate in the Italian population for p.Tyr179Cys (fc=0.00016); (b) age estimate in the German population for p.Tyr179Cys (fc=0.00012); (c) age estimate for p.Tyr179Cys using combined data from both populations (fc=0.00014); (d) age estimate in the Italian population for p.Gly396Asp (fc=0.00004); (e) age estimate in the German population for p.Gly396Asp (fc=0.000047); (f) age estimate for p.Gly396Asp using combined data from both populations (fc=0.000043). The vertical broken lines indicate the 95% credible set values.
Using the same genr value, p.Gly396Asp age estimates were 300 g (95% CS: 270–397), corresponding to 7500 years (95% CS: 6750–9925) and 347 g (95% CS: 306–456), corresponding to 8675 years (95% CS: 7650–11 400), for the Italian and German populations, respectively. Combined data for the two populations gave an estimate of 350 g (95% CS: 313–435), corresponding to 8750 years (95% CS: 7825–10 875) (Figure 2d–f).
The following estimates were obtained for the combined German and Italian populations when the genr was set at 0.0075: 1245 g (95% CS: 1035–1486) for p.Tyr179Cys and 1309 g (95% CS: 1172–1579) for p.Gly396Asp (data not shown); these correspond to 31 125 years (95% CS: 25 875–37 150 years) and 32 725 years (95% CS: 29 300–39 475 years), respectively.
DISCUSSION
Biallelic MUTYH germline mutations contribute to a considerable proportion of attenuated forms of colorectal adenomatous polyposis. To date, around 300 different MUTYH mutations have been identified in MAP patients worldwide (www.lovd.nl/MUTYH). Regional and ethnic differences in the mutation spectrum reported in MUTYH screening projects since 2002, and the presence of specific mutations at higher frequency in some populations suggest the existence of multiple founder effects.28, 34, 58, 59, 60, 61
p.Tyr179Cys and p.Gly396Asp are the most common MAP-linked MUTYH mutations in populations of European origin. The finding of highly conserved core haplotypes for both mutations in German and Italian MAP patients suggests that the p.Tyr179Cys and p.Gly396Asp chromosomes sampled derive from two ancestral chromosomes, on which the mutations arose independently. Although a single common haplotype was associated with p.Tyr179Cys, two distinct haplotypes, sharing the D1S3175 allele, were observed at similar frequencies on p.Gly396Asp chromosomes. The latter finding could be due to an early mutation at marker D1S2677 or to a recombination event. A higher haplotype diversity for p.Gly396Asp is not surprising, as this mutation is more frequent and the age estimates indicate that it is probably older than p.Tyr179Cys.
To investigate the natural history of the two common MUTYH mutations, we calculated their apparent age using the DMLE+2.2 software;54 this is considered to provide the best estimates of gene genealogies and mutation age compared with other methods.62, 63 If it is assumed that g=25 years and that 1960 is the average year of birth of the subjects enlisted in the present study, our overall results would date the age of the MUTYH p.Tyr179Cys and p.Gly396Asp mutations back to 6775–10 450 (or 4775–8450 B.C.) and 7825–10 875 (or 5825–8875 B.C.) years ago, respectively, corresponding to the Neolithic revolution and just before or in concomitance with the development of agriculture and consequent migrations in Europe. The ancient origin of the two mutations is reasonable in light of their even distribution and subpolymorphic frequencies across most European populations.
On the other hand, these results have to be interpreted with some caution, as they are sensitive to changes in the parameters used, namely g and genr. For instance, use of lower genr values, such as 0.0075, corresponding to the average genr estimated for Europe in the last 20 000 years,57 would date the mutations back to around 30 000 years ago, toward the end of the last Pleistocene glaciation in the Paleolithic. However, this result is less compatible with the absence or very low frequency of these mutations in non European populations, as other genetic variants, such as factor V Leiden, that likely date back to that period are widely spread in other ethnic groups.64
The relationship between the age of a mutation and its frequency in the population from which the identical-by-descent MB chromosomes were sampled is influenced by both historical population growth and natural selection. Generally, allelic coalescence models assumed for estimating mutation age are influenced by population growth and selection through a common parameter which is the sum of the two effects.65 Thus, for any mutation, the effect of selection, if any, cannot be separated from that of demography in the shaping of the genealogy of MB chromosomes. The frequency of the p.Gly396Asp mutation is usually higher than that of p.Tyr179Cys in the general population of European countries; specifically, in the largest series investigated so far, involving a total of 5064 control individuals, p.Tyr179Cys and p.Gly396Asp allele frequencies were 0.3 and 0.7%, respectively.35 Genetic drift and/or selection might account for the differences in frequencies and estimated ages between the two mutations.
Considering the general late onset of both adenoma and CRC formation in MAP patients,4, 6, 8, 11, 23 negative selection would be expected to be limited. Nevertheless, a robust correlation between certain MUTYH genotypes and the colorectal phenotype has recently been established.66 Both p.Gly396Asp homozygotes and p.Gly396Asp/p.Tyr179Cys compound heterozygotes show a later presentation and have a significantly lower CRC hazard than p.Tyr179Cys homozygotes (P<0.001). Interestingly, while the allele frequency in large samples of healthy controls is clearly skewed towards p.Gly396Asp, p.Tyr179Cys dominates in MAP patients (Table 1). In addition, the frequencies of these two mutations in population-based CRC patients are similar to those observed in controls of corresponding ethnicity; this is likely due to the milder phenotype in MAP patients not selected for the presence of polyposis.44
These observations indicate that the two mutations have uneven effects on clinical phenotype, and suggest that they might be associated with slightly different selection coefficients. This implies that caution is required in using cumulative allele frequencies (combined samples of general population and patients) when a deleterious mutation is investigated for its natural history.
In conclusion, our findings and the analysis of data from the literature support the hypothesis that the high frequency of the p.Tyr179Cys and p.Gly396Asp mutations in MAP patients from the present German and Italian populations is attributable to founder events that occurred during early European history. The possibility that selection may have had a major role in shaping the allelic genealogy of the two mutations and in determining their present day frequencies cannot be excluded without further evidence. Comparative haplotype studies in different populations of chromosomes bearing the same MUTYH mutations will refine our knowledge on their origin and spread into the European continent. In addition, the different distribution of MUTYH mutations among populations have implications for mutation detection strategies. A screening approach restricted to p.Tyr179Cys and p.Gly396Asp does not seem to be appropriate for patients with a non European ethnic background.
Acknowledgments
This study was supported by grants from Istituto Toscano Tumori and Ente Cassa Risparmio Firenze (to MG) and from the German Cancer Aid (Deutsche Krebshilfe e.V. Bonn, grant no. 108421, to SA).
The authors declare no conflict of interest.
References
- Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Morreau H, Vasen HF, Hes FJ. MUTYH-associated polyposis (MAP) Crit Rev Oncol Hematol. 2011;79:1–16. doi: 10.1016/j.critrevonc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Kanter-Smoler G, Björk J, Fritzell K, et al. Novel findings in Swedish patients with MYH-associated polyposis: mutation detection and clinical characterization. Clin Gastroenterol Hepatol. 2006;4:499–506. doi: 10.1016/j.cgh.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Sieber OM, Lipton L, Crabtree M, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- Out AA, Tops CM, Nielsen M, et al. Leiden Open Variation Database of the MUTYH gene. Hum Mutat. 2010;31:1205–1215. doi: 10.1002/humu.21343. [DOI] [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Goergens H, et al. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer. 2006;119:807–814. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- Morak M, Laner A, Bacher U, et al. MUTYH-associated polyposis–variability of the clinical phenotype in patients with biallelic and monoallelic MUTYH mutations and report on novel mutations. Clin Genet. 2010;78:353–363. doi: 10.1111/j.1399-0004.2010.01478.x. [DOI] [PubMed] [Google Scholar]
- Gismondi V, Meta M, Bonelli L, et al. Prevalence of the Y165C, G382D and 1395delGGA germline mutations of the MYH gene in Italian patients with adenomatous polyposis coli and colorectal adenomas. Int J Cancer. 2004;109:680–684. doi: 10.1002/ijc.20054. [DOI] [PubMed] [Google Scholar]
- Venesio T, Molatore S, Cattaneo F, Arrigoni A, Risio M, Ranzani GN. High frequency of MYH gene mutations in a subset of patients with familial adenomatous polyposis. Gastroenterology. 2004;126:1681–1685. doi: 10.1053/j.gastro.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Aceto G, Curia MC, Veschi S, et al. Mutations of APC and MYH in unrelated Italian patients with adenomatous polyposis coli. Hum Mutat. 2005;26:394. doi: 10.1002/humu.9370. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Franken PF, Reinards TH, et al. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP) J Med Genet. 2005;42:e54. doi: 10.1136/jmg.2005.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulová M, Zídková K, Kleibl Z, et al. Mutation analysis of the MYH gene in unrelated Czech APC mutation-negativepolyposis patients. Eur J Cancer. 2007;43:1617–1621. doi: 10.1016/j.ejca.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Vandrovcová J, Stekrová J, Kebrdlová V, Kohoutová M. Molecular analysis of the APC and MYH genes in Czech families affected by FAP or multiple adenomas: 13 novel mutations. Hum Mutat. 2004;23:397. doi: 10.1002/humu.9224. [DOI] [PubMed] [Google Scholar]
- Lefevre JH, Rodrigue CM, Mourra N, et al. Implication of MYH in colorectal polyposis. Ann Surg. 2006;244:874–879. doi: 10.1097/01.sla.0000246937.54435.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguen G, Manfredi S, Blayau M, et al. Colorectal adenomatous polyposis associated with MYH mutations: genotype and phenotype characteristics. Dis Colon Rectum. 2007;50:1612–1617. doi: 10.1007/s10350-007-9027-0. [DOI] [PubMed] [Google Scholar]
- Russell AM, Zhang J, Luz J, et al. Prevalence of MYH germline mutations in Swiss APC mutation-negative polyposis patients. Int J Cancer. 2006;118:1937–1940. doi: 10.1002/ijc.21470. [DOI] [PubMed] [Google Scholar]
- Isidro G, Laranjeira F, Pires A, et al. Germline MUTYH (MYH) mutations in Portuguese individuals with multiple colorectal adenomas. Hum Mutat. 2004;24:353–354. doi: 10.1002/humu.9282. [DOI] [PubMed] [Google Scholar]
- Filipe B, Baltazar C, Albuquerque C, et al. APC or MUTYH mutations account for the majority of clinically well-characterized families with FAP and AFAP phenotype and patients with more than 30 adenomas. Clin Genet. 2009;76:242–255. doi: 10.1111/j.1399-0004.2009.01241.x. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Dolwani S, Jones S, et al. Autosomal recessive colorectal adenomatous polyposis due to inherited mutation of MYH. Lancet. 2003;362:39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- Gómez-Fernández N, Castellví-Bel S, Fernández-Rozadilla C, et al. Molecular analysis of the APC and MUTYH genes in Galician and Catalonian FAP families: a different spectrum of mutations. BMC Med Genet. 2009;10:57. doi: 10.1186/1471-2350-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera B, González S, Sánchez-Tomé E, et al. Clinical and genetic characterization of classical forms of familial adenomatous polyposis: a Spanish population study. Ann Oncol. 2011;22:903–909. doi: 10.1093/annonc/mdq465. [DOI] [PubMed] [Google Scholar]
- Croitoru ME, Cleary SP, Berk T, et al. Germline MYH mutations in a clinic-based series of Canadian multiple colorectal adenoma patients. J Surg Oncol. 2007;95:499–506. doi: 10.1002/jso.20724. [DOI] [PubMed] [Google Scholar]
- Wang L, Baudhuin LM, Boardman LA, et al. MYH mutations in patients with attenuated and classic polyposis and with young-onset colorectal cancer without polyps. Gastroenterology. 2004;127:9–16. doi: 10.1053/j.gastro.2004.03.070. [DOI] [PubMed] [Google Scholar]
- Eliason K, Hendrickson BC, Judkins T, et al. The potential for increased clinical sensitivity in genetic testing for polyposis colorectal cancer through the analysis of MYH mutations in North American patients. J Med Genet. 2005;42:95–96. doi: 10.1136/jmg.2004.025973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitroski CE, Cossio SL, Koehler-Santos P, et al. Frequency of the common germline MUTYH mutations p.G396D and p.Y179C in patients diagnosed with colorectal cancer in Southern Brazil. Int J Colorectal Dis. 2011;26:841–846. doi: 10.1007/s00384-011-1172-1. [DOI] [PubMed] [Google Scholar]
- Kairupan CF, Meldrum CJ, Crooks R, et al. Mutation analysis of the MYH gene in an Australian series of colorectal polyposis patients with or without germline APC mutations. Int J Cancer. 2005;116:73–77. doi: 10.1002/ijc.20983. [DOI] [PubMed] [Google Scholar]
- Chiang JM, Chen HW, Tang RP, et al. Mutation analysis of the APC gene in Taiwanese FAP families: low incidence of APC germline mutation in a distinct subgroup of FAP families. Fam Cancer. 2010;9:117–124. doi: 10.1007/s10689-009-9292-2. [DOI] [PubMed] [Google Scholar]
- Kim DW, Kim IJ, Kang HC, et al. Germline mutations of the MYH gene in Korean patients with multiple colorectal adenomas. Int J Colorectal Dis. 2007;22:1173–1178. doi: 10.1007/s00384-007-0289-8. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Yamaguchi T, et al. Germline mutations of the MYH gene in Japanese patients with multiple colorectal adenomas. Mutat Res. 2005;578:430–433. doi: 10.1016/j.mrfmmm.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Yanaru-Fujisawa R, Matsumoto T, Ushijima Y, et al. Genomic and functional analyses of MUTYH in Japanese patients with adenomatous polyposis. Clin Genet. 2008;73:545–553. doi: 10.1111/j.1399-0004.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- Ajith Kumar VK, Gold JA, Mallon E, Thomas S, Hodgson SV. Sebaceous adenomas in an MYH associated polyposis patient of Indian (Gujarati) origin. Fam Cancer. 2008;7:187–189. doi: 10.1007/s10689-007-9161-9. [DOI] [PubMed] [Google Scholar]
- Cao X, Hong Y, Eu KW, Loi C, Cheah PY. Singapore familial adenomatous polyposis (FAP) patients with classical adenomatous polyposis but undetectable APC mutations have accelerated cancer progression. Am J Gastroenterol. 2006;101:2810–2817. doi: 10.1111/j.1572-0241.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- Laarabi FZ, Cherkaoui Jaouad I, et al. The first mutations in the MYH gene reported in Moroccan colon cancer patients. Gene. 2012;496:55–58. doi: 10.1016/j.gene.2011.12.024. [DOI] [PubMed] [Google Scholar]
- Abdelmaksoud-Dammak R, Miladi-Abdennadher I, Amouri A, et al. High prevalence of the c.1227-1228dup (p.Glu410GlyfsX43) mutation in Tunisian families with MUTYH-associated polyposis. Fam Cancer. 2012;11:503–508. doi: 10.1007/s10689-012-9543-5. [DOI] [PubMed] [Google Scholar]
- Lubbe SJ, Di Bernardo MC, Chandler IP, et al. Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Oncol. 2009;27:3975–3980. doi: 10.1200/JCO.2008.21.6853. [DOI] [PubMed] [Google Scholar]
- Balaguer F, Castellví-Bel S, Castells A, et al. Identification of MYH mutation carriers in colorectal cancer: a multicenter, case-control, population-based study. Clin Gastroenterol Hepatol. 2007;5:379–387. doi: 10.1016/j.cgh.2006.12.025. [DOI] [PubMed] [Google Scholar]
- Cleary SP, Cotterchio M, Jenkins MA, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology. 2009;136:1251–1260. doi: 10.1053/j.gastro.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz F, Cohen I, Barnett-Griness O, et al. Common MUTYH mutations and colorectal cancer risk in multiethnic populations. Fam Cancer. 2012;11:329–335. doi: 10.1007/s10689-012-9516-8. [DOI] [PubMed] [Google Scholar]
- Avezzù A, Agostini M, Pucciarelli S, et al. The role of MYH gene in genetic predisposition to colorectal cancer: another piece of the puzzle. Cancer Lett. 2008;268:308–313. doi: 10.1016/j.canlet.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Zhou XL, Djureinovic T, Werelius B, et al. Germline mutations in the MYH gene in Swedish familial and sporadic colorectal cancer. Genet Test. 2005;9:147–151. doi: 10.1089/gte.2005.9.147. [DOI] [PubMed] [Google Scholar]
- Küry S, Buecher B, Robiou-du-Pont S, et al. The thorough screening of the MUTYH gene in a large French cohort of sporadic colorectal cancers. Genet Test. 2007;11:373–379. doi: 10.1089/gte.2007.0029. [DOI] [PubMed] [Google Scholar]
- Webb EL, Rudd MF, Houlston RS. Colorectal cancer risk in monoallelic carriers of MYH variants. Am J Hum Genet. 2006;79:768–771. doi: 10.1086/507912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann C, Peto J, Cheadle J, Shah B, Sampson J, Houlston RS. Comprehensive analysis of the contribution of germline MYH variation to early-onset colorectal cancer. Int J Cancer. 2004;109:554–558. doi: 10.1002/ijc.20020. [DOI] [PubMed] [Google Scholar]
- Farrington SM, Tenesa A, Barnetson R, et al. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am J Hum Genet. 2005;77:112–119. doi: 10.1086/431213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Campbell H, Barnetson R, Porteous M, Dunlop M, Farrington SM. Association of MUTYH and colorectal cancer. Br J Cancer. 2006;95:239–242. doi: 10.1038/sj.bjc.6603239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enholm S, Hienonen T, Suomalainen A, et al. Proportion and phenotype of MYH-associated colorectal neoplasia in a population-based series of Finnish colorectal cancer patients. Am J Pathol. 2003;163:827–832. doi: 10.1016/S0002-9440(10)63443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno V, Gemignani F, Landi S, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- Beiner ME, Zhang WW, Zhang S, Gallinger S, Sun P, Narod SA. Mutations of the MYH gene do not substantially contribute to the risk of breast cancer. Breast Cancer Res Treat. 2009;114:575–578. doi: 10.1007/s10549-008-0042-1. [DOI] [PubMed] [Google Scholar]
- Peterlongo P, Mitra N, Chuai S, et al. Colorectal cancer risk in individuals with biallelic or monoallelic mutations of MYH. Int J Cancer. 2005;114:505–507. doi: 10.1002/ijc.20767. [DOI] [PubMed] [Google Scholar]
- Colebatch A, Hitchins M, Williams R, Meagher A, Hawkins NJ, Ward RL. The role of MYH and microsatellite instability in the development of sporadic colorectal cancer. Br J Cancer. 2006;95:1239–1243. doi: 10.1038/sj.bjc.6603421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu X, Fan Y, et al. Germline mutations and polymorphic variants in MMR, E-cadherin and MYH genes associated with familial gastric cancer in Jiangsu of China. Int J Cancer. 2006;119:2592–2596. doi: 10.1002/ijc.22206. [DOI] [PubMed] [Google Scholar]
- Laarabi FZ, Cherkaoui Jaouad I, Benazzouz A, Squalli D, Sefiani A. Prevalence of MYH-associated polyposis related to three recurrent mutations in Morocco. Ann Hum Biol. 2011;38:360–363. doi: 10.3109/03014460.2010.521520. [DOI] [PubMed] [Google Scholar]
- Rennert G, Lejbkowicz F, Cohen I, Pinchev M, Rennert HS, Barnett-Griness O. MutYH mutation carriers have increased breast cancer risk. Cancer. 2012;118:1989–1993. doi: 10.1002/cncr.26506. [DOI] [PubMed] [Google Scholar]
- Rannala B, Reeve JP. High-resolution multipoint linkage-disequilibrium mapping in the context of a human genome sequence. Am J Hum Genet. 2001;69:159–178. doi: 10.1086/321279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Panta L, Rettaroli R. Introduzione Alla Demografia Storica. Rome: Editori Laterza; 1994. [Google Scholar]
- Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Seielstad MT, Perez-Lezaun A, et al. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- Wasielewski M, Out AA, Vermeulen J, et al. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat. 2010;124:635–641. doi: 10.1007/s10549-010-0801-7. [DOI] [PubMed] [Google Scholar]
- Dolwani S, Williams GT, West KP, et al. Analysis of inherited MYH/(MutYH) mutations in British Asian patients with colorectal cancer. Gut. 2007;56:593. doi: 10.1136/gut.2006.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Shinmura K, Hanaoka T, et al. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis. 2004;25:1859–1866. doi: 10.1093/carcin/bgh206. [DOI] [PubMed] [Google Scholar]
- Pin E, Pastrello C, Tricarico R, et al. MUTYH c.933+3A>C, associated with a severely impaired gene expression, is the first Italian founder mutation in MUTYH-Associated Polyposis. Int J Cancer. 2013;132:1060–1069. doi: 10.1002/ijc.27761. [DOI] [PubMed] [Google Scholar]
- Clendenning M, Baze ME, Sun S, et al. Origins and prevalence of the American Founder Mutation of MSH2. Cancer Res. 2008;68:2145–2153. doi: 10.1158/0008-5472.CAN-07-6599. [DOI] [PubMed] [Google Scholar]
- Greenwood CM, Sun S, Veenstra J, et al. How old is this mutation?–a study of three Ashkenazi Jewish founder mutations. BMC Genet. 2010;14:39. doi: 10.1186/1471-2156-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivelin A, Mor-Cohen R, Kovalsky V, et al. Prothrombin 20210G>A is an ancestral prothrombotic mutation that occurred in whites approximately 24000 years ago. Blood. 2006;107:4666–4668. doi: 10.1182/blood-2005-12-5158. [DOI] [PubMed] [Google Scholar]
- Rannala B, Bertorelle G. Using linked markers to infer the age of a mutation. Hum Mutat. 2001;18:87–100. doi: 10.1002/humu.1158. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Joerink-van de Beld MC, Jones N, et al. Analysis of MUTYH genotypes and colorectal phenotypes in patients with MUTYH-associated polyposis. Gastroenterology. 2009;136:471–476. doi: 10.1053/j.gastro.2008.10.056. [DOI] [PubMed] [Google Scholar]