Abstract
Rationale
Antioxidant enzymes play an important role in the defense against oxidative stress in the lung and in the pathogenesis of chronic obstructive pulmonary disease (COPD). Sequence variation in genes encoding antioxidant enzymes may alter susceptibility to COPD by affecting longitudinal change in lung function in adults.
Methods
We genotyped 384 sequence variants in 56 candidate genes in 1,281 African-American and 1,794 European-American elderly adults of the Health, Aging, and Body Composition study. Single-marker associations and gene-by-smoking interactions with rate of change in FEV1 and FEV1/ FVC were evaluated using linear mixed effects models, stratified by race/ethnicity.
Results
In European-Americans, rs17883901 in GCLC was statistically significantly associated with rate of change in FEV1/FVC; the recessive genotype (TT) was associated with a 0.9% per year steeper decline (P = 4.50 × 10−5). Statistically significant gene-by-smoking interactions were observed for variants in two genes in European-Americans: the minor allele of rs2297765 in mGST3 attenuated the accelerated decline in FEV1/FVC in smokers by 0.45% per year (P = 1.13 × 10−4); for participants with greater baseline smoking pack-years, the minor allele of rs2073192 in IDH3B was associated with an accelerated decline in FEV1/FVC (P = 2.10 × 10−4). For both genes, nominally significant interactions (P < 0.01) were observed at the gene-level in African-Americans (P = 0.007 and 4.60 × 10−4, respectively). Nominally significant evidence of association was observed for variants in SOD3 and GLRX2 in multiple analyses.
Conclusions
This study identifies two novel genes associated with longitudinal lung function phenotypes in both African- and European-Americans, and confirms a prior finding for GCLC. These findings suggest novel mechanisms and molecular targets for future research and advance the understanding of genetic determinants of lung function and COPD risk.
Keywords: Antioxidant enzymes, cigarette smoking, gene by environment interaction, genetic association, longitudinal change, lung function, oxidative stress
INTRODUCTION
Lung function is an important predictor of morbidity and mortality in the general population [1]. Spirometric measures of lung function, such as forced expiratory volume in the first second (FEV1) and the ratio of FEV1/forced vital capacity (FEV1/FVC) are easily measured and reliable indicators of the physiological state of the lungs and airways and provide the basis for diagnosing and staging chronic obstructive pulmonary disease (COPD) [2]. Decline in lung function occurs naturally with aging, but accelerated decline can be caused by exposures such as cigarette smoking and can lead to low lung function that characterizes COPD [3, 4]. Therefore, longitudinal changes in lung function are informative predictors of COPD risk, and studies of these outcomes provide important insights for understanding disease pathogenesis [3–6].
The imbalance between chronic oxidative stress and antioxidant protection is postulated to play a key role in accelerated lung function loss [7, 8]. Cigarette smoke, a major source of exogenous oxidants, exposes the lung to elevated levels of oxidative stress, whereas dietary antioxidants and endogenous antioxidant enzymes are the two major forms of antioxidant defense that counteract these processes. The observation that only a subset of smokers develop COPD [9] and that a substantial proportion of COPD cases cannot be explained by smoking [10] led to the hypothesis that dietary intake of antioxidants and genetic variation in genes encoding antioxidant enzymes both play an important role in modifying antioxidant defense against cigarette smoke in the lung with ultimate effects on COPD risk.
In support of this hypothesis, observational epidemiologic studies have provided evidence of a positive association between dietary antioxidant intake and lung function, with stronger effects in cigarette smokers [11–15]. Genetic variation in antioxidant enzymes has also been studied in candidate gene association studies using population data, but published studies have limitations [16]. First, most studies considered only limited numbers of candidate genes, leaving many biologically relevant genes unstudied. Second, very few studies considered longitudinal lung function phenotypes [17–19]. Third, despite compelling evidence for their importance [20, 21], gene-by-smoking interactions are rarely investigated. Finally, very few studies include individuals of non-European ancestry, limiting inference to individuals of European descent. While recent, large-scale genome-wide association studies (GWAS) of lung function phenotypes together identified numerous novel genetic loci, these studies are limited in that they only consider European ancestry and cross-sectional phenotypes [22–24].
We hypothesized that common single nucleotide polymorphisms (SNPs) in genes encoding antioxidant enzymes affect longitudinal decline in lung function. We further hypothesized that gene-by-smoking interactions are present such that some genetic variants affect lung function decline contingent on exposure to cigarette smoke. To investigate these hypotheses, we selected 56 candidate genes that either had putative functional relevance to antioxidant defense in the lung or were previously investigated in relation to COPD-related phenotypes. Functional and tagging SNPs in these genes were genotyped and tested for single-marker associations and gene-by-smoking interactions with rate of change in FEV1 and FEV1/FVC in a population of African-American and European-American elderly adults from the Health, Aging, and Body Composition (Health ABC) study.
MATERIAL AND METHODS
Subjects
The Health ABC study is a longitudinal, prospective cohort study comprising 1,281 African-American and 1,794 European-American community-dwelling men and women, aged 70–79 years at baseline (1996–1997) and residing in the metropolitan areas of Pittsburgh, PA and Memphis, TN [25]. Participants reported self-proclaimed race initially as “Black” or “White”, but the terms “African-American” and “European-American” are used herein. To be eligible, participants were required to be ambulatory at baseline as confirmed by self-report of no difficulty walking one-quarter of a mile or climbing 10 steps without resting, no difficulty performing basic activities of daily living, and no use of a cane, walker, crutches or other special equipment to ambulate. In addition, participants were required to have no history of active treatment for cancer in the prior 3 years, and no plan to move out of the area in the subsequent 3 years. The Health ABC study was approved by the Institutional Review Boards of the University of Pittsburgh and the University of Tennessee, and the work reported herein was approved by the Institutional Review Board for Human Participants at Cornell University.
Pulmonary Function Testing
Spirometry was completed at four time points (baseline, years 4, 7 and 9) in accordance with standardized guidelines of the American Thoracic Society (ATS), as previously reported [25]. The study used a horizontal, dry rolling seal HF6 Spirometer (Sensor Medics Corporation, Yorba Linda, CA, USA) during clinical visits, and the EasyOne Model 2001 diagnostic spirometer (ndd Medizintechnik AG, Zurich, Switzerland) during home visits starting in year 8. The two devices were evaluated for comparability and provided virtually identical values. Consistent with the quality control standard used in recent lung function GWAS [22–24], all FEV1 (ml) and FEV1/FVC (%) measures meeting the ATS criteria for acceptability were included in the current study.
Cigarette Smoking
Participants were classified based on their long-term smoking status during the study follow-up as: (1) never smokers (never smoker at all spirometry time points), who were considered as the reference group in analyses, (2) persistent smokers (current smoker at all time points), (3) former smokers (former smoker at all time points), and (4) intermittent smokers (changing smoking status at different time points). Lifetime smoking dose was quantified as pack-years and calculated at study baseline for current and former smokers.
Candidate Gene Selection and Genotyping
Based on a previous systematic review of genetic association studies and gene expression studies investigating antioxidant enzymes and COPD-related phenotypes [16], we identified 56 candidate genes encoding antioxidant enzymes known to be expressed in lung tissue and postulated to affect the balance of antioxidants/oxidants. 384 functional and tagging SNPs were selected to capture variation across each gene and its regulatory regions (2 kilobases upstream and downstream). Details of the SNP selection strategy are provided elsewhere [26]. Separate consideration was given to African-Americans and European-Americans in SNP selection to maximize coverage in both populations, given differences in linkage disequilibrium (LD) structure and allele frequencies. Details of DNA extraction and genotyping quality, which were excellent, are provided elsewhere [26].
Four genes (GGT2, GSTK1, GSTM1, and GSTT1) were excluded from subsequent analyses due to low genotyping quality or atypical clustering of assayed SNPs. For the remaining SNPs with successful genotyping, Hardy-Weinberg equilibrium (HWE) was tested using the chi-squared goodness-of-fit test, stratified by race. After removing SNPs with genotyping call rate < 95%, minor allele frequency (MAF) < 1%, or p-value < 0.005 for the HWE test, the study included 314 SNPs in 52 genes in the African-American analyses and 284 SNPs in the same 52 genes in the European-American analyses (Supplementary Table 1).
Statistical Analysis
Linear mixed effects models were used to investigate single-marker associations and gene-by-smoking interactions with rate of change in FEV1 and rate of change in FEV1/FVC; all analyses were stratified by race/ethnicity. A continuous time variable quantified the time elapsed between each spirometry test and the study baseline. Random intercept and time effects were included at the individual level to differentiate between- and within-individual variation. All models were adjusted for gender, study site, height at each time point, age and smoking pack-years (both at study baseline), smoking status and smoking status × time. To address potential confounding by population substructure, the first two principal component variables for genetic ancestry [27] (computed separately by race/ethnicity; based on data from GWAS completed in Health ABC) were included in all models.
Single-marker associations with change in pulmonary function were tested by evaluating the product term of SNP × time. Gene-by-smoking interactions were tested by evaluating the three-way product term of SNP × smoking × time. Two smoking variables, smoking status during follow-up and baseline smoking pack-years, were tested separately for interactions, with smoking status during follow-up collapsed into two categories, as follows: smokers (persistent + intermittent) and non-smokers (former + never, which comprised the reference group).
Each SNP was coded by the minor allele and analyzed using an additive genetic model. SNPs with a nominal P < 0.05 were further tested using the dominant and recessive genetic models to refine estimates of the underlying genetic effect. The effect estimates for the genetic model with the most significant association were reported. To maintain statistical validity, we presented findings only for SNPs with a participant count ≥ 10 for the least frequent genotype category in the single-marker analyses and for the least frequent genotype-smoking status category in the interaction analyses.
In genetic association studies, the risk of false positives must be minimized without ruling out true associations. GWAS-scale multiple testing adjustments are not appropriate for the hypothesis-based investigation of candidate genes reported herein. Given the presence of LD among analyzed SNPs, we controlled for multiple testing using a Bonferroni adjustment based on the effective number of independent tests (Meff) [28, 29]. Meff was computed based on the correlation matrix of genotypes of all analyzed SNPs, and then used in a Bonferroni adjustment at the experiment-wise α level of 0.05. Given the difference in LD patterns, the adjustment was performed separately for each race/ethnicity. For African-Americans (Meff = 223), the Bonferroni-corrected significance threshold was P < 2.3 × 10−4; for European-Americans (Meff = 171), the analogous threshold of P < 3.0 × 10−4 was used. In addition, nominally significant associations were defined using P < 0.005 for single-marker analyses and P < 0.01 for gene-by-smoking interaction analyses.
All statistical analyses were conducted using SAS software version 9.1 (SAS Institute, Cary, NC, USA). LD in the Health ABC population was evaluated using Haploview 4.2 [30].
RESULTS
Population Characteristics
After exclusion for missing covariate data, 1,022 African-Americans with 2,432 FEV1 measurements and 1,487 European-Americans with 4,157 FEV1 measurements were included in the FEV1 analysis (Table 1). Similarly, 979 African-Americans with 2,244 FEV1/FVC measurements and 1,469 European-Americans with 4,018 FEV1/FVC measurements were included in the FEV1/FVC analysis.
Table 1.
Characteristics of Study Participants, by Phenotype and Race/Ethnicity, for the Health ABC Studya
| Characteristic | FEV1 Phenotype | FEV1/FVC Phenotype | ||
|---|---|---|---|---|
| African Americans |
European Americans |
African Americans |
European Americans |
|
| No. of participants | 1,022 | 1,487 | 979 | 1,469 |
| No. spirometry measurements | 2,432 | 4,157 | 2,244 | 4,018 |
| Males | 441 (43.2) | 773 (52.0) | 431 (44.02) | 766 (52.1) |
| Age at baseline (yr) | 73.4 (2.9) | 73.8 (2.9) | 73.4 (2.9) | 73.8 (2.9) |
| Height at baseline (cm) | 165.3 (9.4) | 166.7 (9.2) | 165.5 (9.5) | 166.7 (9.2) |
| Study site | ||||
| Memphis, TN | 469 (45.9) | 727 (48.9) | 439 (44.8) | 714 (48.6) |
| Pittsburgh, PA | 553 (54.1) | 760 (51.1) | 540 (55.2) | 755 (51.4) |
| Smoking status during follow-up | ||||
| Never smokers | 448 (43.8) | 649 (43 6) | 423 (43.2) | 638 (43.4) |
| Persistent smokers | 112 (11.0) | 48 (3.2) | 107 (10.9) | 48 (3.3) |
| Intermittent smokers | 62 (6.1) | 57 (3.8) | 60 (6.1) | 57 (3.9) |
| Former smokers | 400 (39.1) | 733 (49.3) | 389 (39.7) | 726 (49.4) |
| Pack-years at baselineb | 22.0 (1 – 126) | 28.5 (1 – 192) | 22.0 (1 – 126) | 29.0 (1 – 192) |
| FEV1 at baseline (ml) | 1924.4 (565.4) | 2288.6 (645.0) | - | - |
| FEV1/FVC at baseline (%) | - | - | 75.1 (8.3) | 74.2 (7.5) |
Abbreviations: FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity.
Data are presented as n, n (%), mean (standard deviation), or median (range).
Data for ever-smokers only.
We observed statistically significant annual decline in FEV1 and statistically significant annual decline in FEV1/FVC in both African-Americans and European-Americans (Table 2). For never smokers, the estimated rate of decline in FEV1/FVC was about 0.5% per year in both African-Americans and European-Americans, while the estimated annual decline in FEV1 was greater in European-Americans (about 40 versus 32 ml per year). In general, the effects of smoking on lung function were stronger in European-Americans compared to African-Americans, consistent with greater smoking doses observed in the former group. Thus, for FEV1/FVC, while persistent and intermittent smokers had significantly faster declines in both groups compared to never smokers, the effect size of persistent smoking in European-Americans was about twice that in African-Americans. While the difference in FEV1/FVC decline between former smokers and never smokers was not significant in African-Americans, it was borderline significant in European-Americans. Similar patterns were observed for annual decline in FEV1, although not all associations were statistically significant; the P value for the persistent smoking association was 0.06, and all associations followed expectations for magnitude and size of effect.
Table 2.
The Association of Smoking with Rate of Change in Spirometry Phenotypes by Race/Ethnicity, for Participants in the Health ABC Studya
| Population and Variable | Rate of Change in FEV1 | Rate of Change in FEV1/FVC | ||||
|---|---|---|---|---|---|---|
| Effect Estimate (ml/yr) |
P Value |
P Value for set |
Effect Estimate (%/yr) |
P Value |
P Value for set |
|
| African Americans | ||||||
| Timeb | −32.21 ± 1.71 | < 0.0001 | −0.50 ± 0.04 | < 0.0001 | ||
| Smoking statusc | ||||||
| Never smokers | Reference | Reference | ||||
| Persistent smokers | −5.96 ± 4.84 | 0.219 | 0.273 | −0.30 ± 0.13 | 0.019 | 0.005 |
| Intermittent smokers | −0.45 ± 4.78 | 0.925 | −0.42 ± 0.14 | 0.004 | ||
| Former smokers | 2.97 ± 2.47 | 0.229 | −0.05 ± 0.06 | 0.440 | ||
| European Americans | ||||||
| Timeb | −39.77 ± 1.35 | < 0.0001 | −0.52 ± 0.02 | < 0.0001 | ||
| Smoking statusc | ||||||
| Never smokers | Reference | Reference | ||||
| Persistent smokers | −12.05 ± 6.47 | 0.063 | 0.120 | −0.62 ± 0.15 | < 0.0001 | <0.0001 |
| Intermittent smokers | −6.51 ± 4.66 | 0.163 | −0.34 ± 0.11 | 0.001 | ||
| Former smokers | −2.27 ± 1.75 | 0.194 | −0.07 ± 0.04 | 0.051 | ||
Abbreviations: FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity.
Data presented are from the linear mixed effects models for the indicated phenotype and race/ethnicity analyses; all models included the following predictors: gender, study site, height at each time point, age and smoking pack-years (both at study baseline), time, smoking status, and smoking status × time.
Effect estimate for time corresponds to the estimated annual rate of change in the phenotype for never smokers; negative values represent declines in the phenotype.
Effect estimate for each smoking category is for the smoking status × time product term; negative values represent accelerations and positive values represent attenuations of decline, relative to never smokers.
Single-Marker Associations
Three genes showed nominal evidence of associations with rate of change in FEV1 in African-Americans and one gene was associated with FEV1 decline in European-Americans, although no associations survived the adjustment for multiple testing (Table 3). However, a SNP in superoxide dismutase 3 (SOD3), rs8192287, was marginally statistically significantly associated with a 20 ml per year faster decline in FEV1 per copy of the minor allele (T) in African-Americans (P = 2.45 × 10−4).
Table 3.
The Most Statistically Significant Associations for Single Genetic Variants with Rate of Change in FEV1 by Race/Ethnicity, for Participants in the Health ABC Studya
| Population and Gene |
SNP | Chr | Base Pair Position |
Minor Allele |
MAF | β (ml/yr)b | P Value | Genetic Modelc |
|---|---|---|---|---|---|---|---|---|
| African Americans | ||||||||
| mGST3 | rs10800120 | 1 | 163871934 | A | 0.20 | −5.8 ± 2.0 | 0.004 | Additive |
| SOD3 | rs8192287 | 4 | 24405666 | T | 0.02 | −19.8 ± 5.4 | 2.45 × 10−4 | Additive |
| GSR | rs8190996 | 8 | 30673548 | T | 0.32 | 12.3 ± 3.8 | 0.001 | Recessive |
| European Americans | ||||||||
| GSTA4 | rs6904771 | 6 | 52964138 | G | 0.02 | −13.2 ± 4.2 | 0.002 | Additive |
Abbreviations: FEV1, forced expiratory volume in 1 s; SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; β, regression coefficient; mGST3, microsomal glutathione S-transferase 3; SOD3, superoxide dismutase 3; GSR, glutathione reductase; GSTA4, glutathione S-transferase A4.
Data shown for associations with P < 0.005 for the SNP single-marker effect on rate of change in FEV1, sorted by race/ethnicity, chromosome and base pair position. Statistically significant associations satisfying the Bonferroni-adjusted threshold (African Americans: P < 2.3 × 10−4; European Americans: P < 3.0 × 10−4) are bolded.
Regression coefficient and standard error for the SNP × time product term in the corresponding mixed effects model.
Genetic model is defined in reference to the minor allele for each SNP.
Four genes were nominally associated with rate of change in FEV1/FVC in African-Americans (Table 4). The most statistically significant association, which did not pass the Bonferroni-adjusted threshold, was for a glutaredoxin 2 (GLRX2) SNP, rs35358794; each copy of the minor allele (A) was associated with a 0.3% per year slower decline. In European-Americans, two genes showed evidence of associations with rate of change in FEV1/FVC. The most statistically significant association, which survived the Bonferroni adjustment for multiple testing, was for the glutamate-cysteine ligase catalytic subunit (GCLC) SNP rs17883901 (P = 4.50 × 10−5). The recessive genotype (TT) was associated with a 0.9% per year steeper decline compared with the reference genotypes (CC/CT).
Table 4.
The Most Statistically Significant Associations for Single Genetic Variants with Rate of Change in FEV1/FVC by Race/Ethnicity, for Participants in the Health ABC Studya
| Population and Gene |
SNP | Chr | Base Pair Position |
Minor Allele |
MAF | β (%/yr)b | P Value | Genetic Modelc |
|---|---|---|---|---|---|---|---|---|
| African Americans | ||||||||
| GLRX2 | rs35358794 | 1 | 191336492 | A | 0.06 | 0.29 ± 0.09 | 0.001 | Additive |
| SOD2 | rs4342445 | 6 | 160018212 | A | 0.15 | −0.53 ± 0.17 | 0.002 | Recessive |
| TXN2 | rs2267337 | 22 | 35200417 | T | 0.22 | −0.16 ± 0.05 | 0.002 | Additive |
| TXN2 | rs2281082 | 22 | 35202696 | T | 0.22 | −0.15 ± 0.05 | 0.004 | Additive |
| PRDX4 | rs528960 | 23 | 23601182 | C | 0.24 | 0.19 ± 0.06 | 0.003 | Dominant |
| European Americans | ||||||||
| GCLC | rs17883901 | 6 | 53517996 | T | 0.09 | −0.86 ± 0.21 | 4.50 × 10−5 | Recessive |
| GSTO2 | rs157077 | 10 | 106027884 | C | 0.46 | −0.09 ± 0.03 | 3.68 × 10−4 | Additive |
Abbreviations: FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; β, regression coefficient; GLRX2, glutaredoxin 2; SOD2, superoxide dismutase 2; TXN2, thioredoxin 2; PRDX4, peroxiredoxin 4; GCLC, glutamate-cysteine ligase (catalytic subunit); GSTO2, glutathione S-transferase O2.
Data shown for associations with P < 0.005 for the SNP single-marker effect on rate of change in FEV1/FVC, sorted by race/ethnicity, chromosome and base pair position. Statistically significant associations satisfying the Bonferroni-adjusted threshold (African Americans: P < 2.3 × 10−4; European Americans: P < 3.0 × 10−4) are bolded.
Regression coefficient and standard error for the SNP × time product term in the corresponding mixed effects model.
Genetic model is defined in reference to the minor allele for each SNP.
Gene-by-Smoking Interactions
Potential interactions between SNPs and cigarette smoking were investigated in relation to rate of change in FEV1 and FEV1/FVC separately in African-Americans and European-Americans. Two smoking variables, smoking status during follow-up and baseline smoking pack-years, were investigated separately for gene-by-smoking interactions (Table 5; also Supplementary Tables 2 to 9).
Table 5.
The Most Statistically Significant Gene-by-Smoking Interactions with Rate of Change in FEV1 and FEV1/FVC by Race/Ethnicity, for Participants in the Health ABC Studya
| Phenotype | Population and Gene |
SNP-Smoking Interactionc |
Chr | Base Pair Position |
Minor Allele |
MAF | Interaction Effectd |
P Value | Genetic Modele |
|---|---|---|---|---|---|---|---|---|---|
| FEV1 | African Americans | ||||||||
| GLRX2 | rs34552619 × smoking status | 1 | 191331738 | C | 0.08 | −32.4 ± 8.8 | 2.74 × 10−4 | Dominant | |
| IDH1 | rs1437410 × smoking pack-years | 2 | 208825562 | C | 0.22 | −1.2 ± 0.4 | 6.27 × 10−4 | Recessive | |
| European Americans | |||||||||
| SOD3 | rs1007991 × smoking status | 4 | 24409783 | C | 0.34 | 17.9 ± 5.7 | 0.002 | Additive | |
| GSTZ1 | rs2111699 × smoking pack-years | 14 | 76858350 | G | 0.32 | −0.4 ± 0.1 | 9.55 × 10−4 | Recessive | |
| FEV1/FVC | African Americans | ||||||||
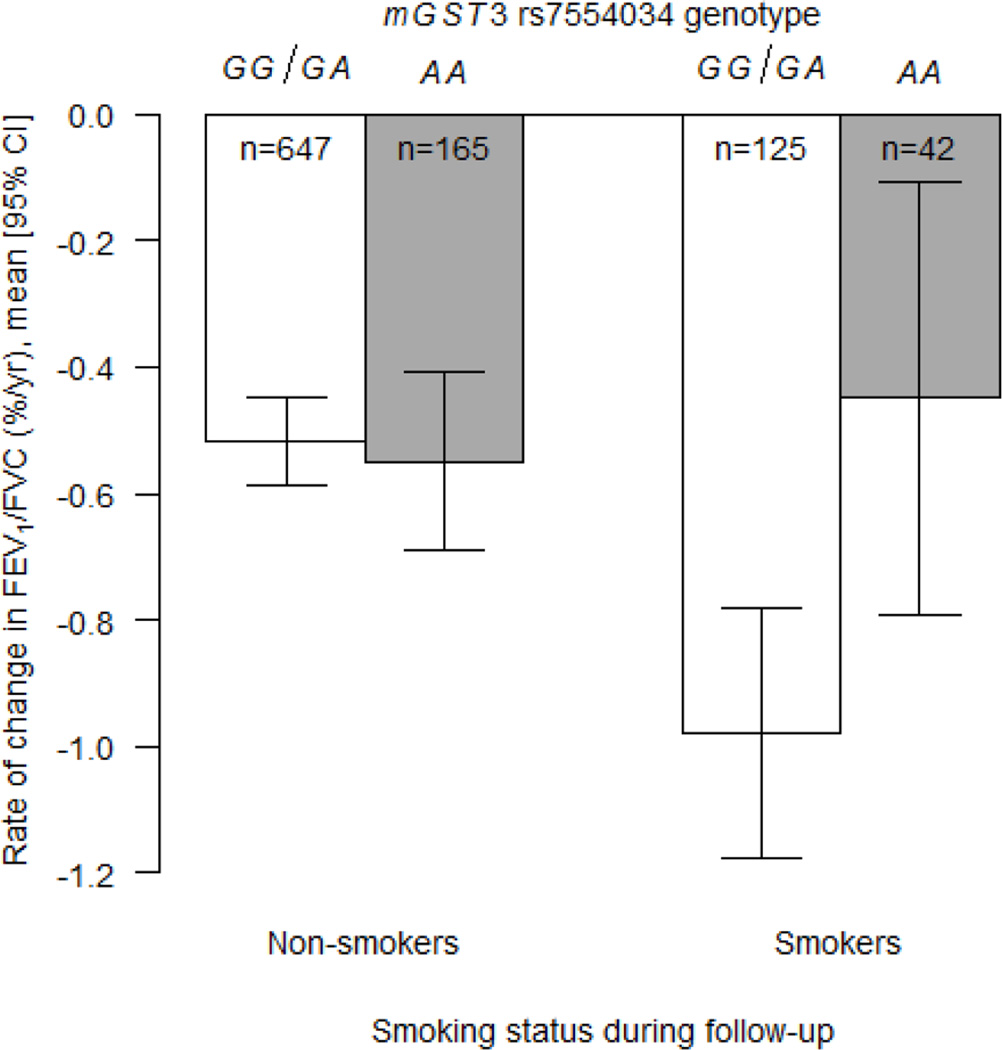
| mGST3b | rs7554034 × smoking status | 1 | 163877088 | A | 0.46 | 0.56 ± 0.21 | 0.007 | Recessive | |
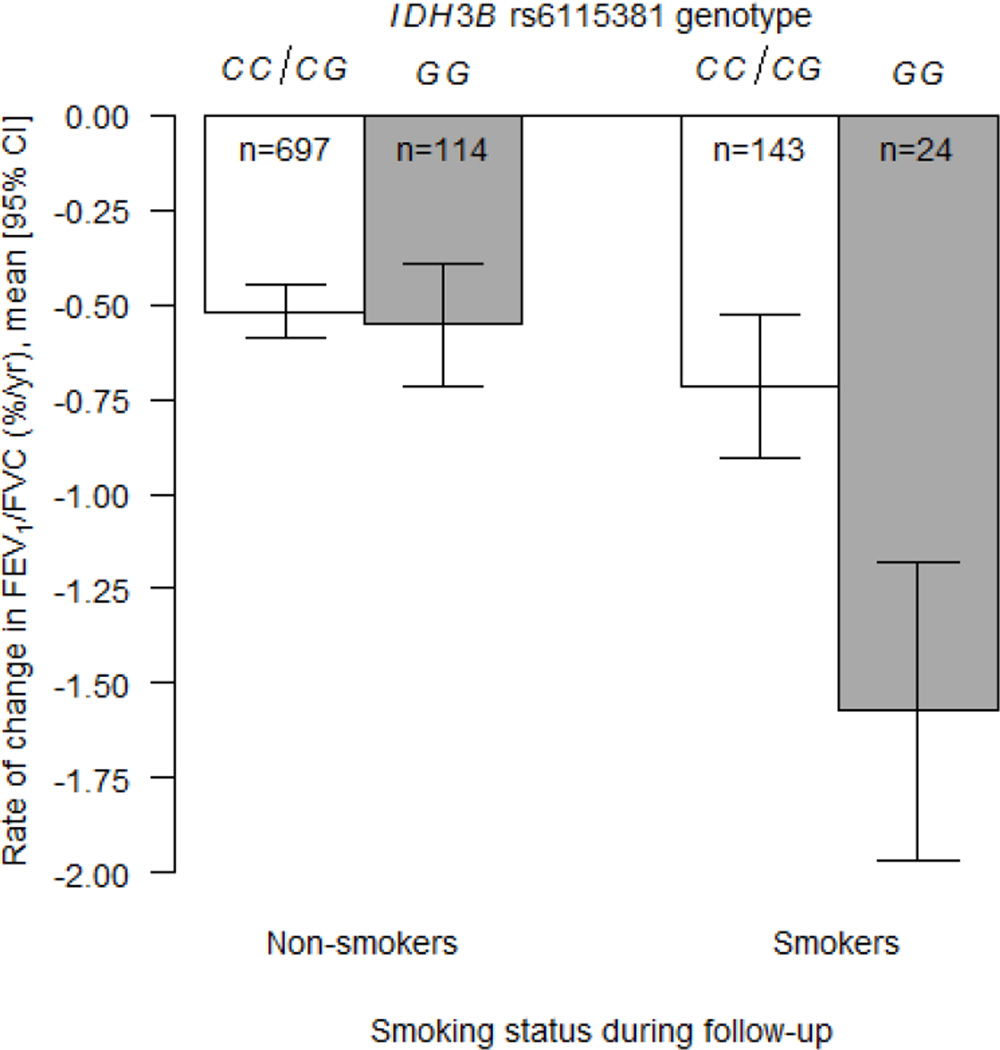
| IDH3Bb | rs6115381 × smoking status | 20 | 2590376 | G | 0.37 | −0.82 ± 0.23 | 4.60 × 10−4 | Recessive | |
| SOD1 | rs2070424 × smoking status | 21 | 31961191 | G | 0.19 | 0.68 ± 0.19 | 3.62 × 10−4 | Dominant | |
| G6PD | rs2472394 × smoking pack-years | 23 | 153424545 | A | 0.13 | −0.012 ± 0.003 | 7.75 × 10−4 | Dominant | |
| European Americans | |||||||||
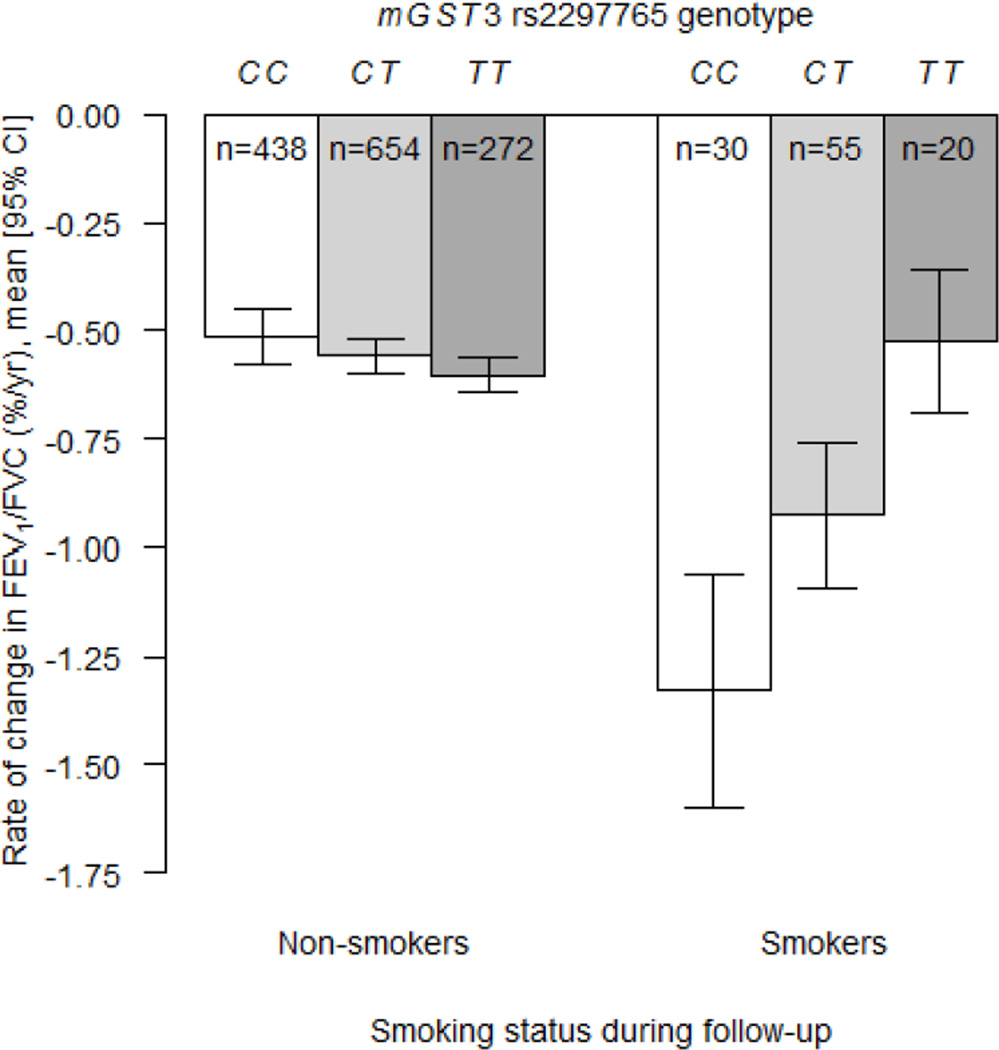
| mGST3 | rs2297765 × smoking status | 1 | 163888831 | T | 0.44 | 0.45 ± 0.12 | 1.13 × 10−4 | Additive | |
| SOD3b | rs2284659 × smoking status | 4 | 24403895 | T | 0.37 | 0.64 ± 0.22 | 0.004 | Recessive | |
| IDH3B | rs6115381 × smoking pack-years | 20 | 2590376 | G | 0.07 | −0.008 ± 0.002 | 3.82 × 10−4 | Additive | |
| IDH3B | rs6107100 × smoking pack-years | 20 | 2592685 | A | 0.07 | −0.008 ± 0.002 | 3.68 × 10−4 | Additive | |
| IDH3B | rs2073192 × smoking pack-years | 20 | 2592996 | A | 0.07 | −0.008 ± 0.002 | 2.10 × 10−4 | Additive | |
Abbreviations: FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; GLRX2, glutaredoxin 2; IDH1, isocitrate dehydrogenase 1; SOD3, superoxide dismutase 3; GSTZ1, glutathione S-transferase Z1; mGST3, microsomal glutathione S-transferase 3; IDH3B, isocitrate dehydrogenase 3B; SOD1, superoxide dismutase 1; G6PD, glucose-6-phosphate dehydrogenase.
Data shown are for the most statistically significant gene-by-smoking interactions for each phenotype and race/ethnicity analysis. Statistically significant interactions satisfying the Bonferroni-adjusted threshold (African Americans: P < 2.3 × 10−4; European Americans: P < 3.0 × 10−4) are bolded.
These nominally significant (P < 0.01) interactions were selectively presented since they represent gene-level replications of most statistically significant interactions in another phenotype and race/ethnicity analysis.
Smoking status was defined as a two-level categorical variable: smokers vs. non-smokers (reference group) during follow-up; smoking pack-years was modeled as a continuous variable.
Beta coefficient and standard error for the SNP × smoking × time product term in the corresponding mixed effects model; ml per year for the effect on rate of change in FEV1 and % per year for the effect on rate of change in FEV1/FVC.
Genetic model is defined in reference to the minor allele for each SNP.
For rate of change in FEV1, in African-Americans, a nominally significant interaction was identified between rs34552619 in GLRX2 and smoking status. African-American smokers with at least one copy of the minor allele (C) had a 32.4 ml per year steeper decline (P = 2.74 × 10−4) than smokers without the minor allele. In European-Americans, rs1007991 in SOD3 had a nominally significant interaction with smoking status; each copy of the minor allele (C) attenuated the accelerated decline in FEV1 in smokers by 17.9 ml per year (P = 0.002). In contrast, neither SNP was associated with rate of change in FEV1 in non-smokers during follow-up.
For rate of change in FEV1/FVC, two genes had statistically significant interactions with smoking that passed the Bonferroni adjustment for multiple testing in European-Americans and gene-level replications were observed for both genes in African-Americans. In European-Americans, the association between rs2297765 in microsomal glutathione S-transferase 3 (mGST3) and rate of change in FEV1/FVC differed by smoking status such that each copy of the minor allele (T) attenuated the decline in smokers by 0.45% per year, but the SNP had no effect on decline in non-smokers (Table 5; P = 1.13 × 10−4; Figure 1). In African-Americans, a different mGST3 variant, rs7554034, had a nominally significant interaction with smoking status; compared to the reference genotype, the recessive genotype (AA) attenuated the decline in smokers by 0.56% per year, but genotype had no effect on rate of decline in non-smokers (Table 5; P = 0.007; Figure 2). These mGST3 SNPs were not in LD in either group (r2 = 0.002 and 0.01 for African-Americans and European-Americans, respectively). In European-Americans, the association between rs2073192 in isocitrate dehydrogenase 3 beta (IDH3B) and rate of change in FEV1/FVC differed by smoking pack-years; in participants with higher smoking dose, the minor allele (A) was associated with a faster decline in FEV1/FVC (Table 5, P = 2.10 × 10−4). Two other IDH3B SNPs, which were in strong LD with rs2073192 in European-Americans (r2 ≥ 0.92), showed similar, but less statistically significant, evidence of interaction with smoking pack-years. In African-Americans, a nominally significant interaction was observed for one of the IDH3B SNPs (rs6115381) and smoking status (Table 5; P = 4.60 × 10−4) such that the recessive genotype (GG) was associated with a 0.82% per year greater decline (compared to the reference genotype) in smokers only; no such difference was observed across genotypes in non-smokers (Figure 3). In African-Americans, the rs6115381 and rs6107100 SNPs in IDH3B were in moderate LD (r2 = 0.63), whereas rs2073192 was not in LD with rs6115381 and rs6107100 (r2 = 0.09 and 0.10, respectively). A nominally significant interaction was also observed between rs2284659 in SOD3 and smoking status in European-Americans for rate of change in FEV1/FVC (P = 0.004).
Figure 1.
Estimated rates of change in FEV1/FVC (% per year) according to smoking status during follow-up and mGST3 rs2297765 genotype for European American participants in the Health ABC study. Open bars represent the CC genotype, light shaded bars represent the CT genotype and dark shaded bars represent the TT genotype. The estimates were computed from the linear mixed effects model that was adjusted for all covariates and included the SNP × smoking status × time product term (P = 1.13 × 10−4 following an additive genetic effect model). CI = confidence interval.
Figure 2.
Estimated rates of change in FEV1/FVC (% per year) according to smoking status during follow-up and mGST3 rs7554034 genotype for African American participants in the Health ABC study. Open bars represent the GG/GA genotypes and shaded bars represent the AA genotype. The estimates were computed from the linear mixed effects model that was adjusted for all covariates and included the SNP × smoking status × time product term (P = 0.007 following a recessive genetic effect model). CI = confidence interval.
Figure 3.
Estimated rates of change in FEV1/FVC (% per year) according to smoking status during follow-up and IDH3B rs6115381 genotype for African American participants in the Health ABC study. Open bars represent the CC/CG genotypes and shaded bars represent the GG genotype. The estimates were computed from the linear mixed effects model that was adjusted for all covariates and included the SNP × smoking status × time product term (P = 4.60 × 10−4 following a recessive genetic effect model). CI = confidence interval.
DISCUSSION
This study was designed to investigate the hypothesis that genetic variation in candidate genes encoding antioxidant enzymes, which is expected to affect antioxidant defense in the lung, is associated with rate of change in lung function phenotypes, FEV1 and FEV1/FVC, and thus contributes to COPD susceptibility, especially in individuals with elevated oxidative stress due to cigarette smoking. Consistent with several recent GWAS of lung phenotypes there were more findings overall for rate of change in FEV1/FVC compared to rate of change in FEV1, although the reasons for this are not yet clear [22–24].
A novel gene, mGST3, was associated with rate of change in FEV1/FVC, with evidence of gene-by-smoking interactions in both European- and African-Americans. In European-Americans, the effect of rs2297765 differed significantly by smoking status, and rs7554034 had a similar interaction in African-Americans. The two SNPs are common in both groups, thus the effects on lung function in smokers are of public health interest. MGST3 is a membrane-bound antioxidant enzyme in the microsomal GST family with close links to antioxidant defense. In microarray studies of gene expression, the mouse Mgst3 gene was up-regulated in the small intestine and liver in response to oxidative stress [31]. MGST3 also catalyzes the conjugation reaction that produces leukotriene C4, an important inflammation mediator with a role in allergy and asthma [32, 33]. The association of mGST3 with lung function phenotypes is novel, but microsomal enzymes, as a class, have been linked to lung health in prior studies. Epoxide hydrolase 1 (EPHX1), another microsomal enzyme, detoxifies xenobiotics including products in cigarette-smoke, and genetic variation in EPHX1 was associated with pulmonary phenotypes including childhood asthma, lung cancer and COPD [34–36].
IDH3B was implicated in a prior study of cross-sectional lung function phenotypes in the Health ABC cohort (gene-by-smoking interactions in African-Americans) [26]. In the current study of longitudinal lung function phenotypes in the same cohort, SNPs in IDH3B had a statistically significant interaction with smoking pack-years in European-Americans and a nominally significant interaction with smoking status in African-Americans in relation to rate of change in FEV1/FVC. The IDH enzymes, the majority of which localize to the mitochondrial matrix, supply the reducing equivalents for the antioxidant activity of the many members of the glutathione and thioredoxin systems. In fibroblasts, decreased expression of IDH genes led to higher lipid peroxidation, oxidative DNA damage, intracellular peroxide generation, and increased senescence, indicating an important regulatory role for these genes in the defense against oxidative stress [37].
In the present study, the rs17883901 SNP in GCLC was associated with rate of change in FEV1/FVC in European-Americans, but the gene-by-smoking interactions for rs17883901 could not be investigated given the limited number of smokers carrying the minor allele. Although no association was detected in African-Americans, there was a considerably lower MAF in African-Americans (1%). These findings are consistent with prior reports. The rs17883901 SNP was associated with an increased risk of COPD in a Chinese population [38], and two variants (rs17883901 and a GAG repeat variant (TNR)) were investigated jointly in relation to several pulmonary phenotypes, including change in FEV1, in two Dutch cohorts [18]. Using a nominal significance threshold of P < 0.05, the Dutch study reported associations for both variants, including an interaction between TNR and smoking pack-years in relation to rate of change in FEV1. GCLC encodes the catalytic subunit of the heterodimeric enzyme glutamatecysteine ligase, which catalyzes the de novo synthesis of glutathione. GCLC is predominantly expressed in lung epithelium [39], and rs17883901 was associated with lower expression of GCLC in endothelial cells in vitro [40], suggesting a potential mechanism for the population-level association. Overall, these findings support a role for GCLC in longitudinal change in lung function.
SNPs in SOD3 and GLRX2 showed nominally significant evidence of associations in multiple analyses, and the GLRX2 findings are novel. SOD3 is a major extracellular antioxidant enzyme highly expressed in the lung; SOD3 binds lung matrix components (collagen I, hyaluronan and heparin sulfate) to protect them against oxidative fragmentation and plays a central role in antioxidant defense in lung tissue [41]. Genetic variation in SOD3 has been extensively studied in relation to pulmonary phenotypes at the population level, although primarily in individuals of European descent. Rs1799895, a rare functional SNP in SOD3, was associated with lower COPD risk and slower FEV1 decline in never smokers [42–44], and rs8192287 and rs8192288, which are in strong LD in individuals of European descent, were associated with reduced lung function and increased emphysema risk [45, 46]. Rs1799895 was analyzed in the present study for single-marker associations in European-Americans, but no statistically significant associations emerged. Rs8192287 was associated with a faster decline in FEV1 in African-Americans, providing novel evidence for an association of rs8192287 with lung function phenotypes in this under-studied group. The observed gene-by-smoking interactions involving other SOD3 SNPs support effect modification by smoking in this genotype—phenotype association. Novel findings emerged for GLRX2, which encodes a mitochondrial antioxidant enzyme in the glutaredoxin family and has been recognized as an important redox regulator [47]. GLRX2 is ubiquitously expressed in various tissues including lung [47], and its over-expression was shown to prevent H2O2-induced apoptosis in human lens epithelial cells and to reduce myocardial cell death by preventing apoptosis and necrosis in mice [48, 49].
The study has several strengths. First, this large, epidemiologic cohort study with longitudinal follow-up data on pulmonary function assessed by high-quality spirometry is a unique resource. The long duration allows the estimation of meaningful decline in lung function, making the investigation of the difference in rate of change among individuals possible. The use of up to 4 repeated measurements per individual provides the data to accurately capture the true trajectory of lung function change over time. Second, the study had high-quality data on important risk and confounding factors, including cigarette smoking and principle component variables for genetic ancestry. The adjustment for genetic ancestry avoids potential confounding due to population substructure in each racial/ethnic group. Multiple forms of smoking data were available, allowing the consideration of both long-term smoking status and lifetime smoking dose in the single-marker analyses and to investigate potential interactions of genotype with these two different aspects of smoking exposure. Third, the Health ABC study includes a sufficiently large sample of African-Americans, allowing race-specific analyses to be performed. This is important because African-Americans have lower lung function compared with their European-American counterparts and they are understudied in pulmonary and genetic epidemiology. Finally, despite the heterogeneity in the frequency and pattern of genetic variation and the challenges in the replication of genetic associations across racial/ethnic groups, this study provides compelling evidence of gene-by-smoking interactions consistent on the gene level between African-Americans and European-Americans for two novel candidate genes.
A few limitations should be considered when evaluating the study findings. First, despite the goal to comprehensively include genes encoding enzymes in relevant antioxidant pathways in the lung, a few genes did not pass genotyping quality control, and other enzymes with antioxidant activities may have been omitted inadvertently. Second, although the Health ABC study recorded extensive data on smoking behaviors, the statistical modeling of smoking in the study may not fully capture the effect of smoke exposure, possibly due to inaccuracy in participants’ self-reports and uncertainty in defining the most relevant aspects of smoking in affecting pulmonary function. Despite these limitations, and limited power due to sample size, we were able to identify meaningful gene-by-smoking interactions. Third, the analyzed SNPs admittedly provide imperfect coverage of genetic variation in the candidate genes. The SNPs showing significant results are therefore likely “proxies” of the true causal variants. Considering the incomplete linkage between these variants, the true associations of causal variants with the corresponding phenotypes are expected to be greater than what was observed. While the current study focused on an elderly population, given their disproportionately high risk of accelerated lung function loss and consequent morbidity and mortality, the findings may or may not generalize to younger populations, and additional studies are needed to test the reported associations in populations with different characteristics. Finally, due to the risk of false discovery inherent in genetic association studies, we adopted a conservative significance threshold that may be overly conservative.
In conclusion, this study explored genetic variation in candidate genes encoding antioxidant enzymes, cigarette smoking, and longitudinal change in two lung function phenotypes in African-American and European-American elderly adults. Evidence of association was observed for several novel genes. Of particular importance are the novel findings of gene-by-smoking interactions for mGST3 and IDH3B, which were observed consistently at the gene level in both African-Americans and European-Americans. The findings for GCLC and SOD3 strengthen existing knowledge and extend the evidence base by the novel consideration of longitudinal phenotypes and African-Americans. Future research, especially in the understudied African-American population, is warranted to further validate these findings and to elucidate the underlying molecular mechanisms.
Supplementary Material
Highlights.
We investigated the association of genetic variants in a network of antioxidant enzymes with change in lung function
A longitudinal cohort study of African- and European-American elderly adults was used for the study
A highly significant genetic main effect was identified for a variant in GCLC in European Americans
Significant gene-by-smoking interactions were identified for mGST3 and IDH3B in both races
These findings suggest novel directions for future research on the genetic basis of lung function
Acknowledgements
none
Funding: This research was supported by NIH R01HL071022 (PAC), and by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University, contract number HHSN268200782096C. Genotyping services specific to this study were provided by the Johns Hopkins University under the U.S. Federal Government contract number N01HV48195. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (ARB).
Abbreviations
- COPD
chronic obstructive pulmonary disease
- FEV1
forced expiratory volume in the first second
- FVC
forced vital capacity
- CAT
catalase
- G6PD
glucose-6-phosphate dehydrogenase
- GCLC
glutamate–cysteine ligase(catalytic subunit)
- GCLM
glutamate-cysteine ligase (modulatory subunit)
- GGT1
γ-glutamyl transferase 1
- GLRX
glutaredoxin
- GPX
glutathione peroxidase
- GSR
glutathione reductase
- GSS
glutathione synthetase
- GST
glutathione S-transferase
- HMOX
hemeoxygenase
- IDH
isocitrate dehydrogenase
- mGST
microsomal glutathione S-transferase
- PRDX
peroxiredoxin
- SEP
selenoprotein
- SOD
superoxide dismutase
- TXN
thioredoxin
- TXNRD
thioredoxin reductase.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest: No competing interests.
Study Approval and Participant Consent: The Health ABC study was approved by Institutional Review Boards at the University of Pittsburgh (Pittsburgh, PA) and the University of Tennessee (Memphis, TN) and participants provided written informed consent for the study. The Institutional Review Board at Cornell University (Ithaca, NY) approved the current study.
Contributor Statement: WT, ARB and PAC designed this study and TH, SK, ABN, DCB, and BM designed the Health ABC study. WT and PAC analyzed data and wrote the manuscript. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Schunemann HJ, Dorn J, Grant BJ, Winkelstein W, Jr, Trevisan M. Pulmonary function is a long-term predictor of mortality in the general population: 29-year follow-up of the Buffalo Health Study. Chest. 2000;118:656–664. doi: 10.1378/chest.118.3.656. [DOI] [PubMed] [Google Scholar]
- 2.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher C. The natural history of chronic bronchitis and emphysema: an eight-year study of early chronic obstructive lung disease in working men in London. New York: Oxford University Press; 1976. [Google Scholar]
- 4.Rennard SI, Vestbo J. Natural Histories of Chronic Obstructive Pulmonary Disease. Proceedings of the American Thoracic Society. 2008;5:878–883. doi: 10.1513/pats.200804-035QC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise R. The value of forced expiratory volume in 1 second decline in the assessment of chronic obstructive pulmonary disease progression. Am J Med. 2006;119:4–11. doi: 10.1016/j.amjmed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Han MK, Agusti A, Calverley PM, Celli BR, Criner G, Curtis JL, Fabbri LM, Goldin JG, Jones PW, MacNee W, et al. Chronic obstructive pulmonary disease phenotypes: The future of COPD. Am J Respir Crit Care Med. 2010;182:598–604. doi: 10.1164/rccm.200912-1843CC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahman I, Adcock IM. Oxidative stress and redox regulation of lung inflammation in COPD. Eur Respir J. 2006;28:219–242. doi: 10.1183/09031936.06.00053805. [DOI] [PubMed] [Google Scholar]
- 8.Rahman I, Biswas S, Kode A. Oxidant and antioxidant balance in the airways and airway diseases. Eur J Pharmacol. 2006;533:222–239. doi: 10.1016/j.ejphar.2005.12.087. [DOI] [PubMed] [Google Scholar]
- 9.Halbert RJ, Natoli JL, Gano A, Badamgarav E, Buist AS, Mannino DM. Global burden of COPD: systematic review and meta-analysis. Eur Respir J. 2006;28:523–532. doi: 10.1183/09031936.06.00124605. [DOI] [PubMed] [Google Scholar]
- 10.Eisner MD, Anthonisen N, Coultas D, Kuenzli N, Perez-Padilla R, Postma D, Romieu I, Silverman EK, Balmes JR. An official American Thoracic Society public policy statement: novel risk factors and the global burden of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:693–718. doi: 10.1164/rccm.200811-1757ST. [DOI] [PubMed] [Google Scholar]
- 11.Hu G, Zhang X, Chen J, Peto R, Campbell TC, Cassano PA. Dietary vitamin C intake and lung function in rural China. Am J Epidemiol. 1998;148:594–599. doi: 10.1093/oxfordjournals.aje.a009685. [DOI] [PubMed] [Google Scholar]
- 12.Butland BK, Fehily AM, Elwood PC. Diet, lung function, and lung function decline in a cohort of 2512 middle aged men. Thorax. 2000;55:102–108. doi: 10.1136/thorax.55.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKeever TM, Scrivener S, Broadfield E, Jones Z, Britton J, Lewis SA. Prospective study of diet and decline in lung function in a general population. Am J Respir Crit Care Med. 2002;165:1299–1303. doi: 10.1164/rccm.2109030. [DOI] [PubMed] [Google Scholar]
- 14.Ochs-Balcom HM, Grant BJ, Muti P, Sempos CT, Freudenheim JL, Browne RW, McCann SE, Trevisan M, Cassano PA, Iacoviello L, et al. Antioxidants, oxidative stress, and pulmonary function in individuals diagnosed with asthma or COPD. Eur J Clin Nutr. 2006;60:991–999. doi: 10.1038/sj.ejcn.1602410. [DOI] [PubMed] [Google Scholar]
- 15.Bentley AR, Kritchevsky SB, Harris TB, Holvoet P, Jensen RL, Newman AB, Lee JS, Yende S, Bauer D, Cassano PA. Dietary antioxidants and forced expiratory volume in 1 s decline: the Health, Aging and Body Composition study. Eur Respir J. 2012;39:979–984. doi: 10.1183/09031936.00190010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentley AR, Emrani P, Cassano PA. Genetic variation and gene expression in antioxidant related enzymes and risk of COPD: a systematic review. Thorax. 2008;63:956–961. doi: 10.1136/thx.2007.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He JQ, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med. 2002;166:323–328. doi: 10.1164/rccm.2111059. [DOI] [PubMed] [Google Scholar]
- 18.Siedlinski M, Postma DS, van Diemen CC, Blokstra A, Smit HA, Boezen HM. Lung function loss, smoking, vitamin C intake, and polymorphisms of the glutamate-cysteine ligase genes. Am J Respir Crit Care Med. 2008;178:13–19. doi: 10.1164/rccm.200711-1749OC. [DOI] [PubMed] [Google Scholar]
- 19.Curjuric I, Imboden M, Schindler C, Downs SH, Hersberger M, Liu SL, Matyas G, Russi EW, Schwartz J, Thun GA, et al. HMOX1 and GST variants modify attenuation of FEF25–75% decline due to PM10 reduction. Eur Respir J. 2010;35:505–514. doi: 10.1183/09031936.00044309. [DOI] [PubMed] [Google Scholar]
- 20.Zhai G, Valdes AM, Cherkas L, Clement G, Strachan D, Spector TD. The interaction of genes and smoking on forced expiratory volume: a classic twin study. Chest. 2007;132:1772–1777. doi: 10.1378/chest.07-1438. [DOI] [PubMed] [Google Scholar]
- 21.Gottlieb DJ, Wilk JB, Harmon M, Evans JC, Joost O, Levy D, O'Connor GT, Myers RH. Heritability of longitudinal change in lung function. The Framingham study. Am J Respir Crit Care Med. 2001;164:1655–1659. doi: 10.1164/ajrccm.164.9.2010122. [DOI] [PubMed] [Google Scholar]
- 22.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YMTA, Chen T-h, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2009;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat Me, Zhao JH, Ramasamy A, Zhai G, Vitart V, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2009;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterer GW, Wan JY, Kritchevsky SB, Wunderink RG, Satterfield S, Bauer DC, Newman AB, Taaffe DR, Jensen RL, Crapo RO. Airflow limitation is underrecognized in well-functioning older people. J Am Geriatr Soc. 2001;49:1032–1038. doi: 10.1046/j.1532-5415.2001.49205.x. [DOI] [PubMed] [Google Scholar]
- 26.Bentley AR, Kritchevsky SB, Harris TB, Newman AB, Bauer DC, Meibohm B, Clark AG, Cassano PA. Genetic variation in antioxidant enzymes and lung function. Free Radic Biol Med. 2012 doi: 10.1016/j.freeradbiomed.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Higgins LG, Hayes JD. Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents. Drug Metab Rev. 2011;43:92–137. doi: 10.3109/03602532.2011.567391. [DOI] [PubMed] [Google Scholar]
- 32.Jakobsson PJ, Mancini JA, Riendeau D, Ford-Hutchinson AW. Identification and characterization of a novel microsomal enzyme with glutathione-dependent transferase and peroxidase activities. J Biol Chem. 1997;272:22934–22939. doi: 10.1074/jbc.272.36.22934. [DOI] [PubMed] [Google Scholar]
- 33.Duroudier NP, Tulah AS, Sayers I. Leukotriene pathway genetics and pharmacogenetics in allergy. Allergy. 2009;64:823–839. doi: 10.1111/j.1398-9995.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 34.Hu G, Shi Z, Hu J, Zou G, Peng G, Ran P. Association between polymorphisms of microsomal epoxide hydrolase and COPD: results from meta-analyses. Respirology. 2008;13:837–850. doi: 10.1111/j.1440-1843.2008.01356.x. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Hu Z, Qu X, Zhu J, Li L, Ring BZ, Su L. Putative EPHX1 enzyme activity is related with risk of lung and upper aerodigestive tract cancers: a comprehensive meta-analysis. PLoS ONE. 2011;6:e14749. doi: 10.1371/journal.pone.0014749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tung KY, Tsai CH, Lee YL. Microsomal epoxide hydroxylase genotypes/diplotypes, traffic air pollution, and childhood asthma. Chest. 2011;139:839–848. doi: 10.1378/chest.10-2479. [DOI] [PubMed] [Google Scholar]
- 37.Kil IS, Huh TL, Lee YS, Lee YM, Park JW. Regulation of replicative senescence by NADP+ -dependent isocitrate dehydrogenase. Free Radic Biol Med. 2006;40:110–119. doi: 10.1016/j.freeradbiomed.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Li B, Zhou Y, Zhong N, Ran P. Genetic analysis of CC16, OGG1 and GCLC polymorphisms and susceptibility to COPD. Respirology. 2007;12:29–33. doi: 10.1111/j.1440-1843.2006.00958.x. [DOI] [PubMed] [Google Scholar]
- 39.Pierrou S, Broberg P, O'Donnell RA, Pawlowski K, Virtala R, Lindqvist E, Richter A, Wilson SJ, Angco G, Moller S, et al. Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175:577–586. doi: 10.1164/rccm.200607-931OC. [DOI] [PubMed] [Google Scholar]
- 40.Koide S, Kugiyama K, Sugiyama S, Nakamura S, Fukushima H, Honda O, Yoshimura M, Ogawa H. Association of polymorphism in glutamate-cysteine ligase catalytic subunit gene with coronary vasomotor dysfunction and myocardial infarction. J Am Coll Cardiol. 2003;41:539–545. doi: 10.1016/s0735-1097(02)02866-8. [DOI] [PubMed] [Google Scholar]
- 41.Petersen SV, Oury TD, Ostergaard L, Valnickova Z, Wegrzyn J, Thogersen IB, Jacobsen C, Bowler RP, Fattman CL, Crapo JD, et al. Extracellular superoxide dismutase (ECSOD) binds to type i collagen and protects against oxidative fragmentation. J Biol Chem. 2004;279:13705–13710. doi: 10.1074/jbc.M310217200. [DOI] [PubMed] [Google Scholar]
- 42.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:858–864. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- 43.Young RP, Hopkins R, Black PN, Eddy C, Wu L, Gamble GD, Mills GD, Garrett JE, Eaton TE, Rees MI. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax. 2006;61:394–399. doi: 10.1136/thx.2005.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siedlinski M, van Diemen CC, Postma DS, Vonk JM, Boezen HM. Superoxide dismutases, lung function and bronchial responsiveness in a general population. Eur Respir J. 2009;33:986–992. doi: 10.1183/09031936.00171507. [DOI] [PubMed] [Google Scholar]
- 45.Dahl M, Bowler RP, Juul K, Crapo JD, Levy S, Nordestgaard BG. Superoxide dismutase 3 polymorphism associated with reduced lung function in two large populations. Am J Respir Crit Care Med. 2008;178:906–912. doi: 10.1164/rccm.200804-549OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorheim IC, DeMeo DL, Washko G, Litonjua A, Sparrow D, Bowler R, Bakke P, Pillai SG, Coxson HO, Lomas DA, et al. Polymorphisms in the superoxide dismutase-3 gene are associated with emphysema in COPD. COPD. 2010;7:262–268. doi: 10.3109/15412555.2010.496821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, Johansson M, Holmgren A. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276:26269–26275. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 48.Nagy N, Malik G, Tosaki A, Ho YS, Maulik N, Das DK. Overexpression of glutaredoxin-2 reduces myocardial cell death by preventing both apoptosis and necrosis. J Mol Cell Cardiol. 2008;44:252–260. doi: 10.1016/j.yjmcc.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, Xing K, Lou MF. Glutaredoxin 2 prevents H(2)O(2)-induced cell apoptosis by protecting complex I activity in the mitochondria. Biochim Biophys Acta. 2010;1797:1705–1715. doi: 10.1016/j.bbabio.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.