Abstract
Glutaredoxin 2 is a vertebrate specific oxidoreductase of the thioredoxin family of proteins modulating the intracellular thiol pool. Thereby, glutaredoxin 2 is important for specific redox signaling and regulates embryonic development of brain and vasculature via reversible oxidative posttranslational thiol modifications. Here, we describe that glutaredoxin 2 is also required for successful heart formation. Knock-down of glutaredoxin 2 in zebrafish embryos inhibits the invasion of cardiac neural crest cells into the primary heart field. This leads to impaired heart looping and subsequent obstructed blood flow. Glutaredoxin 2 specificity of the observed phenotype was confirmed by rescue experiments. Active site variants of glutaredoxin 2 revealed that the (de)-glutathionylation activity is required for proper heart formation. Our data suggest that actin might be one target during glutaredoxin 2 regulated cardiac neural crest cell migration and embryonic heart development. In summary, this work represents further evidence for the general importance of redox signaling in embryonic development and highlights additionally the importance of glutaredoxin 2 during embryogenesis.
Keywords: S-glutathionylation, Zebrafish, Glutaredoxin, Cardiac development, Migration
Abbreviations: A, atrium; CNC, cardiac neural crest; CCV, common cardinal vein; GSH, glutathione; Grx, glutaredoxin; NC, neural crest; V, ventricle; zf, zebrafish
Highlights
-
•
Reversible redox regulation, S-glutathionylation, regulates heart formation.
-
•
Glutaredoxin 2 controls migration of neural crest cells.
-
•
Loss of glutaredoxin 2 impairs heart looping and subsequently heart functionality.
Graphical Abstract
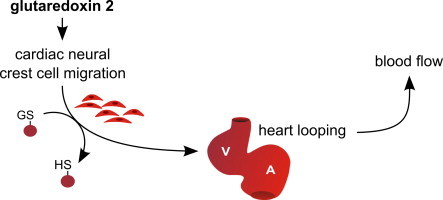
Introduction
About 2.5 billion years ago, photosynthesis evolved and the raising oxygen levels changed radically the composition of the Earth's atmosphere. Rapidly, organisms evolved strategies to exploit this new resource, but also to defend themselves against the unavoidable toxic side products of aerobic metabolism: reactive oxygen species (ROS). ROS have long been considered to be only deleterious to organisms, damaging DNA, proteins, and lipids, however the last two decades of extensive research have established a fundamental role of ROS as second messengers [1]. Today, redox signaling is known to be essential for many cellular processes [2] and pivotal for proper embryonic development [3]. The major targets for redox signaling events are protein thiol groups, which can regulate protein activity through reversible oxidative modifications [4].
The redox state of these cysteine residues is regulated by members of the thioredoxin family of proteins including glutaredoxins (Grxs) [5]. Grxs can be subdivided in dithiol Grxs harboring a Cys-X-X-Cys active site, and monothiol Grxs that are characterized by a Cys-X-X-Ser motif. Whereas the catalytic activity of the latter is yet to be characterized, it is well established that dithiol Grxs reduce protein disulfides via the dithiol mechanism that is dependent on both active site cysteines [6]. Additionally, dithiol Grxs are able to reduce protein-GSH-mixed disulfides (de-glutathionylation) for which solely the N-terminal cysteine is required [6]. After reducing disulfides or glutathionylated cysteine residues, Grxs get recycled on the expense of GSH and NADPH as final electron donor [6].
Grxs are among the few proteins that are able to reversible (de)-glutathionylate substrates and therefore likely to be prime regulators of redox signaling via protein S-glutathionylation [7]. Grx2 is characterized as vertebrate specific oxidoreductase by two conserved additional cysteine residues forming an intramolecular disulfide. Isoforms are localized in the mitochondria (Grx2a) or cytosol/nucleus (Grx2b/c) [8]. Our group has unraveled that vertebrate Grx2 is essential for successful embryogenesis. Using the zebrafish as a model system, we found that Grx2 regulates vertebrate neuronal survival and axon growth via a thiol–disulfide mechanism [9] as well as vertebrate angiogenesis through S-glutathionylation of the histone-deacetylase sirtuin1 [10]. The zebrafish is a powerful model that offers multiple advantages for in vivo studies and it has played a fundamental role in refining the knowledge on vertebrate embryonic development including the formation of the heart [11].
Here, we report that zfGrx2 is required for the formation of the zebrafish heart. Loss of zfGrx2 leads to migratory defects of the cardiac neural crest (CNC) which results in heart looping defects that causes impaired heart functionality.
Material and methods
Zebrafish husbandry
Zebrafish were kept in standard conditions, obtained by mass mating and raised in an E3 medium. For precise age-matching of the different embryo groups, early embryos were staged by counting the number of somites and embryos older 24 h by hours post fertilization (hpf) following the protocol of Kimmel [12]. The 12 somite stage corresponds to ~15 hpf, 16 somite stage to ~17 hpf and the 19 somite stage to ~18.5 hpf. To prevent pigmentation of stages older than 24 (hpf), Phenyl-2-thiourea (Sigma) was added to the E3 medium. All experiments were reviewed and granted by the North Stockholm Ethical Council.
Morpholino and mRNA injections
The morpholino knocking down zfGrx2 was designed and obtained from Genetools (http://www.gene-tools.com) and injected as described before [9,10]. Capped mRNA was synthesized with the mMessage/Machine Kit (Ambion) using constructs described elsewhere [9]. Morpholino and mRNA were injected into 1 cell embryos to ensure ubiquitous distribution.
In situ hybridization and acridine orange staining
The generation of riboprobes and in situ hybridization was carried out according to standard protocols [13]. As marker genes, we used cmlc2 (demarks cardiac mesoderm [14]), foxD3 as well as crestin (both demarking neural crest [15,16]). To detect cell death in living embryos, we immersed them in 0.002% acridine orange (Sigma) solution for 45 min followed by several washing steps with PBS and imaging directly afterwards.
Microscopy, image processing, and statistics
Fixed specimens were mounted in glycerol, life embryos were mounted in low-melting agarose and bright field pictures were taken with a Leica MZ16 microscope equipped with a Leica DFC300FX camera. Images were processed with Gimp (http://www.gimp.org) without obstructing any original data. Movies were captured with a Zeiss Axiovert 40 equipped with a Zeiss Axiocam ICM1 and angiograms were calculated with ImageJ using a previously published protocol [17]. To avoid unspecific effects, tricaine was not added for angiogram experiments. Data are expressed as mean ± SD. Statistical significance was revealed using two-tailed Student’s T-test. *p < 0.05; **p < 0.005; and ***p < 0.0005.
Mass spectrometry and mass spectrometric data analysis
Rabbit muscle actin (Sigma) (0.2 µg/µl) was incubated with 5 mM GSSG for 1 h. After removal of GSSG (Zeba Spin Desalting column, Thermo Scientific) actin solution was divided into three parts. First part was incubated with 10 mM NEM (glutathionylated sample), 2nd part was incubated with 10 µM zfGrx2/100 µM GSH, and 3rd part was incubated with 5 mM DTT. After 45 min 2nd and 3rd parts were incubated with 10 mM NEM and subsequently, actin protein was separated on SDS polyacrylamide gels, stained with coomassie brilliant blue, destained, alkylated with iodoacetamide (55 mM in an aqueous solution containing 50 mM ammonium bicarbonate) and digested with chymotrypsin. Peptides were extracted from the gel and finally resuspended in 0.1% trifluoroacetic acid and subjected to liquid chromatography. Mass spectrometry parameters were if not otherwise stated as described earlier [18].
Briefly, peptides were separated over a 54 min LC-gradient using an Ultimate 3000 Rapid Separation liquid chromatography system (Dionex/Thermo Scientific) equipped with an Acclaim PepMapRSLC 100 C18 column (2 µm C18 particle size, 100 Å pore size, 75 µm inner diameter, 25 cm length, Dionex/Thermo Scientific, Dreieich, Germany). Mass spectrometry was carried out on an Orbitrap Elite high resolution instrument (Thermo Scientific, Bremen, Germany) operated in positive mode and equipped with a nanoelectrospray ionization source. Survey scans were carried out in the Orbitrap analyzer over a mass range from 350 to 1700 m/z at a resolution of 60,000 (at 400 m/z). The target value for the automatic gain control was 1,000,000 and the maximum fill time 200 ms. The 15 = 2+ charged peptide ions (minimal signal intensity 500) were isolated, transferred to the linear ion trap (LTQ) part of the instrument and fragmented using collision induced dissociation (CID) or in some runs fragmented by higher-energy collisional dissociation (HCD) in the HCD cell of the mass spectrometer and transferred back to the orbitrap analyzer. Already fragmented ions were excluded from fragmentation for 30 s.
Raw files were further processed for protein and peptide identification with the MaxQuant software suite version 1.4.1.2 (Max Planck Institute of Biochemistry, Planegg, Germany) if not otherwise stated with default parameters. Within the software suite database searches were carried out within 888 Oryctolagus cuniculus sequences from the UniProtKB/Swiss-Prot database (downloaded on 2014-01-30) using the following parameters: mass tolerance Fourier transformed mass spectra (Orbitrap) first/second search: 20 ppm/4.5 ppm, mass tolerance fragment spectra (linear ion trap): 0.5 Da, variable modification: glutathione, carbamidomethyl (C), methionine oxidation, acetylation at protein N-termini, chymotryptic (FWYML) digestion. Quantification of 2+ and 3+ charged peptide ions (VCGlutathioneDNGSGL: m/z 535.1999, RCGlutathionePETLF: m/z 390.8355 and m/z 585.7495, VGDEAQSKRGIL: m/z 636.8494, ESAGIHETTY: m/z 554.2513, and QKEITAL: m/z 401.7371) was carried out by the integration of extracted ion chromatogram (10 ppm mass window, 3 min time window within one experiment) areas using Xcalibur 2.2 SP1.48 Qual Browser (Thermo Scientific, Bremen, Germany). Summed signals of the detectable charge states were used as quantitative correlate for relative peptide amounts.
Results
Knock-down of zfGrx2 impairs cardiac functionality
Using antisense morpholinos targeting the translation-initiation codon of zfGrx2, we are able to knock-down specifically zfGrx2 protein synthesis in zebrafish embryos by 75% as described and verified before [9,10]. Examining embryos at 48 hpf, we observed a reduced blood flow in the dorsal aorta as well as in the common cardinal vein in 93% (n = 249) of embryos lacking zfGrx2, whereas no obstructed blood flow was observed in age-matched controls (n = 150) (Fig. 1, supplementary movies). Performing rescue experiments, i.e. injecting the anti-zfGrx2 morpholino together with capped mRNA encoding zfGrx2, we were able to reduce the phenotypic penetrance to 35% (n = 57), verifying specificity of the observed phenotype. We additionally observed a reduced pumping efficiency of hearts in zfGrx2 knock-down embryos, which likely caused the reduced perfusion in the main axial vessels (Fig. 1A, Movie S2).
Fig. 1.
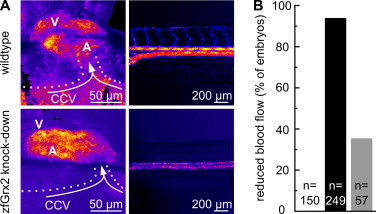
Knock-down of zfGrx2 impairs cardiac functionality and perfusion. (A) Angiograms based on supplementary movies demonstrate lower heart pumping capability (left panel, Movie S2) and subsequently reduced blood flow in the pre-cardiac region of the common cardinal vein (CCV) (left panel) and the main axial vessels (right panel, Movie S1) upon loss of zfGrx2 compared to wildtype embryos 48 hpf (V: ventricle, A: atrium). (B) Quantification of A, right panel shows that none of 150 examined wildtype embryos displayed reduced blood flow, whereas 93% of embryos lacking zfGrx2 (n = 249, black bar) showed reduced blood flow. Phenotypic penetrance was reduced to 35% (n = 57, gray bar) when capped zfGrx2 mRNA was injected in parallel with the morpholino.
Knock-down of zfGrx2 leads to cardiac looping defects
Since a reduced pumping efficiency was likely the cause for the obstructed blood flow in the trunk of zfGrx2 morpholino injected embryos, we examined their heart morphology at 48 hpf in detail. In situ hybridization detecting cmlc2 transcripts, a cardiomyocyte marker gene [14], revealed that the hearts of embryos lacking zfGrx2 do not undergo proper heart looping in contrast to controls (Fig. 2). In control embryos we measured an angle between atrium and ventricle of 54 ± 17° (n = 29), whereas age-matched morpholino injected embryos had a significantly more stretched angle of 112 ± 24° (n = 35). As reduced perfusion itself, also defects in heart looping were specific for the lack of zfGrx2, as rescue experiments restored the angle between atrium and ventricle to 89 ± 21° (n = 27).
Fig. 2.
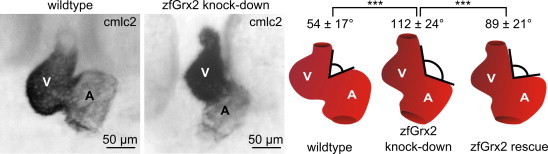
Knock-down of zfGrx2 leads to cardiac looping failure. In situ hybridization detecting transcripts of the cardiac-myocyte specific gene cmlc2 revealed that embryos lacking zfGrx2 suffer from heart looping defects at 48 hpf. Whereas the angle between atrium and ventricle in wildtype embryos was 54 ± 17° (n = 29), we measured an angle of 112 ± 24° in zfGrx2 morpholino injected embryos (n = 35). Cardiac looping could be restored to an angle of 89 ± 21° when zfGrx2 capped mRNA was injected simultaneously with the morpholino (n = 27). A: Atrium, V: ventricle. Presented are mean ± SD (***p < 0.0005).
Knock-down of zfGrx2 impairs migration of cardiac neural crest cells
To unravel the processes during cardiac development which are affected by the loss of zfGrx2, we dissected early heart formation in detail. Cardiomyocytes are specified in the lateral mesoderm and detectable as bilateral, cmlc2 positive stripes at the 16 somite stage [19]. Determining size and shape of these structures, we did not find any significant difference between embryos lacking zfGrx2 and controls (n = 25 and 26 respectively, Fig. 3). However, at the 19 somite stage, we found the cardiac fields of embryos with reduced levels of zfGrx2 to be significantly smaller (81 ± 11 µm, n = 37) compared to age-matched controls (100 ± 9 µm, n = 28) (Fig. 3). Cardiac neural crest (CNC) cells migrate to the primary heart field between the 16 and 19 somite stage and contribute to proper cardiac looping in zebrafish and mammals [20–22]. We followed the movement of neural crest (NC) cells during early embryonic development by making use of their specific crestin expression [16]. At 12 somites, we observed a shorter distance between the lateral edges of NC cells, indicating a slower lateral movement of this cell population. We found the distance between the lateral NC fields in embryos lacking zfGrx2 78 ± 15 µm (n = 69) compared to 109 ± 10 µm (n = 34) in age-matched controls (Fig. 3). Later in development, at the 16 somite stage, the width of the caudal NC stream was significantly smaller in embryos lacking zfGrx2 (62 ± 12 µm, n = 17) compared to controls (98 ± 16 µm, n = 22) and additionally the middle NC stream was markedly reduced in 78% of Grx2 knock-down embryos (n = 22) (Fig. 3). This phenotype remained at the 19 somite stage with the caudal NC stream being thinner in fish lacking Grx2 (98 ± 20 µm, n = 18) compared to controls (147 ± 17 µm, n = 10) and the middle stream was almost undetectable in 83% of embryos lacking zfGrx2 (n = 18).
Fig. 3.
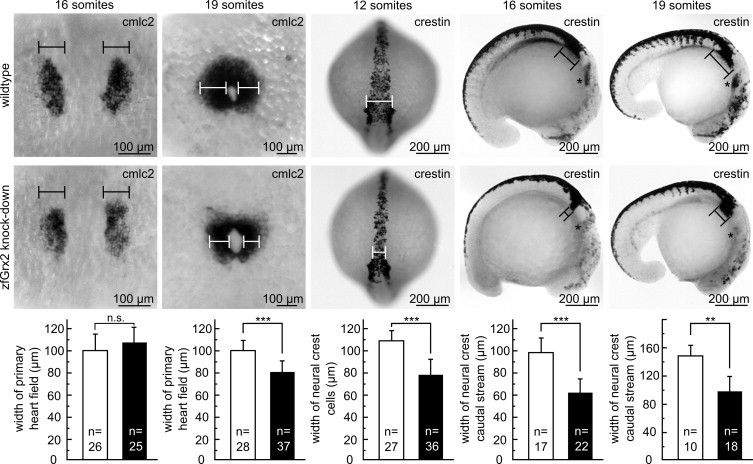
Knock-down of zfGrx2 impairs migration of neural crest cells. Using in situ hybridization detecting cmlc2 transcripts, the width of the primary cardiac field was measured in wildtype and morpholino injected embryos at the 16, and the 19 somite stage. Impaired migration of neural crest cells was demonstrated by anti-crestin in situ hybridization at 12, 16, and 19 somites. Quantification shows the width of the bilateral edges at 12 somites and the width of the caudal stream at later stages. Asterisks mark the middle stream. Presented are mean ± SD (**p < 0.005, ***p < 0.0005).
Monothiol mechanism of Grx2 is important for heart formation
To gain insight into the molecular mechanism by which zfGrx2 is involved in cardiac formation, we generated variants of zfGrx2 that were either enzymatically inactive (zfGrx2C37S) or only able to de-glutathionylate substrates via the monothiol mechanism (zfGrx2C40S). The mRNA variants were injected together with the morpholino and rescue efficiencies were compared to that of injection of mRNA coding unmodified zfGrx2 (Fig. 4A). The enzymatically inactive variant of zfGrx2 was not able to rescue the cardiac looping phenotype (rescue efficacy of 22 ± 1.4% compared to wildtype mRNA, n = 12). However, zfGrx2C40S rescued the heart looping defect as efficiently as unmodified zfGrx2 (rescue efficacy of 106 ± 2.7% compared to wildtype mRNA, n = 19), indicating that the biochemical mechanism involves (de)-glutathionylation of target proteins. It is well established that posttranslational modifications of components of the cytoskeleton is tightly controlled and a prerequisite for cellular migration [23]. Actin is a crucial protein required for cytoskeletal reorganization and migration and its polymerization is regulated via reversible S-glutathionylation [23,24]. Therefore, we tested the ability of zfGrx2 to modulate the glutathionylation state of actin in vitro. Recombinant muscle rabbit actin was glutathionylated and afterwards incubated with zfGrx2/GSH or DTT and analyzed by mass spectrometry. Two peptides harboring cysteines 12 (VCDNGSGL) and 259 (RCPETLF) were identified as targets for S-glutathionylation (Figs. S1 and S2 ). To determine relative amount of glutathionylated peptides, summed signals of the detectable charge states of three non-modified peptides (VGDEAQSKRGIL, ESAGIHETTY, QKEITAL) (Fig. S1) were used as quantitative correlates (Fig. 4B and C). Compared to glutathionylated samples (VCDNGSGL: 100 ± 30%, RCPETLF: 100 ± 14%), treatment with both zfGrx2 (VCDNGSGL: 19 ± 11%, RCPETLF: 9 ± 0.7%) and DTT (VCDNGSGL: 0.7 ± 0.7%, RCPETLF: 2 ± 1%) significantly reduced the amount of glutathionylated peptides indicating that actin is a potential target for zfGrx2 mediated de-glutathionylation during CNC migration and heart formation.
Fig. 4.
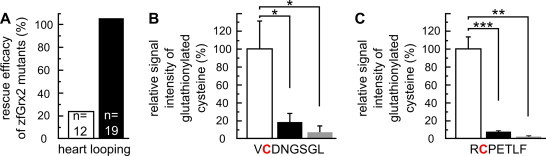
Heart development depends on reversible S-glutathionylation. (A) Rescue efficacy of heart looping defect using active site cysteine variants of zfGrx2 (white bar: zfGrx2C37S, black bar: zfGrx2C40S) related to efficacy of rescue with unmodified zfGrx2 mRNA. (B,C) S-glutathionylation state of recombinant rabbit actin was determined by mass spectrometry after incubation with GSSG (white bars) following incubation with zfGrx2 (black bars) or DTT (gray bars). Two cysteines were identified as S-glutathionylated (Cys12 (B) and Cys259 (C)). Relative signal intensity was calculated in relation to 3 peptides without cysteines and compared to GSSG treated samples.
Knock-down of zfGrx2 leads to cell death in the region of neural crest cells
It has been reported before that NC cells which fail to migrate undergo cell death [25], which is also suggested in our experimental setup by the reduced staining for crestin positive cells in zfGrx2 lacking embryos (Fig. 3). By acridine orange staining, we confirmed that loss of zfGrx2 induces massive cell death during the time and in the region of migrating NC cells in 83% of embryos analyzed (n = 37) compared to only very few acridine orange positive cells in controls (n = 20) (Fig. 5). This observation was further supported by detecting apoptotic cells using TUNEL staining at 24 hpf (Fig. 5) and by the loss of foxD3 demarked NC cells in morpholino injected 24 hpf embryos (Fig. 5).
Fig. 5.
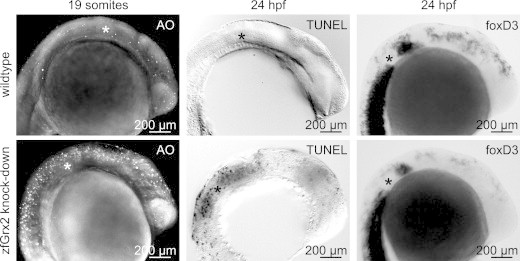
Knock-down of glutaredoxin 2 induces cell death in the areas of formation and migration of neural crest cells. Using acridine orange (AO) staining in 19 somite stage embryos, we detected cell death in the area of specification and migration of NC cells in zfGrx2 knock-down embryos, whereas wildtype controls showed only few acridine orange positive cells. Apoptotic cell death was confirmed by TUNEL staining at 24 hpf. Detecting neural crest cells using the specific marker foxD3 shows less staining rostral to the otic vesicle as well as in the rostral brain of embryos lacking zfGrx2 compared to wildtypes, indicating loss of NC cells. Asterisks mark the otic vesicle.
Discussion
Glutaredoxins are oxidoreductases that control the cellular thiol redox pool [6]. Our previous research has unraveled that Grx2 is a crucial mediator for redox signaling during embryonic development [9,10]. Members of the thioredoxin family of proteins including glutaredoxins are known to be expressed in the mammalian heart and involved in cardiovascular physiology and pathology [26]. However, our report presents the first evidence that Grx2 is required for the development of the heart itself. Importantly, the heart is the first organ to develop and function during vertebrate embryonic development and therefore the prerequisite for the successful formation of a new organism. Knock-down of zfGrx2 in zebrafish embryos leads to a reduced pumping efficacy of hearts and obstructed blood flow in the main axial vessel due to disrupted heart looping. Looping of the cardiac tube is not only the first manifestation of embryonic laterality which is only poorly understood, but also an essential process during cardiac development [27]. In mammals and lower vertebrates cardiac looping depends, like outflow tract septation and cushion formation, on the tightly orchestrated migration and invasion of CNC cells into the cardiac field [20,22,28]. Those migratory cells represent a specialized subgroup of NC cells that originate, together with the other NC cells, from the neuroepithelium [29].In line with these reports, we found that migration of CNC cells was severely impaired in embryos lacking zfGrx2. Modulating the levels of zfGrx2 reduced the size of cardiac fields only after the presumptive arrival of CNC cells [30], indicating a reduced migration speed of this cell population if zfGrx2 is absent. Moreover, during early development the lateral migration of NC cells is inhibited and during the course of development the number of NC cells is severely reduced, most prominently in the caudal and middle streams. Interestingly, those NC streams are recognized as the origin of CSC cells in zebrafish [31]. If NC cells are disturbed in their migration and terminal differentiation, they undergo cell death [25,32]. In line with these previous studies, we have observed massive cell death in the region where NC cells are located. Cell migration, including migration of NC cells, is based on continuous remodeling of the cytoskeleton [33–35]. Posttranslational modifications, including reversible redox modifications of actin and other proteins of the cytoskeleton, are involved in cytoskeleton remodeling and cell migration [24,36–39]. Several proteins of the cytoskeleton have been recently identified as potential interaction partners of Grx2 [40]. Using an in vitro approach in combination with mass spectrometry analysis, we present evidence that the observed phenotype might be connected to reversible S-glutathionylation of actin by zfGrx2. Nevertheless, zfGrx2 might also regulate additional proteins or pathways which have to be elucidated in future.We have previously identified collapsin-response-mediator protein 2 (CRMP2) as substrate for Grx2 [9]. CRMP2 is involved in semaphorin signaling which has been implicated in cardiogenesis and CNC migration [41,42]. However, the Grx2-CRMP2 signaling axis seems not to regulate cardiac looping and CNC migration in our setting as (i) we were unable to rescue the phenotype by overexpression of CRMP2 (data not shown), (ii) we previously found that Grx2 regulates CRMP2 activity via reduction of a disulfide, and (iii) we and others were unable to detect CRMP2 transcripts in the heart [43] (Fig. S3 ). Moreover, semaphorin signaling in cardiac development seems to be mostly independent of CRMP2 [41].Here, we have described that zfGrx2 is crucial for successful heart formation by controlling the invasion of cardiac neural crest cells into the primary heart field (Fig. S4). Together with our recent work on the role of glutaredoxin 2 in brain [9] and vascular development [10], this work represents further evidence for the general importance of redox signaling in embryonic development and highlights additionally the importance of Grx2 during embryogenesis.
Acknowledgments
We thank Lena Ringdén for administrative assistance, the Karolinska Institutet zebrafish core facility for excellent service, and Ulrich Rüther (Heinrich Heine University) for helpful discussions. Templates for foxD3 and crestin riboprobes were generously provided by the Yost laboratory (University of Utah, USA) and Bo Zhang (Peking University, P.R. China). This work was supported by Karolinska Institutet (postdoctoral fellowships to CB, PhD fellowship (KID) Dnr. 2379/07-225 to LB), German Research Foundation (DFG priority program 1710, BE 3259/5-1 to CB), the Swedish Society for Medical Research (fellowship to L.B.) the K.A. Wallenberg Foundation (KAW 2006.0192 to AH), the Swedish Cancer Society (961 to AH), and the Swedish Research Council (K2012-68X-03529-41-3 to AH).
Contributor Information
Carsten Berndt, Email: carsten.berndt@med.uni-duesseldorf.de.
Lars Bräutigam, Email: lars.braeutigam@ki.se.
Appendix A. Supplemental Materials
Supplementary material associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.redox.2014.04.012
Appendix A. Supplementary Materials
Spectra of glutathionylated peptides of rabbit actin (upper spectra, S-glutathionylated cystein residues are marked red), and three non-modified spectra used as quantitative correlates for relative peptide amounts.
Alignment of rabbit actin and zebrafish actin. Peptides described within this work are marked in gray. S-glutathionylated cysteines are highlighted in green, identified non-modified cysteines in yellow, and not detected cysteines in red.
In situ hybridization of CRMP2 transcripts in zebrafish embryos 48 hpf.
The morpholino knocking-down zfGrx2 was injected in fertilized eggs. From 15 hpf onwards, we observed impaired NC migration that manifested in cell death and a smaller primary heart field at 18.5 hpf. Impaired NC migration resulted in faulty cardiac looping that was assessed at 48 hpf.
Blood flow in zebrafish embryos was recorded by video microscopy. A: Anterior, P: posterior.
The heart beat was recorded by video microscopy.
References
- 1.Winterbourn C.C. Reconciling the chemistry and biology of reactive oxygen species. Nature Chemical Biology. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 2.Brigelius-Flohé R., Flohé L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxidants and Redox Signaling. 2011;15:2335–2381. doi: 10.1089/ars.2010.3534. 21194351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ufer C., Wang C.C., Borchert A., Heydeck D., Kuhn H. Redox control in mammalian embryo development. Antioxidants and Redox Signaling. 2010;13:833–875. doi: 10.1089/ars.2009.3044. 20367257 [DOI] [PubMed] [Google Scholar]
- 4.Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radical Biology and Medicine. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. 18544350 [DOI] [PubMed] [Google Scholar]
- 5.Holmgren A. Thiol redox control via thioredoxin and glutaredoxin systems. Biochemical Society Transactions. 2005;33:1375–1377. doi: 10.1042/BST0331375. 16246122 [DOI] [PubMed] [Google Scholar]
- 6.Lillig C.H., Berndt C. Glutaredoxins in thiol/disulfide exchange. Antioxidants and Redox Signaling. 2013;18(13):1654–1665. doi: 10.1089/ars.2012.5007. 23231445 [DOI] [PubMed] [Google Scholar]
- 7.Mieyal J.J., Chock B. Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on S-glutathionylation. Antioxidants and Redox Signaling. 2012;16:471–475. doi: 10.1089/ars.2011.4454. 22136616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lillig C.H., Berndt C., Holmgren A. Glutaredoxin systems. Biochimica et Biophysica Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. 18621099 [DOI] [PubMed] [Google Scholar]
- 9.Bräutigam L. Vertebrate-specific glutaredoxin is essential for brain development. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20532–20537. doi: 10.1073/pnas.1110085108. 22139372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bräutigam L. Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20057–20062. doi: 10.1073/pnas.1313753110. 24277839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staudt D., Stainier D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annual Review of Genetics. 2012;46:397–418. doi: 10.1146/annurev-genet-110711-155646. 22974299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 13.Thisse C., Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. 18193022 [DOI] [PubMed] [Google Scholar]
- 14.Yelon D., Horne S.A., Stainier D.Y.R. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Developmental Biology. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. 10491254 [DOI] [PubMed] [Google Scholar]
- 15.Stewart R.A. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Developmental Biology. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. 16499899 [DOI] [PubMed] [Google Scholar]
- 16.Luo R., An M., Arduini B.L., Henion P.D. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 2001;220:169–174. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. 11169850 [DOI] [PubMed] [Google Scholar]
- 17.Chico T.J.A., Ingham P.W., Crossman D.C. Modeling cardiovascular disease in the zebrafish. Trends in Cardiovascular Medicine. 2008;18:150–155. doi: 10.1016/j.tcm.2008.04.002. 18555188 [DOI] [PubMed] [Google Scholar]
- 18.Poschmann G. High-fat diet induced isoform changes of the Parkinson's disease Protein DJ-1. Journal of Proteome Research. 2014 doi: 10.1021/pr401157k. [DOI] [PubMed] [Google Scholar]
- 19.Yelon D. Cardiac patterning and morphogenesis in zebrafish. Developmental Dynamics : An Official Publication of the American Association of Anatomists. 2001;222:552–563. doi: 10.1002/dvdy.1243. [DOI] [PubMed] [Google Scholar]
- 20.Li Y.-X. Cardiac neural crest in zebrafish embryos contributes to myocardial cell lineage and early heart function. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 2003;226:540–550. doi: 10.1002/dvdy.10264. 12619138 [DOI] [PubMed] [Google Scholar]
- 21.Creazzo T.L., Godt R.E., Leatherbury L., Conway S.J., Kirby M.L. Role of cardiac neural crest cells in cardiovascular development. Annual Review of Physiology. 1998;60:267–286. doi: 10.1146/annurev.physiol.60.1.267. 9558464 [DOI] [PubMed] [Google Scholar]
- 22.Yelbuz T.M. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation. 2002;106:504–510. doi: 10.1161/01.cir.0000023044.44974.8a. 12135953 [DOI] [PubMed] [Google Scholar]
- 23.Sakai J. Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity. 2012;37:1037–1049. doi: 10.1016/j.immuni.2012.08.017. 23159440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J. Reversible glutathionylation regulates actin polymerization in A431 cells. Journal of Biological Chemistry. 2001;276:47763–47766. doi: 10.1074/jbc.C100415200. 11684673 [DOI] [PubMed] [Google Scholar]
- 25.Barrallo-Gimeno A., Holzschuh J., Driever W., Knapik E.W. Neural crest survival and differentiation in zebrafish depends on Mont Blanc/tfap2a gene function. Development (Cambridge, England) 2004;131:1463–1477. doi: 10.1242/dev.01033. 14985255 [DOI] [PubMed] [Google Scholar]
- 26.Berndt C., Lillig C.H., Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. American Journal of Physiology: Heart and Circulatory Physiology. 2007;292:H1227–H1236. doi: 10.1152/ajpheart.01162.2006. 17172268 [DOI] [PubMed] [Google Scholar]
- 27.Miquerol L., Kelly R.G. Organogenesis of the vertebrate heart. Wiley Interdisciplinary Reviews: Developmental Biology. 2013;2:17–29. doi: 10.1002/wdev.68. 23799628 [DOI] [PubMed] [Google Scholar]
- 28.Waldo K.L. Cardiac neural crest is necessary for normal addition of the myocardium to the arterial pole from the secondary heart field. Developmental Biology. 2005;281:66–77. doi: 10.1016/j.ydbio.2005.02.011. 15848389 [DOI] [PubMed] [Google Scholar]
- 29.Snider P., Olaopa M., Firulli A.B., Conway S.J. Cardiovascular development and the colonizing cardiac neural crest lineage. The Scientific World Journal. 2007;7:1090–1113. doi: 10.1100/tsw.2007.189. 17619792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato M., Tsai H.-J., Yost H.J. Semaphorin3D regulates invasion of cardiac neural crest cells into the primary heart field. Developmental Biology. 2006;298:12–21. doi: 10.1016/j.ydbio.2006.05.033. 16860789 [DOI] [PubMed] [Google Scholar]
- 31.Sato M., Yost H.J. Cardiac neural crest contributes to cardiomyogenesis in zebrafish. Developmental Biology. 2003;257:127–139. doi: 10.1016/s0012-1606(03)00037-x. 12710962 [DOI] [PubMed] [Google Scholar]
- 32.Hinoue A. Disruption of actin cytoskeleton and anchorage-dependent cell spreading induces apoptotic death of mouse neural crest cells cultured in vitro. Anatomical Record Part A: Discoveries in Molecular, Cellular, and Evolutionary Biology. 2005;282:130–137. doi: 10.1002/ar.a.20150. 15627983 [DOI] [PubMed] [Google Scholar]
- 33.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. 12600310 [DOI] [PubMed] [Google Scholar]
- 34.Terman J.R., Kashina A. Post-translational modification and regulation of actin. Current Opinion in Cell Biology. 2013;25:30–38. doi: 10.1016/j.ceb.2012.10.009. 23195437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stossel T.P. On the crawling of animal cells. Science (New York, N.Y.) 1993;260:1086–1094. doi: 10.1126/science.8493552. 8493552 [DOI] [PubMed] [Google Scholar]
- 36.Hou X. Coactosin accelerates cell dynamism by promoting actin polymerization. Developmental Biology. 2013;379:53–63. doi: 10.1016/j.ydbio.2013.04.006. 23603493 [DOI] [PubMed] [Google Scholar]
- 37.Nie S., Kee Y., Bronner-Fraser M. Caldesmon regulates actin dynamics to influence cranial neural crest migration in Xenopus. Molecular Biology of the Cell. 2011;22:3355–3365. doi: 10.1091/mbc.E11-02-0165. 21795398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X., Francis R., Wei C.J., Linask K.L., Lo C.W. Connexin 43-mediated modulation of polarized cell movement and the directional migration of cardiac neural crest cells. Development (Cambridge, England) 2006;133:3629–3639. doi: 10.1242/dev.02543. 16914489 [DOI] [PubMed] [Google Scholar]
- 39.Landino L.M., Moynihan K.L., Todd J.V., Kennett K.L. Modulation of the redox state of tubulin by the glutathione/glutaredoxin reductase system. Biochemical and Biophysical Research Communications. 2004;314:555–560. doi: 10.1016/j.bbrc.2003.12.126. 14733943 [DOI] [PubMed] [Google Scholar]
- 40.Schütte L.D. Identification of potential protein dithiol–disulfide substrates of mammalian Grx2. Biochimica et Biophysica Acta. 2013;1830:4999–5005. doi: 10.1016/j.bbagen.2013.07.009. 23872354 [DOI] [PubMed] [Google Scholar]
- 41.Toyofuku T., Kikutani H. Semaphorin signaling during cardiac development. Advances in Experimental Medicine and Biology. 2007;600:109–117. doi: 10.1007/978-0-387-70956-7_9. 17607950 [DOI] [PubMed] [Google Scholar]
- 42.Scholl A.M., Kirby M.L. Signals controlling neural crest contributions to the heart. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2009;1:220–227. doi: 10.1002/wsbm.8. 20490374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christie T.L., Starovic-Subota O., Childs S. Zebrafish collapsin response mediator protein (CRMP)-2 is expressed in developing neurons. Gene Expression Patterns. 2006;6:193–200. doi: 10.1016/j.modgep.2005.06.004. 16168718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spectra of glutathionylated peptides of rabbit actin (upper spectra, S-glutathionylated cystein residues are marked red), and three non-modified spectra used as quantitative correlates for relative peptide amounts.
Alignment of rabbit actin and zebrafish actin. Peptides described within this work are marked in gray. S-glutathionylated cysteines are highlighted in green, identified non-modified cysteines in yellow, and not detected cysteines in red.
In situ hybridization of CRMP2 transcripts in zebrafish embryos 48 hpf.
The morpholino knocking-down zfGrx2 was injected in fertilized eggs. From 15 hpf onwards, we observed impaired NC migration that manifested in cell death and a smaller primary heart field at 18.5 hpf. Impaired NC migration resulted in faulty cardiac looping that was assessed at 48 hpf.
Blood flow in zebrafish embryos was recorded by video microscopy. A: Anterior, P: posterior.
The heart beat was recorded by video microscopy.


