Summary
N-glycans modify the great majority of all secreted and plasma membrane proteins, which themselves constitute one-third to one-half of the proteome. The ultimate definition of the glycoproteome would be the identification of all the N-glycans attached to all the modified asparaginyl sites of all the proteins, but glycosylation heterogeneity makes this an unachievable goal. However, mass spectrometry in combination with other methods does have the power to deeply mine the N-glycome of Dictyostelium, and characterize glycan profiles at individual sites of glycoproteins. Recent studies from our laboratories using mass spectrometry-based methods have confirmed basic precepts of the N-glycome based on prior classical methods using radiotracer methods, and have extended the scope of glycan diversity and the distribution of glycan types across specific glycoprotein attachment sites. The protocols described here simplify studies of the N-glycome and -glycoproteome, which should prove useful for interpreting mutant phenotypes, conducting interstrain and interspecies comparisons, and investigating glycan functions in glycoproteins of interest.
Keywords: Dictyostelium, glycosylation, glycome, glycoproteome, mass spectrometry, phosphoglycans, sulfated glycans
1. Introduction
The many attractions for studying the social soil amoebae include their amoebal lifestyle and multicellular developmental cycle elicited by starvation, and their tractability for molecular genetics, biochemistry and cell biology. In development, the amoebae chemotactically assemble a migrating slug which, in response to appropriate environmental signals, emerges from the soil to culminate into a fruiting body whose aerial spores can be dispersed to new locations. The amoebae are surrounded by a rich glycocalyx whose glycan composition changes in concert with the developmental cycle.
Dictyostelium discoideum, like most other eukaryotes (1), carries out extensive N-glycosylation of its membrane and secreted proteins. During protein translocation into the rER, Glc3Man9GlcNAc2- is transferred from the canonical lipid-linked oligosaccharide (LLO) precursor (2) by oligosaccharyltransferase to the amide of an Asn residing in an N-sequon consisting of Asn-x-Ser/Thr (x ≠ Pro). As described in other organisms, removal of the peripheral glucose (Glc) and certain peripheral mannose (Man) residues are likely to be involved in ERQC and ERAD (endoplasmic reticulum quality control and associated degradation) pathways that contribute to nascent protein folding and complex assembly (3), and consequential dislocation of unfolded proteins into the cytoplasm. Subsequent remodeling of the N-glycans produces a spectrum of N-glycans that contribute to post rER functions in the life of the carrier protein.
Early studies suggested that the types of N- and O-glycans formed are developmentally regulated based on studies with lectins including wheat germ agglutinin (WGA) and Concanavalin A (ConA), and anticarbohydrate monoclonal antibodies (4). Subsequently, metabolic labeling with 3H-sugars, 35SO4, and 32PO4, coupled with chromatographic methods, exoglycosidase treatments and NMR, indicated that the N-glycome is dominated by high-mannose structures with variable processing and a variety of substitutions including CH3-PO4- and SO4- on peripheral Man residues, βGlcNAc at bisecting and intersecting positions, and core α-fucosylation (5–13). Developmental progression was accompanied by increased Man-trimming with decreased or increased peripheral modifications (2, 6, 14). Genetic analyses have implicated several genes responsible for precursor assembly and specific processing steps. Genomic searches identified 77 candidate glycosyltransferase and glycophosphotransferase genes, and many glycosidase genes, that are expected to contribute to the assembly and remodeling of the N- and other types of glycans (15, 16). This enumeration implies that structural heterogeneity is greater than could be resolved by previous chromatographic methods.
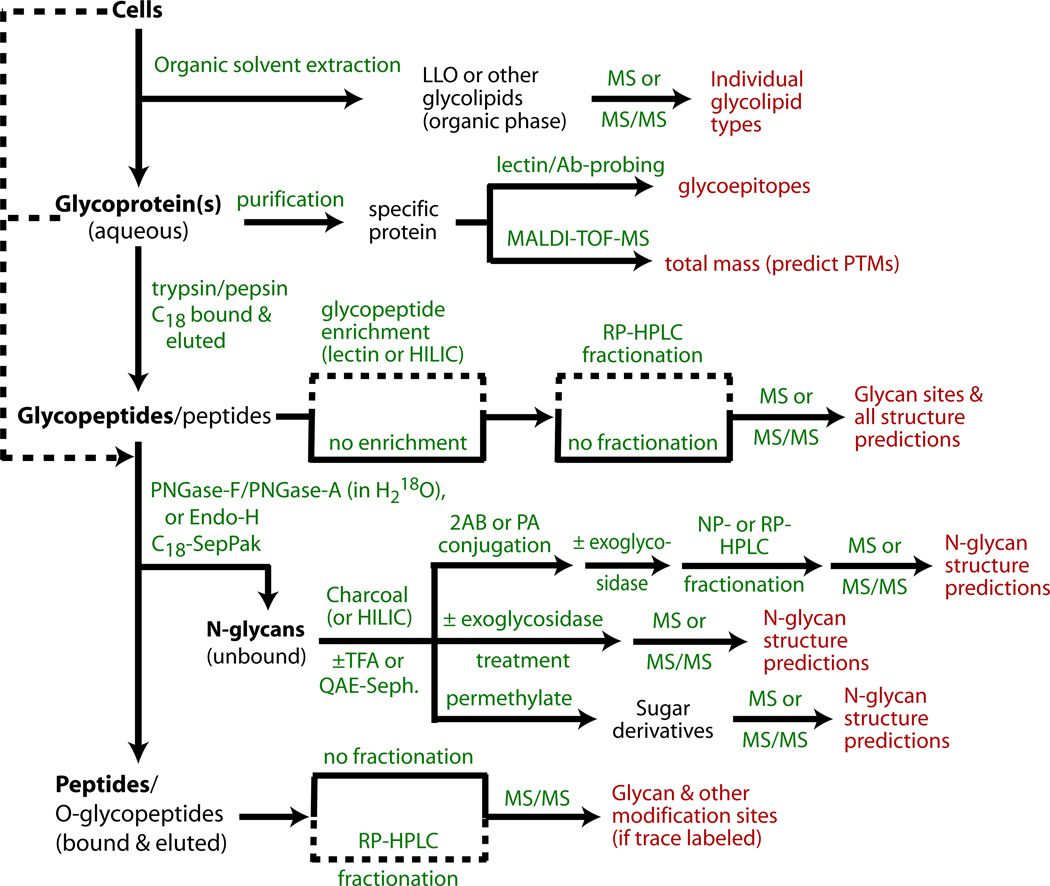
Recent studies using mass spectrometry and other high-resolution methods have re-investigated the overall N-glycomes of total cells (17, 16) and a single cell surface glycoprotein gp130 implicated in cell adhesion (18). The basic findings of the classical studies have been confirmed and, not surprisingly, the detailed diversity of the N-glycome has been considerably expanded. Studies of other social amoebae are beginning to reveal substantial differences in peripheral processing of the high-mannose structures (16). Interestingly, glycoproteomic studies of the heavily N-glycosylated gp130 revealed that nearly the entire diversity of the N-glycome is expressed on this single cell surface glycophosphatidylinositol (GPI)-anchored protein. The application of mass spectrometry (MS) and complementary strategies for defining the N-glycome and site-specific representation on glycoproteins is the subject of this article. We describe how to characterize the dolichol-PP-glycan (lipid-linked oligosaccharide or LLO) precursor, and alternative methods for enzymatically liberating and recovering different N-glycan classes from glycoproteins. An overview is presented in the flow chart in Fig. 1. Methods for MS profiling of the native N-glycans themselves or after conjugation via their reducing ends to fluorophores or permethylation are then described. Though MS methods are excellent at determining glycan compositions based on proportions of hexoses, HexNAcs, deoxyhexoses, etc., additional approaches are required to determine sugar identities and linkage types. Fortunately, so-called biosynthetic rules based on prior studies, and MS-MS fragmentation methods, can frequently permit accurate predictions of structural models for N-glycans in Dictyostelium and additional methods that assist in these determinations are included. Orthogonal separation and enrichment strategies to resolve glycan subclasses, including normal and reversed phase and ionic exchange methods, as well as the use of anticarbohydrate antibodies to differentiate core α1,3-fucosylation, are also described. Finally, we show how modifications of the approaches can be applied to analysis of glycans prior to release from their Asn anchors, i.e., as glycopeptides, in order to map their locations on proteins.
Figure 1. Flow chart for N-glycomic and N-glycoproteomic studies of cells and glycoproteins.
Cells are optionally initially extracted by organic solvent combinations to remove lipids and other small molecules (Subheading 3.1), and LLOs can be recovered. The remaining material can then be degraded by proteolysis to generate glycopeptides that can be analyzed for site-specific N-glycosylation (Subheading 3.13). Alternatively, purified glycoproteins can be analyzed for total mass and probed for lectin/Ab binding to query for discrete glycan structures (Subheading 3.16), and then introduced into the processing pipeline. Glycopeptides, which are more susceptible than intact glycoproteins to enzymatic de-N-glycosylation, are treated with PNGase F, PNGase A, or Endo-H to release N-glycans which are separated from peptides on a C18-SepPak (Subheading 3.3). The unbound glycans can be analyzed by MS in their native forms, before or after treatment with α-mannosidase or other exoglycosidases to probe the structure or non-reducing termini (Subheading 3.8). Alternatively, N-glycans are derivatized at their reducing termini with PA or 2AB (Subheading 3.6.1), and characterized chromatographically (Subheading 3.7), and/or analyzed by MS and MS/MS (Subheading 3.10) before or after permethylation (Subheading 3.6.2). If enzymatic de-N-glycosylation is performed in the presence of H218O, the peptides can be analyzed by MS for characteristic mass changes that allow confirmation of N-glycosylation (Subheading 3.15). Dashed lines indicate alternative paths or potential bypasses.
2. Materials
High quality source of deionized water, to dissolve reagents and samples and dilute organic solvents.
2.1. Reagents, Buffers and Columns
2.1.1. Cell Lysis, Extraction and Proteolytic Digestion
0.1 M iodoacetamide (IAM) (Sigma-Aldrich) in water, prepare fresh.
1 M dithiothreitol (DTT) (EMD Millipore) in water. Store at −20°C.
Urea, ultrapure (MP Biomedicals). Use only freshly prepared solutions.
Sodium dodecyl sulfate (SDS), high purity (Thermo 28312).
10% NP40, protein grade (e.g., Calbiochem/EMD Millipore). Store at 5°C.
Reagent grade TPCK-treated trypsin, 4 mg/mL in 50 mM acetic acid; store at −80°C (Sigma-Aldrich).
Sequencing grade modified trypsin, 1 mg/mL in 50 mM acetic acid; store at −80°C (Promega).
Proteomics grade pepsin (Amresco, M142), 3 mg/mL prepared fresh in 5% formic acid or pepsin from porcine gastric mucosa (Sigma), 4.22 U/mg protein.
Phosphate buffered saline (PBS): 150 mM NaCl, 20 mM Na phosphate, pH 7.4. Adjust pH by addition of 1 M NaOH to a solution of NaH2PO4 (monobasic) prior to bringing to a final volume to achieve 20 mM.
Tight fitting glass homogenizer, e.g., glass Dounce-type with B-pestle (custom-adjusted).
8-mL polypropylene tubes with screw caps (e.g., Sarstedt 60.542).
2.1.2. Glycan Release
NH4HCO3, certified grade (Fisher), 25–200 mM as needed, at its natural pH of 7.8–8.0.
N-glycosidase F (PNGase F), glycerol free (New England Biolabs), supplied at 500,000 U/mL in 50 mM NaCl, 5 mM Na2EDTA, 20 mM Tris-HCl (pH 7.5). Prepare intermediate stock solutions by dilution in 25 mM NH4HCO3, pH 7.8 (for glycopeptides) or 50 mM sodium phosphate, pH 7.5 (for glycoproteins). Alternatively, use PNGase F from Roche (250 U in 0.25 mL) as indicated.
N-glycosidase A (PNGase A) from almond meal (Roche Applied Science), supplied at 5 mU/100 µL in 50 mM citrate-phosphate buffer (pH 5.0), 50% glycerol.
Endoglycosidase Hf (Endo Hf) (New England Biolabs), supplied at 1,000,000 U/mL in 50 mM NaCl, 5 mM Na2EDTA, 20 mM Tris-HCl (pH 7.5). Prepare intermediate stock solutions by dilution in 50 mM sodium citrate, pH 5.5.
Alkaline borohydride solution: 50 mM NaOH (diluted from Fisher 50% (w/w) stock solution), 1 M NaBH4 (Sigma, 71320). Store NaBH4 powder in a desiccator. Prepare 100 mM NaOH from a 50% (w/w) stock solution (Fisher), and 2 M NaBH4 from the powder, immediately before use. Allow gas evolution when adding the NaBH4 to the water. Prepare final solution by mixing equal volumes of the NaOH and NaBH4 solutions.
40–48% hydrofluoric acid (Mallinckrodt or Sigma-Aldrich).
H216O and H218O, 97% isotope purity (Cambridge Isotope Laboratories, Andover, MA).
Dowex 50W×8, 200–400 mesh, H+ form (Sigma-Aldrich), pre-equilibrated in 5% acetic acid solution.
Sephadex™ G15 and G25 (GE Healthcare).
Orcinol monohydrate (Sigma), 200 mg dissolved in 100 mL of 20% (v/v) H2SO4, suitable for spraying.
Thin-layer chromatography (TLC) sprayer, e.g., Merck.
TLC Silica gel 60 on aluminum plate (Merck).
96 F black plates (Nunc).
2.1.3. Glycan and Glycopeptide Recovery
Acetonitrile (ACN) (HPLC grade), dried over 3-Å molecular sieves (Sigma-Aldrich) (see Note 1).
C18-SepPak (100 mg) cartridge (Waters). Pre-equilibrate by sequential application of methanol (MeOH), water, 50% ACN in water, 0.1% trifluoroacetic acid (TFA) in water, 0.1% TFA in water.
Syringe mounted on an adaptor for expelling solutions from SepPak (Grace/Alltech, 210705).
Carbograph cartridge: 400 mg Carbograph cartridge (Grace). Pre-equilibrate by sequential application of ethanol (EtOH), water, 50% ACN in water, 0.1% TFA in water.
Hydrophilic interaction chromatography (HILIC): polyHYDROXYETHYL A TopTips (PolyLC). Pre-cycle by sequential application of 0.5% formic acid in ACN; 0.5% formic acid in 80% ACN/water; 0.5% formic acid/water; 80% ACN/water (see Note 2).
SupelClean™ENVI™ CarbSPE Tubes 6 mL/0.25 g (Sigma-Aldrich). Pre-equilibrate by sequential application of 100% ACN and water.
Non-Porous graphitized carbon (NPGC) column: 25 mg ENVI™ Carb bulk material (Sigma-Aldrich) per 1-mL SPE (solid-phase extraction) tube. Pre-equilibrate by sequential application of 100% ACN, 40% ACN, and water.
2-mL polypropylene Bio-Spin column (Bio-Rad).
2.1.4. Glycan Derivatization or Modification
2-aminopyridine (2AP or PA) (Sigma-Aldrich).
2-aminobenzamide (2AB) (Sigma-Aldrich).
Sodium cyanoborohydride (Sigma-Aldrich).
Dimethylsulfoxide (DMSO), dried over CaH2 (see Note 1).
ACN: see Subheading 2.1.3, item 1.
NaOH, 20–40 mesh beads (sealed under N2).
Chloroform (CHCl3), certified ACS (Fisher) (see Note 1).
Isopropanol, reagent grade (Fisher) (see Note 1).
n-butanol, reagent grade (Fisher) (see Note 1).
Methyl iodide (CH3I) (Sigma-Aldrich), stored at 4°C (see Note 1).
Partial dextran hydrolysate, 2–20 glucose units (Sigma), PA-labeled in-house (see Subheading 3.6.1.2).
Jack bean α-mannosidase (Sigma-Aldrich) (see Note 3).
Jack bean β-N-acetylhexosaminidase (Prozyme).
2.1.5. Glycan and Glycopeptide Fractionation and Enrichment
Lectins: Agarose-bound ConA, 10 mg ConA/mL resin (Vector Laboratories); agarose-bound WGA, 7 mg/mL (Vector Labs).
ConA binding buffer: 10 mM HEPES-NaOH, pH 7.5, 1 mM CaCl2, 1 mM MnCl2.
General lectin elution buffers: 100 mM acetic acid (pH ~3).
α-methyl mannoside (Sigma).
QAE-Sephadex A-25 (Sigma-Aldrich, 5 g): Rehydrate in 25 mL of 0.5 M NaCl, 2 mM Trizma base (pH 9.6), equilibrate with 100 mL of 1 mM Trizma base (natural pH of 9.6), and pack into a 2-mL Bio-Spin column to a final bed volume of 0.5 mL.
QAE-Sephadex solutions: 1 mM Trizma base (pH 9.6) with 0, 70, or 140 mM NaCl.
SupelClean™ENVI™ CarbSPE Tubes, 6 mL/0.25 g, equilibrated in 40% ACN followed by water.
PepMap100 C18 column, 3-µm, 100-Å pore size (Acclaim), or an equivalent alternative. Pre-equilibrate in 0.09% formic acid, 0.01% TFA, 2% ACN, in water.
Phenomenex Jupiter C18 resin, 3 µm (Jupiter), self-packed (19) into an 8 cm × 50 µm i.d. PicoTip (New Objective), or a commercially available alternative. Pre-equilibrate in 0.1% formic acid, 2% (v/v) ACN, in water.
Micro Spin columns (Harvard Apparatus, 74–4420).
Tosoh Amide-80 column (4.6 × 250 mm), for normal phase (NP)-HPLC. Pre-equilibrate in a 1:3 mixture of 10 mM ammonium formate (take the appropriate volume to achieve 10 mM formic acid in the final volume and adjust with ammonia to pH 7.0; buffer A) and 95% ACN (buffer B) in water.
Agilent Hypersil ODS (4 mm × 250 mm, 5 µm; stored in 30% MeOH), for reverse phase (RP)-HPLC. Pre-equilibrate in 0.1 M ammonium acetate (buffer C; take the appropriate volume of concentrated acetic acid to achieve 0.1 M in the final volume and adjust to pH 4.0 with NH3).
Sephadex G15: see Subheading 2.1.2, item 9.
2.1.6. Mass Spectrometry
- MALDI matrices (see Note 4)
- 20 mg/mL 2,5-dihydroxybenzoic acid (DHB), recrystallized, in 30% (v/v) ACN/water (Sigma-Aldrich, 85707).
- 2–10 mg/mL 2',4',6'-trihydroxyacetophenone monohydrate (THAP) in 50% (v/v) ACN/water (Sigma-Aldrich).
- 5–10 mg/mL 6-aza-2-thiothymine (ATT) in water or 50% (v/v) ACN/water (Sigma-Aldrich).
- 10 mg/mL α-cyano-4-hydroxycinnamic acid in 0.1% TFA, 50% ACN/water (Sigma-Aldrich).
Dextran Mr ladder, Mr 500–3000 (V-labs; Covington, LA).
Seven-component peptide Mr standard mixture (Bruker Daltonics).
2.1.7. Western Blotting
Standard SDS-PAGE electrophoresis apparatus (e.g., BioRad) including standard buffers.
Trans blot SD-semi dry transfer cell (e.g., BioRad) and associated materials and reagents.
Antibody dilution and blocking solution (also used for antibody dilution): 0.5% (w/v) crystalline bovine serum albumin (Roth 8076.3) in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.05% (v/v) Tween-20.
Anti-horseradish peroxidase (anti-HRP) from rabbit; use at 1 µg/mL (Sigma-Aldrich) in blocking solution.
Goat anti-rabbit IgG, conjugated with alkaline phosphatase (Vector Labs); use at 1:2000 in blocking solution.
Biotinylated Wheat germ agglutinin (WGA) (Vector Labs); use at 2.5 µg/mL in blocking solution.
Anti-Biotin from goat conjugated with alkaline phosphatase (Sigma); use at 1:10,000 in blocking solution.
2.2. Equipment
Vacuum centrifuge (e.g., Speedvac, Thermo), attached to an Edwards pump (EB1M18) capable of achieving a vacuum of 50 torr. The pump is protected by a −105°C glass cold trap (Thermo RVT4104) and a Thermo acid trap (Thermo disposable cartridge SCT120 and housing).
MALDI-TOF-TOF-MS: Ultraflex II MALDI-TOF-TOF (Bruker Daltonics, Billerica, MA). Viable alternatives are available commercially.
NanoLC (Ultimate 3000 nanoLC system; Dionex, Sunnyvale CA) with direct infusion into a QSTAR Elite (Applied Biosystems). Viable alternatives are available commercially.
Nanoelectrospray ion trap MS: Eskigent nanoLC 1D (Dublin, CA) with direct splitless infusion into a LTQ XL (Thermo-Electron, Waltham, MA). Viable alternatives are available commercially.
Liquid chromatograph LC-10 AD with system controller SCL-10A, two pumps FCV- 10 AL and fluorescence detector RF 10 AXL (Shimadzu).
Fraction collector, such as BioRad model 2110.
Lyophilizer with manifold for standard flasks, e.g., Martin Christ GmbH.
Multifunctional microplate reader, excitation/emission: 320/400 nm (such as Infinite M200 monochromator based instrument; Tecan).
Micro-centrifuge, such as Hereaus (Thermo).
Probe sonifier, e.g., model 250 (Branson).
Source of dry N2.
Upright compound microscope equipped with a 20× phase contrast objective (any model).
Hand-held UV illuminator (long wave-length), e.g., Mineralight lamp from Ultra-violet Products.
2.3. Analysis Software
Glycoworkbench 2.1 (free download at www.glycoworkbench.org/).
flexAnalysis (Bruker Daltonics).
Xcalibur (Thermo).
Analyst (ABI/Sciex).
3. Methods (see flow chart in Fig. 1)
3.1. Samples
Cellular slime mold samples, ranging from whole cells to cell fractions to purified proteins, can be analyzed for N-glycans. Potential pitfalls to avoid include contamination from nutrient sources and environment (see Note 5), and artifactual degradation from endogenous enzymes (see Note 6).
3.2. Lipid-linked Oligosaccharide Extraction (20)
Wash 600 mg (wet weight) of cells by centrifugation, at 2000 g for 10 min, and resuspension by gentle pipetting in 5 mL of ice-cold PBS. Repeat the centrifugation. Sonicate the final pellet in 5 mL of MeOH in an 8-mL Sarstedt tube (suggested parameters: duty cycle 30%, output control 2–3 for 2 × 10 sec). Remove the majority of MeOH under a stream of N2.
Add 5 mL of CHCl3:MeOH;2:1 (v/v) at room temperature to the residue and probe sonicate twice until well dispersed. Centrifuge the suspension at 2500 g for 10 min. Discard the supernatant and repeat. Add 2 mL of MeOH followed by another round of sonication and air drying. Next, add 5 mL of water and sonicate the pellet twice with occasional vortexing to ensure complete resuspension. Centrifuge again, discard the supernatant, and repeat. Resuspend the pellet and sonicate with 2 mL MeOH, and remove the MeOH under N2.
Extract the LLOs twice with 5 mL of CHCl3, MeOH and water (10:10:3 [v/v/v]). Sonicate and centrifuge as in step 2. Repeat with an additional 5 mL. Dry the final supernatant (containing LLOs) under N2.
Cleave the oligosaccharides from the dolichol pyrophosphate using 2 mL of 0.1 M HCl in 50% isopropanol, for 1 h at 50°C. Lyophilize and resuspend in 0.2 mL water.
Apply the resuspended sample to a pre-equilibrated 1-mL NPGC column (e.g., ENVI™ Carb), wash with water (5 × 1 mL aliquots), and elute the oligosaccharides with 1 mL of 40% ACN. After lyophilization and dissolving in 10 µL water, analyze the oligosaccharides by MALDI-TOF-MS (see Subheading 3.10).
3.3. Release of N-Glycans (see Note 7)
3.3.1. Release by PNGase F
-
1
Resuspend cell pellet (107 cells) in 200 µL of 25 mM NH4HCO3 (pH 7.8) and lyse by probe sonication until visibly dispersed (sonicator setting of 4, 2–4 s bursts with 5 s pauses for 30–60 s). Snap-freeze at −80°C or process immediately. Delipidated samples from Subheading 3.2, step 3 (pellet) are suitable for analysis. Purified glycoproteins (1–10 µg) should be dissolved in 20 µL of compatible buffer.
-
2
Add ultrapure urea and DTT to final concentrations of 6 M and 10 mM, respectively, and incubate at 37°C for 45 min.
-
3
Alkylate proteins (see Note 8) by adding IAM to 25 mM and incubating at room temperature for 1 h in the darkness. Alkylation is terminated by addition of DTT to a final concentration of 30 mM.
3.3.1.1. Option 1 (see Note 9)
-
4
Dilute urea to 0.8 M with 25 mM NH4HCO3, pH 7.8.
-
5
Add 20 µg of reagent grade trypsin (or at a 1:50 weight ratio) by addition of trypsin (Sigma) stock solution. Digest samples for 18 h at 37°C and terminate by addition of 10 µL of 30% (v/v) acetic acid solution.
-
6
Purify tryptic peptides by C18-SepPak (18). Vacuum filter the sample over the pre-equilibrated SepPak, wash with 3 × 1 mL aliquots of water and elute with 50% ACN, 0.1% TFA. Dry samples by vacuum centrifugation to remove ACN.
-
7
De-N-glycosylate peptides by resuspension in 100 µL of 25 mM NH4HCO3, pH 7.8 containing 25–50 U/µL of PNGase F and incubate at 37°C for 18 h (see Note 10).
3.3.1.2. Option 2 (see Note 9)
-
4
Dilute sample in 0.5% (w/v) SDS in 50 mM sodium phosphate (pH 7.5), boil for 3 min, dilute 5-fold with 1% (v/v) NP40, and treat with 50 U PNGase F/µg protein for 18 h at 37°C.
-
5
Alternatively, dilute alkylated protein to 0.8 M urea with 25 mM NH4HCO3, pH 7.8, and treat with PNGase F.
3.3.2. Release by PNGase A (see Notes 6, 7 and 11)
Resuspend cell pellet (107 cells) in 200 µL of 5% formic acid and lyse by probe sonication as above. Add 50 µg Amresco pepsin (i.e., dilute stock solution to a final concentration of 250 µg/mL; or 1:20 (w/w;enzyme:protein) and incubate at 37°C for 18 h.
Lyophilize to remove formic acid, resuspend in 0.1% TFA, purify on C18-SepPak as in Subheading 3.3.1.1, step 6, and remove ACN under a stream of N2.
Redissolve the sample in PNGase A buffer containing 1–3 µU/µL of PNGase A and incubate at 37°C for 18 h.
3.3.3. Sequential Release by PNGase F and PNGase A (see Note 11)
Suspend axenically grown cells (1–6 g wet weight) in 10 mL of boiling water for 5 min.
After cooling, disperse cells using a tight fitting glass homogenizer or probe sonifier, using sufficient force to generate subcellular particles as determined using phase contrast microscopy.
Add formic acid [up to 5% (v/v)] and 1 mg of pepsin (Sigma). Incubate for 1 day at 37°C and centrifuge to remove insoluble material.
Incubate the supernatant with 10 packed mL of prewashed Dowex-50 for 1 h at 23°C. Pour into a column and reapply the flow-through fraction. Wash the column with 2% (v/v) acetic acid to remove unbound material, and elute glycopeptides with 0.5 M ammonium acetate (pH 6.0). Lyophilize and resuspend in 3 mL of water.
Subject the sample to gel filtration on an 80-mL Sephadex G25 column in 0.5% (v/v) acetic acid. Elute 2 ml fractions and spot 2 µL of each fraction onto a TLC plate and spray with the orcinol reagent. Develop the plate for 5 min at 90°C.
Pool the orcinol positive fractions and lyophilize.
Suspend in 250 µL of water, heat for 5 min at 95°C, bring to a final concentration of 50 mM NH4HCO3 (pH 8.0), add 3 µL PNGase F (Roche), and subject to digestion overnight at 37°C. The final volume should not exceed 500 µL.
Repeat the Dowex-50 chromatography step (step 4), and lyophilize the unbound (free N-glycans, lacking core α1,3-fucose, ready for further purification) and the bound fractions (remaining glycopeptides).
Desalt the glycopeptides on Sephadex G25 (as in step 5). Lyophilize orcinol-positive fractions and dissolve together in 50 mM ammonium acetate, pH 5.0. Incubate with 3 µL of PNGase A (Roche) overnight at 37°C.
Repeat the Dowex-50 chromatography step (step 4). The unbound fraction contains core α1,3-fucosylated glycans which are also ready for further purification below.
3.3.4. Release by Endo-H
Treat samples prepared as for PNGase F digestion (see Subheading 3.3.1.1, step 6) by resuspending glycopeptides in 10 µL of 250 U of Endo H in 50 mM sodium citrate (pH 5.5) at 37°C for 18 h.
3.3.5. Release by Reductive Alkaline Cleavage (see Note 12)
Resuspend washed cells, delipidated samples, or purified proteins in a mixture of 100 µL of 100 mM NaOH and 100 µL of 2 M NaBH4 and incubate at 45°C for 16 h.
Add 10-µL aliquots of 30% acetic acid until evolution of H2 (bubbling) subsides. Pass over a 1-mL Dowex-50 column, followed by 3 ml of 5% acetic acid, and subject the pooled flow-through fractions to vacuum centrifugation.
Remove borates with the addition of 200 µL of MeOH and evaporation under a stream of N2. Repeat this step two more times, or until the remaining residue no longer diminishes in amount.
3.4. N-glycan Recovery
Dilute glycan samples with an equal volume of 0.1% TFA (v/v) and apply to a pre-equilibrated C18-SepPak. Wash twice with 1.5 mL of 0.1% TFA and pool the eluates.
Apply to a Carbograph cartridge and wash with 10 mL of 0.1% TFA.
Elute N-glycans sequentially with 3 mL of 50% ACN, 0.1% TFA in water followed by 0.1% TFA in 80% ACN/water.
Dry by vacuum centrifugation before further derivatization or analysis.
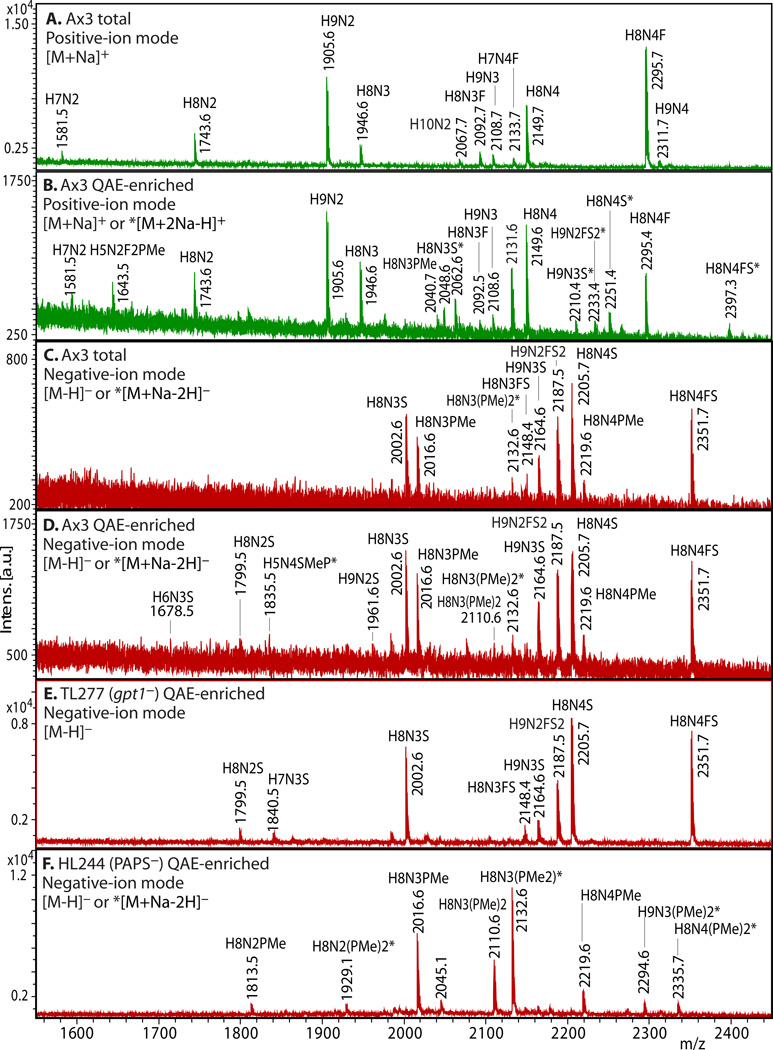
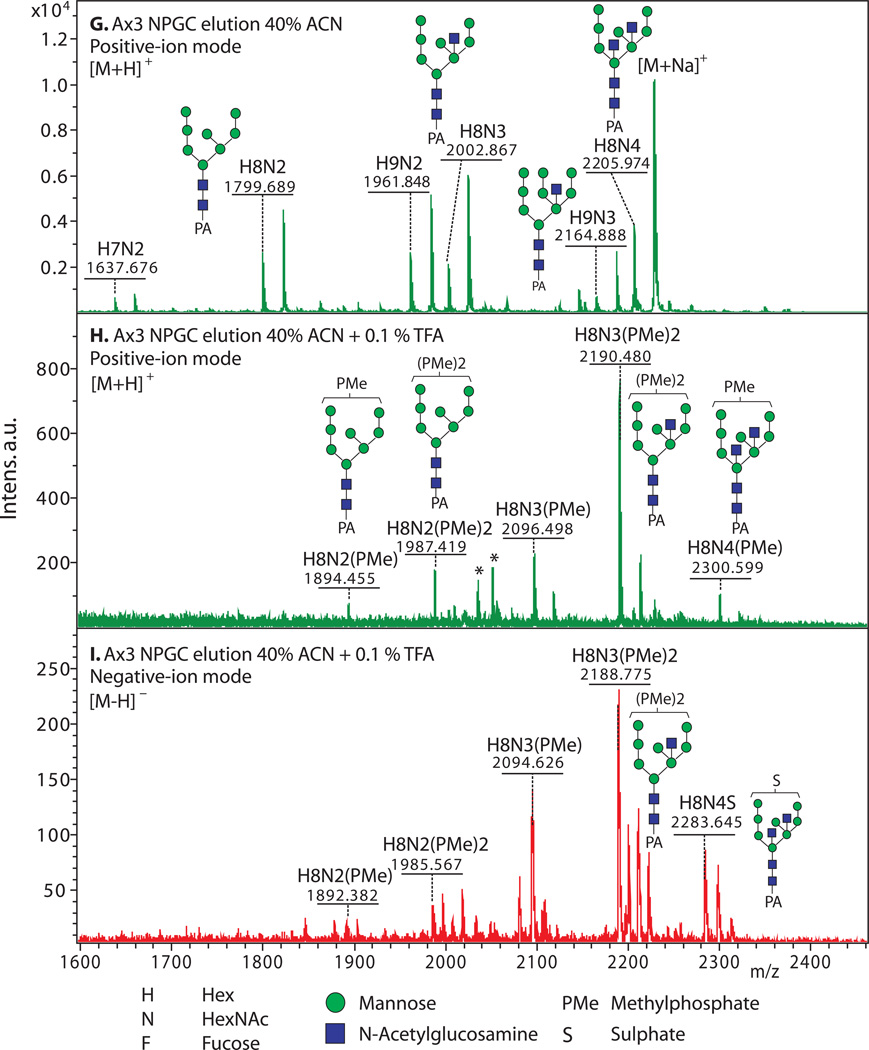
3.5. Enrichment of Anionic N-Glycans (examples shown in Fig. 2)
Figure 2. Enrichment of anionic N-glycans and analysis by MALDI-TOF-MS.
Enrichment based on anionic exchange (A–F) and porous graphitized carbon (G–I) are compared. (A–F) Total N-glycans were released from cells growing logarithmically in HL-5 axenic medium using PNGase A according to Subheading 3.3.2, and recovered according to Subheading 3.4. Anionic glycans were enriched using 70 mM NaCl according to Subheading 3.5.1, and MS-analyzed according to Subheading 3.10 using DHB. The underivatized native glycans were analyzed in standard positive-ion mode (green traces in panels A, B), or negative-ion mode (red traces in panels C-F) to enhance detection of anionic glycans. Glycan compositions (H=Hexose (Man); N=HexNAc or N-acetylhexosamine (GlcNAc); F=fucose(deoxyhexose); S=sulfate; PMe=methylphosphate) are assigned based on mass matching to glycan models (Subheading 3.11), with accuracy of 50 ppm, and confirmed (not shown) by MS/MS, α-mannosidase digestion (Subheading 3.8), and/or detection as their permethylated derivatives (Subheading 3.6.2.2). Structural models are illustrated in G–I. A) A mixture of high-mannose, β-GlcNAc-bisected and/or -intersected, ± core α3-fucosylated neutral species are revealed in the positive ion mode. Glycans ionize in their [M+Na]+ form. B) After anion exchange enrichment, mono-sulfated and mono-methylphosphorylated species are observed in the positive ion mode, in addition to carryover neutral species seen in panel A. Sulfated species appear in their [M+2Na-H]+ form under these conditions. C) Anionic species are selectively detected in negative ion mode analysis, as illustrated in this analysis of total glycans. Most ions occur in their [M–H]− forms, but di-methylphosphorylated glycans appear as their [M+Na–2H]− forms. D) Anion exchange enrichment typically enhances detection of anionic glycans in the negative ion mode, as suggested by evidence for additional low abundance species in this example. E) Similar analysis of strain TL277 (10), which is unable to form the methyl-phosphate substituent owing to the absence of gpt1, confirms assignments of the sulfated species in panel D. F) Similar analysis of strain HL244 (11), unable to form the sulfate substituent owing to a deficiency in PAPS (12), confirms assignments of the mono- and di-methylphosphorylated species. It is not known whether enhanced detection of the methylphosphorylated species is due to increased production in the absence of sulfation, or enhanced sensitivity in the absence of suppression by the sulfated species. (G–I) N-glycans were released using PNGase F from pepsin-glycopeptides from strain Ax3 (unicellular stage) (Subheading 3.3.3, steps 1–5). After adsorption to ENVI™ Carb NPGC material (Subheading 3.5.2), the neutral N-glycans (G) were eluted using ACN, and the acidic N-glycans with TFA/ACN (H, I). After separation, N-glycans were labeled with 2-aminopyridine (PA) (Subheading 3.6.1.2), and the glycans analyzed using MALDI-TOF-MS in positive- (G, H; green) or negative- (I; red) reflectron ion mode. The [M+H]+ ions are annotated with putative compositions and structures based on biosynthetic rules and MS/MS studies (not shown); sodium adducts [M+Na]+ are also present in the spectra. Peaks annotated with (*) are putatively not N-glycans. Further studies are needed to understand the reason for preferential detection of sulfated or methylphosphorylated N-glycans using the two methods, which were conducted on separate samples in different laboratories.
3.5.1. Enrichment by Ion-Exchange
Resuspend samples in 100 µL of 5 mM Trizma base (pH 9.6), check to ensure alkaline pH, and apply to QAE-Sephadex (0.5 mL bed volume prepared in a Bio-Spin column) pre-equilibrated in 1 mM Trizma base.
Wash neutral glycans from the resin with 2 mL 1 mM Trizma base. Elute glycans of increasing negative charge with salt steps of 70 mM NaCl and 140 mM NaCl in 1 mM Trizma base.
Desalt on a Carbograph cartridge as above (Subheading 3.4).
3.5.2. Enrichment by Porous Graphitized Carbon
Dissolve glycans in water and adsorb them to pre-equilibrated NPGC.
Elute neutral N-glycans with 40% ACN, and elute acidic glycans with 0.1 % TFA in 40% ACN. Dry by vacuum centrifugation or lyophilization.
3.6. Derivatization of N-glycans (see Notes 11 and 13)
3.6.1. Reductive Amination of Glycans
Fluorophores are conjugated at the reducing terminus of the glycan, which is uniquely constituted by a reactive carbonyl moiety. Many fluorophores are available including 2-aminobenzamide (2AB) and 2-aminopyridine (2AP or PA) described here.
3.6.1.1. Option 1: 2-aminobenzamide, 2AB (18)
Dry 2 µg of glycans in a 1.5-mL polypropylene microcentrifuge tube.
Add 80 µL of 0.8 M 2AB, 1 M sodium cyanoborohydride, pre-mixed in DMSO:acetic acid;7:3 (v/v) at 80°C for 2 h.
Remove excess 2AB by spotting onto Whatman 3MM chromatography paper. After drying, subject the sample to ascending chromatography by placing in closed jar containing n-butanol:EtOH:water;4:1:1 (v/v) to a depth of several mm.
Elute labeled glycans, which remain at the origin and can be visualized with a hand-held UV illuminator, with 5% acetic acid in water, and dry by vacuum centrifugation.
3.6.1.2. Option 2: 2-Aminopyridine, PA (21, 22)
Dissolve 100 mg of PA in a mixture of 76 µL of concentrated HCl and 152 µL of water.
Transfer 80 µL to the glycan sample dried in a 1.5-mL polypropylene microcentrifuge tube, and incubate in boiling water for 15 min.
Add 4.4 mg of sodium cyanoborohydride to a mixture of 9 µL of the PA solution and 13 µL of water.
Continue the reaction by transferring 4 µL of the cyanoborohydride/PA solution to the sample and incubating overnight at 90°C.
Dilute the sample in 1.5 mL of 0.5% acetic acid, apply to a 50 mL Sephadex G15 column equilibrated in 0.5% acetic acid and collect 2.0 mL fractions. Transfer an aliquot of 80 µL from each fraction into a 96 F black plate and detect fluorescence in a microtiter plate reader. Pool fluorescent glycans depleted of excess labeling reagent and lyophilize.
3.6.2. Permethylation of Glycans (see Note 14)
Two options exist for this procedure. Option 2 may result in improved signal:noise ratio, and permits better fractionation. All procedures are performed at room temperature.
3.6.2.1. Option 1: Solution phase permethylation and liquid/liquid extraction (23)
Prepare a fresh DMSO:NaOH slurry (e.g., 300 µL:100 µg NaOH) in a 1.5-ml polypropylene conical microcentrifuge tube using a glass rod.
Dry the native (or 2AB-labeled) glycan sample into a separate tube. Sequentially add 70 µL of the slurry and 70 µL of CH3I. Vortex continuously for 5 min.
Lightly spin the sample to the bottom, add 300 µL of 200 mM NaCl in water and 300 µL of CHCl3, vortex for 5 min, and spin at 10,000 g × 1 min. Remove the upper aqueous layer with standard pipetteman and discard.
Wash the CHCl3 sample with 300 µL of water, repeating the process three times. Remove the CHCl3 under a stream of dry N2.
3.6.2.2. Option 2: Spin column permethylation and solid-phase extraction (24)
Suspend NaOH in dried ACN (same ratio as in Subheading 3.6.2.1, step 1), and transfer a sufficient quantity to a Micro Spin column to fill to 90% maximal volume. Wash the bed with 100 µL of dry ACN, then 100 µL of dry DMSO.
Redissolve the dried glycan sample in 70 µL of dry DMSO and add 70 µL of CH3I. After continuous vortexing for 5 min, apply the sample to the spin column and allow to percolate by gravity pressure. Reapply the flow-through three times.
Collect sample by centrifuging for 1 min at 800 g, dilute with an equal volume of water, and apply to a C18 SepPak. Push the sample through using an air-filled syringe mounted on an adaptor. Sequentially elute with four 250-µL aliquots of 0.1% TFA in 5% ACN into the same recipient tube.
Successively elute the SepPak in the same way (4 × 250 µL aliquots) with 0.1% TFA in 15%, 35%, 50%, and 80% ACN, into separate tubes. Dry the samples and resuspend in 10 µL of 50% MeOH for MS analysis. N-glycans usually appear in the 35 and 50% fractions.
3.7. HPLC Separation Methods
Pyridylaminated N-glycans can be analyzed by either NP- or RP-HPLC using a HPLC system equipped with a fluorescence detector. Columns are calibrated daily in terms of glucose units, using PA-labeled forms of partial dextran hydrolysates (see Note 15).
3.7.1. Option 1: NP-HPLC
Dissolve dried sample in 50 µL of a 1:3 mixture of buffer A (10 mM ammonium formate, pH 7.0) and buffer B (95% ACN).
Inject sample into a Tosoh-80 column equilibrated in the same 1:3 mixture.
Elute column at 1 mL/min as follows: 0–5 mins, 75% buffer B (from step 1, with the balance made up with buffer A); 5–15 mins, 75–65% B; 15–40 mins, 65% B; 40–55 mins, 65–57% B; followed by a return to the starting conditions.
Detect glycans by fluorescence using excitation at 320 nm and emission at 400 nm. Collect fractions for later analyses.
3.7.2. Option 2: RP-HPLC
Dissolve dried sample in 50 µL of water.
Inject sample onto a pre-equilibrated Hypersil ODS column.
Starting with 100% buffer C, elute at 1.5 mL/min using a linear gradient of 30% (v/v) MeOH in water, increasing at 1% per min. Collect fractions and save for later analyses.
3.7.3. Option 3: 2-D HPLC
First fractionate by NP-HPLC (see Subheading 3.7.1).
Collect fractions, lyophilize and identify fractions of interest by MALDI-TOF-MS (see Subheading 3.10).
Subject desired fractions to RP-HPLC (see Subheading 3.7.2) and analyze by MALDI-TOF-MS (see Subheading 3.10).
3.8. Exoglycosidase Digestions (see Note 16)
Resuspend pmol quantities of 2AB- or PA-labeled glycans in 50% MeOH and spot directly onto MALDI target plates with an equal volume of DHB or ATT matrix solution.
Dry spots in a vacuum desiccator.
Resuspend in 1 µL of 50 mM ammonium acetate (pH 5.0) containing 10 U/mL jack bean β-N-acetylhexosaminidase or α-mannosidase. Incubate plates at 37°C in a humidified box for 2, 4 or 24 h.
3.9. Phosphodiester Bond Cleavage
Incubate dried glycan fractions with 40–48% hydrofluoric acid on ice overnight, and then dry under a stream of dry N2 gas.
3.10. MALDI-TOF and TOF-TOF Analysis
Dry native or derivatized samples completely under vacuum centrifugation and resuspend in 20 µL of 50% (v/v) ACN.
Prepare a method blank identically to experimental samples to differentiate signals from background contamination.
Spot 0.5 µL of the sample onto a polished steel MALDI target plate with an equal volume of DHB MALDI matrix and vacuum dry for co-crystallization. Anionic glycans are routinely screened also using THAP or ATT matrices. Spot a dextran ladder as an external mass calibrant.
Analyze glycans in an Ultraflex II MALDI-TOF-TOF in reflectron positive ion mode with an accelerating voltage of 23 kV, or negative ion mode for anionic glycans. Acquire MALDI spectra at laser frequency of 66 Hz and sum 1000–2000 individual spectra or each sample. Precursor ions are accelerated to 8 kV and selected by a timed ion gate.
For glycan fragmentation, perform TOF-TOF MS/MS experiments by accelerating fragment ions generated by laser-induced dissociation at 19 kV using the incorporated LIFT™ device in Bruker Ultraflex instruments. High energy collision-induced decay is performed using Ar as the collision gas; alternatively, laser-induced dissociation (post source decay) can also be performed.
3.11. Glycan Data Analysis
Analyze MS spectra using flexAnalysis software. Ions are typically singly charged under the MALDI conditions described, and exact monoisotopic m/z values (i.e., the 12C-only ion) should be examined. Select either [M+H]+ or [M+Na]+ ions for peak picking in positive ion mode, or [M-H]− in negative ion mode. For anionic native glycans, variations occur as shown in Fig. 2B–F.
Predict glycan compositions based on mass/charge (m/z) matching with potential glycan compositions using manual calculations or GlycoWorkbench software. Typical parameters are as follows: must match +/− 0.3 daltons and contain Hex (2–15), HexNAc (2–10), deoxyHex (0–4), sulfate (0–3), methylphosphate (0–3). Apply corrections for reducing terminal derivatization or permethylation. Prior studies of Dictyostelium N-glycans indicate that Hex=Man, HexNAc=GlcNAc, and deoxyHex=Fuc (18, 17).
Examples of native and pyridylaminated species in standard positive ion mode are shown in Fig. 2A, G. Other panels show variations on the method that permit detection of anionic glycans, which are more difficult to detect under standard conditions. The different approaches selectively emphasize the abundance of sulfated or methylphosphorylated N-glycans, which illustrates a challenge for sample preparation and MS that has yet to be resolved. See Note 17 for samples that contain a background of poly-Hex species.
Predict structural models based on known biosynthetic rules, and confirm using exoglycosidase digestions (Subheading 3.8), chemical cleavage (Subheading 3.9), and MS/MS studies (Subheading 3.10). These models can be converged with findings from elution times in 2-D HPLC studies (Subheading 3.7).
Estimate relative abundance in permethylated samples by dividing the ion abundance for individual glycans by the total ion abundance of all identified N-glycans.
3.12. Glycopeptide Enrichment (see Note 18)
Generate tryptic glycopeptides as in Subheading 3.3.1, steps 1–5, except use sequencing grade trypsin. Glycopeptides from proteolytic digests can then be enriched by HILIC or by lectin ConA selection.
3.12.1. Option 1: HILIC
Pre-equilibrate polyHYDROXYETHYL A TopTips with loading buffer (0.5% formic acid in 80% ACN).
Resuspend lyophilized peptides in 10 µL of loading buffer, and gently apply to the upper surface of the TopTip. Wash out unbound peptides with 5 column volumes of loading buffer.
Elute glycopeptides with 3 column volumes of 0.5% formic acid and dry in vacuo.
3.12.2. Option 2: ConA
Dissolve peptides in 100 µL of ConA binding buffer and incubate with 200 µL of ConA agarose beads in a 1.5-mL microcentrifuge tube for 1 h at 22°C with rocking.
Transfer the slurry to a Bio-Rad Bio-spin column, and elute non-glycosylated peptides with 10 bed volumes of ConA binding buffer.
Elute glycopeptides with 3 bed volumes of 0.2 M α-methyl mannoside in ConA binding buffer, and desalt on a C18-SepPak as above (see Subheading 3.3.1.1, step 6).
3.13. MS Analysis of Glycopeptides (see Note 19)
3.13.1. Option 1: MALDI-TOF-TOF-MS
Spot the samples with an equal volume of α-cyano-4-hydroxycinnamic acid matrix solution and vacuum dry.
Spot a seven-component peptide mixture as an external mass calibrant.
3.13.2. Option 2: nanoLC-MS/MS
Load 2 µg of total trypsin digest onto a pre-equilibrated PepMap100 C18 column, and elute, at a flow rate of 200 nL/min over a period of 40 min, with a linear gradient of 10–70% of a buffer made of 0.09% formic acid, 0.0085% TFA, 95% ACN, in water.
Directly infuse the effluent via the Dionex nanoLC pump into the QSTAR Elite MS (ion spray voltage: 2500 V, ion source gas: 22 psi, curtain gas: 20 psi), such that the peak width is ~30 s.
Conduct data-dependent MS/MS fragmentation of precursor ions on the top 3 ions with a 250 s exclusion time (survey: 620–3300 m/z with charge 2 to 5 if signal exceeds 10 cts).
Externally calibrate the instrument daily with the peptide mixture.
3.13.3. Option 3: nanoLC-MSn
Load 2 pmol (~2 µL) of a pepsin or trypsin digest (see Note 20) into a pre-equilibrated, self-packed C18 column mounted on the Eskigent nanoLC pump.
Elute peptides with a 0–68% linear gradient of a buffer composed of 2% water, 0.1% formic acid in ACN, over 45 min directly infused into the LTQ XL MS at a rate of 200 nL/min.
Conduct a MS survey scan of the range from 300–1800 m/z every 1–2 s, followed by data-dependent MS/MS fragmentation at 35% collision energy on the top three ions, with a 120 s exclusion time.
3.14. Glycopeptide Data Analysis (see Note 16)
Select glycopeptide elution intervals from an extracted ion chromatogram of the MS/MS spectra based on the presence of glycan oxonium ions at m/z 146 (dHex), 162 (Hex), 203 (HexNAc) or disaccharide combinations. Program the detection using Analyst software for the QSTAR, or Xcalibur software for the LTQ.
Examine regions of the chromatogram containing signature glyconium ions for candidate glycopeptide ions consisting of peptide fragment ions (b- and y-type ions) decorated with glycan types from the glycomic analysis above (Subheadings 3.10 and 3.11).
Expand the search by manually probing the entire LC/MS-MS run for mass matches against the predicted glycopeptides.
Estimate the relative abundance of each glycoform on a single glycosylation site by dividing its ion intensity by the sum of ion intensities for all of the glycoforms within a glycopeptide family over the entire elution window. Confirm by averaging over separate LC/MS-MS experiments. The assumption that each glycoform exhibits equal ionization efficiency is likely valid for neutral glycans.
3.15. N-glycan Site Mapping (see Note 21)
Perform de-N-glycosylation in a 1:1 mixture of H216O and H218O. PNGases convert the glycosylated asparagine residue into an aspartate and incorporate either an 16O or an 18O atom, resulting in a 1 or 3-Da increase in mass of the residue relative to unmodified asparagines.
Detect peptides bearing N-glycosylated sites based on appearance of signal doublets separated by 2 mass units when analyzed by MALDI-TOF or nLC-electrospray MS.
Infer partial site occupancy by presence of unmodified peptides.
Identify peptides in MS/MS fragmentation experiments to sequence the peptides and confirm the mass difference at the N-sequon site.
3.16. Western blotting (see Note 22)
Analyze crude whole cell extracts (25 µg total protein material) or purified glycoproteins of interest by Western blotting after separation by SDS-PAGE and transfer to nitrocellulose membrane using a semi-dry blotting apparatus. Use standard Western blot procedures to probe the membrane with anti-carbohydrate antibody or lectin, such as rabbit anti-HRP for α3-linked core Fuc, or biotin-conjugated wheat germ agglutinin lectin followed by either alkaline phosphatase–conjugated goat anti-rabbit antibody or alkaline phosphatase conjugated anti-biotin antibody, followed by subsequent color detection with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium.
Acknowledgments
Studies in the OUHSC OCMG Core Lab were supported by the OUHSC Dept. of Biochemistry & Molecular Biology, the OUHSC VP Office for Research, and NIH grants R01-GM037539 and R01-GM084383 to C.M.W. This work was also supported by a grant to I.B.H.W. from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF) [grant number P19615].
Footnotes
Hazardous reagent. Observe MSDS sheet for proper use and safety precautions.
Tap the TopTip device gently to ensure that the resin is positioned at the bottom, and remove the cap by twisting. Introduce liquid using a long 10-µL tip, and induce flow by applying positive pressure to the tip using a syringe or pipettor, ensuring that the resin is fully wetted with no visible bubbles.
Jack bean α-mannosidase (Sigma-Aldrich) can be purified, to remove MS-interfering material, by chromatographing 50 µL (310 µg protein, 6.8 Units) on a Superdex 75 column (3.2/30 cm/mm) equilibrated in 10 mM ammonium acetate (pH 7.25), 0.1 mM zinc acetate. The α-mannosidase activity elutes in a single peak (~75 µL total volume).
If high purity matrix is not available, it is advisable to re-crystallize. For example, heat a saturated solution of the matrix in 70% ACN/30% water to the boiling point and allow the solid to dissolve completely (filter to remove remaining particulate if necessary). Cool to room temperature, then on ice. Collect the resulting precipitate by suction filtration on Whatman paper (any grade) and dry.
The methodology is highly sensitive and can detect contaminating glycans from Dictyostelium food sources or residue from previously used plasticware. Cells should be well washed and, though not routinely required, can be allowed to grow on defined medium or to digest food sources by starvation-induced development. It is important to include separate control samples starting from sources containing medium or buffer only.
Avoid N-glycan release in extracts by endogenous Endo S (25), which rapidly cleaves N-glycans between the two core GlcNAc residues- the same position as Endo H. This activity, which can result in appearance of glycans lacking the reducing terminal GlcNAc, is a candidate for inhibition by NAG-thiazoline (26).
PNGase F is the standard N-glycosidase for releasing N-glycans, but does not release N-glycans modified by Fuc α3-linked to the core GlcNAc. PNGase A has broader specificity to include core α3-fucosylated N-glycans, but requires small peptides for optimal activity and is more expensive. Endo Hf is inhibited by sulfation (12) and bisecting αGlcNAc (27). The latter has not been reported in D. discoideum.
Alkylation of Cys-residues is desirable to prevent formation of disulfide bonds which can interfere with accessibility of enzymes to N-glycans.
Enzymatic deglycosylation is most effective after proteolysis, especially for PNGase A. However, satisfactory results may be obtained with PNGase F when intact proteins are denatured by SDS or urea, if SDS is first diluted in the presence of NP40, or urea is sufficiently diluted.
In event of failure, confirm proper pH by dispensing 1 µL on a strip of appropriate pH indicator paper.
Different procedures were developed for historical reasons and are presented separately.
This chemical release method is typically reserved for release of O-glycans but also releases N-glycans. Higher concentrations of NaOH have been reported but may result in enhanced peeling (degradation at the reducing terminus). Note that reduction of the reducing terminus precludes future fluorescence derivatization. Hydrazinolysis is an alternative chemical release method (6).
It is simplest to analyze released N-glycans directly by MS. However, derivatization of the reducing terminus with a fluorophore improves MS sensitivity, and allows for detection of glycans by RP or NP chromatography for identification based on co-chromatography with known standards (28). Either form can be analyzed after exoglycosidase digestion. PA and 2AB are commonly used fluorophores.
Derivatization by permethylation attaches a methyl group to all free hydroxyls including the reducing terminus, and offers several advantages: improvement of sensitivity, removal of contaminants, and stabilization that inhibits glycoside isomerization due to branch migration. In addition, the derivatization makes various glycans more chemically similar which permits reasonable quantitation of relative amounts based on ion current values. If m/z values obtained after permethylation yield compositions that match compositions inferred from analysis of native or 2AB/PA-conjugated glycans, this usually constitutes proof of identity even in the absence of additional information such as MS-MS or glycosidase digestions.
Calibrated HPLC separations can allow structure prediction based on elution times compared to known standards, because compositional isoforms may be differentially retained on select stationary phases.
Similar results are obtained when enzyme reactions are conducted in microtubes in the absence of matrix. Although jack bean α-mannosidase is considered non-specific with respect to the linkage position on the underlying sugar, reaction rates vary considerably and thus times need to be extended to achieve removal of sterically constrained linkages. Other exoglycosidases useful for Dictyostelium N-glycans include α-glucosidase and α1,2-mannosidase from Aspergillus saitoi.
Contamination by a Hexn series can reportedly be removed by addition of rice α-glucosidase (Sigma). Alternatively, samples (e.g., glycopeptides) can be pre-reduced with 50 µL of 1% (w/v) sodium borohydride at room temperature for 2 h followed by addition of 2 µL of glacial acetic acid and lyophilization. The reduced material will then be inert to derivatization by reductive amination (29, 30).
Glycopeptides are difficult to detect in mixtures with peptides owing to ion suppression. Therefore it is advantageous to enrich for glycopeptides prior to analysis to reduce levels of competing peptides. ConA is specific for high mannose N-glycans typical for Dictyostelium, and HILIC, while less selective, captures a broader range of N-glycans. Other lectins or antibodies might be used for selective enrichment of glycan subsets.
MALDI-TOF-MS instruments have high resolution and are relatively easy to set up and operate. However, suppression of glycopeptide signals by non-glycosylated peptides often requires further purification such as can be conveniently achieved by RP-HPLC mated directly to an electrospray input as in the QSTAR Elite and LTQ XL instruments. The QSTAR Elite MS consists of a quadrupole front end for ion separation, an intermediate collision chamber for fragmentation, and a TOF stage for detection. This design has high resolution (5–10 ppm) and a relatively wide mass range. The LTQ XL (linear ion trap) MS has the ability to perform MSn experiments and has greater sensitivity, but lower mass resolution.
Trypsin is the standard enzyme for proteolysis because it is robust, and the cleavage on the C-terminal side of basic residues results in peptides with two positively charged sites (including the N-terminus) which tends to ionize well in positive ion mode. However, if tryptic peptides are too large for downstream applications chymotrypsin can be included for additional cleavages after select hydrophobic residues. Pepsin also results in smaller peptides and its use at low pH can be desirable to inhibit other enzyme activities. Finally, proteins that are difficult to trypsinize might be sensitive to treatment with Endo Lys-C (Wako) dissolved in 200 mM Tris-HCl (pH 9.2), 2 M urea.
N-glycosylation sites can be inferred by detection of peptides bearing the N-sequon (Nx[≠P]S/T) based on loss of a single mass unit owing to conversion of Asn to Asp during enzymatic de-N-glycosylation. However, inclusion of a 1:1 mixture of H216O and H218O avoids false positive detection owing to spontaneous deamidation, and facilitates scanning of complex spectra owing to the characteristic m/z-doublet formed. An alternative method is to de-N-glycosylate using Endo H, which leaves the core GlcNAc providing a 203 m/z mass tag.
Antibodies and lectins can help differentiate glycoforms, such as core α3- vs. α6-linked Fuc when using anti-HRP, at the glycoprotein level. The Western blotting method is readily adapted for any lectin or antibody, and any enzyme-linked or fluorescence detection method. Concentrations should be optimized according to the reagents used by comparison with appropriate negative and positive controls.
Contributor Information
Christa L. Feasley, Email: christa-feasley@ouhsc.edu.
Alba Hykollari, Email: alba.hykollari@boku.ac.at.
Katharina Paschinger, Email: katharina.paschinger@boku.ac.at.
Iain B. H. Wilson, Email: iain.wilson@boku.ac.at.
Christopher M. West, Email: Cwest2@ouhsc.edu.
References
- 1.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivatt RL, Das OP, Henderson EJ, Robbins PW. Glycoprotein biosynthesis in Dictyostelium discoideum: developmental regulation of the protein-linked glycans. Cell. 1984;38:561–567. doi: 10.1016/0092-8674(84)90510-5. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West CM, Erdos GW, Davis R. Glycoantigen expression is regulated both temporally and spatially during development in the cellular slime molds Dictyostelium discoideum and D. mucoroides. Mol Cell Biochem. 1986;72:121–140. doi: 10.1007/BF00230640. [DOI] [PubMed] [Google Scholar]
- 5.Couso R, van Halbeek H, Reinhold V, Kornfeld S. The high mannose oligosaccharides of Dictyostelium discoideum glycoproteins contain a novel intersecting N-acetylglucosamine residue. J Biol Chem. 1987;262:4521–4527. [PubMed] [Google Scholar]
- 6.Amatayakul-Chantler S, Ferguson MAJ, Dwek RA, Rademacher TW, Parekh RB, Crandall IE, Newell PC. Cell surface oligosaccharides in Dictyostelium during development. J Cell Sci. 1991;99:485–495. [Google Scholar]
- 7.Nakagawa M, Tojo H, Fujii S. A glycan of Psi-factor from Dictyostelium discoideum contains a bisecting-GlcNAc, an intersecting-GlcNAc, and a core α-1,6-fucose. Biosci Biotechnol Biochem. 2011;75:1964–1970. doi: 10.1271/bbb.110369. [DOI] [PubMed] [Google Scholar]
- 8.Freeze HH. Dictyostelium discoideum glycoproteins: using a model system for organismic glycobiology. In: Montreuil J, Vliegenthart JFG, Schachter H, editors. Glycoproteins II. Elsevier Science BV; 1997. pp. 89–121. [Google Scholar]
- 9.Srikrishna G, Wang L, Freeze HH. Fucoseβ-1-P-Ser is a new type of glycosylation: using antibodies to identify a novel structure in Dictyostelium discoideum and study multiple types of fucosylation during growth and development. Glycobiology. 1998;8:799–811. doi: 10.1093/glycob/8.8.799. [DOI] [PubMed] [Google Scholar]
- 10.Qian Y, West CM, Kornfeld S. UDP-GlcNAc:Glycoprotein N-acetylglucosamine-1-phosphotransferase mediates the initial step in the formation of the methylphosphomannosyl residues on the high mannose oligosaccharides of Dictyostelium discoideum glycoproteins. Biochem Biophys Res Commun. 2011;393:678–681. doi: 10.1016/j.bbrc.2010.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knecht DA, Dimond RL, Wheeler S, Loomis WF. Antigenic determinants shared by lysosomal proteins of Dictyostelium discoideum Characterization using monoclonal antibodies and isolation of mutations affecting the determinant. J Biol Chem. 1984;259:10633–10640. [PubMed] [Google Scholar]
- 12.Lacoste CH, Freeze HH, Jones JA, Kaplan A. Characteristics of the sulfation of N-linked oligosaccharides in vesicles from Dictyostelium discoideum: in vitro sulfation of lysosomal enzymes. Arch Biochem Biophys. 1989;273:505–515. doi: 10.1016/0003-9861(89)90510-9. [DOI] [PubMed] [Google Scholar]
- 13.Freeze HH, Hindsgaul O, Ichikawa M. A novel pathway for phosphorylated oligosaccharide biosynthesis. Identification of an oligosaccharide-specific phosphate methyltransferase in Dictyostelium discoideum. J Biol Chem. 1992;267:4431–4439. [PubMed] [Google Scholar]
- 14.Sharkey DJ, Kornfeld R. Developmental regulation of processing α-mannosidases and ‘intersecting’ N-acetylglucosaminyltransferase in Dictyostelium discoideum. J Biol Chem. 1991;266:18477–18484. [PubMed] [Google Scholar]
- 15.West CM, van der Wel H, Coutinho PM, Henrissat B. Glycosyltransferase genomics in Dictyostelium discoideum. In: Loomis WF, Kuspa A, editors. Dictyostelium Genomics. Norfolk, UK: Horizon Scientific Press; 2005. pp. 235–264. [Google Scholar]
- 16.Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, Lindquist E, Shapiro H, Bruce D, Schmutz J, Salamov A, Fey P, Gaudet P, Anjard C, Babu MM, Basu S, Bushmanova Y, van der Wel H, Katoh-Kurasawa M, Dinh C, Coutinho PM, Saito T, Elias M, Schaap P, Kay RR, Henrissat B, Eichinger L, Rivero F, Putnam NH, West CM, Loomis WF, Chisholm RL, Shaulsky G, Strassmann JE, Queller DC, Kuspa A, Grigoriev IV. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 2011;12:R20. doi: 10.1186/gb-2011-12-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiller B, Hykollari A, Voglmeir J, Pöltl G, Hummel K, Razzazi-Fazeli E, Geyer R, Wilson IBH. Development of Dictyostelium discoideum is associated with alteration of fucosylated N-glycan structures. Biochem J. 2009;423:41–52. doi: 10.1042/BJ20090786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feasley CL, Johnson JM, West CM, Chia CP. Glycopeptidome of a heavily N-glycosylated cell surface glycoprotein of Dictyostelium implicated in cell adhesion. J Proteome Res. 2010;9:3495–3510. doi: 10.1021/pr901195c. [DOI] [PubMed] [Google Scholar]
- 19.Swanson SK, Florens L, Washburn MP. Generation and analysis of multidimensional protein identification technology datasets. Methods Mol Biol. 2009;492:1–20. doi: 10.1007/978-1-59745-493-3_1. [DOI] [PubMed] [Google Scholar]
- 20.Gao N. Fluorophore-assisted carbohydrate electrophoresis: a sensitive and accurate method for the direct analysis of dolichol pyrophosphate-linked oligosaccharides in cell cultures and tissues. Methods. 2005;35:323–327. doi: 10.1016/j.ymeth.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Hase ST, Ibuki T, Ikenaka T. Reexamination of the pyridylamination used for fluorescence labeling of oligosaccharides and its application to glycoproteins. J Biochem. 1984;95:197–203. doi: 10.1093/oxfordjournals.jbchem.a134585. [DOI] [PubMed] [Google Scholar]
- 22.Paschinger K, Hykollari A, Razzazi-Fazeli E, Greenwell P, Leitsch D, Walochnik J, Wilson IBH. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology. 2012;22:300–313. doi: 10.1093/glycob/cwr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciucanu I, Costello CE. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J Am Chem Soc. 2003;125:16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 24.Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- 25.Freeze HH, Etchison JR. Presence of a nonlysosomal endo-β-N-acetylglucosaminidase in the cellular slime mold Dictyostelium discoideum. Arch Biochem Biophys. 1984;232:414–421. doi: 10.1016/0003-9861(84)90557-5. [DOI] [PubMed] [Google Scholar]
- 26.Abbott DW, Macauley MS, Vocadlo DJ, Boraston AB. Streptococcus pneumoniae endohexosaminidase D, structural and mechanistic insight into substrate-assisted catalysis in family 85 glycoside hydrolases. J Biol Chem. 2009;284:11676–11689. doi: 10.1074/jbc.M809663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buser R, Lazar Z, Käser S, Künzler M, Aebi M. Identification, characterization, and biosynthesis of a novel N-glycan modification in the fruiting body of the basidiomycete Coprinopsis cinerea. J Biol Chem. 2010;285:10715–10723. doi: 10.1074/jbc.M109.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomiya N, Awaya J, Kurono M, Endo S, Arata Y, Takahashi N. Anal Biochem. 1988;171:73–90. doi: 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- 29.Paschinger K, Razzazi-Fazeli E, Furukawa K, Wilson IBH. Presence of galactosylated core fucose on N-glycans in the planaria Dugesia japonica. J Mass Spectrom. 2011;46:561–567. doi: 10.1002/jms.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pöltl G, Kerner D, Paschinger K, Wilson IBH. N-glycans of the porcine nematode parasite Ascaris suum are modified with phosphorylcholine and core fucose residues. FEBS J. 2007;274:714–726. doi: 10.1111/j.1742-4658.2006.05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]