Abstract
Methionine is an essential proteogenic amino acid. In addition, it is a methyl donor for DNA and protein methylation and a propylamine donor for polyamine biosyn-thesis. Both the methyl and propylamine donation pathways involve metabolic cycles, and methods are needed to quantitate these cycles. Here, we describe an analytical approach for quantifying methionine metabolic fluxes that accounts for the mixing of intracellular and extracellular methionine pools. We observe that such mixing prevents isotope tracing experiments from reaching the steady state due to the large size of the media pools and hence precludes the use of standard stationary metabolic flux analysis. Our approach is based on feeding cells with 13C methionine and measuring the isotope-labeling kinetics of both intracellular and extracellular methionine by liquid chromatography−mass spectrometry (LC-MS). We apply this method to quantify methionine metabolism in a human fibrosarcoma cell line and study how methionine salvage pathway enzyme methylthioadenosine phosphorylase (MTAP), frequently deleted in cancer, affects methionine metabolism. We find that both transmethylation and propylamine transfer fluxes amount to roughly 15% of the net methionine uptake, with no major changes due to MTAP deletion. Our method further enables the quantification of flux through the pro-tumorigenic enzyme ornithine decarboxylase, and this flux increases 2-fold following MTAP deletion. The analytical approach used to quantify methionine metabolic fluxes is applicable for other metabolic systems affected by mixing of intracellular and extracellular metabolite pools.
13C-Metabolic Flux Analysis (MFA) is a commonly used approach to determine cellular metabolic fluxes, which has significantly contributed to biotechnology and medicine.1−5 It is based on feeding cells with isotope-labeled nutrients, measuring the labeling patterns of intracellular metabolites, and applying computational methods to analyze metabolite labeling data to estimate fluxes.2,6,7 The most standard MFA approach requires an isotopic steady state, resolving flux through alternative pathways by their characteristic scrambling of 13C from labeled media substrates.
A factor that complicates flux inference in some cases is exchange of isotope label between intracellular and media metabolite pools. This may lead to gradual change in labeling pattern of both intracellular and media metabolite pools due to the large size of metabolite pools in media that prevents reaching an isotopic steady state within the experimental time frame. Simply increasing the experimental duration is not feasible due to depletion of nutrient(s) from the media. To address this problem, variants of MFA rely on isotopic label kinetics without requiring reaching an isotopic steady state.8−11 However, these methods are experimentally and computationally intensive, involving repeated cell sampling, extensive metabolite labeling analysis, and subsequent computational simulation of large systems of differential equations.
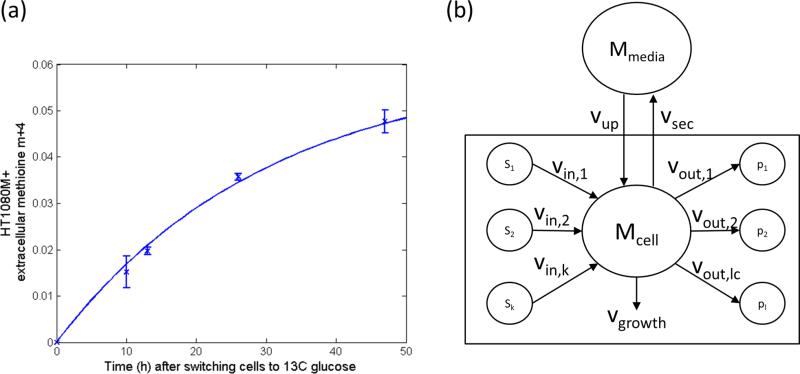
Here, we describe a more streamlined analytical approach for quantifying metabolic fluxes that is robust to mixing of intracellular and extracellular metabolite pools. We use this approach to study methionine metabolism, focusing on the transmethylation and the propylamine transfer cycles, both of which have a known tumorigenic role.12−15 Feeding fibrosarcoma cells with 13C glucose, we observe mixing of intracellular and media methionine pools, as evident by the gradual increase of labeled methionine in media over time (Figure 1a). We then derive closed-form expressions for flux through transmethylation and propylamine transfer cycles based on experimental measurements of isotope labeling kinetics by mass spectrometry. The application of this approach is demonstrated in human fibrosarcoma cells, which have a deletion of the methionine salvage pathway enzyme methylthioadenosine phosphorylase (MTAP). MTAP is commonly deleted in cancer, likely due to its chromosomal location near the known tumor suppressor p16.16 To further study the effect of MTAP deletion on methionine fluxes, we also apply this approach to an isogenic cell line in which MTAP was reintroduced.
Figure 1.
(a) Labeling kinetics of media methionine in HT1080 cells in which MTAP was reintroduced after switching cells into [U−13C]-glucose. (b) A schematic representation of metabolic reactions consuming and producing metabolite M whose intracellular and extracellular pools are mixed.
METHODS
Cell Lines and Culture Conditions
We used a human fibrosarcoma cell line with a homozygous deletion in MTAP (HT1080M−) and an isogenic cell line in which MTAP was reintroduced (HT1080M+).17 The cell lines were grown in Dulbecco's modified eagle media (DMEM) supplemented with 2 mM glutamine, 10% dialyzed fetal bovine serum (HyClone), 100 μg/mL of penicillin, 100 μg/mL of streptomycin, and 250 μg/mL of G418. Cells were grown in an incubator containing 5% CO2 and ambient oxygen at 37 °C. For labeling experiments, the medium was prepared from DMEM without glucose or methionine, with the desired isotopic form of glucose and/or methionine added at the same concentration as in standard DMEM media. Short-term experiments (e.g., nutrient uptake and kinetic labeling) were conducted at 70− 80% confluency. Cellular growth rate was determined by measuring packed cell volume at five different time points and fitting an exponential growth function to the obtained curve.
Metabolomic Experiments and LC-MS Analysis
For all metabolomic and isotope-tracer experiments, the metabolite extraction procedure consisted of aspirating media, washing as quickly as possible by pipetting on 4 °C PBS and immediately aspirating the wash solution, and quenching metabolism directly thereafter by adding a −80 °C 80:20 methanol/water extraction solution. Samples were analyzed by multiple LC-MS systems (each from Thermo Scientific and fed by electrospray ionization), as described previously.18−20 Briefly, a stand-alone orbitrap mass spectrometer (Exactive) operating in negative ion mode was coupled to reversed-phase ion-pairing chromatography and used to scan from m/z 85 to 1000 at 1 Hz and 100 000 resolution. ATSQ Quantum Ultra triple-quadrupole mass spectrometer operating in positive ion mode was coupled to hydrophilic interaction chromatography on an aminopropyl column and used to analyze selected compounds by multiple reaction monitoring, and a TSQ Quantum Discovery triple− quadrupole mass spectrometer operating in negative ion mode was coupled to reverse-phase ion-pairing chromatography and used to analyze selected compounds by multiple reaction monitoring. Amino acids were derivatized before analysis to improve sensitivity and selectivity by the following procedure: 10 μL of triethylamine was added and mixed into a 100 μL cell extract in 80% methanol, and then 2 μL of benzyl chloroformate was added to the sample. After incubation at room temperature for 30 min, the resulted derivatized amino acids were quantified by negative-mode LC-ESI-tandem mass spectrometry (MS/MS).21,22 To detect polyamines, we utilized dansyl derivatization.23 Data were analyzed using the MAVEN software suite.24 Measured metabolite levels were normalized by packed cell volume. The results are adjusted for natural 13C abundance and enrichment impurity of labeled substrate supplied to cells.
Absolute metabolite levels were quantified via an isotope-ratio approach, as described previously.25−27 Specifically, cells are fed with 13C methionine to near-complete isotopic enrichment and then extracted and analyzed by LC-MS twice: once with the same extraction solvent described above (to measure the ratio of labeled-to-unlabeled endogenous metabolite, R1) and once with the extraction solvent with unlabeled internal standards added in known concentrations (S, to measure the ratio of labeled endogenous metabolite to unlabeled standard + unlabeled endogenous metabolite, R2). The ratio of endogenous metabolite to internal standard (in the second cell extract) is determined by (R2 + R2/R1)/(1 − R2/ R1). The product of this ratio and the unlabeled standard amount is equal to the amount of endogenous metabolite present in the cells. The cellular concentration of the metabolite is then calculated based on the packed volume of the extracted cells.
RESULTS AND DISCUSSION
A Generic Analytical Approach for Quantifying Metabolic Fluxes Accounting for Metabolite Exchange with Media
Figure 1b shows a schematic representation of metabolic reactions consuming, producing, and transporting metabolite (M). Intracellular metabolite M, denoted Mcell, is synthesized via k reactions with rates vin,1...vin,k (in nmol/μL-cells/h) from substrates S1, ..., Sk, consumed via l reactions with rates vout,1, ..., vout,l, taken up from a media pool Mmedia at a rate of vup, and simultaneously secreted to the media at rate vsec. The growth dilution of metabolite M is denoted vgrowth. The transient change in the amount (in nmol) of a specific labeling pattern of intracellular M in time t, denoted can be described as follows:
| (1) |
where denotes the total amount of intracellular M, V(t) denotes the total cellular volume at time t (μL cells; representing pack-volume), denotes the intracellular amount (in nmol) of a specific labeling pattern of Si synthesizing , and denotes the total amount of Si. Considering that mass balance considerations for intracellular M entail that
| (2) |
Equation 1 can be expressed as
| (3) |
The transient change in the amount of labeled M in media, denoted is
| (4) |
where denotes the total amount of media M. Plugging eq 4 into eq 3 gives
| (5) |
Taking the integral of eq 5 over time (from time zero, when nonlabeled media are switched with a labeled one, until T hours later), we get the following:
| (6) |
This can be simplified using the following definitions:
We refer to the resulting equation as the isotopomer cumulative balance equation for M*:
| (7) |
Note that the β and M* parameters can be calculated directly from experimentally measured isotope labeling data, and vnet_up from the time-dependent change in the media concentration of M. Thus, eq 7 can be used to constrain, or in many cases directly calculate, vin,I, i.e., intracellular fluxes. This utility will be exemplified below for the case of methionine metabolism.
Quantifying Propylamine Transfer Flux in a Fibrosarcoma Cell Line
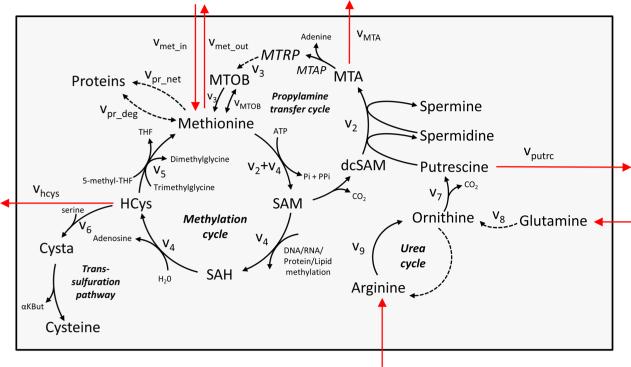
Propylamine transfer is facilitated by a cyclic pathway, in which methionine and ATP react to make S-adenosylmethionine (SAM; Figure 2). SAM is decarboxylated to form S-adenosylmethioninamine. S-adenosylmethioninamine serves as the propylamine donor for polyamine biosynthesis, producing methylthioadenosine (MTA) as a byproduct. MTA is cleaved by methyl-thio-adenosine-phosphorylase (MTAP) to adenine and 5-methylthioribose-1-phosphate, and the latter is recycled back to methionine via the methionine salvage pathway.
Figure 2.
Methionine metabolism, including transmethylation cycle, polyamine biosynthesis, and methionine salvage cycle.
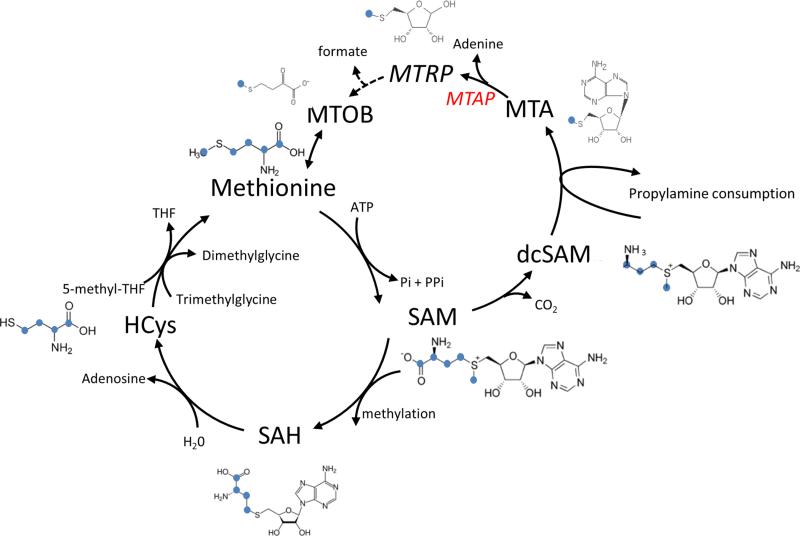
We use the above method to derive an analytical expression for the flux through the propylamine transfer cycle, given metabolite labeling data following a switch from a nonlabeled medium to [U−13C]-methionine. Tracking methionine carbons through the propylamine transfer cycle shows that a methionine molecule that goes through the cycle gets four carbons replaced by ribose carbons (from an ATP), while the methyl group remains from the original methionine molecule (Figure 3). Hence, a methionine m+5 going through the propylamine transfer cycle would make methionine m+1. We denote the net methionine salvage flux by v3 and the bidirectional transamination flux from 4-methylthio-2-oxobutanoate (MTOB) to methionine and back by vMTOB (Figure 2). Methionine m+1 can be further consumed by protein biosynthesis and then mixed back with the methionine pool via proteolysis (Figure 2). We denote the rate of methionine production through protein degradation by vpr_deg. Hence, the isotopomer cumulative balance equation for methionine m+1 is
| (8) |
where PrMet denotes methionine within a cellular protein pool. MTOB is synthesized from 5-methylthioribose-phosphate (MTRP) and the isotopomer cumulative balance equation for MTOB m+1 is
| (9) |
Combining the balance equations in eqs 8 and 9 gives
| (10) |
To quantitate the terms in this equation, we measured methionine and MTOB using an isotope ratio-based approach. This revealed that the amount of intracellular labeled methionine and MTOB is negligible compared to the amount of extracellular labeled methionine as the intracellular concentration of both methionine and MTOB is less than 200 μM and the cellular volume is less than 10 μL, whereas the media concentration of methionine is 200 μM and the media volume is 3 mL. Hence, methionine salvage flux can be approximated by
| (11) |
To compute βMet,m+1, we utilized a numerical integration method in Matlab based on the measured cellular growth curve and the measured labeling kinetics of intracellular methionine.
Figure 3.
Tracking of methionine carbons in the propylamine transfer and transmethylation cycles.
To compute βPrMet,m+1, we simulated the labeling kinetics of methionine in proteins, considering that the transient change in the amount of PrMet that is m+1 is
| (12) |
The total concentration of methionine in proteins (which is assumed to remain fixed during the experiment due to the metabolic steady-state) was calculated based on the net consumption of methionine for protein biosynthesis (vpr_net) and cellular doubling time (μ):
| (13) |
Net methionine consumption for protein biosynthesis is calculated via measured net uptake and secretion of methionine, MTA, and homocysteine, as well as cysteine biosynthesis rate, considering mass-balance considerations:
| (14) |
The rate of protein degradation is calculated considering that the average half-life of a human protein (denoted γ) is 6.9 h:28
| (15) |
To calculate the standard deviation for the methionine salvage flux calculated via eq 11, we performed Monte Carlo simulations considering the experimental noise in the measurement of metabolite labeling, metabolite concentrations, and growth rate (i.e., repeatedly applying eq 11 on parameters drawn from a normal distribution whose mean and standard deviation are derived from the experimental data). To further account for uncertainty in assumed average protein half-life, this parameter was drawn from a normal distribution considering a standard deviation of 50% of the experimental value.
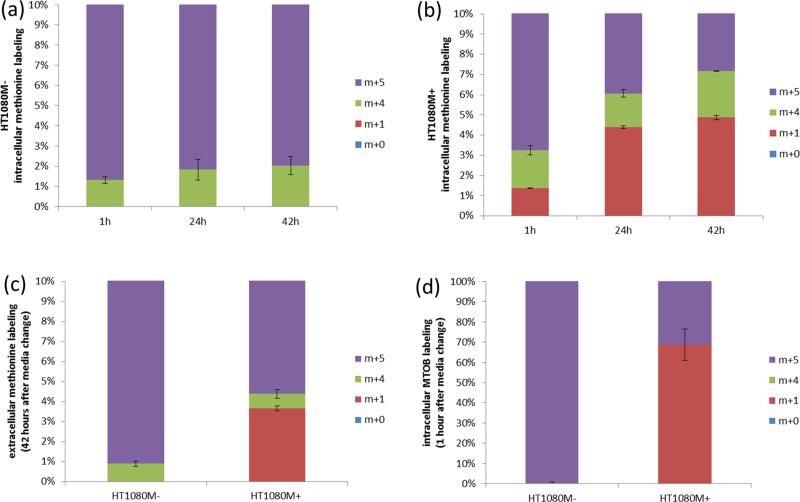
Next, we applied this method to quantify the propylamine transfer flux in a human fibrosarcoma-derived cell line that lacks MTAP expression (HT1080M−) and in an isogenic cell line in which MTAP was reintroduced via stable transfection (HT1080M+).29 Toward this goal, we measured cellular growth rate, finding that the doubling time of both cell lines was on the order of 23 h (Methods). Utilizing LC-MS to measure net methionine uptake from media, we find an uptake of ~0.8 ± 0.1 nmol/μL-cells/h in both cell lines (Table 1). To quantify the net consumption of methionine for protein biosynthesis via eq 14, we measured MTA and homocysteine secretion rates and determined that there is no cysteine biosynthesis from homocysteine (based on feeding cells with [U−13C] glucose and measuring the labeling pattern of serine and cysteine; data not shown). We find a net consumption of methionine for protein biosynthesis of 0.71 and 0.65 nmol/uL-cells/h for HT1080M+ and HT1080M−, respectively (Table 1). Next, we measured the kinetics of intracellular methionine labeling for 42 h after switching to a medium containing [U−13C]-methionine (Figure 4a,b), as well as the absolute amount and labeling of extracellular labeled methionine after 42 h (Figure 4c).
Table 1.
Inferred Fluxes in nmol/μL-cells/h in Both HT1080M– and HT1080M+ Based on Isotope Labeling Coupled with Computational Modelinga
| reaction | HT1080M+ | HT1080M– |
|---|---|---|
| v1 (vmet_in – vmet_out) | 0.8 ± 0.1 | 0.9 ± 0.1 |
| v 2 | 0.17 ± 0.02 | 0.11 ± 0.01 |
| v 3 | 0.17 ± 0.02 | 0 |
| v 4 | 0.12 ± 0.02 | 0.16 ± 0.02 |
| v 5 | 0.03 ± 0.02 | 0.02 ± 0.01 |
| v 6 | 0 | 0 |
| v 7 | 0.18 ± 0.01 | 0.33 ± 0.03 |
| v 8 | 0.07 ± 0.01 | 0.13 ± 0.01 |
| v 9 | 0.11 ± 0.01 | 0.2 ± 0.02 |
| v MTA | <0.005 | 0.11 ± 0.01 |
| v hyc | 0.09 ± 0.01 | 0.14 ± 0.02 |
| v putr | 0.01 ± 0 | 0.21 ± 0.02 |
| v pr_net | 0.71 ± 0.1 | 0.65 ± 0.02 |
Net methionine uptake (v1) MTA secretion (v_MTA), homocysteine secretion (vhyc), and putrescine secretion (vputr) were quantified based on the accumulation or depletion of these metabolites in media, as detected by LC-MS. Net consumption rate of methionine for protein biosynthesis (vpr_net) is equal to v1 – vMTA – vhyc – v6, based on mass-balance considerations. Methionine salvage flux (v3) was calculated via eq 11. Propylamine transfer flux (v2) is equal to v3 + Vmta, based on mass-balance considerations. Methionine synthase flux (v5) is calculated via eq 20. Cysteine biosynthesis flux (v6)is determined via an auxiliary experiment with glucose labeling. Transmethylation flux (v4) is equal to v5 + vhcys + v6, due to mass-balance considerations. ODC flux (v7) is equal to v2 + v_putr, due to mass-balance considerations. Ornithine biosynthesis via glutamine pathway (v8) and arginine pathway (v9) are calculated based on ODC flux (v7), and glucose labeling experiments showing that arginine and glutamate pathways contribute 60% and 40%, respectively, to ornithine production (Figure 5d).
Figure 4.
Quantifying propylamine production and methionine salvage flux via [U−13C] methionine isotope tracing. (a) Labeling kinetics of intracellular methionine in HT1080M−. (b) Labeling kinetics of intracellular methionine in HT1080M+. (c) Labeling of extracellular methionine 42 h after cells were switched into [U−13C]-methionine. (d) Labeling of intracellular MTOB 1 h after cells were switched into [U−13C]-methionine.
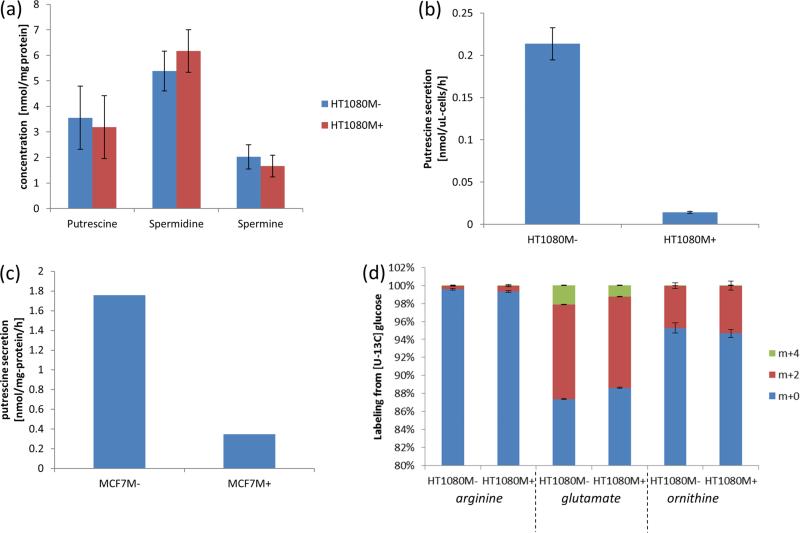
To apply eq 11 to quantify methionine salvage flux (v3) in HTM1080M+, we measured MTOB labeling one hour after switching the medium to 13C methionine and discovered that it is already fully labeled by that time (Figure 4d). This suggests that the pool of MTOB's upstream precursor in the methionine salvage pathway, MTRP, is already fully m+1 labeled one hour after the medium switch, and hence βMTRP,m+1 roughly equals . Note that the observed rapid labeling of MTOB (relative to the time scale of the experiment) obviates the need for more detailed kinetic measurements of MTOB labeling, saving experimental effort. Applying eq 11, we get that the methionine salvage flux (v3) in HT1080M+ is 0.17 ± 0.02 nmol/u-cells/h. As no MTA secretion was found in this cell line (v_MTA < 0.005 nmol/uL-cells/h, considering MTA limit of detection), the methionine salvage flux is equal to the propylamine transfer flux (v2; Table 1). Considering cellular doubling time, our estimated propylamine transfer rate for polyamine biosynthesis suggests that the total spermidine and spermine pools are no larger than a few mM, which is consistent with previous measurements of polyamines concentrations performed in human lung cancer cell lines.30
In HT1080M−, methionine salvage flux is zero, as cells lacking MTAP do not produce methionine m+1 or MTOB m+1 (Figure 4a,c,d). Propylamine transfer (v2) is hence equal to the rate of MTA secretion into the medium, which is 0.11 ± 0.015 nmol/μL-cells/h (Table 1). Consistent with the fact that we do not observe a major drop in propylamine transfer following MTAP deletion, we do not see a change in steady-state concentration of polyamines following MTAP deletion (Figure 5a). Notably, in agreement with previous studies, we find a 7-fold increase in intracellular MTA level following MTAP deletion. While MTA was previously shown to inhibit spermidine and spermine biosynthesis in vitro,31 our results show that the increased MTA level due to MTAP deletion does not impair spermidine and spermine synthesis in vivo.
Figure 5.
Changes in putrescine biosynthesis and secretion due to MTAP deletion. (a) Concentration of intracellular polyamines. (b) Secretion rate of putrescine to the medium in the HTM1080 cell line. (c) Secretion rate of putrescine to the medium in the MCF7 cell line. (d) Labeling of intracellular arginine, glutamine, and ornithine from [U−13C] glucose.
From a methodological perspective, it is worth noting that ignoring the mixing of the intracellular and extracellular methionine pools (i.e., assuming no secretion of labeled methionine to the medium) and applying standard steady-state MFA to quantify the propylamine transfer flux in HT1080M+ would have led to an erroneous flux estimate. Specifically, under an isotopic steady-state, the balance equation for intracellular methionine m+1 is
| (16) |
where HCY denotes homocysteine. The balance equation for MTOB m+1 is
| (17) |
Additionally, mass-balance for intracellular methionine entails
| (18) |
Combining eqs 16−18 and considering that under an isotopic steady state, the fractional labeling of homocysteine m+1 and methionine m+1 within protein is equal to that of free methionine m+1 and that the entire MTRP pool is in m+1 form, gives the following expression for the methionine salvage flux:
| (19) |
Utilizing eq 19 to quantify v3 leads to an incorrect estimation of 0.04 nmol/μL-cells/h, which is ~4-fold lower than the true flux as inferred above.
Our method for quantifying propylamine transfer flux can also be used to quantify the flux through ornithine decarboxylase (ODC; v7 in Figure 2), which is upregulated in a wide variety of cancers and is known to promote tumorigenesis.32 Specifically, ODC flux should be equal to propylamine transfer flux plus additional putrescine outflux. In our case, we detect ~15-fold higher putrescine secretion in the MTAP negative cells, reaching a rate of 0.21 nmol/μL-cells/h (Figure 5b). Hence, considering that total propylamine transfer flux is not significantly affected by MTAP deletion, the flux through ODC roughly doubles following MTAP deletion (Table 1). To assess whether MTAP deletion is associated with putrescine secretion also in another cell type, we analyzed media samples from another pair of isogenic MTAP positive and negative cell lines: the breast cancer cell line MCF-7 (denoted MCF7M−), which lacks MTAP expression, and its isogenic pair in which MTAP was reintroduced (MCF7M+).33 We find that also here, the MTAP negative cell line shows a markedly higher putrescine secretion rate, with a 5-fold increase compared to the MTAP positive cells (Figure 5c). Notably, ODC activity in MCF-7 cells was previously associated with the capacity for cellular growth in soft agar.33 This study provides the first direct evidence for an increase in flux through ODC in live cells due to MTAP deletion. A potential mechanistic explanation for the observed increase in flux through ODC following MTAP comes from yeast, where the downstream intermediate in the methionine salvage pathway, MTOB, negatively regulates ODC, and hence its depletion following MTAP deletion may induce ODC flux.34 In support of this hypothesis, we find a roughly 2-fold higher concentration of MTOB in HT1080M+ versus HTM1080−. Further research is needed to test this hypothesis, and more generally to confirm ODC induction following MTAP deletion and to explore other potential underlying mechanisms.
To examine whether the biosynthetic route of ODC's substrate, ornithine, may also depend on MTAP, we fed cells with [U−13C]-glucose. Ornithine can be made via either arginine or glutamate. In both HT1080M+ and HT1080M−, 10% of the glutamate pool is m+2 (labeled from glucose), while less than 1% of the arginine pool gets labeled from glucose (Figure 5d). In both the cell lines, ~4% of the ornithine pool is m+2; thus, MTAP deletion does not change the relative contribution of each of these pathways to ornithine biosyn-thesis, with the arginine and glutamate pathways contributing 60% and 40%, respectively, to ornithine biosynthesis (v8 and v9 in Table 1).
Quantifying Transmethylation Flux
Transmethylation is facilitated by another cyclic pathway, which partially overlaps the propylamine transfer cycle (Figure 2). In transmethylation, SAM serves as the methyl donor yielding S-adenosylhomocysteine (SAH) as a byproduct. SAH loses the adenosine group to make homocysteine, which is then remethylated by methionine synthase to make methionine (with a methyl group from tetrahydrofolate or the choline degradation pathway).
We use a similar approach as above to derive an analytical expression for the flux through the transmethylation cycle. Tracking methionine carbons in transmethylation, a methio-nine m+5 going through this cycle would make methionine m+4, with the methyl group replaced (Figure 3). Notably, a methionine molecule going through both cycles would hence make methionine m+0, though this event is highly unlikely as evident by the undetected level of methionine m+0 (Figure 4a,b). The isotopomer cumulative balance equation for methionine m+4 can hence be used to calculate the methionine synthase flux (v5) as follows:
| (20) |
Due to mass-balance considerations, the transmethylation flux (v4) is equal to the methionine synthase flux (v5) plus homocysteine secretion to the medium (vhcys) and cysteine biosynthesis flux (v6; which is zero as specified above). In the fibrosarcoma cell lines, the homocysteine secretion rate was measured to be ~0.1 nmol/μL-cells/h (Table 1). As the concentration of all four metabolites in the transmethylation cycle was found to be ~50 μM, and transmethylation flux is at least 100 μM/h (based on homocysteine secretion rate), the m+4 labeling of the homocysteine pool is rapid (<2 h; determined via simulation) and hence
Using eq 20 to quantify methionine synthase flux (v5), we find that it is 0.03 ± 0.02 and 0.02 ± 0.01 nmol/μL-cells/h in the MTAP positive and negative cell lines, respectively (Table 1). Hence, quite surprisingly, most of the transmethylation flux (v4) in these cell lines ends with the secretion of homocysteine, instead of the remethylation of homocysteine back to methionine. The low methionine synthase flux is consistent with previous experiments showing the inability of HT1080M− and HT1080M+ to grow in methionine-free media supplemented with homocysteine.35 Notably, while previous studies have shown that MTA inhibits transmethylation in vitro,36,37 our results show that its accumulation due to MTAP deletion does not lead to a major change in transmethylation rate in vivo.
CONCLUSION
This paper presents an analytical method for quantifying methionine metabolic fluxes in mammalian cells that accounts for the exchange of intracellular and media methionine pools, which prevents isotope labeling experiments from reaching an isotopic steady state. Specifically, we provide analytical expressions for fluxes through the propylamine transfer and transmethylation cycles, which depend on straightforward experimental measurements. In marked contrast to existing nonstationary MFA methods that infer flux via complex differential equation models, our method provides closed form expressions for calculating fluxes. To the best of our knowledge, this study represents the first attempt to comprehensively quantify metabolic flux through the multiple methionine-related pathways that are physiologically important in growing cells, including in human cancer. Our results include the first direct evidence that MTAP deletion in cancer directly promotes an increase in flux through ornithine decarboxylase (ODC). Notably, mixing of intracellular and extracellular metabolite pools is not unique to methionine and has been previously demonstrated for other amino acids, such as serine.38 While the applicability of our approach to other metabolic systems depends on the ability to measure required parameters according to the isotopomer cumulative balance equations, we expect that it will prove useful also in many other metabolic systems.
ACKNOWLEDGMENTS
We would like to thank Michel Nofal and Naama Tepper for commenting on this paper.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Sauer U. Mol. Syst. Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Metab. Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boghigian BA, Seth G, Kiss R, Pfeifer BA. Metab. Eng. 2010;12:81–95. doi: 10.1016/j.ymben.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Jin ES. Anal. Biochem. 2004;327:149–155. doi: 10.1016/j.ab.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 5.Sillers R, Al-Hinai MA, Papoutsakis ET. Biotechnol. Bioeng. 2009;102:38–49. doi: 10.1002/bit.22058. [DOI] [PubMed] [Google Scholar]
- 6.Wiechert W, Mollney M, Isermann N, Wurzel M, de Graaf AA. Biotechnol. Bioeng. 1999;66:69–85. [PubMed] [Google Scholar]
- 7.Zamboni N. Curr. Opin. Biotechnol. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Jazmin LJ, Young JD. Methods Mol. Biol. 2013;985:367–390. doi: 10.1007/978-1-62703-299-5_18. [DOI] [PubMed] [Google Scholar]
- 9.Antoniewicz MR. Metab. Eng. 2007;9:277–292. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noh K, Wiechert W. Biotechnol. Bioeng. 2006;94:234–251. doi: 10.1002/bit.20803. [DOI] [PubMed] [Google Scholar]
- 11.Yuan J, Bennett BD, Rabinowitz JD. Nat. Protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baylin SB. Nat. Clin. Pract. Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 13.Xu W. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerner EW, Meyskens FL., Jr. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 15.Laird PW, Jaenisch R. Annu. Rev. Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 16.Bertino JR, Waud WR, Parker WB, Lubin M. Cancer Biol. Ther. 2011;11:627–632. doi: 10.4161/cbt.11.7.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang B, Testa JR, Kruger WD. Cancer Biol. Ther. 2012;13:1082–1090. doi: 10.4161/cbt.21115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munger J. Nat. Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemons JM. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu W. Anal. Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraml CM, Zhou D, Byrne N, McConnell O. J. Chromatogr. A. 2005;1100:108–115. doi: 10.1016/j.chroma.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Lu W, Bennett BD, Rabinowitz JDJ, Chromatogr B. Anal. Technol. Biomed. Life Sci. 2008;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducros V. Anal. Biochem. 2009;390:46–51. doi: 10.1016/j.ab.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Melamud E, Vastag L, Rabinowitz JD. Anal. Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Nat. Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mashego MR. Biotechnol. Bioeng. 2004;85:620–628. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- 27.Birkemeyer C, Luedemann A, Wagner C, Erban A, Kopka J. Trends Biotechnol. 2005;23:28–33. doi: 10.1016/j.tibtech.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Eden E. Science. 2011;331:764–768. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 29.Kadariya Y. J. Biomol. Screening. 2011;16:44–52. doi: 10.1177/1087057110386371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu I. J. Biol. Chem. 2011;286:4902–4911. doi: 10.1074/jbc.M110.198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pajula RL, Raina A. FEBS Lett. 1979;99:343–345. doi: 10.1016/0014-5793(79)80988-6. [DOI] [PubMed] [Google Scholar]
- 32.Bello-Fernandez C, Packham G, Cleveland JL. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christopher SA, Diegelman P, Porter CW, Kruger WD. Cancer Res. 2002;62:6639–6644. [PubMed] [Google Scholar]
- 34.Subhi AL. J. Biol. Chem. 2003;278:49868–49873. doi: 10.1074/jbc.M308451200. [DOI] [PubMed] [Google Scholar]
- 35.Tang B, Li YN, Kruger WD. Cancer Res. 2000;60:5543–5547. [PubMed] [Google Scholar]
- 36.Della Ragione F, Pegg AE. Biochem. J. 1983;210:429–435. doi: 10.1042/bj2100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferro AJ, Vandenbark AA, MacDonald MR. Biochem. Biophys. Res. Commun. 1981;100:523–531. doi: 10.1016/s0006-291x(81)80208-2. [DOI] [PubMed] [Google Scholar]
- 38.Possemato R. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]