Abstract
Single particle cryo-electron microscopy (cryo-EM) is an emerging powerful tool for structural studies of macromolecular assemblies (i.e., protein complexes and viruses). Although single particle cryo-EM requires less concentrated and smaller amounts of samples than X-ray crystallography, it remains challenging to study specimens that are low-abundance, low-yield, or short-lived. The recent development of affinity grid techniques can potentially further extend single particle cryo-EM to these challenging samples by combining sample purification and cryo-EM grid preparation into a single step. Here we report a new design of affinity cryo-EM approach, cryo-SPIEM, that applies a traditional pathogen diagnosis tool Solid Phase Immune Electron Microscopy (SPIEM) to the single particle cryo-EM method. This approach provides an alternative, largely simplified and easier to use affinity grid that directly works with most native macromolecular complexes with established antibodies, and enables cryo-EM studies of native samples directly from cell cultures. In the present work, we extensively tested the feasibility of cryo-SPIEM with multiple samples including those of high or low molecular weight, macromolecules with low or high symmetry, His-tagged or native particles, and high- or low-yield macromolecules. Results for all these samples (nonpurified His-tagged bacteriophage T7, His-tagged E. coli ribosomes, native Sindbis virus, and purified but low-concentration native Tulane virus) demonstrated the capability of cryo-SPIEM approach in specifically trapping and concentrating target particles on TEM grids with minimal view constraints for cryo-EM imaging and determination of 3D structures.
Keywords: affinity cryo-EM, affinity grid, antibodies, single particle cryo-EM, 3D reconstruction, bacteriophage
Introduction
Solid Phase Immune Electron Microscopy (SPIEM) was first developed as a quantitative assay of plant viruses by Derrick in 1973 (Derrick, 1973a). By precoating a TEM sample grid with a particular antiserum, target virus particles were specifically trapped and concentrated on the grid surface. This method that combines serology and electron microscopy was commonly used in the detection and rapid diagnosis of many plant viruses (Derrick, 1973b; Paliwal, 1977) and mammalian viruses (Berthiaume et al., 1981; Lewis, 1991; Nicolaieff et al., 1980). This technique was later modified by introducing Protein A (Shukla and Gough, 1979), a protein that was initially found in the cell wall of Staphylococcus aureus and can specifically bind to the Fc region of IgG antibodies. The treatment of TEM grids with Protein A prior to antiserum coating allowed specific extraction of IgGs from the non-purified antiserums, presented them in optimal orientations for pathogen binding and therefore increased the sensitivity of this technique (Gough and Shukla, 1980; Shukla and Gough, 1979). Protein A SPIEM was an essential tool for rapid virus detection (Berthiaume et al., 1981; Wu et al., 1990), serology and antigenic studies of enteric viruses (Gerna et al., 1984; Lewis et al., 1995; Lewis, 1990; Lewis et al., 1988). This was especially true during the eighties and early nineties. Although viral diagnosis is now performed mainly using the Polymerase Chain Reaction and sequencing techniques, the SPIEM technique remains a useful, complementary tool for pathogen diagnosis.
Single particle cryo-electron microscopy (cryo-EM) and 3D reconstruction is a rising powerful tool to determine structures of macromolecules with achievable resolution now often at near-atomic resolutions (3–4 Å) (Bai et al., 2013; Jiang et al., 2008; Liao et al., 2013; Liu et al., 2010; Yu et al., 2013b; Zhang et al., 2010). Although single particle cryo-EM and 3D reconstruction technique eliminates the need of sample crystallization and consequently saves a significant amount of efforts, highly purified samples at appropriate concentrations (i.e., ~1 mg/ml) are still necessary for efficient imaging to obtain a large number of particle images for high resolution 3D reconstructions. In recent years, efforts have been made to simplify the sample preparation of cryo-EM and make it a routine, high throughput structural determination approach for most biological systems. One of the directions being explored is to skip the large-scale, time-consuming sample purification step by using the “Affinity Grid” technique (Kelly et al., 2008a) that combines sample purification and grid preparation for cryo-EM imaging into a single step. Currently, multiple related approaches, including the Ni-NTA functionalized lipid monolayer purification methods (Kelly et al., 2008a; Kelly et al., 2010), the streptavidin 2D crystal-based approach (Han et al., 2012), and the recent functionalized carbon surface approach (Llaguno et al., 2014), have been reported. However, a significant amount of efforts are still required for either (genetic or chemical) modification of specimens or generation of the affinity layers on TEM grids. For example, both the lipid monolayer generation and the transfer of lipid monolayer to the TEM grid in the Ni-NTA functionalized lipid monolayer purification method are complicated procedures requiring special tools and training.
Considering the capabilities of “purification” and “concentration” of targets allowed by the SPIEM technique, we combined the traditional SPIEM technique with the emerging structural biology tool cryo-EM and established a new antibody-based affinity grid approach, cryo-SPIEM, by which crude extracts (e.g., patient isolates, cell cultures or lysates) without any modification and extensive purification can be directly used for cryo-EM imaging and 3D reconstruction. Such a cryo-SPIEM technique provides an alternative affinity grid technique that uses a much simpler affinity layer preparation protocol than other approaches. There are multiple advantages for using the cryo-SPIEM technique. First, akin to other affinity grid approaches, it will significantly simplify and expedite the sample preparation of single particle cryo-EM by reducing or even eliminating the need of traditional, relatively large-scale purification of samples. In addition, because of this substantially simplified cryo-EM sample preparation protocol, there would be better chances to capture and image some transient intermediates that have been neglected with traditional purification approaches. Finally, the cryo-SPIEM technique coupled with serologically specific viral particle-affinity will also benefit the serotype-specific structural studies of patient-isolated viruses. This will be especially helpful in the case of viruses with many serotypes, e.g., the Rhinovirus with 99 serotypes (Palmenberg et al., 2009). Moreover, the application of cryo-SPIEM could possibly resolve the structure of the pathogens directly using samples from outbreaks, even before the establishment of laboratory cell culture systems. This aspect is important, given that some non-cultivatable human viruses are already known to be of serious health concern (e.g., human noroviruses (Duizer et al., 2004)). In this work, we extensively demonstrated the feasibility of cryo-SPIEM in imaging non-purified viruses in cell cultures (including viral assembly intermediates), non-purified protein complexes and purified yet low-concentration sample.
Materials and Methods
Antibodies and Antiserums
Monoclonal anti-His IgG antibody (Sigma Aldrich) was diluted to 10 µg/ml in phosphate-buffered saline (PBS), pH 7.4 for the coating of TEM grids. The monoclonal sv127 IgG antibody (Snyder et al., 2011) against the envelope protein 2 (E2) of Sindbis virus was used to trap the virions from infected cell culture supernatants. Anti-TV antiserum was produced by immunization of mouse with purified Tulane virus. The TV antiserum was diluted by 1:100 in PBS, pH 7.4 for grid coating.
Protein A
Protein A (Sigma Aldrich) was dissolved in filter-sterile distilled water to 1 mg/ml. For grid coating, the stock was diluted to 50 µg/ml in PBS, pH 7.4.
Preparation of antibody-based affinity grids
TEM grids, coated with holey/continuous carbon (Ted Pella Inc.) or Formvar-carbon film (homemade), were glow-discharged for about 30 s (EMITECH K950X). 3 µl of purified antibodies at ~1–10 µg/ml (may vary for different antibodies) was directly applied to the carbon side of the TEM grid. After 5 min incubation at room temperature, excess liquids on the grid were removed with filter paper and the grid was ready for applying to specimens. For the non-purified antibodies in the supernatant of hypodermal cell culture (e.g., anti-E2 used in this work) or in antiserum (e.g., anti-TV used here), a Protein A coating step was added to enrich IgG antibodies on the grid surface. The procedure was that of Protein A SPIEM technique (Shukla and Gough, 1979) with some modifications. Briefly, 5 µl of Protein A (50 µg/ml) was applied to the grid surface and incubated for 10 min. After blotting away the excess liquids on grid, 3 µl of non-purified antibodies (may vary for different samples) was applied onto the grid and incubated in a humid chamber for 15 min. After washing three times with PBS, pH 7.4 to remove unbound antibodies and other contaminants in the cell culture supernatant or the antiserum, the antibody-based affinity grid was ready for use.
Data collection and image processing of His-tagged bacteriophage T7 in a cell lysate
(1) Preparing T7 cell lysate. E. coli BL21 cells at an optical density (600 nm) of 0.6 were infected with the His-tagged bacteriophage T7 at a Multiplicity of Infection (MOI) of 10. Incubation was continued at 37 °C with aeration. When visible cell lysis was complete (about 50 min), the cell lysate (titer≈1010/ml) was collected and directly used to prepare EM sample grids. (2) Negative-stain TEM. The Formvar-carbon grid coated with anti-His antibody (1 µg/ml) floating on 50 µl His-T7 cell lysate was incubated in a humid chamber for 30 min at room temperature, after which the grid was washed with PBS (twice) and distilled water (once), and then stained with 2% phosphotungstic acid (PTA), pH 7. TEM images were taken using a Philips CM200 TEM with a 1K×1K CCD. (3) Preparing the frozen-hydrated sample and data collection. The holey/continuous grid coated with anti-His antibody was incubated with 50 µl His-T7 cell lysate at room temperature in a humid chamber for 30 min to 1 hour. After 3 steps of washing with PBS, the grid was blotted with filter paper, plunge-frozen in liquid ethane using the Vitrobot cryo-plunger (FEI Co.) and then imaged using a Philips CM200 TEM operated at 200 KV, liquid nitrogen temperature. Images were recorded on Kodak SO-163 photographic films at 27.5 K nominal magnification with a total dose of 20 e/Å2 and digitized using a Nikon Super CoolScan 9000 scanner. The final sampling of the image was 2.24 Å/pixel. (4) Image processing. The images were 2× binned for further image processing. The three states of T7 phage: procapsid (CI) (300 particles), empty capsid II (CII) (258 particles) and mature phage (900 particles), were selected and grouped visually using the e2boxer.py program of EMAN2 (Tang et al., 2007). The “truly independent reconstruction” strategy (Guo and Jiang, 2014) was adopted for all three datasets. Briefly, after estimating the contrast transfer function (CTF) parameters using the fitctf2.py program (Jiang et al., 2012), individual datasets were split into two halves, whereby de novo initial 3D models were built separately using the “random model” method by assigning random orientations to subsets of randomly picked particles from individual half datasets and iteratively refining until convergence. The 2D alignment for each half dataset was performed independently with the “projection matching” method that compares the CTF-corrected 2D images with the projections of the models, and 3D models were built using the “direct Fourier inversion” approach. All the steps including the initial model building and the iterative 2D alignment and 3D reconstruction were carried out using the in-house developed program jspr.py (Guo and Jiang, 2014) based on EMAN/EMAN2 libraries (Ludtke et al., 1999; Tang et al., 2007). Resolutions of the reconstructions were evaluated using the gold-standard Fourier shell correlation (FSC) criterion, i.e. the independently refined reconstructions built from the two halves were calculated to evaluate the resolution based on the 0.143 cutoff criterion (Rosenthal and Henderson, 2003; Scheres and Chen, 2012).
Data collection and image processing of His-tagged ribosomes in a cell lysate
(1) Preparation of His-tagged RplL ribosome cell lysate. His-tagged ribosome large subunit protein RplL strain in the ASKA(+) library was obtained from NBRP-E.coli at NIG (Kitagawa et al., 2005). The bacterial strain was cultured and induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside for overexpression at 37 °C for 3 hours. Cells were then pelleted by centrifugation and washed with distilled water. The cell pellets were resuspended in buffer containing 20 mM Tris-HCl pH 7.5, 150 mM KCl, 10 mM MgCl2 and 0.2 mg/ml lysozyme, and lysed by sonication. The lysate was diluted by 20-fold for preparation of EM specimen. (2) Preparation of frozen-hydrated sample for cryo-EM data collection. 5 µl diluted cell lysate was applied to the holey/continuous grid coated with anti-His antibody (5 µg/ml) and incubated for 5 min at room temperature in a humid chamber. The grid was washed 3 times with the lysis buffer without lysozyme and plunge-frozen in liquid ethane. Data was collected using a Philips CM200 TEM operated at 200 KV, liquid nitrogen temperature. 50K nominal magnification and a total dose of 20 e/Å2 were used for imaging on the Kodak SO163 photographic films. (3) Image processing. The digitized images were binned by 4×4 to a final sampling of 4.98 Å/pixel. The CTF parameters of each micrograph were estimated using CTFFIND3 (Mindell and Grigorieff, 2003). About 4,800 particles were selected using e2boxer.py program of EMAN2 (Tang et al., 2007). The overall image processing, including reference-free 2D classification, 3D classification and refinement, was performed using RELION (Scheres, 2012). After 2D classification using a regularization number of T=2, the whole data set was separated into 50 classes, from which sub-datasets of the 70S (1663 particles), 50S (2213 particles) and 30S (270 particles) ribosome particles were selected based on size and morphology and bad particles were removed. 3D classification of individual subsets was performed to further select homogenous particles. The 3D classification reference map of 70S ribosome was downloaded from electron microscopy data bank (EMDB-1657) and low-pass filtered to 60 Å. References for the 50S and 30S ribosomal subunits were generated from the X-ray crystallography structures, PDB-3E1B and PDB-4B3M, using e2pdb2mrc.py of EMAN2 and low-pass filtered to 60 Å and 80 Å, respectively. The final refinement of individual relatively homogenous subsets with an angular sampling rate of 3.7° and a regularization number of 3 was performed till convergence. The final resolutions were calculated using the gold standard FSC with 0.143 cutoff criterion (Rosenthal and Henderson, 2003).
Data collection and image processing of Sindbis virus in cell culture supernatant
(1) Cell culture and Sindbis virus (SINV) propagation. The SINV (TE12) strain (Lustig et al., 1988) was propagated in BHK-21 cells at 37 °C. Supernatant containing the progeny SINV virions (titer≈108/ml) was collected at 16 hours post infection. (2) Negative staining TEM. Formvar-carbon grid coated with protein A and anti-E2 was incubated with 50 µl SINV supernatant for about 1 hour at room temperature, and washed and stained with 2% PTA, pH 7. (3) Cryo-EM data collection and image processing. The original SINV supernatant was concentrated by ~8-fold via low speed centrifuge in a Millipore filter unit with 50 KDa cutoff. The holey/continuous grid coated with anti-E2 antibody via protein A was incubated with 50 µl concentrated SINV supernatant at room temperature in a humid chamber for 1–1.5 hour, washed with phosphate buffer and plunge-frozen in liquid ethane. Images were collected and processed using the same conditions and methods adopted by His-T7 bacteriophage.
Data collection and image processing of low-concentration Tulane virus
(1) Purification of Tulane virus (TV). TV sample was purified using the same approach as described elsewhere (Farkas et al., 2008; Yu et al., 2013a). (2) Negative staining TEM. 6–10 µl of purified TV sample was applied to a Formvar-carbon grid coated with protein A and anti-TV antiserum and incubated for 15–30 min in a humid chamber. After washing with PBS buffer and distilled water, the grid was stained with 2% PTA pH 7 and imaged using TEM. (3) Cryo-EM data collection and image processing. Sample was prepared in the same way as for negative staining TEM, except that the Formvar-carbon grid was replaced with a holey/continuous carbon grid and the grid was plunge-frozen into liquid ethane instead of being stained. The imaging conditions, reconstruction, and resolution assessment methods were those used for His-T7.
Results
Cryo-SPIEM could successfully capture His-tagged macromolecules with high or no symmetry from crude cell extracts
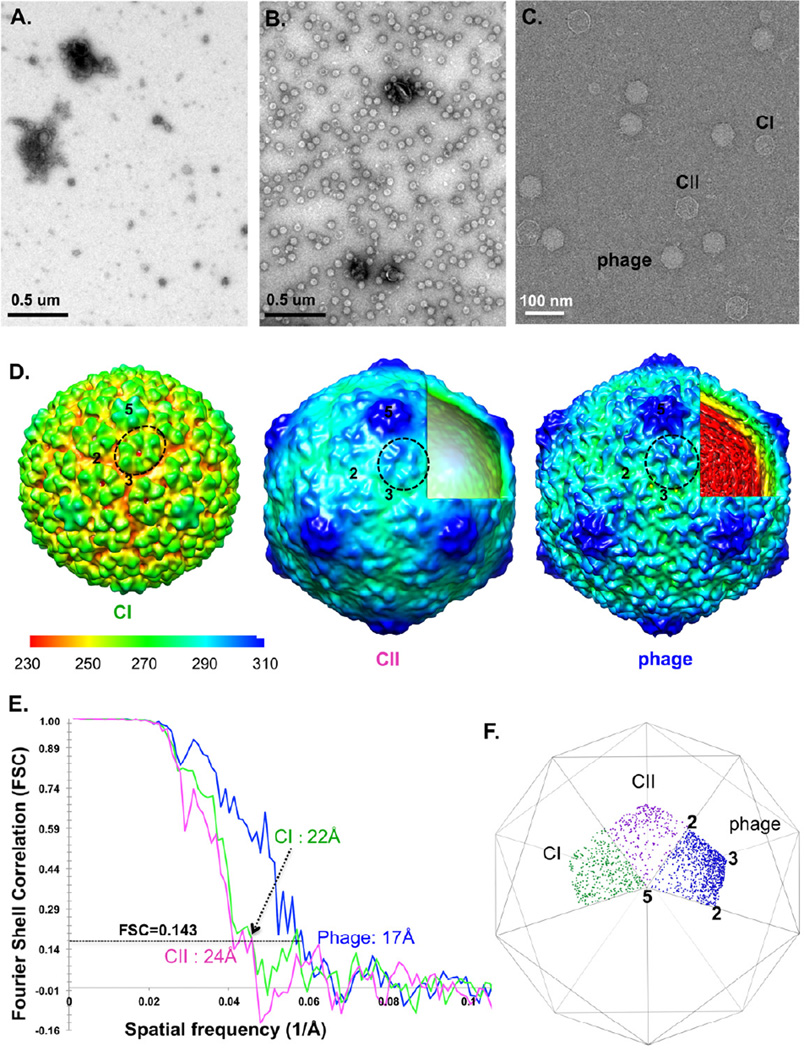
Among the most commonly used affinity tags in biological studies is the His-tag. To test the cryo-SPIEM technique with the anti-His antibody, the purified anti-His IgG monoclonal antibodies were directly coated onto the carbon film with random orientations as illustrated in Figure 1A, and two His-tagged macromolecules, His-tagged bacteriophage T7 with high symmetry and His-tagged ribosome without symmetry, were tested. Bacteriophage T7 in the Podoviridae family is a dsDNA virus that infects most of the E. coli bacterial strains (Studier, 1972). The 6×His-tag was constructed at the surface-facing C-terminus of its major capsid protein gp10 so that the tags are exposed on the T7 particle surfaces without interfering the assembly and maturation of the particles. As shown in Figure 2, His-T7 particles were specifically trapped and concentrated on the anti-His antibody coated grid (Fig. 2B), but not on the control grid without anti-His antibodies (Fig. 2A), which demonstrated the “purification” and “concentration” functions of the anti-His SPIEM grid. Particles in three, known major states of phage T7 were captured simultaneously from the cell lysate: the initial procapsid (CI), the empty capsid II (CII) and the genome DNA-filled mature phage (Fig. 2C). 3D reconstructions were performed for all three states. CI, CII and mature phage were reconstructed to 22 Å, 24 Å and 17 Å resolutions, respectively, using the gold-standard FSC=0.143 criterion (Fig. 2D–E). All three maps reveal T=7 icosahedral shells with different sizes. As previously described (Cerritelli et al., 2003; Cerritelli et al., 1997), the diameter of CI was significantly smaller than the other two expanded states (supplementary Fig.1). However, we are surprised to find that the empty CII capsid was ~8 Å larger than the mature phage in rotationally averaged diameter (supplementary Fig.1). The fact that all the particles were captured on the same TEM grid and imaged simultaneously in the same dataset eliminates the possibility of size variance caused by TEM magnification fluctuation. The molecular mechanism of this newly observed size reduction from CII to mature phage and its potential association with T7 DNA packaging (Serwer et al., 2010), remain to be elucidated. All other structural features, including the elliptical hexamers of CI, the L-shaped ridge of individual gp10 subunits, and the nearly equally-spaced concentric rings for genomic DNA of the mature phage (Fig. 2D), were consistent with previously reported observations on T7 (Agirrezabala et al., 2005; Cerritelli et al., 2003; Cerritelli et al., 1997; Guo et al., 2013).
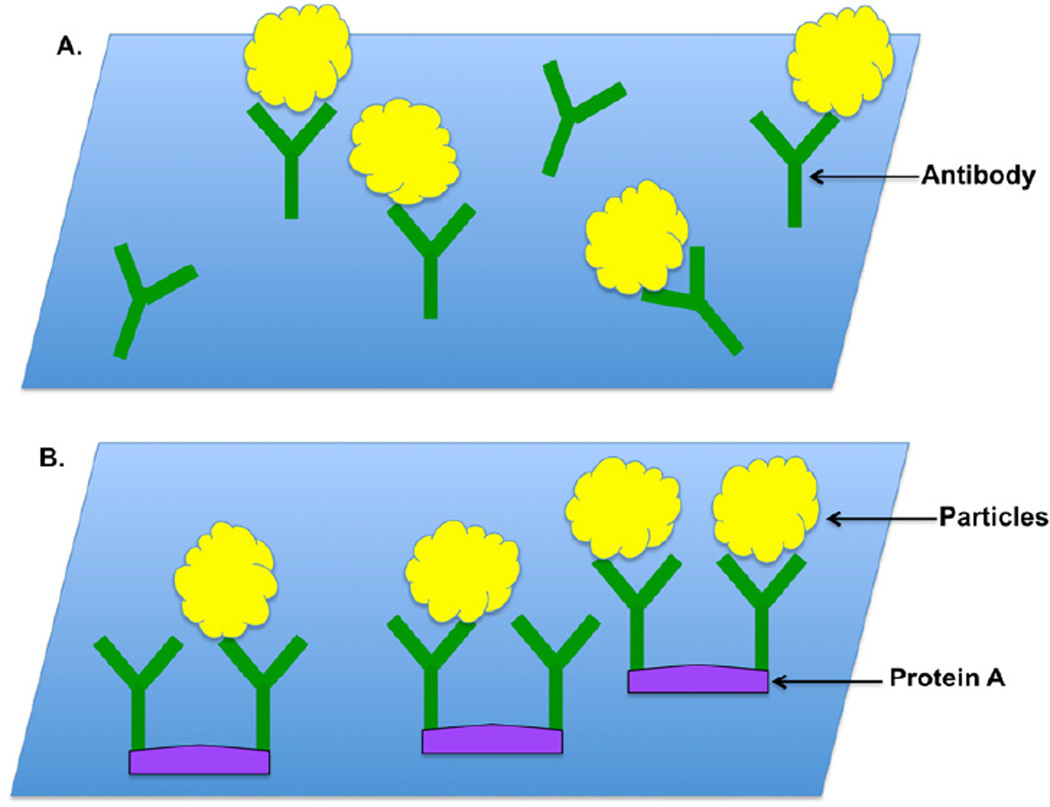
Figure 1. Schematic diagrams of antibody-coated grids.
(A) Grids directly coated with purified antibodies. (B) Non-purified antibody coated grid with the help of protein A.
Figure 2. His-T7 bacteriophages directly captured from a non-purified cell lysate by SPIEM grids coated with anti-His antibodies.
Negative staining TEM images of a control grid (A) and anti-His antibodies coated grid (B) show the specific “trapping” and “concentrating” of target particles. (C) cryo-SPIEM images of His-T7 particles directly from a cell lysate. Three states of T7 phages: initial procapsid (CI), the empty capsid II (CII) and the DNA-filled mature phage. Single particle cryo-EM 3D reconstruction of the three states of His-T7 (D) and the estimated resolutions using gold-standard FSC=0.143 criterion (E). The figures: “2”, “3” and “5”, indicate the icosahedral two-, three- and five-fold axes, respectively. One capsid protein hexamer of each map was circled. (F) shows the Euler angle distributions of CI, CII and mature phage particles on cryo-SPIEM grids.
Euler angle distribution analyses indicated relatively uniform orientation distribution of CI particles, but some bias toward the icosahedral 3-fold axis was observed for the empty CII particles and mature phages (Fig. 2F). The major cause of this bias is probably the angular shape of the CII and mature phage particles, as similar levels of bias were also seen for phages imaged using regular cryo-EM grids (Jiang et al., 2003; Jiang et al., 2006; Jiang et al., 2008). Taken together, this work on His-T7 bacteriophage strongly supports the feasibility of cryo-SPIEM as a significantly simplified sample preparation method for single particle cryo-EM.
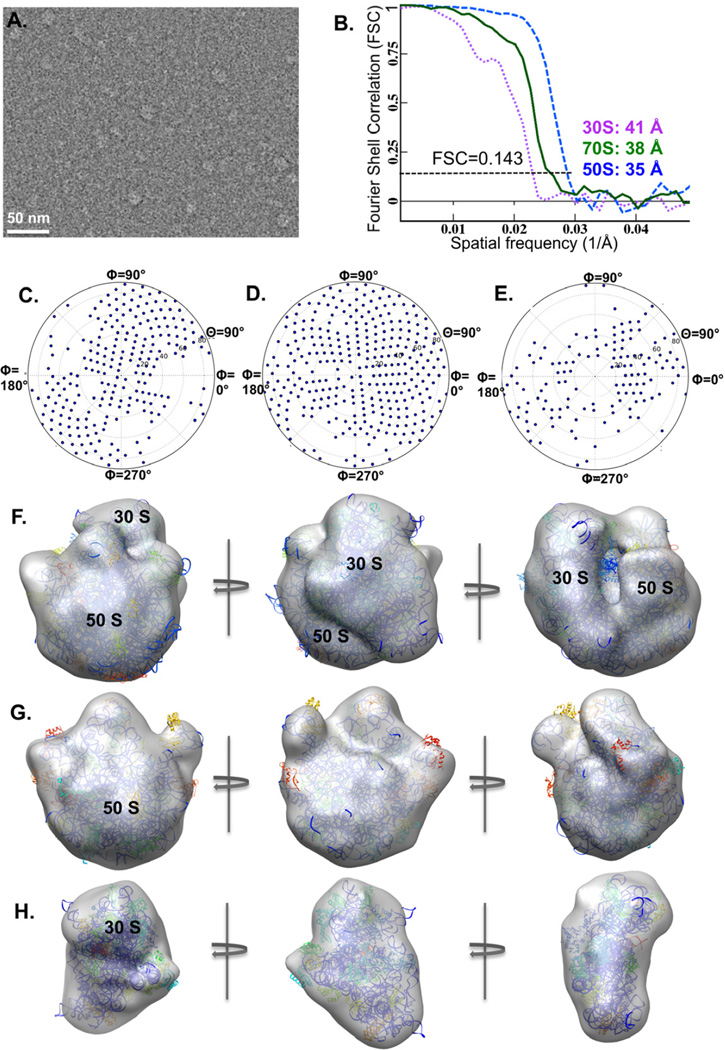
The anti-His affinity grid also worked well in purifying the His-tagged E. coli ribosomes from an unfractionated cell lysate for cryo-EM imaging and structure determination (Fig. 3A). Since the His-tag is located in the 50S ribosomal subunit, the captured particles were dominated by the 50S (2213 particles) and 70S (1663 particles) ribosomes as expected. Meanwhile, a relatively small number of 30S ribosomes (270 particles) that were probably captured through binding to the His-tagged 70S-bound mRNA molecules (Kelly et al., 2008b) or due to the dissociation of His-tagged 70S prior to sample freezing, were also identified after 2D classification using RELION (Scheres, 2012). The 70S, 50S and 30S ribosomes were reconstructed to 38 Å, 35Å and 41Å resolutions, respectively (Fig. 3B). Plots of the Euler angles showed that the particle orientations of each reconstruction covered most of the tilt angle space (θ=0°−90°), although the azimuthal rotation angle space (Φ) was not fully covered possibly due to the limited number of particles (Fig. 3C–E). The reconstructed structures were verified by fitting the atomic structures of 70S (PDB-1ML5), 50S (PDB-3E1B) and 30S (PDB-4B3M) ribosomes into the corresponding cryo-EM densities (Fig. 3F–H). This preliminary success with non-purified ribosome samples demonstrated the feasibility of cryo-SPIEM in studying low-symmetry protein complexes.
Figure 3. His-tagged E. coli ribosomes directly imaged using the cryo-SPIEM approach with anti-His antibodies.
(A) An anti-His antibody based cryo-SPIEM image of E. coli ribosomes from a crude cell lysate. (B) Fourier shell correlations curves of the 70S, 50S and 30S ribosomes captured on the cryo-SPIEM grid. And the Euler angle distributions of the 70S (C), 50S (D) and 30S (E) are plotted. The 3D reconstructions of the 70S, 50S and 30S ribosomes fitted with corresponding crystallographic structures (PDB-1ml5, 3e1b, 4b3m) are shown in (F), (G) and (H), respectively.
Cryo-SPIEM could directly capture enveloped, non-tagged virions from infected cell cultures
To evaluate the feasibility of cryo-SPIEM for enveloped viruses, a Sindbis virus sample was also tested. Sindbis virus (SINV), the prototypical alphavirus in the Togaviridae family, is an enveloped, positive-sense, single-stranded RNA virus that infects both animals and human beings (Kurkela et al., 2008). We bypassed the traditional, extensive purification procedure of SINV (Hernandez et al., 2010; Pletnev et al., 2001) and directly used the supernatants of infected cell cultures for TEM studies. Non-purified, monoclonal antibodies against the envelope protein E2 of SINV were coated on the TEM grids via protein A, as illustrated in Figure 1B. Similar to the case of His-T7, SINV virions were also successfully purified from the original, crude cell culture supernatant by the anti-E2 SPIEM grid (Fig. 4A–B). For cryo-SPIEM grid preparation, the original supernatant was concentrated by ~8-fold via low speed centrifuge in a filter unit. This is to ensure an adequate amount of particles on the cryo-SPIEM grid for efficient cryo-EM imaging. As demonstrated in Figure 4C, a desirable amount of SINV particles were trapped on the grid after plunge-frozen in liquid ethane. From the reconstructed map at 25 Å resolution using about 1,600 particles, the characteristic 80 trimetric “spikes” of SINV envelope proteins are clearly resolved (Fig. 4D–E). The Euler angles of SINV particles displayed a uniform coverage of the angle space (Fig. 4F), indicating that the binding of SINV particles to the anti-E2 antibody on grid surface did not introduce preferred orientations. Altogether, this result suggested that cryo-SPIEM is a promising method for structural studies of non-purified enveloped viruses in a condition more close to their native environments.
Figure 4. Native enveloped Sindbis virus directly captured from infected BHK-21 cell culture supernatant for single particle cryo-EM 3D reconstruction.
Negative staining images show that Sindbis particles specifically bind to the grid coated with anti-E2 antibodies (B) but not to the grid without anti-E2 (A). (C) shows one representative cryo-SPIEM images of Sindbis particles directly from the 8-fold concentrated cell culture supernatant. (D) Single particle 3D reconstruction of the captured particles. “2”, “3” and “5” represent the icosahedral two-, three- and five-fold axes, respectively. (E) Fourier shell correction of the reconstruction. (F) plots the Euler angles of the Sindbis particles captured on the cryo-SPIEM grid.
Cryo-SPIEM could also benefit structural studies of low-yield samples
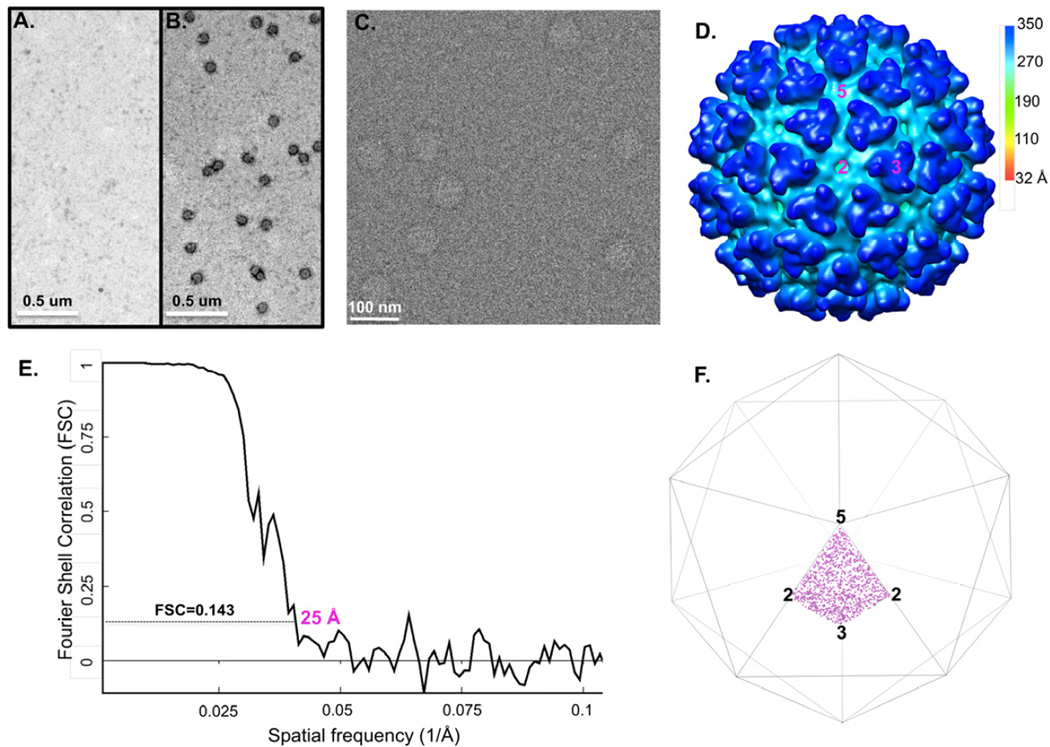
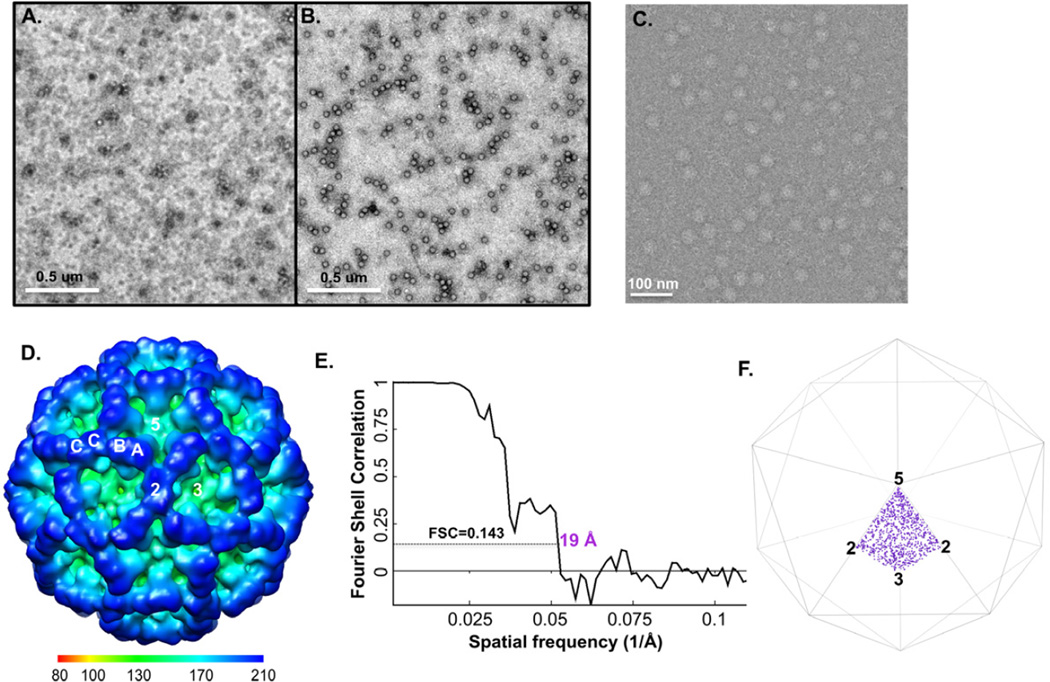
For some low-yield systems or some low-abundance macromolecular species, it remains challenging to obtain samples at the required concentrations for cryo-EM studies. To test the ability of cryo-SPIEM approach in dealing with such challenging samples, a batch of purified Tulane virus (TV) sample of low concentration was used. As shown in Figure 5A, the TV sample was very dilute and not suitable for cryo-EM imaging. By using a larger sample volume (i.e., 10 µl instead of 3 µl) and an affinity surface generated with protein A and anti-TV antiserum, a significant improvement of particle density on the anti-TV grid was obtained (Fig. 5B). Cryo-SPIEM images with desirable particle densities were also collected (Fig. 5C), and a 19 Å TV structure was obtained with about 900 particles using the single particle cryo-EM and 3D reconstruction technique (Fig. 5D–E). The overall structural features of the reconstruction, such as the shape and the arrangement of TV capsid dimers A/B and C/C (Fig. 5D), agreed well with our previous observation (Yu et al., 2013a). The orientation distribution plot showed that TV particles were uniformly distributed in the Euler angle space (Fig. 5F). In summary, this work with a low-concentration TV sample proved that the cryo-SPIEM approach potentially could provide a generic solution to the low sample concentration issue often encountered in the single particle cryo-EM field.
Figure 5. Low-concentration TV sample enriched on the SPIEM grid coated with anti-TV antiserum.
(A) Original TV sample used for the experiment. (B) The sample was concentrated on the TEM grid coated with anti-TV serum with the help of protein A by ~100 fold. (C) Cryo-SPIEM image of TV showing desirable particle density. (D) Single particle cryo-EM 3D reconstruction of the on-grid concentrated TV particles. “2”, “3” and “5”, indicate the icosahedral two-, three- and five-fold axes, respectively. Letters “A”, “B” and “C” represent three types of capsid subunits of TV. (E) Fourier shell correlation curve of the reconstruction indicates a resolution of 19 Å with FSC=0.143 cutoff. (F) Euler angles plot of the TV particles bound on the cryo-SPIEM grid.
Discussion and Conclusion
In this work, we report an alternative affinity grid method, cryo-SPIEM, to immobilize and concentrate particles of interest onto the TEM sample grid surfaces. Neither genome engineering (e.g., 6×His tagging) nor chemical modification (e.g., biotinylation) of particles of interest is required, although they can be used. Omitting of these modification steps substantially reduces the amount of work required and also enables the study of native macromolecules. This is different from other affinity grid approaches, such as the Ni-NTA lipid monolayer-based affinity grid (Kelly et al., 2008a), streptavidin crystal based grid (Han et al., 2012) and the Ni-NTA functionalized carbon surface approach (Llaguno et al., 2014), for which sample modifications are necessary in most cases. Additionally, the cryo-SPIEM approach is much simpler and easier to use compared with other affinity grid approaches in terms of affinity layer generation on surface of TEM grids (see Materials and Methods). Given the broad usage and availability of antibodies already developed for other biological studies, cryo-SPIEM can be easily applicable to most biological systems.
Similar to other affinity grid approaches, the cryo-SPIEM approach could simplify and expedite the cryo-EM sample preparation by directly using crude cell extracts as demonstrated by our work. In addition, in this simplified protocols of cryo-EM sample preparation, particles of interest would be largely maintained in their native environments, which would be favorable for those labile macromolecular complexes that cannot survive extensive purifications.
Despite the aforementioned advantages, one potential concern about affinity grids, in general, is the increased background noise arising from the addition of affinity layers, such as lipids, streptavidin crystal or antibodies (protein A-antibodies for some cases), which probably could limit the achievable resolution. This is possibly less of a problem for affinity grids layered with streptavidin crystals, as the background from streptavidin 2D crystals can be removed by subtracting the diffraction spots of streptavidin crystal lattices from the images (Wang et al., 2008). For both the lipid- and the antibody-based affinity grid approaches, however, it is currently impossible to remove the unwanted background and therefore is more likely to limit the resolution of 3D reconstruction. Therefore, to alleviate this concern and minimize the amount of biomass introduced to the carbon films as much as possible, for the purified antibodies (e.g., the anti-His used in this work), we directly coated them to TEM grids without the intermediate layer of protein A. On the other hand, the immobilization of macromolecular particles onto a thin affinity layer could reduce the Brownian motion and the damage of particles caused by interactions with the air-water interface (Taylor and Glaeser, 2008), and therefore allow for thinner vitreous ice and less background noise to balance out part of the noise from the affinity layer. The resolution limit of the cryo-SPIEM approach and its potential for near-atomic resolution remain to be investigated.
It has been shown that the protein A SPIEM approach can concentrate target viral particles by about 2 orders of magnitude (Shukla and Gough, 1979). Therefore, with the help of this approach and the possibility of applying a larger sample volume to the grid, it could significantly abate the required sample concentration for cryo-EM studies, in general. As shown here, crude samples with titers of 109–1010/ml yielded cryo-EM grids with sufficient amounts of target particles for imaging and 3D reconstruction. For different specimens and different antibodies, however, the required sample titer may vary significantly as the antibody-antigen binding affinities range from 105 M−1 to 1012 M−1. It remains challenging to work with highly dilute specimens (i.e. <107/ml), as the low titer of the specimen would limit the number of bound particles at equilibrium. As we have done with the SINV sample, preconcentration of specimens using low-speed centrifugation is one of the potential solutions. Nevertheless, an interesting direction that deserves exploration is increasing the local concentration of target macromolecules nearby the affinity surface using concentration methods like dielectrophoresis (Nakano and Ros, 2013). In the cryo-SPIEM technique, the antibodies (or protein A) were coated on the TEM grid via hydrophobic interactions between the non-polar residues of antibodies (or protein A) and the carbon substrate, similar to the enzyme-linked immunosorbent assay (ELISA) (Loizou et al., 1985; Yoshizawa et al., 2004). Although ligands coating could be optimized by varying the conditions (e.g., buffers) as shown in ELLSAs, the immobilization efficiency of such a passive adsorption process is usually limited and largely uncontrollable. Therefore, quantitatively and qualitatively controlled covalent linking of antibodies to the supporting substrates (i.e., carbon, graphene/graphene oxide) probably will further improve the particle capturing and concentration capability of the SPIEM grid and further lower the concentration limit of cryo-EM specimens. These potential improvements will be further investigated in follow-up works.
In conclusion, our extensive exploration of the cryo-SPIEM technique, using a diverse set of specimens (low- and high-symmetry, large and small, native and tagged), suggests that cryo-SPIEM can be an alternative, much less time-consuming, easy to use and broadly applicable approach for cryo-EM sample preparation. Particles with uniformly distributed orientations were obtained in the test samples, presumably due to the intrinsic flexibility of the Fab of IgG antibodies. For the case in which purified IgG antibodies were directly coated onto the carbon substrate, the random orientations of coated IgGs possibly also contribute to the uniform Euler angle space coverage of bound particles.
Supplementary Material
Rotationally averaged radial density plots of His-T7 CI (green), CII (purple) and mature phage (blue). Peak positions for the capsid shells of CI (244 Å), CII (281 Å) and mature phage (277 Å) are marked.
Acknowledgement
This work was supported in part by NIH grant R01AI072035. The negative staining TEM and cryo-EM images were taken at the Purdue Biological Electron Microscopy Facility. We thank Melissa Mikolaj and Thomas J. Edwards for providing some of the Sindbis virus test samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. Embo J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiaume L, Alain R, McLaughlin B, Payment P, Trepanier P. Rapid detection of human viruses in faeces by a simple and routine immune electron microscopy technique. The Journal of general virology. 1981;55:223–227. doi: 10.1099/0022-1317-55-1-223. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem. 2003;64:301–323. doi: 10.1016/s0065-3233(03)01008-8. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- Derrick KS. Quantitative Assay for Plant Viruses Using Serologically Specific Electron-Microscopy. Virology. 1973a;56:652–653. doi: 10.1016/0042-6822(73)90068-8. [DOI] [PubMed] [Google Scholar]
- Derrick KS. Detection and Identification of Plant Viruses by Serologically Specific Electron-Microscopy. Phytopathology. 1973b;63:441–441. [Google Scholar]
- Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. The Journal of general virology. 2004;85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- Farkas T, Sestak K, Wei C, Jiang X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J Virol. 2008;82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G, Passarani N, Battaglia M, Percivalle E. Rapid serotyping of human rotavirus strains by solid-phase immune electron microscopy. J Clin Microbiol. 1984;19:273–278. doi: 10.1128/jcm.19.2.273-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough KH, Shukla DD. Further-Studies on the Use of Protein-a in Immune Electron-Microscopy for Detecting Virus-Particles. J Gen Virol. 1980;51:415–419. doi: 10.1099/0022-1317-51-2-415. [DOI] [PubMed] [Google Scholar]
- Guo F, Jiang W. Single Particle Cryo-electron Microscopy and 3-D Reconstruction of Viruses. Methods Mol Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. P Natl Acad Sci USA. 2013;110:6811–6816. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BG, Walton RW, Song A, Hwu P, Stubbs MT, Yannone SM, Arbelaez P, Dong M, Glaeser RM. Electron microscopy of biotinylated protein complexes bound to streptavidin monolayer crystals. J Struct Biol. 2012;180:249–253. doi: 10.1016/j.jsb.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Hernandez R, Sinodis C, Brown DT. Sindbis virus: propagation, quantification, and storage. Curr Protoc Microbiol Chapter. 2010;15(Unit15B):11. doi: 10.1002/9780471729259.mc15b01s16. [DOI] [PubMed] [Google Scholar]
- Jiang W, Guo F, Liu Z. A graph theory method for determination of cryo-EM image focuses. J Struct Biol. 2012;180:343–351. doi: 10.1016/j.jsb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Monolayer purification: a rapid method for isolating protein complexes for single-particle electron microscopy. P Natl Acad Sci USA. 2008a;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kelly DF, Dukovski D, Walz T. Strategy for the Use of Affinity Grids to Prepare Non-His-Tagged Macromolecular Complexes for Single-Particle Electron Microscopy. J Mol Biol. 2010;400:675–681. doi: 10.1016/j.jmb.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The Affinity Grid: a prefabricated EM grid for monolayer purification. J Mol Biol. 2008b;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kurkela S, Ratti O, Huhtamo E, Uzcategui NY, Nuorti JP, Laakkonen J, Manni T, Helle P, Vaheri A, Vapalahti O. Sindbis virus infection in resident birds, migratory birds, and humans, Finland. Emerg Infect Dis. 2008;14:41–47. doi: 10.3201/eid1401.070510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D. Norwalk agent and other small-round structured viruses in the U.K. J Infect. 1991;23:220–222. doi: 10.1016/0163-4453(91)92567-o. [DOI] [PubMed] [Google Scholar]
- Lewis D, Ando T, Humphrey CD, Monroe SS, Glass RI. Use of Solid-Phase Immune Electron-Microscopy for Classification of Norwalk-Like Viruses into 6 Antigenic Groups from 10 Outbreaks of Gastroenteritis in the United-States. J Clin Microbiol. 1995;33:501–504. doi: 10.1128/jcm.33.2.501-504.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DC. Three serotypes of Norwalk-like virus demonstrated by solid-phase immune electron microscopy. J Med Virol. 1990;30:77–81. doi: 10.1002/jmv.1890300117. [DOI] [PubMed] [Google Scholar]
- Lewis DC, Lightfoot NF, Pether JVS. Solid-Phase Immune Electron-Microscopy with Human Immunoglobulin-M for Serotyping of Norwalk-Like Viruses. J Clin Microbiol. 1988;26:938–942. doi: 10.1128/jcm.26.5.938-942.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HR, Jin L, Koh SBS, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic Structure of Human Adenovirus by Cryo-EM Reveals Interactions Among Protein Networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaguno MC, Xu H, Shi L, Huang N, Zhang H, Liu Q, Jiang QX. Chemically functionalized carbon films for single molecule imaging. J Struct Biol. 2014 doi: 10.1016/j.jsb.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizou S, McCrea JD, Rudge AC, Reynolds R, Boyle CC, Harris EN. Measurement of anti-cardiolipin antibodies by an enzyme-linked immunosorbent assay (ELISA): standardization and quantitation of results. Clin Exp Immunol. 1985;62:738–745. [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Lustig S, Jackson AC, Hahn CS, Griffin DE, Strauss EG, Strauss JH. Molecular basis of Sindbis virus neurovirulence in mice. J Virol. 1988;62:2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Nakano A, Ros A. Protein dielectrophoresis: advances, challenges, and applications. Electrophoresis. 2013;34:1085–1096. doi: 10.1002/elps.201200482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaieff A, Obert G, van Regenmortel MH. Detection of rotavirus by serological trapping on antibody-coated electron microscope grids. J Clin Microbiol. 1980;12:101–104. doi: 10.1128/jcm.12.1.101-104.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal YC. Rapid Diagnosis of Barley Yellow Dwarf Virus in Plants Using Serologically Specific Electron-Microscopy. Phytopathol Z. 1977;89:25–36. [Google Scholar]
- Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev SV, Zhang W, Mukhopadhyay S, Fisher BR, Hernandez R, Brown DT, Baker TS, Rossmann MG, Kuhn RJ. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell. 2001;105:127–136. doi: 10.1016/s0092-8674(01)00302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P, Wright ET, Hakala K, Weintraub ST, Su M, Jiang W. DNA packaging-associated hyper-capsid expansion of bacteriophage t3. J Mol Biol. 2010;397:361–374. doi: 10.1016/j.jmb.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla DD, Gough KH. Use of Protein-a, from Staphylococcus-Aureus, in Immune Electron-Microscopy for Detecting Plant-Virus Particles. J Gen Virol. 1979;45:533–536. doi: 10.1099/0022-1317-51-2-415. [DOI] [PubMed] [Google Scholar]
- Snyder JE, Azizgolshani O, Wu B, He Y, Lee AC, Jose J, Suter DM, Knobler CM, Gelbart WM, Kuhn RJ. Rescue of infectious particles from preassembled alphavirus nucleocapsid cores. J Virol. 2011;85:5773–5781. doi: 10.1128/JVI.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Bacteriophage T7. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J Struct Biol. 2008;163:214–223. doi: 10.1016/j.jsb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LG, Ounjai P, Sigworth FJ. Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection. J Struct Biol. 2008;164:190–198. doi: 10.1016/j.jsb.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BG, Mahony JB, Simon G, Chernesky MA. Sensitive Solid-Phase Immune Electron-Microscopy Double-Antibody Technique with Gold-Immunoglobulin-G Complexes for Detecting Rotavirus in Cell-Culture and Feces. J Clin Microbiol. 1990;28:864–868. doi: 10.1128/jcm.28.5.864-868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Kohno H, Ikeda K, Shinoda T, Yokohama H, Morita K, Kusada O, Kobayashi Y. A practical method for measuring deoxynivalenol, nivalenol and T-2+HT-2 toxin in foods by an enzyme-linked immunosorbent assay using monoclonal antibodies. Biosci Biotech Bioch. 2004;68:2076–2085. doi: 10.1271/bbb.68.2076. [DOI] [PubMed] [Google Scholar]
- Yu G, Zhang D, Guo F, Tan M, Jiang X, Jiang W. Cryo-EM structure of a novel calicivirus, Tulane virus. Plos One. 2013a;8:e59817. doi: 10.1371/journal.pone.0059817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XK, Jin L, Jih J, Shih CH, Zhou ZH. 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein. Plos One. 2013b;8 doi: 10.1371/journal.pone.0069729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 angstrom Cryo-EM Structure of a Nonenveloped Virus Reveals a Priming Mechanism for Cell Entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rotationally averaged radial density plots of His-T7 CI (green), CII (purple) and mature phage (blue). Peak positions for the capsid shells of CI (244 Å), CII (281 Å) and mature phage (277 Å) are marked.