Summary
Languishing antibiotic discovery and flourishing antibiotic resistance have prompted development of alternative untapped sources for antibiotic discovery, including previously uncultured bacteria. Here, we screen extracts from uncultured species against M. tuberculosis and identify lassomycin, an antibiotic that exhibits potent bactericidal activity against both growing and dormant mycobacteria, including drug-resistant forms of M. tuberculosis, but little activity against other bacteria or mammalian cells. Lassomycin is a highly basic, ribosomally-encoded cyclic peptide with an unusual structural fold that only partially resembles that of other lasso peptides. We show that lassomycin binds to a highly acidic region of the ClpC1 ATPase complex and markedly stimulates its ATPase activity without stimulating ClpP1P2 catalyzed protein breakdown, which is essential for viability of mycobacteria. This mechanism, uncoupling ATPase from proteolytic activity, accounts for lassomycin's bacteriocidal activity.
Keywords: Drug discovery, Mycobacterium tuberculosis, Lassomycin, Species specific, Lasso peptide
INTRODUCTION
The scarcity of suitable lead compounds is now the major bottleneck in the development of novel antimicrobial drugs (Lewis, 2013; Payne et al., 2007). In the absence of new therapeutic agents, the rise and spread of multidrug resistant pathogens will continue unchecked. Most antibiotics in use today resulted from screening of soil actinomycetes for active compounds. However, repeated exploitation of this limited resource has led to diminishing returns, and most current efforts result in rediscovering known compounds (Lewis, 2012). Consequently, there has been a general elimination of natural product-based discovery in most pharmaceutical companies. Using new microbial sources of compounds could reduce the problem of rediscovery and lead to novel antimicrobials. Uncultured species of bacteria account for 99% of all microbial diversity and by definition represent an unexploited source of secondary metabolites (Staley and Konopka, 1985). We have developed general methods to grow previously uncultured bacteria, based on cultivation in diffusion chambers in their natural environments (Kaeberlein et al., 2002) and on prolonged incubation in vitro (Buerger et al., 2012). This approach results in cultivation of up to 40% of cells from environmental samples. However, even with this new source, most of the effort is expended on rediscovery of known compounds or generally toxic ones. We reasoned that this problem could be addressed by a species-selective approach, whereby compounds with broad antibiotic spectra are eliminated, and only compounds active against a specific species are considered. We chose Mycobacterium tuberculosis as a target organism for this approach, since few natural products are known to act specifically against this pathogen, and therefore most of the specific hits obtained should be new agents. There is also a considerable medical need for novel anti-TB compounds (Sacchettini et al., 2008; Zumla et al., 2013) to stem the spread of extremely- and totally drug-resistant strains of the pathogen.
We screened extracts from a collection of soil bacteria, obtained by in situ cultivation and by prolonged incubation, against M. tuberculosis and counterscreened against S. aureus. A novel antimicrobial, lassomycin, was purified from an extract of Lentzea kentuckyensis sp. Lassomycin is a potent bactericidal compound that we show targets the ClpC1 ATPase, an essential enzyme in mycobacteria, which normally functions in protein degradation together with the ClpP1P2 proteolytic complex (Akopian et al., 2012). This agent is a highly basic lasso peptide antibiotic that is encoded in the genome and is unusual in its specificity for mycobacteria and its mode of action.
RESULTS AND DISCUSSION
Isolation of lassomycin
A library of extracts from soil actinomycetes was screened against M. tuberculosis. To shorten the duration of screens, we constructed a strain constitutively expressing mCherry, and used bacterial fluorescence as the readout. This method allowed for reliable detection of growth inhibition in five days. The screen had a hit rate of 10% against M. tuberculosis. A counterscreen against S. aureus had a hit rate of 30%, and the hit rate for extracts specifically acting against M. tuberculosis was 2%. One of the first extracts identified that acted specifically against M. tuberculosis was from isolate IS009804, a Lentzea kentuckyensis sp. (99.7% identical to L. kentuckyensis, accession number: DQ291145, by 16S rDNA). The extract was fractionated by HPLC, and a single active fraction was identified by bioassay-guided purification. This fraction was lyophilized, leaving a white powder. Analysis of this fraction by LC-MS indicated that a single major compound was present ([M+H]+ = 1880).
Structural elucidation of lassomycin
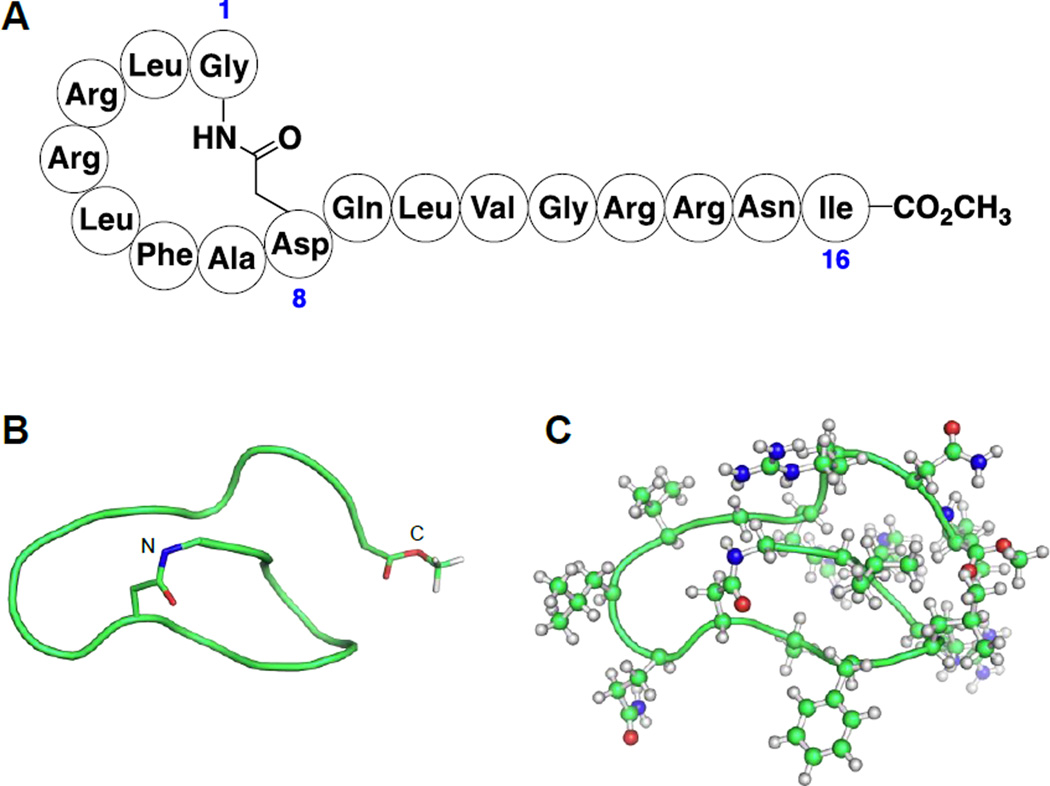
Preliminary NMR studies indicated that the active compound was a peptide, and further analysis revealed an Asp-Gln-Leu-Val-Gly pentapeptide sequence. Elucidation of the entire structure proved to be quite challenging, and multiple approaches were employed. The producing strain was cultured in a medium supplemented with D-glucose-U-13C6, CELTONE base powder-U-13C, U-15N and l-proline-U-13C5, 15N (Cambridge Isotope Laboratories, USA) to produce a uniformly labeled compound for further analysis by three-dimensional NMR techniques; the pentapeptide sequence was used as a search fragment in the producing strain’s genome to identify the biosynthetic genes; and MS/MS was employed to experimentally identify the peptide’s sequence. These combined approaches revealed that the active compound, which we have named lassomycin, consists of 16 amino acids in which the N-terminal residues form an 8-residue ring through formation of an amide bond between the N-terminal amine and the side chain carboxyl group of Asp8. The overall structure resembles a lasso in which the 8-residue ring forms the loop, and residues 9–16 form the spoke. In addition, the C-terminal carboxyl is converted to a methyl ester (Fig. 1A). Acid hydrolysis of lassomycin, followed by derivatization with Marfey’s reagent, and LC/MS analysis established that all of the residues are l-amino acids.
Figure 1.
(A) The amino acid sequence and post-translational modifications of lassomycin. Blue numbering indicates the positions of residues 1, 8 and 16. (B) The backbone structure of lassomycin. The N- and C-termini are labeled. (C) The structure of lassomycin with its side chains shown.
The three-dimensional solution structure of lassomycin was deduced from the NOE distance restraints obtained from three-dimensional NMR data using CYANA 2.1 (Fig. 1B). Surprisingly, the solution structure of lassomycin lacks the characteristic knot structure reported for other homologous lasso peptides like lariatin A and microcin J25 (Arnison et al., 2013) as the C-terminal end packs tightly against the N-terminal ring instead of passing through the macrolactam (Fig. 1B, Fig. S1, Table S1). Lassomycin has four positively charged arginine side chains and no negatively charged groups as the terminal carboxyl is esterified.
Biosynthetic gene cluster of lassomycin
The resulting structure was consistent with the putative biosynthetic genes identified in the producing strain’s genome. The structural gene itself shows highest homology by BLAST to larA in the larABCDE operon which codes for lariatin A. This antibiotic is a prototypical member of the lasso peptide family (Fig. 1A), which is produced by Rhodococcus jostii and inhibits cell wall biosynthesis (Iwatsuki et al., 2006). Lasso peptides consist of 16–21 amino acids and feature a macrolactam ring formed from a connection between the N-terminus and a sidechain carboxyl group. Lasso peptides are produced by both actinobacteria (Streptomyces, Rhodococcus) and proteobacteria (Escherichia, Burkholderia). Lariatin A is an 18-amino acid peptide with an 8-residue ring that is formed between the N-terminal glycine amino group and a glutamic acid side chain at position 8. The larA gene encodes a precursor peptide that is believed to be cleaved by LarD and enzymatically converted to the mature lasso structure by LarB to produce the active peptide. LarE exports the mature peptide. LarC’s function is unknown, but it is necessary for antibiotic activity (Inokoshi et al., 2012). The lariatin A precursor peptide shares only 53% homology with the predicted lassomycin precursor, LasA. In contrast, four other genes in the putative lassomycin operon (Fig. 2), lasB, lasC, lasD and lasE, share high (84–97%) homology with the corresponding genes in the lariatin operon. The last gene product of the lassomycin biosynthetic cluster, LasF, is a putative O-methyltransferase that is likely responsible for the formation of the methyl ester at the C-terminus of the mature peptide.
Figure 2.

The putative lassomycin biosynthetic operon.
Lassomycin bioactivity
Lassomycin had a minimum inhibitory concentration (MIC) of 0.8–3 μg/ml, fairly potent for a peptide, against a variety of M. tuberculosis strains, including MDR (multidrug resistant) and XDR (extremely drug resistant) isolates (Table 1).
Table 1.
Lassomycin spectrum of activity.
| Strains | MIC, μg/ml | MIC, μM |
|---|---|---|
| M. tuberculosis | ||
| H37Rv | 0.78–1.56 | 0.41–0.83 |
| 186, susceptible clinical isolate | 1.56 | 0.83 |
| 83, susceptible clinical isolate | 1.56–3.1 | 0.83–1.65 |
| 84, resistant to INH, STR | 1.56–3.1 | 0.83–1.65 |
| 85, resistant to INH, RIF | 1.56–3.1 | 0.83–1.65 |
| 7, resistant to INH, RIF | 1.56 | 0.83 |
| 86, resistant to INH, RIF, STR | 1.56 | 0.83 |
| 136, resistant to INH, RIF, STR, FQ | 0.78 | 0.41 |
| 133, resistant to INH, RIF, STR, FQ | 0.78 | 0.41 |
| 189, resistant to INH, RIF, STR, FQ | 3.1 | 1.65 |
| 3, resistant to INH, RIF, EMB, FQ | 3.1 | 1.65 |
| 30, resistant to INH, RIF, EMB, PZA, FQ | 0.78 | 0.41 |
| 181, resistant to INH, RIF, EMB, PZA, FQ | 0.78 | 0.41 |
| 183, resistant to INH, RIF, STR, EMB, PZA, FQ | 3.1 | 1.65 |
| 188, resistant to INH, RIF, STR, EMB, PZA, FQ | 3.1 | 1.65 |
| mc26020 | 0.39 – 0.78 | 0.21–0.41 |
| Other Mycobacteria | ||
| M. avium subsp. Paratuberculosis | 0.125 – 0.25 | 0.07–0.13 |
| M. smegmatis | 0.78 – 2 | 0.41–1.06 |
| Other Actinobacteria | ||
| Propionibacterium acnes | 12.5 – 25 | 6.7–13 |
| Bifidobacterium longum | 25 – 50 | 13–27 |
| Other Gram-positive bacteria | ||
| Clostridium difficile | >50 | >27 |
| Clostridium perfringens | >50 | >27 |
| Lactobacillus reuteri | >50 | >27 |
| Lactobacillus casei | >50 | >27 |
| Streptococcus mutans | >50 | >27 |
| Enterococcus faecalis | >50 | >27 |
| Enterococcus faecalis VRE | >50 | >27 |
| Bacillus anthracis Sterne | >50 | >27 |
| Staphylococcus aureus | >50 | >27 |
| Gram-negative bacteria | ||
| Bacteroides fragilis | >50 | >27 |
| Escherichia coli K12 | >50 | >27 |
| Klebsiella pneumoniae | >50 | >27 |
INH, isoniazid; RIF, rifampicin; STR, streptomycin; EMB, ethambutol; PZA, pyrazinamide; FQ, resistant to at least one fluoroquinolone.
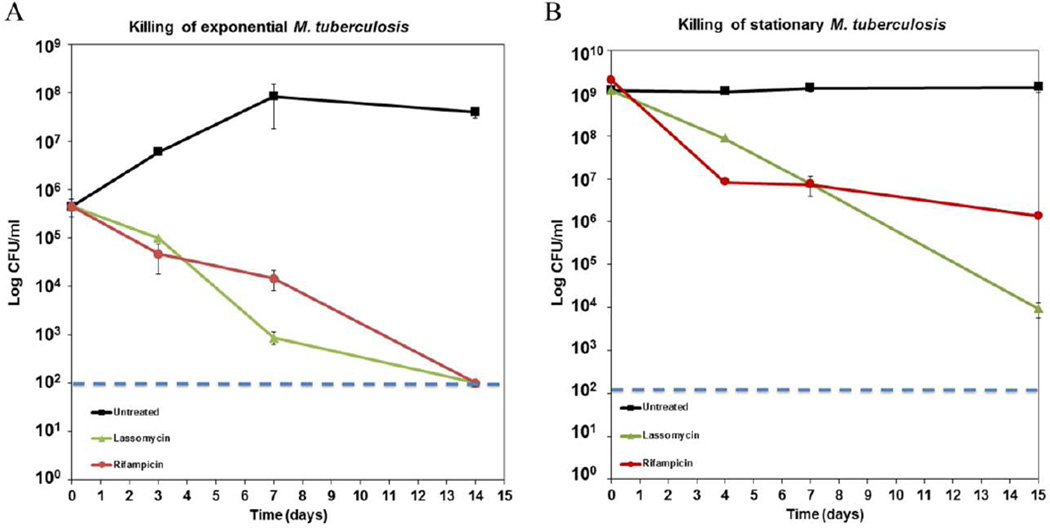
Lassomycin was discovered in a screen designed to identify compounds acting specifically against M. tuberculosis. We therefore tested the compound against a panel of diverse bacterial species (Table 1). Lassomycin was highly active against M. tuberculosis; Mycobacterium avium subsp. paratuberculosis that is a gastrointestinal pathogen of cattle and a suspected pathogen in Crohn’s disease; and Mycobacterium smegmatis, a soil microorganism. The compound was less active against other actinobacteria tested, and had no activity against other microorganisms tested (Table 1). Importantly, lassomycin was inactive against symbionts of the human microbiota that are suppressed by conventional non-specific antibiotics (Dethlefsen and Relman, 2011). The lassomycin MIC was unchanged in the presence of 10% fetal bovine serum, indicating resistance to serum proteases and the lack of significant protein binding. The compound caused no lysis of erythrocytes and had low cytotoxicity (IC50, 350 μg/ml) against human NIH 3T3 and HepG2 cells, perhaps due to poor penetration into mammalian cells. Lassomycin has a minimum bactericidal concentration (MBC) of 1–4 μg/ml against M. tuberculosis and M. avium subsp. paratuberculosis. Lassomycin showed excellent killing activity in a time-dependent assay against exponentially growing cells of M. tuberculosis (Fig. 3A). Thus, its potency against Mtb is similar to that of the best existing bactericidal agent, rifampicin (Fig. 3A). Stationary phase cells of M. tuberculosis are highly tolerant to most antibiotics, for example, rifampicin showed a characteristic biphasic killing (Fig. 3B) and a significant number of persister cells survive exposure (Keren et al., 2011). By contrast, lassomycin had greater killing activity against stationary M. tuberculosis than rifampicin without an obvious presence of surviving persisters.
Figure 3.
Time-dependent killing of M. tuberculosis mc26020 by lassomycin. All drugs were administered at 10 ×MIC. Each point represents the average of three biological replicates. Rifampicin (red circles), lassomycin (green triangles), or untreated (black squares) of exponential (A) and stationary (B) M. tuberculosis. Dashed blue line indicates the limit of detection. The error bars represent standard deviation.
Target identification and mechanism of action of lassomycin
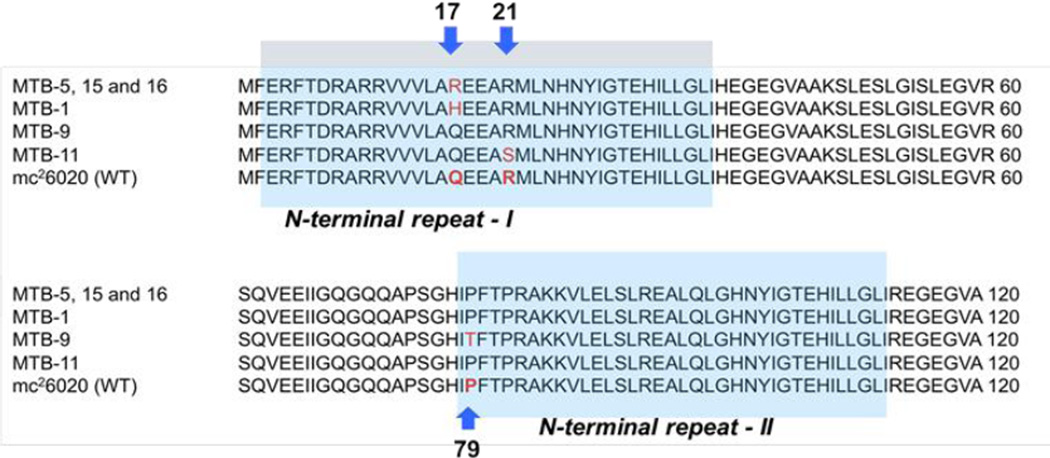
In order to determine the target of lassomycin, resistant mutants of M. tuberculosis were obtained by selecting colonies on nutrient agar plates containing the compound. Mutants resistant to 16 μg/ml of lassomycin were obtained at a frequency of 3×10−7. Genome sequencing of six resistant mutants (Fig. 4) showed mutations in the clpC1 gene, which encodes the subunit of the hexameric ATPase complex, ClpC1. It functions in protein degradation with the two-ring protease complex, ClpP1P2, in mycobacteria (Akopian et al., 2012; Raju et al., 2012). Together, they form the large (26-subunit) ATP-dependent protease complex, ClpC16P17P27C16, in which the ClpC1 ATPase binds protein substrates, unfolds them, and translocates them into the ClpP1P2 central chamber for proteolysis (Akopian et al., 2012).
Figure 4.
Sequences of the two N-terminal repeat regions of ClpC1 mutants resistant to lassomycin. Amino acid changes are indicated in red.
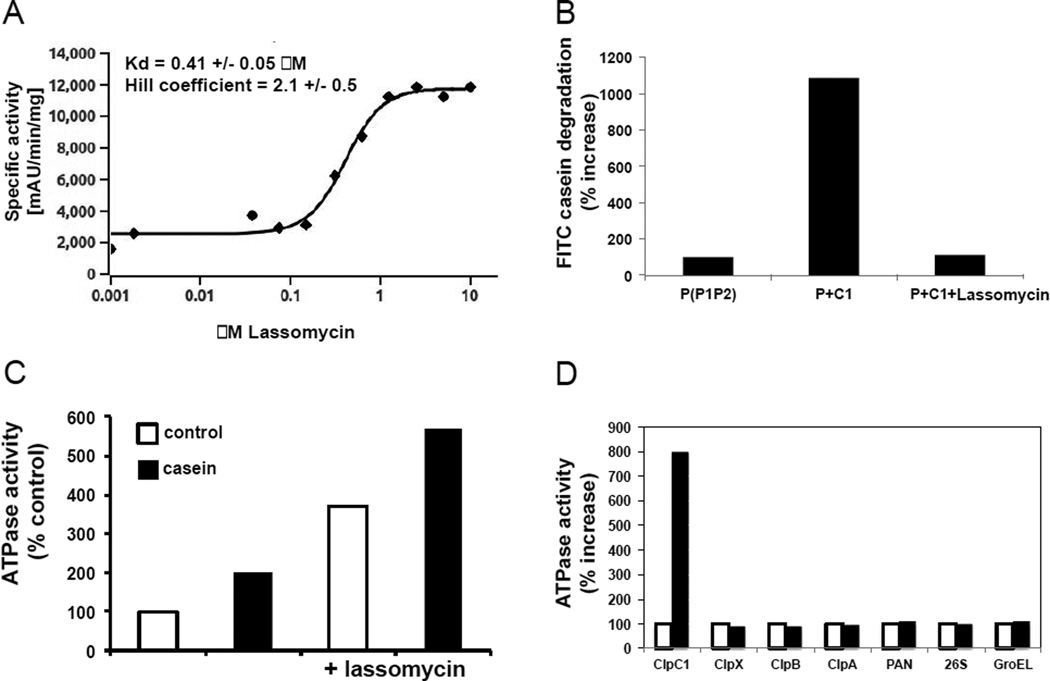
Because all the resistant mutants were mapped to the clpC1 gene, we tested directly if lassomycin altered either of ClpC1’s enzymatic activities: its ability to hydrolyze ATP and to support ATP-dependent protein breakdown by ClpP1P2 (Akopian et al., 2012; Raju et al., 2012), a novel member of the ClpP family of compartmentalized proteases (Lupas et al., 1997; Yu and Houry, 2007). The cloned His6-tagged M. tuberculosis ClpC1 was expressed in M. smegmatis and purified to near-homogeneity, as described previously (Akopian et al., 2012). As expected, ClpC1 exhibited ATPase activity and stimulated the ATP-dependent degradation of the model protein substrate, casein, by the pure ClpP1P2 protease complex, prepared as we recently described (Akopian et al., 2012). Thus, ClpC1 catalyzes casein translocation into the proteolytic compartment formed by ClpP1P2. Surprisingly, in the presence of low concentrations of lassomycin, ATP hydrolysis by ClpC1 increased up to 7–10 fold. This effect of lassomycin on ATP hydrolysis was highly cooperative and showed a Hill coefficient of 2 (Fig. 5A). This strong activation by lassomycin showed an apparent Kd of 0.41 μM, which resembles the value of its MIC against M. tuberculosis cells. Since killing seems to result from binding to ClpC1 (see below), there is probably little or no barrier to lassomycin’s entry into the bacteria.
Figure 5. Lassomycin effect on ClpC1 ATPase activity and protein degradation by ClpC1P1P2 complex.
A) Lassomycin stimulates ATPase activity of ClpC1. 32 nM of pure ClpC1 were mixed with 100 μl of the assay buffer (50 mM TrisHCl pH 7.8; 100 mM KCl; 10% glycerol; 1 mM phosphoenolpyruvate; 1 mM NADH; 2 units of pyruvate kinase/lactic dehydrogenase (Sigma); 4 mM MgCl2 and 1 mM ATP) and the ATPase activity of ClpC1 was followed by measuring the coupled oxidation of NADH to NAD+ spectrometrically at 340 nm. Similar results were obtained when the ATPase activity was measured with the Malachite Green method (Geladopoulos et al., 1991). The rate of ATPase activity in the absence of lassomycin was taken as 100%. The apparent Kd and Hill coefficient for lassomycin activation of ClpC1 ATPase were determined using curve fitting with classic Hill-kinetic through a Scaled Levenberg-Marquardt algorithm; tolerance 0.0001. B) ClpC1 does not activate degradation of casein by ClpP1P2 in the presence of lassomycin. ClpP1P2 (100 nM) and ClpC1 (100 nM) were mixed in 80µlofreaction buffer containing 50 mM potassium phosphate (pH 7.6), 100 mM KCl, 8 mM MgCl2, 5% glycerol, 2 mM ATP, 5 mM Z-Leu-Leu. Enzymatic activity was measured fluorometrically using FITC-casein as a substrate in the presence or absence of 10 µM of lassomycin. The rate of degradation of ClpP1P2 was taken as 100%. C) Lassomycin does not interfere with casein binding to ClpC1. ATPase activity of ClpC1 (32nM) was measured as in Fig. 5A in the presence or absence of casein (10 µM) and lassomycin (1 µM). The activity of ClpC1 alone (control) was taken as 100%. D) Stimulation of ClpC1 ATPase activity by lassomycin is highly specific. The activities of purified ATPases from bacteria (M. tuberculosis ClpX; E.coli ClpA, ClpB and GroEL), archaea (PAN) and mouse (26S proteasome) were measured in the presence and absence of lassomycin (10 μM). ATPase activity of each ATPase in the absence of lassomycin was taken as 100%.
This unexpected stimulation of ClpC1’s ATPase activity initially suggested that lassomycin also activated protein degradation by the ClpC1P1P2 protease. Such a mechanism appeared likely in light of the discovery that the natural product antibiotics, acyldepsipeptides (Kirstein et al., 2009), are bactericidal by binding to ClpP and inducing excessive breakdown of cell proteins. In fact by activating ClpP these antibiotics can kill persisters and eradicate a chronic biofilm infection (Conlon et al., 2013). Another antibiotic, cyclomarin A, which binds to ClpC1, has also been proposed to cause excessive proteolysis (Schmitt et al., 2011; Vasudevan et al., 2013). Surprisingly, however, we found that lassomycin, while markedly stimulating the ATPase activity of ClpC1, completely eliminated its ability to support ATP-dependent degradation of casein (Fig 5B), and somehow uncoupled the ATPase activity of ClpC1 from proteolysis. The resulting inhibition of proteolysis, while accelerating ATP hydrolysis, represents a totally novel mechanism of antibiotic action that should be highly deleterious to the bacteria, since it prevents the regulated selective destruction of key cell proteins by the ClpC1P1P2 complex.
One potential and easily testable mechanism for uncoupling of ATP hydrolysis from protein degradation would be if lassomycin prevented the binding of protein substrates to ClpC1. This possibility could be easily tested, because protein substrates of ClpC1 are known to enhance its ATPase activity 2–3 fold (Akopian et al., 2012). To determine whether lassomycin blocks casein binding to ClpC1, we tested whether it decreases the activation of ATP hydrolysis by casein. On the contrary, lassomycin and casein had additive effects on ClpC1’s ATPase activity (Fig 5C), and therefore, they probably bind to ClpC1 in distinct places (see below). Therefore, in the presence of the antibiotic, ClpC1 seems to bind protein substrates normally, but is incapable of translocating them into the proteolytic compartment.
ClpC1 is a member of the large AAA family of ATPases that serve a variety of key functions in animal and bacterial cells (Erzberger and Berger, 2006; Hanson and Whiteheart, 2005; White and Lauring, 2007). Use of lassomycin as an anti TB drug would presumably not be advisable if it activated other such ATPases. To determine whether lassomycin also affected the activities of other AAA ATPases, we tested several purified well-characterized bacterial and mammalian homologs. In M. tuberculosis, ClpX, like ClpC1, supports protein degradation by ClpP1P2; nevertheless, lassomycin had no effect on ATP hydrolysis by ClpX. In addition, no stimulation was observed with the E. coli ClpC1 homolog, ClpA, a component of the E. coli ClpAP protease complex, PAN, the proteasomal activating ATPase, from the archaebacteria, Methanococcus jannaschii, and the mammalian 26S proteasome (Fig. 5D). This highly specific activation of ClpC1 is clearly novel and of appreciable mechanistic interest. Somehow, binding of multiple lassomycin molecules must lead to an accelerated ATP-ADP exchange cycle (Smith et al., 2011) and much more rapid ATP hydrolysis by its six subunits.
Docking of lassomycin to ClpC1
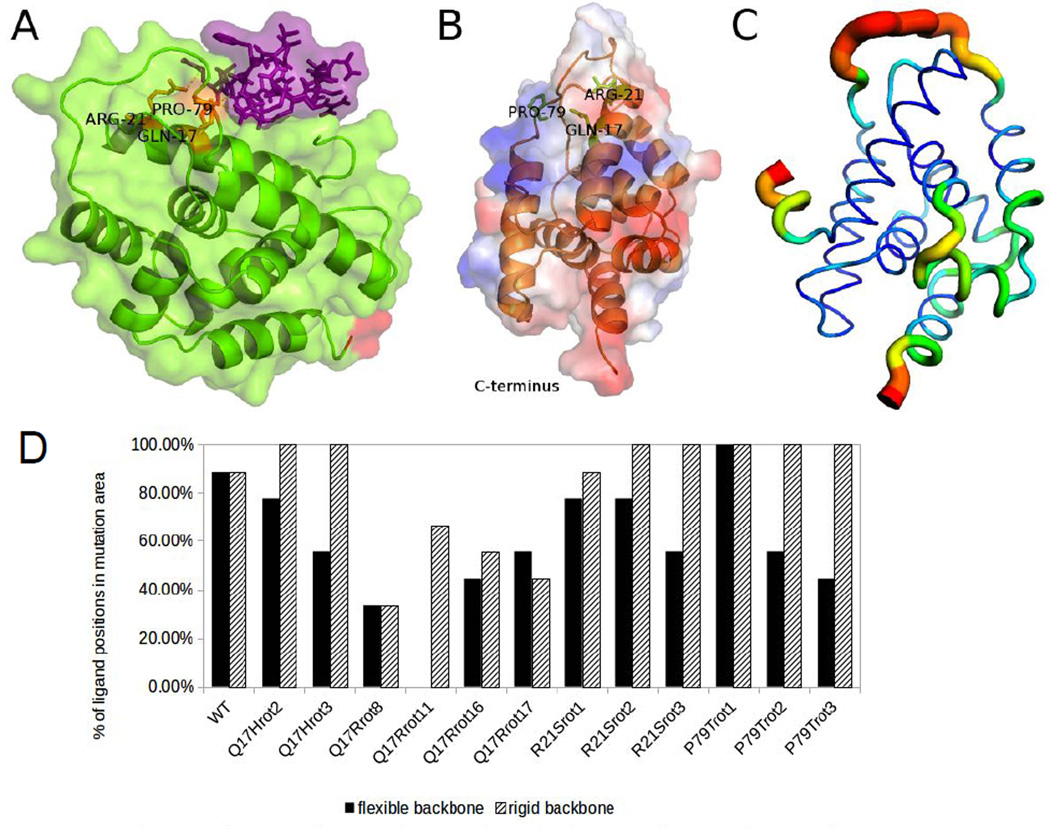
In order to gain further insight into the interaction of lassomycin with ClpC1, an in silico approach was utilized. We focused on the N-terminal region where all the lassomycin-resistant mutations were localized. Recently, the X-ray structure of the ClpC1 N-terminus was solved by Vasudevan et al. (Vasudevan et al., 2013). Visualization of the mutation sites in this model revealed them to be close to each other on ClpC1 (Fig. 6A). Furthermore, they were all located in a highly acidic region, which is likely to be the binding site for lassomycin based on its four positively charged guanidinium groups Docking (Trott and Olson, 2010) of lassomycin onto this structure for ClpC1’s N-terminus showed that eight of the nine obtained binding sites were in the same vicinity (Fig. 6A,B). Analysis of the residues contributing to binding indicated that Gln17, which was altered in four of six resistant mutants (Fig. 6A), is the major interacting residue through hydrogen bonding. In the resistant strains, Gln17 was mutated to an Arg or His, and the resulting reversal of the charge should markedly reduce the tendency for lassomycin binding. The other mutation sites, Arg21 and particularly Pro79 are located, in most of the binding states, on the rim of the surface area contacting the peptide (Fig. 6A).
Figure 6. Images of a docking result and structure evaluations.
(A) Lassomycin (purple) binds at the mutation sites in the M. tuberculosis ClpC1 N-domain (green; C-terminus in red). Mutation sites are labeled and shown in orange. (B) Transparent overlapping surface map shows electrostatics, making positively charged regions (blue) and negatively charged regions (red) visible. Protein backbone in orange, mutant residues in green. (C) The temperature-factor value of the top loop of the ClpC1N-terminus crystal structure 3WDB indicates a very flexible part (red) of the molecule. The color changes from very flexible in red to very rigid in blue. (D) Mutant ClpC1 model variants yield fewer binding positions for the region of the lassomycin-resistant mutations than the wild-type model (WT). Docking was performed assuming a flexible (black) or rigid (shaded) backbone of lassomycin.
To clarify the mode of binding of lassomycin, models of the resistant mutant forms of ClpC1 were created. Because the actual conformations resulting from the mutations of these various residues is not known, several variants of the models were tested. The docking showed that the Gln17Arg mutation has the largest impact on lassomycin binding, and all of the four Gln17Arg mutant models showed a significant reduction in the number of likely positions for lassomycin (Fig. 6D). Two variants of the Pro79Thr and two variants of the Arg21Ser mutation also reduced binding greatly, considering a flexible backbone of lassomycin. Both results confirmed that Gln17 is critical for antibiotic binding, while Arg21 and Pro79 appear less important and might decrease binding by altering the protein's tertiary structure. This analysis, while useful, has clear limitations. For example, changes in ClpC1’s tertiary structure upon hexamer formation or ATP hydrolysis are not considered. In fact, AAA ATPases are highly dynamic structures (Smith et al., 2011), and the upper loop in the C. glutamicum ClpC N-terminal crystal structure has a large temperature-factor (Debye-Waller) value, indicating a flexible region (Fig. 6C). Also the conformation of lassomycin on the enzyme and possible influence of water on its binding are not known. Nevertheless, the approach indicates clear differences between WT and mutant structures in the likely binding site for lassomycin (Fig. 6D). In the future it will be valuable to analyze the crystal structure of ClpC1 with lassomycin bound in order to understand the basis for the large acceleration of ATP hydrolysis and its inability to support ATP-dependent proteolysis. Even though lassomycin and substrates (casein) both activate ATPase, casein must bind to a different site on ClpC1 since casein could stimulate ClpC1 in the presence of lassomycin (Fig 5C). It is also unclear how lassomycin’s binding to this highly acidic N-terminal region can allosterically regulate ClpC1’s ATPase activity and lead to the inhibition of ClpP1P2-dependent proteolysis. Most likely, the conformational changes induced by lassomycin interfere with ClpC1’s association with ClpP1P2. Since mutations in the binding region prevent killing by lassomycin, this novel uncoupling of ATP hydrolysis from protein degradation must be responsible for its bactericidal actions.
SIGNIFICANCE
In multiple respects, lassomycin is a very unusual bactericidal antibiotic with a novel mechanism of action. 1) It kills mycobacteria selectively. This specificity is due to targeting ClpC1 without affecting related hexameric AAA ATPases. ClpC1, like ClpP1P2, is essential for viability in mycobacteria but not in other bacteria. 2) Its structure is generated by cleavage of a ribosomal product, cyclization, and C-terminal esterification. 3) It is an unusually basic, 16-residue lasso peptide, which even at low concentrations, enters M. tuberculosis and binds to an acidic N-terminal pocket on ClpC1. 4) It is unusual for an antibiotic to activate rather than inhibit its target enzyme. An important question for future research is to understand how lassomycin’s binding to ClpC1 stimulates its ATPase activity but uncouples it from ClpP1P2-dependent proteolysis. Normally, AAA ATPases, like ClpC, function by an ordered reaction cycle involving each of its six subunits (Smith et al., 2011), which extend loops into specific pockets on ClpP heptamers. Most likely, this coupling mechanism is disrupted by lassomycin. 5) Although this interaction with ClpC1’s N-terminus is critical for lassomycin’s bactericidal action, it is unclear whether cell death results from this prevention of ClpC1-mediated breakdown of key proteins or from the large increase in ClpC1’s activity (e.g. excessive ATP hydrolysis or excessive protein unfolding). 6) Based on lassomycin’s potency, which resembles or exceeds that of the standard treatments for tuberculosis, agents with similar effects on ClpC1 function represent promising approaches to treat this disease and its drug-resistant forms.
MATERIALS AND METHODS
Bacterial strains and plasmids
M. smegmatis mc2155 were grown at 37 °C in Middlebrook 7H9 broth with 0.05% Tween-80 and ADC (0.5% BSA, 0.2% dextrose, 0.085% NaCl, 0.003 g catalase/1 L medium) with hygromycin (50 μg/ml) and anhydrotetracycline (ATc) (100 ng/ml). M. tuberculosis strains were grown in Middlebrook 7H9 Broth (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (Difco) with additional supplements. The supplements for auxotrophic strain of M. tuberculosis mc26020 (ΔlysA,ΔpanCD) (Sambandamurthy et al., 2005) included 0.5% glycerol, 0.2% casamino acids (Amresco), 0.05% tyloxapol, pantothenic acid (24 μg/ml), and lysine (80 μg/ml). The M. tuberculosis strain mc26020 was transformed with a plasmid pBEN (gift from Dr. L. Ramakrishnan, University of Washington). Before transformation, the gene for green fluorescent protein in the pBEN plasmid was replaced with a gene coding for the red fluorescent protein mCherry (Shaner et al., 2004). The supplements for M. tuberculosis clinical isolates included 0.2% glycerol and 0.05% Tween-80. The clinical isolates were kindly provided by Dr. Clifton Barry III (NIH). C-terminally 6XHis-tagged ClpC1 was expressed in M. smegmatis on pTetOR plasmid, which has an inducible tetracycline promoter.
Long-term incubation setup
1 g of soil sample was vortexed vigorously in 10 ml of deionized H2O for 10 minutes in a 50 ml conical tube. This sample was serially diluted and mixed with molten 2% SMS agar (0.125 g casein, 0.1 g potato starch, 1 g casamino acids, 20 g Bacto-agar in 1 L deionized H2O) for plating at multiple densities of colony forming units. 100 μl of these mixtures were then pipetted into flat bottom 96-well plates. Plates were incubated at room temperature (22 °C) in a humidified chamber and observed weekly under a 50X dissecting microscope.
Fermentation of natural isolates
Colonies of IS009804 were homogenized and transferred into a 250 mL Erlenmeyer flask containing 40 mL of seed broth (15 g glucose, 10 g malt extract, 10 g soluble starch, 2.5 g yeast extract, 5 g casamino acids, and 0.2 g CaCl2•2H2O per 1 L deionized H2O, pH adjusted to 7.0 before autoclaving). The seed broth was incubated for 7 days at 28 °C on a rotary shaker (2.5 inch throw, 200 rpm) prior to production medium inoculation at 2.5% (v/v). IS009804 was screened for antibiotic production after growth in a panel of fermentation production media by removal of aliquots of the crude broths after 4 and 11 days of growth. The active fermentation production medium was R4 broth (10 g glucose, 1 g yeast extract, 0.1 g casamino acids, 3 g proline, 10 g MgCl2-6H2O, 4 g CaCl2-2H2O, 0.2 g K2SO4, 5.6 g TES free acid (2-[[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]amino]ethanesulfonic acid) per 1 L deionized H2O, pH to 7 before autoclaving).The samples were concentrated to dryness and reconstituted in 100% DMSO. Further fermentation was performed in 500 mL aliquots of R4 in 2 L tri-baffled flasks for 5 days at 28 °C on a rotary shaker (2.5 inch throw, 200 rpm).
Production of uniformly labeled [13C, 15N]lassomycin
The spores and mycelia of IS009804 were used to inoculate 30 ml of BioExpress-1000-CN Cell Growth medium (13C, 98%; 15N, 98%, Cambridge Isotope Laboratories, USA). The inoculated medium was incubated for 48 hours at 30 °C on a rotary shaker (200 rpm). Two percent of the inoculum from the BioExpress medium were used to seed the modified R4 broth (10 g D-glucose-U-13C6, 1.1 g CELTONE base powder-U-13C, U-15N, 0.5g l-proline-U-13C5, 15N (Cambridge Isotope Laboratories, USA); 10 g MgCl2•6H2O, 4 g CaCl2•2H2O, 0.2 g K2SO4, 5.6 g TES, 1% Trace element solution (ATCC), 1 L H2O, the pH was adjusted to 7.5 before autoclaving). The culture was incubated for 8 days at 30 °C on a rotary shaker (200 rpm).
Strain identification
16S rDNA sequence analysis was utilized to determine the taxonomic identity of isolate IS009804. Chromosomal DNA was isolated from approximately 106 cells after a five minute vigorous agitation in the presence of 50 mg of glass beads and 100 μl of H2O in a 0.5 ml Eppendorf tube. PCR amplification of nucleotide bases 20 through 710 of the gene encoding the 16S rRNA was carried out using IS009804 chromosomal DNA, GoTaq Green Master Mix (Promega M7122), and universal primers Bac8F and 782R (Baker 2003). PCR thermocycler parameters included 30 cycles of 95 °C for 30 s, 45 °C for 30 s, and 72 °C for 105 s. The amplified DNA fragment was sequenced by Macrogen (Rockville, MD) using primer 782R and compared by BLAST alignment to the GenBank nucleotide collection.
Isolation of lassomycin
The fermentation culture (4.0 L) was centrifuged for 20 minutes and the supernatant was decanted and the cell pellet was extracted with acetone (2.0 L) and centrifuged again. The acetone extract was combined with the supernatant and HP-20 resin (75 g). This mixture was then concentrated under reduced pressure on a rotary evaporator until all of the acetone had been removed. The resin was first washed with water, then with 80% aqueous acetone before eluting with acetone. Each sample was tested in a M. smegmatis bioassay. The acetone sample contained all of the activity. The acetone solution was then concentrated under reduced pressure to an orange oil. Hexanes were added and the mixture was then centrifuged. The hexanes were then decanted, tested for activity and discarded because there was no activity. The remaining residue was then reconstituted in 50% aqueous DMSO and the sample was fractionated using an HPLC instrument equipped with a reversed-phase C18 column eluting with a gradient of H2O/Acetonitrile/0.1% trifluoroacetic acid (TFA) over 45 minutes. All fractions were tested for activity against M. smegmatis and only one fraction was active. This fraction was lyophilized, leaving a white powder (80 mg). Analysis of this fraction by LC-MS indicated that a major compound was present ([MH]+ = 1880). Subsequent searches in a natural product database (AntiBase) did not result in any previously reported compound that matched our results.
ESI-LC-MS analysis
ESI-LC-MS data were recorded on a MicroMass Q-Tof-2 spectrometer equipped with an Agilent 1100 solvent delivery system and an online diode array detector using a Phenomenex Gemini-C18 reversed phase column (50 × 2.0 mm, 3.0 μm particle size). Elution was performed with a linear gradient using deionized water with 0.1% formic acid and CH3CN with 0.1% formic acid as solvents A and B, respectively, at a flow rate of 0.2 mL/min. The gradient increased from 25 to 100% of solvent B over 10 minutes followed by an isocratic elution at 100% of solvent B for 7 minutes. Analytical and semi-preparative chromatography was performed on a Zorbax SB-C18 reversed phase column (250 × 9.4 mm, 5.0 μm particle size) using a Shimadzu SCL-10AVP HPLC system including a SPD-M10AVP diode array detector set at 254 nm. Elution was performed with a linear gradient using deionized water with 0.1% trifluoroacetic acid and CH3CN with 0.1% trifluoroacetic acid as solvents A and B, respectively, at a flow rate of 3.0 mL/min. The gradient increased from 10 to 100% of solvent B over 20 minutes followed by an isocratic elution at 100% of solvent B for 8 minutes.
Partial hydrolysis of lassomycin
The peptide was partially hydrolyzed using microwave-assisted acid hydrolysis. In a 1.5 mL polypropylene centrifuge tube, the sample solution contained 0.1 μg/μL peptide in 25% TFA or 3 M HCl. The sample tube was placed in the water bath of a CEM microwave chamber (CEM Discover System, CEM Corporation, Matthews, NC) to perform the hydrolysis. Microwave hydrolysis conditions were as follows: 80°C, 300W, 32 to 70 minutes for TFA hydrolysis and 12 minutes for HCl hydrolysis. After the microwave hydrolysis, samples were either dried using a SpeedVac vacuum centrifuge (Savant, Holbrook, NY), purified or diluted 5 to 10 times by water, followed by MS analysis.
MS/MS sequencing and exact mass determination of partially hydrolyzed lassomycin
The partially hydrolyzed lassomycin peptide was diluted 5 to 10 times and directly spotted (0.5 μL) onto a Bruker Daltonics MTP AnchorChip 800/384 target and air dried. 0.5 μL of α-cyano-4-hydroxy cinnamic acid matrix solution (CHCA) was spotted on top and air dried. The spots were washed 3 times by water to remove excess acids and salts. The matrix solution was prepared by diluting 36 μL of saturated matrix solution in 0.1% TFA in 90:10 ACN:H2O to 800 μL final volume using 0.1% TFA in 85:15 ACN:H2O, containing 1 mM ammonium phosphate. Mass spectra were obtained in the positive reflectron mode of ionization using a Bruker Daltonics (Bremen, Germany) UltrafleXtreme MALDI TOF/TOF mass spectrometer. The MS and MS/MS spectra were obtained in an automated mode of operation; for MS/MS analysis the CID (collision-induced dissociation) gas was turned off. The instrument was calibrated over the mass range 700 to 3500 Da using a mixture of standard peptides.
The high-resolution exact mass MALDI-MS data of the partially hydrolyzed peptide was collected on a Bruker 9.4T Apex-Qe FTICR (Bruker Daltonics, Billerica, MA) using the samples already spotted on the MTP AnchorChipTM 800/384 target. The FTICR was externally calibrated using a mixture of standard peptides (Fig. S2).
In addition, LC-MS/MS was performed as follows: the partially hydrolyzed peptide was ZipTip (Millipore, Billerica, MA) purified and 5 μL of the resultant peptide solution was loaded onto a Waters nanoAcquity UPLC system (Waters, Milford, MA) using a peptide trap (180 μm × 20 mm, Symmetry® C18 nanoAcquity™ column, Waters, Milford, MA) and an analytical column (75 μm × 150 mm, Atlantis™ dC18 nanoAcquity™ column, Waters, Milford, MA). Desalting on the peptide trap was achieved by flushing the trap with 2% acetonitrile, 0.1% formic acid at a flow rate of 10 μL/min for 3 min. Peptides were separated with a gradient of 2–60% solvent B (acetonitrile, 0.1% formic acid) over 35 min at a flow rate of 350 nL/min. The column was connected to a Waters Q-TOF Premier (Waters, Milford, MA) for ESI-MS/MS analysis. Data interpretation was completed manually to provide a proposed peptide sequence.
NMR spectroscopy of [13C, 15N]lassomycin
NMR data were acquired and processed as previously described (Rea et al., 2010; Sit et al., 2011). A Varian Inova 800-MHz spectrometer with a triple-resonance HCN cold probe and pulsed field gradients (PFGs) was used to record spectra. [13C, 15N]lassomycin was dissolved in dimethyl sulfoxide-d6 (Cambridge Isotope Laboratories, Andover, MA), and the sample was heated to 40 °C for data collection. Table S2 lists the experimental parameters used to acquire the NMR spectra for lassomycin. Tables S3–S4 list the proton, backbone nitrogen and carbon chemical shift assignments of the peptide. The 15N-HSQC (Fig. S1A) gave reasonably well-dispersed peaks, with 12 out of 16 unique backbone NH signals observed, indicating that lassomycin holds a defined structure in solution. The backbone NH signals for Arg3, Leu5, Arg14 and Ile16 could not be definitively assigned due to spectral overlap. Most of the proton chemical shift assignments were made based on data from the HCCH-TOCSY, 13C-NOESYHSQC and 15N-TOCSYHSQC experiments. Most of the carbon and nitrogen chemical shift assignments were made based on the backbone experiments HNCACB and CBCA(CO)NH (Sit et al., 2011).
Structure calculations
CYANA 2.1 was used to calculate the structures of all the stereoisomers (Guntert et al., 1997), using NOE restraints measured from the 13C-NOESYHSQC and 15N-NOESYHSQC experiments combined with angle restraints obtained from the HNHA experiment. The automatically assigned NOEs were calibrated within CYANA according to their intensities. The same NOE peak lists were used for the structure calculations of each stereoisomer, following the same procedure as previously reported (Sit et al., 2011). After seven rounds of calculation for lassomycin (10,000 steps per round), a total of 449 cross-peak NOE assignments and 1 3JHNH coupling constant were used in the final calculation. The structural statistics for the 20 lowest energy conformations of lassomycin are summarized in Table S5. These conformations had no constraint violations, a high number of long-range NOEs (61) used in the structure calculation, a low average target function value (0.01), a low backbone rmsd (0.35 ± 0.10 Å), and a low heavy atom rmsd (1.00 ± 0.23 Å). Coordinates for lassomycin have been deposited in the Protein Data Bank (2mai) and chemical shift assignments have been deposited in the BioMagResBank (19355). All other figures were generated using PyMOL.
Compound spectrum, activity
Potency was determined by measuring MIC by broth microdilution. Strains were grown to an optical density of 0.3 at 650 nm and diluted 1:100 into fresh media. Antibiotics were serially diluted two fold in 96-well plates and an equal volume of diluted bacterial culture was added to each well. The plates were sealed with Breathe-Easy® (3M Company) and incubated at 37°C for 14 days after which growth was visually inspected. The MIC was the concentration of compound that resulted in no growth compared to controls wells. MBC was determined by plating cells from wells containing 2X, 4X and 8X MIC concentrations of compound. The plates were incubated for 3–4 weeks at 37°C. The MBC was the concentration of compound that decreased colony count by ≥ 99.9%.
To monitor in vitro cytotoxicity, exponentially growing NIH 3T3, and HepG2 cells in media supplemented with 10% fetal bovine serum were harvested and seeded at 5000 cells per well in a 96-well flat bottom plate (Smee et al., 2002). After 24 hours the medium was replaced with fresh medium containing compounds added at a two-fold serial dilution, in a process similar to an MIC assay. After 24 and 48 hours of incubation, cell viability was measured with the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega), according to manufacturer’s recommendations.
High throughput screen
Frozen aliquot of M. tuberculosis mc26020 pBEN mCherry used to inoculate (0.5–1% inoculum) 50 ml of 7H9 with OADC, glycerol, casamino acids, taloxapol, panthothenic acid, lysin and 50 μg/ml of kanamycin. The culture was incubated at 37 °C with shaking until the optical density (OD600) reached 0.4 to 0.5. The culture was then diluted to have approximately 106 CFU/ml, and transferred to black, clear-bottom 96-well microplates (Corning) and 1–2% of the crude extracts were added to the tested wells. Rifampicin 1 μg/ml (Sigma) was used as a positive control. Last column of the plate was used as negative control (cell with 2 μl of DMSO). Plates were sealed with Breathe-Easy Sealing Membrane (RPI Corp., USA) and were incubated with shaking at 37 °C for 5–7 days. After incubation fluorescence was measured from the bottom using SynergyMX plate reader (BioTek) with excitation 580 nm and emission 610 nm.
An auxotrophic strain of M. tuberculosis mc26020 (ΔlysA,ΔpanCD) (Sambandamurthy et al., 2005) expressing the red fluorescent protein mCherry was screened against 25,600 crude extracts (3200 strains, 4 fermentation media, 2 time points). All crude extracts were counterscreened against S. aureus. The extracts showing activity against S. aureus were excluded from the screening steps, and the resulting hit rate was 2%. To evaluate the quality of our assay the Z’-factor was calculated for several growth conditions. A Z´ factor between 1 and 0.9 is considered an excellent assay, between 0.9 and 0.7 is good, and between 0.7 and 0.5 will benefit significantly from any improvement (Zhang et al., 1999). The highest Z’ factor (0.8 ± 0.06) was obtained for the plates containing 150 μl of cell culture after 5 days of incubation with shaking, so we proceeded with these conditions for the high throughput screening.
Whole-genome sequencing
Genomic DNA from M. tuberculosis mc26020 (ΔlysA ΔpanCD) wild type and resistance mutants were isolated using previously described protocol (Kieser et al., 2000). An Illumina single-end library was constructed and 50 bp sequencing was conducted on Illumina HiSeq2000 following manufacturer's instructions at Biopolymers facility (Harvard Medical School, Boston, MA). Genomes were assembled and mapped using the reference genome of H37Rv with CLC Genomics Workbench (CLC bio, Cambridge, MA). Genomes of resistance mutants were aligned with the genome of the wild type to detect single base-pair polymorphisms (SNPs).
The results were filtered to exclude any SNPs that had low confidence after assembly. The remaining SNPs were verified by PCR amplification and Sanger sequencing.
Purification of M. tuberculosis ClpC1
C-terminally 6XHis-tagged M. tuberculosis ClpC1 subunits were overexpressed in M. smegmatis using an ATc inducible expression system. After overnight induction with ATc (100 ng/ml), cells were resuspended in buffer A (50 mM TrisHCl pH 7.8, 100 mM KCl2, 10% glycerol, 1 mM ATP, 4 mM MgCl2), lysed by French press, and lysates were centrifuged for 1 hour at 100,000 g. ClpC1 was isolated from the supernatant by Ni-NTA affinity chromatography (Qiagen). Eluted fractions containing ClpC1 protein were pooled and further purified by size-exclusion chromatography on Sephacryl S-300 column in buffer A. Fractions containing ClpC1 protein were concentrated and used in the assays of ATPase activity and protein degradation.
Measurement of ATPase activity
2 μg of pure ClpC1 were mixed with 100 µl of the assay buffer (50 mM TrisHCl pH 7.8; 100 mM KCl; 10% glycerol; 1 mM phosphoenolpyruvate; 1 mM NADH; 2 µl pyruvate kinase/lactic dehydrogenase (Sigma); 4 mM MgCl2 and 1 mM ATP) and the ATPase activity of ClpC1 was followed by measuring the coupled oxidization of NADH to NAD spectrometrically at 340 nm.
Measurement of casein degradation
Assay of proteolytic activity was performed at 37 °C in 96 wells plate using Plate Reader SpectraMax M5 (Molecular Devices, USA). Wells contained 3 µg ClpP1P2, 6 µg ClpC1, 5 mM Z-Leu-Leu and 2–5 µg of FITC-casein in 80 µl of 50 mM phosphate buffer (pH 7.6) with 5% glycerol, 100 mM KCl, 8 mM MgCl2 and 2 mM ATP. FITC-casein hydrolysis was continuously monitored at 518 nm (Ex at 492 nm). Deviations in the measurements of FITC-casein were less than 10 %.
Protein structure visualization
The PyMOL Molecular Graphics System, Version 1.5.0.1 Schrödinger, LLC.
Molecule docking
AutoDock Vina v1.1.2 was used to perform the docking runs (Trott and Olson, 2010). The structure of the ClpC1 N-domain was obtained through the Protein Data Bank (www.rcsb.org; (Berman et al., 2000); PDB entry 3WDB) and prepared using PyMol. Solvents and molecules other than the ClpC1 N-domain were removed and in silico mutations were introduced. All structure files were validated with MolProbity 4.02 (Chen et al. 2010). Only rotamers resulting in a clashscore close to the wild-type model (clashscore < 6; wild-type was 5.2) were chosen for the docking. The structure files for the ligand and the receptor were prepared with AutoDockTools v1.5.6 (Morris et al. 2009). Preparation of the receptor molecules (M. tuberculosis ClpC1 N-domain model and mutants) included the following steps: addition of all hydrogens, computation of Gasteiger charges, and merging of non-polar hydrogens. To account for the fact, that the bioactive conformation of lassomycin is not known, its structure file was converted with AutoDockTools in two variants: (1) All bonds were assumed rotatable, except the closed lasso ring backbone. (2) Only side chains were rotatable, but the whole peptide backbone remains rigid. The search space was restricted to the acidic half of the ClpC1 N-domain and the exhaustiveness set to 100 (default is 8) to limit the generation of local minima in the optimization function. Computations of the docking runs were conducted on the Harvard University Faculty of Arts and Science Research Computing Cluster “Odyssey”. Vina calculates a binding energy for each position it finds based on the number of rotatable bonds allowed for the ligand and the environment on the receptor (Trott and Olson, 2010). Hence, in the docking approach here, a comparison of the binding energies between different receptor models is not appropriate, because the mutant models have a different protein sequence. Instead, the results were scored by the position of the docked ligand on the receptor molecule. Human error in this evaluation was eliminated by the fact that positive and negative positions showed a great distance from each other.
Supplementary Material
Highlights.
Lassomycin has potent and selective bactericidal activity against M. tuberculosis.
Lassomycin is a lasso peptide but lacks the family’s characteristic knot.
Lassomycin activates ClpC1 ATP hydrolysis, uncoupling it from ClpP1P2 proteolysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akopian T, Kandror O, Raju RM, Unnikrishnan M, Rubin EJ, Goldberg AL. The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J. 2012;31:1529–1541. doi: 10.1038/emboj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and posttranslationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. Microbial scout hypothesis and microbial discovery. Appl. Environ. Microbiol. 2012;78:3229–3233. doi: 10.1128/AEM.07308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Geladopoulos TP, Sotiroudis TG, Evangelopoulos AE. A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 1991;192:112–116. doi: 10.1016/0003-2697(91)90194-x. [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Inokoshi J, Matsuhama M, Miyake M, Ikeda H, Tomoda H. Molecular cloning of the gene cluster for lariatin biosynthesis of Rhodococcus jostii K01-B0171. Appl. Microbiol. Biotechnol. 2012;95:451–460. doi: 10.1007/s00253-012-3973-8. [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Tomoda H, Uchida R, Gouda H, Hirono S, Omura S. Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J. Am. Chem. Soc. 2006;128:7486–7491. doi: 10.1021/ja056780z. [DOI] [PubMed] [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio. 2011;2:e00100–e00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- Kirstein J, Hoffmann A, Lilie H, Schmidt R, Rubsamen-Waigmann H, Brotz-Oesterhelt H, Mogk A, Turgay K. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol. Med. 2009;1:37–49. doi: 10.1002/emmm.200900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Antibiotics: Recover the lost art of drug discovery. Nature. 2012;485:439–440. doi: 10.1038/485439a. [DOI] [PubMed] [Google Scholar]
- Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- Lupas A, Flanagan JM, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem. Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- Raju RM, Unnikrishnan M, Rubin DH, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 2012;8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. SciU.SA. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2008;6:41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- Sambandamurthy VK, Derrick SC, Jalapathy KV, Chen B, Russell RG, Morris SL, Jacobs WR., Jr Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect. Immun. 2005;73:1196–1203. doi: 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt EK, Riwanto M, Sambandamurthy V, Roggo S, Miault C, Zwingelstein C, Krastel P, Noble C, Beer D, Rao SP, et al. The natural product cyclomarin kills Mycobacterium tuberculosis by targeting the ClpC1 subunit of the caseinolytic protease. Angew. Chem. Int. Ed. 2011;50:5889–5891. doi: 10.1002/anie.201101740. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric, red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sit CS, McKay RT, Hill C, Ross RP, Vederas JC. The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon crosslinks. J. Am. Chem. Soc. 2011;133:7680–7683. doi: 10.1021/ja201802f. [DOI] [PubMed] [Google Scholar]
- Smee DF, Morrison AC, Barnard DL, Sidwell RW. Comparison of colorimetric, fluorometric, and visual methods for determining anti-influenza (H1N1 and H3N2) virus activities and toxicities of compounds. J. Virol. Methods. 2002;106:71–79. doi: 10.1016/s0166-0934(02)00137-4. [DOI] [PubMed] [Google Scholar]
- Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D, Rao SP, Noble CG. Structural basis of mycobacterial inhibition by cyclomarin A. J. Biol. Chem. 2013;288:30883–30891. doi: 10.1074/jbc.M113.493767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- Yu AY, Houry WA. ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 2007;581:3749–3757. doi: 10.1016/j.febslet.2007.04.076. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 2013;12:388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.