Abstract
The contribution of mitochondrial dysfunction to insulin resistance is a contentious issue in metabolic research. Recent evidence implicates mitochondrial dysfunction as contributing to multiple forms of insulin resistance. However, some models of mitochondrial dysfunction fail to induce insulin resistance, suggesting greater complexity describes mitochondrial regulation of insulin action. We report that mitochondrial dysfunction is not necessary for cellular models of insulin resistance. However, impairment of mitochondrial function is sufficient for insulin resistance in a cell type-dependent manner, with impaired mitochondrial function inducing insulin resistance in adipocytes, but having no effect, or insulin sensitising effects in hepatocytes. The mechanism of mitochondrial impairment was important in determining the impact on insulin action, but was independent of mitochondrial ROS production. These data can account for opposing findings on this issue and highlight the complexity of mitochondrial regulation of cell type-specific insulin action, which is not described by current reductionist paradigms.
Keywords: Mitochondria, Insulin action, Reactive oxygen species, Adipocyte, Hepatocyte
Abbreviations: AMPK, AMP-activated protein kinase; AS160, Akt substrate of 160 kDa; BSA, bovine serum albumin; ECAR, extracellular acidification rate; FoxO1, forkhead box protein O1; GP, glucose production; HI-FBS, heat-inactivated foetal bovine serum; IRS1, insulin receptor substrate 1; GLUT4, facilitative glucose transporter isoform 4; G.O., glucose oxidase; LDH, lactate dehydrogenase; MMP, mitochondrial membrane potential; MnTBAP, manganese (III) tetrakis (4-benzoic acid) porphyrin chloride; PI3K, phosphatidylinositol 3-kinase; ROS, reactive oxygen species; SOD, superoxide dismutase; T2D, type 2 diabetes; TNFα, tumour necrosis factor alpha
1. Introduction
Type 2 diabetes (T2D) involves insulin resistance in skeletal muscle, the liver and adipose tissue [1]. One of the most contentious issues in metabolic research is the role of mitochondrial dysfunction in the development of insulin resistance. The term mitochondrial dysfunction can describe impairments in numerous mitochondrial function indices, including respiration, ATP production, membrane potential, proton leak and reactive oxygen species (ROS) production [2]. Impaired mitochondrial function has been observed in skeletal muscle [3–6], the liver [7–9] and adipose tissue [10–12] of T2D patients and animal models of T2D. Similar impairments have been observed in the skeletal muscle of insulin resistant offspring of T2D patients [13]. This could suggest a role for impaired mitochondrial function in the development of insulin resistance. This is supported by observations of insulin resistance in humans with mitochondrial DNA mutations that result in impaired mitochondrial function [14–16]. Mitochondrial dysfunction has been proposed to induce insulin resistance through ectopic lipid accumulation secondary to reduced β-oxidation, which impairs insulin signalling [17,18]. More recently, production of ROS has emerged as a direct link between mitochondrial dysfunction and insulin resistance driven by numerous insults such as saturated fatty acids, inflammatory cytokines and glucocorticoids [19,20]. However, numerous animal models with impaired mitochondrial function have either unchanged or increased insulin sensitivity [21–23], questioning both the causality of this relationship and whether mitochondrial dysfunction is necessary and/or sufficient for insulin resistance. Further adding to controversy on this issue, anti-diabetic agents such as the biguanide and thiazolidinedione family of compounds, which enhance insulin action primarily in the liver and adipose tissue respectively, have been reported to inhibit complex I of the electron transport chain and/or the mitochondrial pyruvate carrier (MPC), which impairs mitochondrial function [24–26]. These counterintuitive findings have been balanced by evidence that biguanides can prevent ROS production by complex I under conditions of electron backflow from complex II, such as during high fat feeding [27]. However, given that most of our knowledge regarding the role of mitochondria in the regulation of insulin action has been generated from studies of skeletal muscle, coupled with the fact that the primary tissues of action of these anti-diabetic drugs are not skeletal muscle, could raise the possibility that there are cell/tissue type-specific responses in this relationship that are not yet fully understood. Indeed, studies of either insulin resistant humans or animal models of mitochondrial dysfunction have not been able to mechanistically dissect this relationship with any certainty. This is due to reasons such as the non-physiological nature of gene ablation, the markedly different mitochondrial respiratory rates and metabolic function of tissues involved in whole body insulin action, the complexity in controlling substrate flux to individual insulin-sensitive tissues and inter-tissue cross-talk in vivo. While these factors are important for the development of the whole body metabolic phenotype in insulin resistance, they are nonetheless confounding variables when examining the fundamental link between mitochondrial dysfunction and insulin action.
The fundamental biology underpinning these paradoxical findings describing the relationship between mitochondria and insulin action is poorly understood. Indeed, the potential importance of specific mitochondrial enzyme impairments and the tissue/cell type in which impairments occur are unknown. Furthermore, it is unknown how alterations in many of the interdependent indices of mitochondrial function impact on cellular insulin action. As in vivo studies have been unable to dissect these mechanisms, fundamental studies in cellular systems that define the biological complexity in the relationship between mitochondrial function and insulin action in multiple cell types are required before our understanding of the physiological role of mitochondria in the development of insulin resistance can advance. Therefore, the aims of this study were to: 1. determine whether mitochondrial dysfunction is necessary and/or sufficient for cellular insulin resistance; 2. establish whether specific mitochondrial enzyme impairment is important for this response in a cell type-dependent manner; and 3. assess whether ROS production is a universal link between impaired mitochondrial function and insulin resistance. We used 3T3L1 adipocytes and FAO hepatoma cells, as models of adipocytes and hepatocytes, respectively, to address these aims, assessing glucose uptake and suppression of glucose production as measures of insulin action. These cell lines have been used extensively to study insulin action and these cell types are characterised by impaired mitochondrial function in insulin resistant states [7,12]. However, few studies have utilised these cell types to mechanistically examine the role of mitochondria in insulin action.
2. Materials and methods
2.1. Cell culture
Mouse immortalised 3T3L1 fibroblasts were cultured in 10% CO2 at 37 °C in growth media consisting of DMEM (4.5 g/L glucose; Invitrogen), 10% heat-inactivated foetal bovine serum (HI-FBS; Thermo Scientific) and antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin; Life Technologies). Cells were induced to differentiate 2 days after reaching confluence (day 0), by supplementing growth media with 3 nM insulin (Humulin R; Eli Lilly), 0.25 μM dexamethasone (Sigma–Aldrich) and 0.5 mM 1-methyl-3-isobutyl-xanthine (Sigma–Aldrich). From day 3 until day 7, cells were maintained in growth media supplemented with 3 nM insulin after which the mature adipocytes were maintained in growth media. All treatments were for 24 h in growth media unless stated otherwise. Rat FAO immortalised hepatoma hepatocytes [28] were cultured in 5% CO2 at 37 °C in growth media consisting of RPMI 1640 medium (2 g/L glucose; Invitrogen) and 10% foetal bovine serum (FBS; Thermo Scientific). Assays were carried out when cells were ∼90% confluent. All treatments of hepatocytes were for 24 h in glucose- and serum-free RPMI 1640 media supplemented with 2 mM sodium pyruvate, 20 mM sodium l-lactate and 0.1% BSA (glucose production media; GP media), except where indicated. For 3T3L1 models of insulin resistance, cells were treated with 25 mU/mL glucose oxidase (G.O.; Sigma–Aldrich), 10 ng/mL tumour necrosis factor-α (TNFα; Peprotech) or 10 nM chronic insulin. For chronic insulin treatments, cells were returned to growth media containing no insulin 2 h before beginning glucose uptake assays or protein collection. Treatment doses for oligomycin (Sigma–Aldrich) and rotenone (Sigma–Aldrich) models of mitochondrial dysfunction as well as Antimycin A (Sigma–Aldrich) and FCCP (Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; Sigma–Aldrich) in both 3T3L1 and FAO cells are as stated in Figures 2 and 3, Figures S2 and S3 . The doses of rosiglitazone and phenformin are stated in Figure 4 and Figure S4. MnTBAP (Manganese (III) tetrakis (4-benzoic acid) porphyrin chloride; Enzo Life Sciences) co-treatments were at a dose of 300 μM and wortmannin (wort; Sigma–Aldrich) co-treatments were at a dose of 100 nM.
Figure 2.
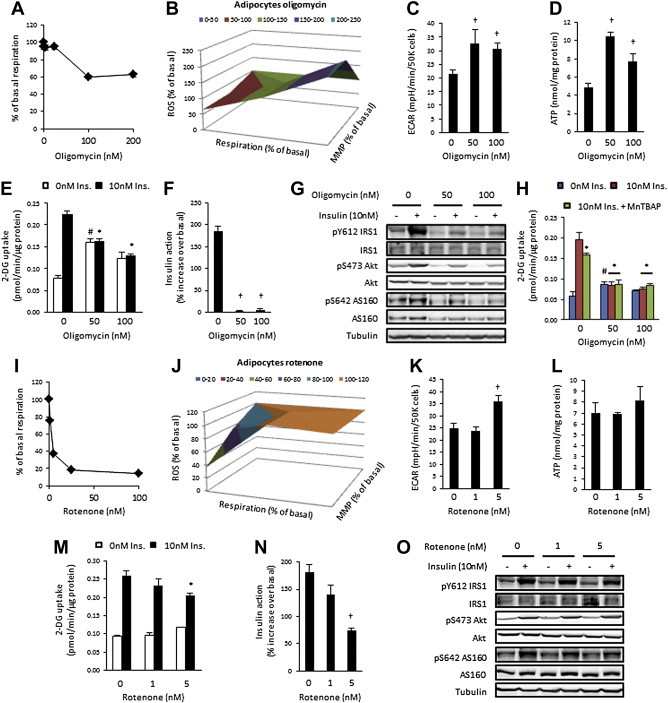
Impaired mitochondrial function induces insulin resistance in 3T3L1 adipocytes. (A) Basal respiration; (B) Mitochondrial function; (C) Extracellular acidification rate (ECAR); (D) ATP concentration; (E) Basal and insulin-stimulated (10 nM) 2-deoxyglucose uptake; (F) Insulin action; (G) Insulin signalling; and (H) Basal and insulin-stimulated (10 nM) 2-deoxyglucose uptake in the presence or absence of MnTBAP in vehicle, 50 nM or 100 nM oligomycin treated (24 h) 3T3L1 adipocytes. (I) Basal respiration; (J) Mitochondrial function; (K) ECAR; (L) ATP concentration; (M) Basal and insulin-stimulated (10 nM) 2-deoxyglucose uptake; (N) Insulin action; and (G) Insulin signalling in vehicle, 1 nM or 5 nM rotenone treated (24 h) 3T3L1 adipocytes. Data presented as mean ± SEM, n = 3–6 biological replicates. Denotes significantly different from; † vehicle-treated cells, # vehicle/non-insulin-treated cells, * vehicle/insulin-treated cells (p < 0.05).
Figure 3.
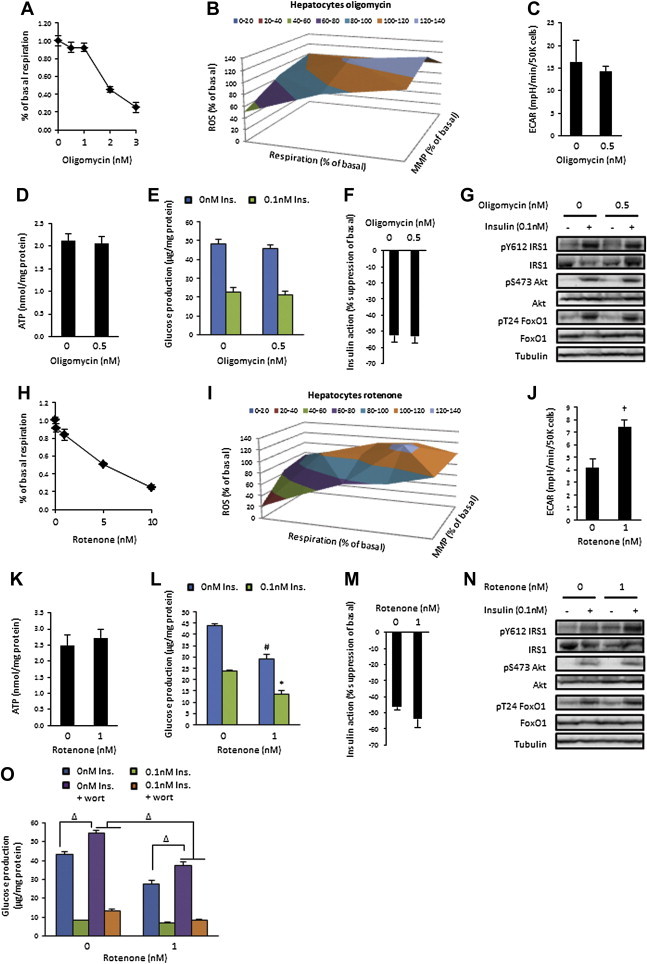
Impaired mitochondrial function has no effect on insulin action in FAO hepatocytes. (A) Basal respiration; (B) Mitochondrial function; (C) Extracellular acidification rate (ECAR); (D) ATP concentration; (E) Basal and insulin-suppressed (0.1 nM) glucose production; (F) Insulin action; and (G) Insulin signalling in vehicle and 0.5 nM oligomycin treated (24 h) FAO hepatocytes. (H) Basal respiration; (I) Mitochondrial function; (J) ECAR; (K) ATP concentration; (L) Basal and insulin-suppressed (0.1 nM) glucose production; (M) Insulin action; (N) Insulin signalling; and (O) Basal and insulin-suppressed (0.1 nM) glucose production in the presence of absence of wortmannin (100 nM) in vehicle and 1 nM rotenone treated FAO hepatocytes. Data presented as mean ± SEM, n = 3–6 biological replicates. Denotes significantly different from; † vehicle-treated cells, # vehicle/non-insulin-treated cells, * vehicle/insulin-treated cells, Δ designated difference (p < 0.05).
Figure 4.
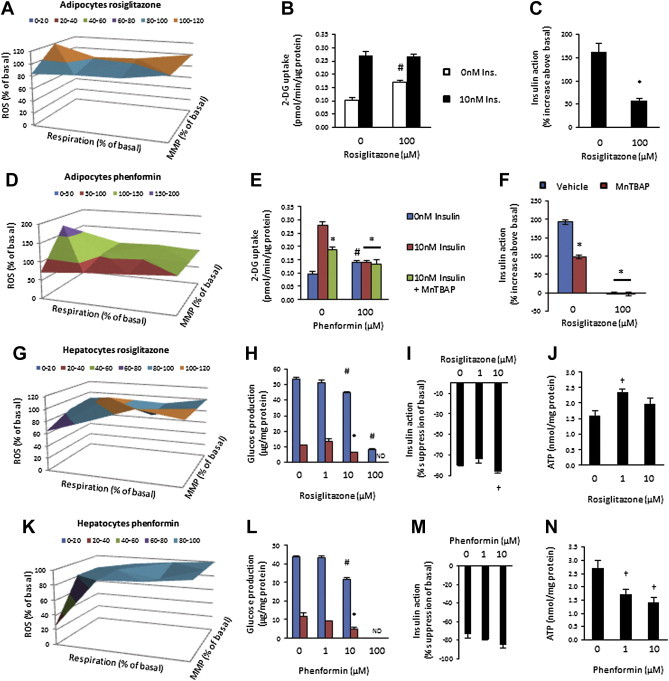
Anti-diabetic agents rosiglitazone and phenformin induce mitochondrial dysfunction and have cell type-dependent effects on insulin action. (A) Mitochondrial function; (B) Basal and insulin-stimulated (10 nM) 2-deoxyglucose uptake; and (C) Insulin action in vehicle and 100μM rosiglitazone treated (24 h) 3T3L1 adipocytes. (D) Mitochondrial function; (E) Basal and insulin-stimulated (10 nM) 2-deoxyglucose uptake; and (F) Insulin action in the presence or absence of MnTBAP in vehicle and 100μM rosiglitazone treated (24 h) 3T3L1 adipocytes. (G) Mitochondrial function; (H) Basal and insulin-suppressed (0.1 nM) glucose production; (I) Insulin action; and (J) ATP levels in FAO hepatocytes treated (24 h) with increasing doses of rosiglitazone. (K) Mitochondrial function; (L) Basal and insulin-suppressed (0.1 nM) glucose production; (M) Insulin action; and (N) ATP levels in FAO hepatocytes treated (24 h) with increasing doses of phenformin. Data presented as mean ± SEM, n = 3–6 biological replicates. Denotes significantly different from; # vehicle/non-insulin-treated cells, * vehicle/insulin-treated cells, † vehicle-treated cells (p < 0.05). ND = not detected.
2.2. Bioenergetics and respiration analyses
The cellular bioenergetics profile of 3T3L1 adipocytes and FAO hepatocytes was assessed using the Seahorse XF24 Flux Analyzer (Seahorse Bioscience). 3T3L1 fibroblasts were seeded into a 24-well XF24 cell culture microplate (Seahorse Bioscience) and were differentiated to maturity, as described above, at which time the cells were treated for 24 h. FAO hepatocytes were also seeded into a XF24 microplate at a density of 50,000 cells per well and 4 h later, 24 h treatments were begun in growth media. Cells were washed and incubated in 600 μl unbuffered DMEM (containing 25 mM glucose, 1 mM pyruvate and 1 mM glutamate) pH 7.4, at 37 °C in a non-CO2 incubator (1 h prior to bioenergetics assessment). Three basal oxygen consumption rate (OCR) measurements were performed using the Seahorse analyzer, and measurements were repeated following injection of oligomycin (1 μM), FCCP (1 μM) and Antimycin A (1 μM). Basal extracellular acidification rate (ECAR) was determined from data collected at basal measurement points. Calculations of respiratory parameters of mitochondrial function were performed as previously described [29] and included subtraction non-mitochondrial respiration from all mitochondrial respiration parameters. Following completion of the assay cell number was determined using the CyQuant® Cell Proliferation Assay kit (Molecular Probes) according to manufacturer's instructions.
2.3. Glucose uptake assay
Mature adipocytes in 24-well plates were treated for 24 h with insults, mitochondrial inhibitors or anti-diabetic agents, as described above, in serum-starve media consisting of DMEM (4.5 g/L glucose) supplemented with 0.2% fatty acid-free bovine serum albumin (BSA; USB Corporation). To begin the assay, cells were washed twice in 1 × Dulbecco's Phosphate-Buffered Saline, pH 7.4 (Life Technologies), containing 0.5 mM MgCl2, 0.5 mM CaCl2 and 0.2% fatty acid-free BSA, and then incubated in the presence or absence of 10 nM insulin at 37 °C for 30 min. Uptake of 50 μM 2-deoxyglucose and 0.2 μCi 2-deoxy-d-[3H]-glucose (PerkinElmer) per well was measured over the final 10 min of this incubation. Cells were washed twice in ice-cold phloretin (80 μg/mL) in PBS and lysed in 0.03% SDS before being analysed by scintillation counting. Protein content per well was measured and results expressed as pmol glucose transported per minute per μg of protein.
2.4. Glucose production assay
FAO hepatocytes in 48-well plates were treated for 24 h with mitochondrial inhibitors or insulin sensitising agents in GP media in the presence or absence of 0.1 nM insulin. To measure the glucose produced by the cells, 40 μl of media was collected from each well and combined with 250 μl of Assay Buffer consisting of 0.12 M NaH2PO4·2H2O pH 7.0, 1 mg/mL phenol, 0.5 mg/mL 4-aminoantipyrine, 1.6 units/mL peroxidise and 10 units/mL glucose oxidase. This was incubated for 25 min at 37 °C after which absorbance was measured at 490 nm. Cells were lysed in 100 μl 0.03% SDS and protein was quantified using a BCA Protein Assay kit (Pierce). Results are expressed as μg glucose per mg protein.
An alternative method was used to measure glucose production in hepatocytes co-treated with MnTBAP as the colour of MnTBAP interferes with any colorimetric measurement. Cells were seeded and treated as in the colorimetric assay and instead glucose production was measured by a fluorometric assay kit (Cayman Chemical Company) following the manufacturer's instructions. Cells were lysed in 100 μl 0.03% SDS and protein was quantified using a BCA Protein Assay kit (Pierce). Results are expressed as μg glucose per mg protein.
2.5. Western blot analysis
Cells in 6-well plates were treated for 24 h as described above, in the absence or presence of insulin for the final 15 min (3T3L1 10 nM insulin in serum-starve media; FAO 0.1 nM insulin). Proteins were extracted using protein lysis buffer (50 mM Tris pH7.5, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% triton X-100, 50 mM NaF, 5 mM Na4P2O7, 1 mM Na3VO4, 1 mM DTT, protease inhibitor cocktail) and protein concentration quantified using a BCA Protein Assay kit (Pierce). 30 μg of protein was separated by SDS-PAGE and transferred onto PVDF membrane using standard protocols. Membranes were blocked for 1 h at room temperature (RT) in TBST (TBS with 0.05% Tween 20, pH 7.4) containing 1% BSA, and incubated overnight at 4 °C with the following primary antibodies, at the dilutions indicated, in TBST: anti-pY612 IRS1 (1:1000, Upstate), anti-IRS1 (1:1000, Upstate), anti-pS473 Akt (1:1000, Cell Signaling), anti-Akt (1:1000, Cell Signaling), anti-pS462 AS160 (1:1000, Cell Signaling), anti-AS160 (1:1000, Upstate), anti-pT24 FoxO1 (1:1000, Cell Signaling), anti-FoxO1 (1:1000, Cell Signaling), anti-α-tubulin (1:1,000; Sigma–Aldrich). Membranes were washed with TBST before being exposed to the appropriate HRP conjugated secondary antibody at a dilution of 1:10,000 in TBST for 1 h at RT. The reaction was developed using chemiluminescence (Invitrogen) and visualised using the ChemiDoc™ XRS+ with Image Lab™ software.
2.6. Lactate dehydrogenase cytotoxicity assay
Lactate dehydrogenase (LDH) is released from cells upon loss of membrane integrity due to apoptosis or necrosis and can therefore be used as a measure of cell viability. Cells in 48-well plates were treated as described above (3T3L1 in serum-starve media). After 24 h the cytotoxic effects of our treatments were assessed using the CytoTox 96® Non-Radioactive Cytotoxicity Assay kit (Promega) as per manufacturer's instructions. Viability was expressed as LDH in the media normalised to total LDH from the media and cell lysate.
2.7. Mitochondrial membrane potential measurement
Mitochondrial transmembrane potential was measured using the membrane-permeable JC-1 dye (Invitrogen). Cells in black-walled 96-well plates were treated for 24 h as described above. In the final 10 min of treatment, cells were incubated with 10 μg/mL JC-1 dye at 37 °C. After three washes with PBS, both green and red fluorescence emissions were measured using an excitation wavelength of 488 nm and emission wavelengths of 522 and 605 nm, respectively.
2.8. Mitochondrial superoxide measurement
MitoSOX™ Red (Molecular Probes) was used to measure mitochondrial specific superoxide. Cells in black-walled 96-well plates were treated for 24 h as described above. In the final 30 min of treatment, cells were incubated with 1μM MitoSOX™ Red at 37 °C. After two washes with PBS, fluorescence was measured using an excitation wavelength of 510 nm and an emission wavelength of 580 nm.
2.9. Statistical analysis
All values are expressed as mean ± SEM. Data were analysed for statistical significance using Minitab 15 Statistical Software. Unpaired t-tests were used where two individual treatments were compared. One-way ANOVA with Tukey post-hoc test was used where more than two groups were compared within a single treatment, and p values less than 0.05 were considered significant.
3. Results
3.1. Mitochondrial dysfunction is not necessary for insulin resistance in adipocytes
In vivo studies have not been able to determine whether mitochondrial dysfunction is necessary for insulin resistance with any certainty. To establish whether mitochondrial dysfunction is necessary for insulin resistance, we assessed mitochondrial function indices in diverse cellular models of insulin resistance in 3T3L1 adipocytes. These included glucose oxidase, TNFα and chronic insulin treatment, which all impaired insulin-stimulated glucose uptake (Figure 1A). Cellular insulin resistance in these models was not attributed to a common defect in insulin signalling at the level of IRS1, Akt or AS160 (Figure 1B). There was a small but significant increase in LDH release with glucose oxidase (Figure 1C), indicating compromised cell viability. As impaired mitochondrial respiration has been linked to the development of insulin resistance, we measured multiple mitochondrial respiration parameters including basal respiration, respiration due to ATP turnover and respiration due to proton leak. Glucose oxidase decreased respiration due to ATP turnover (Figure 1D) and increased mitochondrial ROS production (Figure 1F). Chronic insulin increased basal respiration, respiration for ATP turnover (Figure 1D) and decreased MMP (Figure 1E). As TNFα had no effect on any of the parameters measured and there was no consistent mitochondrial perturbation across all insults, these data suggest that a common defect in mitochondrial function, and mitochondrial dysfunction more generally, is not necessary for all forms of cellular insulin resistance.
Figure 1.
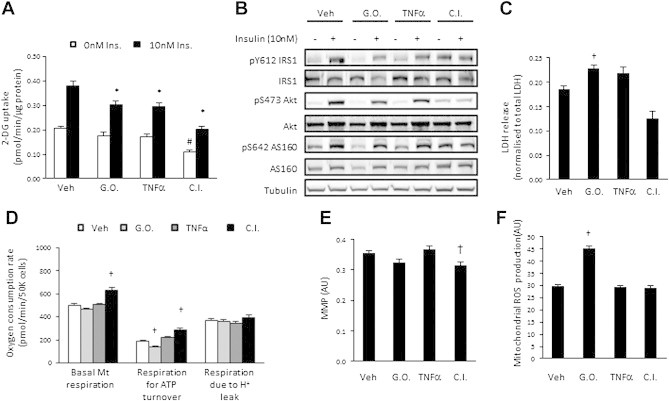
Diverse models of cell-autonomous insulin resistance in 3T3L1 adipocytes is not associated with a common perturbation in mitochondrial function. (A) Basal and insulin-stimulated (10 nM) 2-deoxyglucose uptake; (B) Insulin signalling; (C) LDH release; (D) Oxygen consumption due to basal respiration, ATP turnover and H+ leak; (E) Mitochondrial membrane potential (MMP); and (F) Mitochondrial ROS production, in vehicle (Veh), glucose oxidase (G.O.; 25 mU/mL), TNFα (10 ng/mL) and chronic insulin (10 nM) treated (24 h) cells. Data presented as mean ± SEM, n = 3–6 biological replicates. Denotes significantly different from; # vehicle/non-insulin-treated cells, * vehicle/insulin-treated cells; and † vehicle-treated cells (p < 0.05).
3.2. Mitochondrial dysfunction by inhibition of ATP synthase or complex I is sufficient to induce insulin resistance in adipocytes, independent of ROS production
A number of animal models with mitochondrial impairments have unchanged or enhanced insulin action. We sought to clarify these findings by investigating whether physiological impairment of mitochondrial function was sufficient to induce insulin resistance. As reduced ADP-stimulated respiration has been observed in insulin resistant states [3,4,13], we used the ATP synthase inhibitor oligomycin to mimic aspects of this defect. In 3T3L1 adipocytes, 24 h treatment with 50 nM and 100 nM oligomycin reduced basal respiration by ∼20% and ∼40%, respectively (Figure 2A), similar to that observed in insulin resistant states [13]. Three-variable (respiration, membrane potential and ROS production) surface plots were generated to represent mitochondrial function regulated by oligomycin (Figure 2B). Dose-dependent reductions in respiration due to ATP turnover (Figure S2A), but not proton leak (Figure S2B), were associated with increased MMP and ROS production (Figure 2B, Figure S2C and D). At 100 nM, oligomycin increased LDH release (Figure S2E). The extracellular acidification rate (ECAR), a proxy measure of glycolytic rate, was increased with both doses (Figure 2C) and was associated with an increase in cellular ATP levels (Figure 2D). Insulin-stimulated glucose uptake was decreased with both doses of oligomycin, and at 50 nM, basal glucose uptake was increased (Figure 2E). Insulin action, assessed as the percentage increases in insulin-stimulated glucose uptake over basal, was ablated at both oligomycin doses (Figure 2F). There was also a global reduction in expression and phosphorylation of key components of the insulin signalling pathway (Figure 2G). As mitochondrial ROS production has been implicated as a causative factor in insulin resistance [19,20] and was increased in this model, the SOD mimetic MnTBAP was used to reduce mitochondrial ROS production (Figure S2F). MnTBAP did not restore oligomycin-induced insulin resistance, but did reduce insulin-stimulated glucose uptake in the absence of oligomycin (Figure 2H), suggesting that increased mitochondrial ROS production does not mediate insulin resistance in this model. This was further supported by evidence that antimycin A, a complex III inhibitor that increases ROS production, had no effect on glucose uptake (Figure S2G) at concentrations that reduced respiration by ∼20% and ∼80% respectively (data not shown). These data show that mitochondrial dysfunction, in a model that replicates aspects of mitochondrial dysfunction seen in insulin resistant states in vivo, is sufficient to induce insulin resistance independent of mitochondrial ROS production.
As inhibition of complex I is thought to be the mechanism of action of some anti-diabetic agents, we examined the effect of the complex I inhibitor rotenone on insulin action. In 3T3L1 adipocytes, 1 nM and 5 nM rotenone reduced basal respiration by ∼20% and ∼60% respectively (Figure 2I). Altered mitochondrial function induced by rotenone (Figure 2J) varied from that induced by oligomycin (Figure 2B) and did not include alterations in MMP or mitochondrial ROS production (Figure S2H and I). However, 5 nM but not 1 nM rotenone, significantly reduced respiration associated with ATP turnover (Figure S2J) and proton leak (Figure S2K). While 5 nM rotenone increased ECAR (Figure 2K), both doses of rotenone had no effect on ATP concentration (Figure 2L) or cell viability (Figure S2L). We next tested the effect of rotenone on insulin action in 3T3L1 adipocytes. At 5 nM, but not 1 nM, rotenone significantly reduced insulin-stimulated glucose uptake (Figure 2M) and insulin action (Figure 2N). There were no obvious impairments in insulin signalling through IRS1, Akt and AS160 (Figure 2O). These data suggest that inhibition of complex I and its associated mitochondrial dysfunction is sufficient to induce insulin resistance in 3T3L1 adipocytes. However, this occurred only at supra-physiological impairment of respiration. Nonetheless, these data show that complex I inhibition does not have insulin sensitising effects in 3T3L1 adipocytes. The divergent effects of oligomycin and rotenone on insulin signalling and the magnitude of insulin resistance show that the exact mechanism of mitochondrial impairment is important in predicting subsequent effects on insulin action.
3.3. Impairment of ATP synthase or complex I of the ETC has no effect on the insulin sensitivity of FAO hepatocytes, despite increasing ROS production
Insulin-sensitive cells can differ widely in their oxidative capacity and metabolic functions. Therefore, to determine whether mitochondrial dysfunction is sufficient to induce insulin resistance in multiple cell types, we investigated these effects in FAO hepatocytes. Low doses of oligomycin reduced basal respiration (Figure 3A) and altered mitochondrial function (Figure 3B) in FAO hepatocytes. At 0.5 nM, which reduced respiration by ∼10% (Figure 3A), oligomycin did not significantly alter respiration due to ATP turnover (Figure S3A) and proton leak (Figure S3B), but did increased MMP (Figure S3C) and ROS production (Figure S3D). There was no change in cell viability (Figure S3E). There were no effects on ECAR (Figure 3C) or ATP levels (Figure 3D). We tested whether oligomycin-induced mitochondrial dysfunction was sufficient to reduce insulin sensitivity in FAO hepatocytes by measuring insulin suppression of glucose production (Figure 3E). Oligomycin did not alter basal or insulin suppression of glucose production, or insulin action (Figure 3F). No effect on the insulin signalling pathway was detected (Figure 3G). These data provide evidence that increased mitochondrial ROS production does not induce insulin resistance in these cells. Rather a reduction in mitochondrial ROS (Figure S3F) and alteration of the cellular redox state with MnTBAP completely impaired glucose production (Figure S3G), suggesting a more fundamental role for ROS signalling in the control of glucose production in these cells.
We next assessed the effect of rotenone on respiration in FAO hepatocytes (Figure 3G), which altered mitochondrial function (Figure 3H). At 1 nM, which reduced respiration by ∼20%, this included decreased respiration due to ATP turnover (Figure S3H) and proton leak (Figure S3I) but had no effect on MMP, ROS production (Figure 3H, Figure S3J and K) or cell viability (Figure S3E). ECAR was increased (Figure 3I) and ATP levels were unchanged (Figure 3J). Interestingly, rotenone reduced both basal and insulin-stimulated glucose production (Figure 3M), but had no effect on insulin action (Figure 3N). As rotenone increased both basal and insulin-stimulated tyrosine phosphorylation of IRS1 (Figure 3O), we tested whether inhibiting phosphatidylinositol 3-kinase (PI3K) and subsequent insulin signalling with wortmannin, would eliminate the ability of rotenone to reduce glucose production. Wortmannin significantly increased basal glucose production in vehicle-treated cells, but had no effect on the ability of insulin to suppress glucose production, suggesting that the canonical insulin signalling pathway is not essential for the suppressive effects of insulin on glucose production in this model. In the presence of wortmannin, rotenone was still able to decrease both basal and insulin-stimulated glucose production (Figure 3P), suggesting that rotenone's actions on glucose production are not mediated through insulin signalling pathway sensitisation.
3.4. The anti-diabetic drugs rosiglitazone and phenformin alter mitochondrial function and have cell type-dependent effects on insulin action
Our data show that mitochondrial dysfunction is not a universal initiating factor for insulin resistance. However, inducing mitochondrial dysfunction is sufficient to induce cell type-dependent insulin resistance. As rosiglitazone and phenformin exert their effects, in part, through inhibition of complex I and the mitochondrial pyruvate carrier, resulting in impaired mitochondrial respiration, we examined the acute effects of these agents on mitochondrial function and insulin sensitivity. In 3T3L1 adipocytes, rosiglitazone reduced mitochondrial respiration (Figure S4A) and mitochondrial function (Figure 4A, Figure S4B and C), however the nature of that dysfunction, based on multiple parameter analysis varied from that induced by oligomycin and rotenone in these same cells (Figure 2B and J), reinforcing the importance of measuring multiple parameters of mitochondrial function. Rosiglitazone increased ATP levels (Figure S4D) independent of increases in ECAR (Figure S4E). Consistent with our data for other mitochondrial inhibitors in these cells, insulin action was decreased by rosiglitazone (Figure 4B and C), but this was driven by an increase in basal glucose uptake (Figure 4B). Phenformin induced similar alterations in mitochondrial respiration (Figure S4F) and function (Figure 4A), in adipocytes, albeit with greater inhibitory action on respiration associated with ATP turnover and proton leak (Figures S4G and H). No significant changes in ATP levels (Figure S4I) or ECAR (Figure S4J) were detected with phenformin treatment. Although phenformin increased basal glucose uptake (Figure 4E), insulin action was also completely abrogated (Figure 4F). As phenformin increased mitochondrial ROS production (refer ahead to Figure 5D), we used MnTBAP to counter this increase, with no effect on insulin action (Figure 4E and F). This shows that in this cell type, heterogeneous forms of mitochondrial dysfunction converge to induce insulin resistance, independent of increased mitochondrial ROS production. These drugs also altered mitochondrial function in FAO hepatocytes (Figure 4G and K, Figure S4K–O, Q–U). Rosiglitazone at high doses reduced both basal and insulin suppression of glucose production (Figure 4H) and increased insulin action (Figure 4I). As recent evidence links the efficacy of biguanides on hepatic glucose production to reduced ATP levels [34], we also assessed ATP concentrations. However, rosiglitazone did not reduce ATP levels (Figure 4J), nor ECAR (Figure S4P). Phenformin at higher doses also reduced basal and insulin-stimulated glucose production (Figure 4L), but not insulin action (Figure 4M), despite reduced ATP levels (Figure 4N) and no compensatory change in ECAR (Figure 4SV). These data show that similar to traditional mitochondrial inhibitors, these anti-diabetic agents have cell type-dependent effects on mitochondrial function and insulin action.
Figure 5.
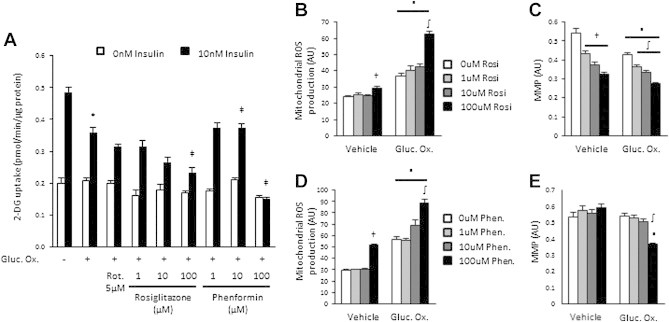
Rosiglitazone and phenformin do not impair mitochondrial ROS production in 3T3L1 adipocytes. (A) Basal and insulin-stimulated (10 nM) glucose uptake in 3T3L1 adipocytes treated with glucose oxidase (Gluc. Ox.; 25 mU/mL) and increasing doses of either rosiglitazone and phenformin; (B) Mitochondrial ROS production via mitosox fluorescence; and (C) Mitochondrial membrane potential (MMP) in 3T3L1 adipocytes treated (24 h) with glucose oxidase and rosiglitazone. (D) Mitochondrial ROS production via mitosox fluorescence; and (E) Mitochondrial membrane potential (MMP) in 3T3L1 adipocytes treated (24 h) with glucose oxidase and phenformin. Data presented as mean ± SEM, n = 3–6 biological replicates. Denotes significantly different from; * vehicle/insulin-treated cells, ‡ glucose oxidase/insulin-treated cells, ∫ glucose oxidase/vehicle-treated cells, ▪ corresponding treatment group/no glucose oxidase treated cells (p < 0.05).
3.5. Rosiglitazone and phenformin do not impair mitochondrial ROS production in 3T3L1 adipocytes
While the exact mechanism linking the effects of rosiglitazone and phenformin on mitochondrial function and insulin action remain unclear, one proposed mechanism is that these compounds can reduce ROS production under conditions of mitochondrial dysfunction [27], suggesting that these drugs could have altered efficacy in stressed and healthy states. While we have not found a role for ROS production linking mitochondrial dysfunction and insulin resistance in our models, we nonetheless explored this possibility. We induced mitochondrial dysfunction in 3T3L1 adipocytes with glucose oxidase, which increased ROS production (Figure 1F), and co-treated with rosiglitazone or phenformin before assessing insulin action (Figure 5A). Glucose oxidase-induced insulin resistance was worsened with increasing doses of both rosiglitazone and phenformin (Figure 5A). ROS production was also increased with co-treatment of glucose oxidase with rosiglitazone (Figure 5B) and phenformin (Figure 5D). This was associated with a loss in MMP with rosiglitazone that was also worsened with glucose oxidase (Figure 5C) and with 100 μM phenformin and glucose oxidase (Figure 5E). These data suggest that these compounds do not reduce ROS production under states of mitochondrial dysfunction in all cell types and that these anti-diabetic drugs can further exacerbate mitochondrial dysfunction and its associated insulin resistance under context-specific conditions.
4. Discussion
These fundamental studies on the relationship between mitochondrial function and cell-autonomous insulin action revealed that acute, physiological impairments in mitochondrial function are sufficient, but not necessary, to induce insulin resistance (Figure 6). Definitive conclusions on this issue have been elusive in human and animal studies. This has been partly due to the difficultly in inducing physiological impairments in mitochondrial function in animal models either through genetic or other means. In addition, interventionist human studies have found it difficult to separate mitochondrial dysfunction with other confounding variables, such as increased fatty acid availability, which is also associated with insulin resistance [30,31]. However the utility of our cell-autonomous system that allows titration of mitochondrial impairment to physiological levels and analysis of insulin action under conditions of constant substrate availability, without the influence of inter-tissue crosstalk, has assisted with mechanistic dissection of this relationship. These findings support data in humans with mitochondrial impairments due to mitochondrial DNA mutations, which can also manifest in insulin resistance in key insulin-sensitive tissues [14–16]. Numerous animal models with impaired mitochondrial function due to genetic or dietary modification of components of the electron transport chain have produced opposing conclusions on this same issue. For example, deletion of the apoptosis initiating factor (AIF) enzyme that is a component of complex I, leads to oxidative phosphorylation defects and protection against insulin resistance in response to high fat feeding [22]. Similar findings are observed with the ablation of cytochrome c oxidase subunit VI peptide 2a (Cox6a2; 23) and iron-containing enzymes of the ETC [21]. However, data from the present study could reconcile these findings. For example, we showed that complex I inhibition decreased insulin-stimulated glucose uptake in adipocytes, but potentiated insulin suppression of glucose production in hepatocytes. These data showing that identical insults have cell type-dependent effects on mitochondrial function and insulin action highlights the complexity in this relationship (Figure 6). Multi-parameter measurement and representation of mitochondrial function in a multi-dimension format in the form of surface plots was also useful in identifying the heterogeneous nature of mitochondrial dysfunction by common insults. Furthermore, the specific mitochondrial enzymes impaired also determine the impact on insulin action. Indeed, oligomycin and rotenone had differential effects on insulin signalling and the magnitude of insulin resistance in 3T3L1 adipocytes, despite similar impairments in respiration. These data suggest that animal models with global defects in mitochondrial function could manifest different phenotypes depending on the site of the mitochondrial impairment and on the specific insulin-sensitive tissue that dominates the whole body phenotype. The impact on whole body insulin action would therefore be difficult to identify and predict. The functional significance of these findings are that any phenotypic investigation into the role of mitochondria in the pathogenesis of insulin resistance in patients or disease models will likely have to establish the exact tissue and enzyme(s) affected to understand the mechanisms involved. Furthermore, the biological complexity in mitochondrial regulation of insulin action identified in our studies suggests that universal and reductionist theories describing this relationship may not be possible.
Figure 6.
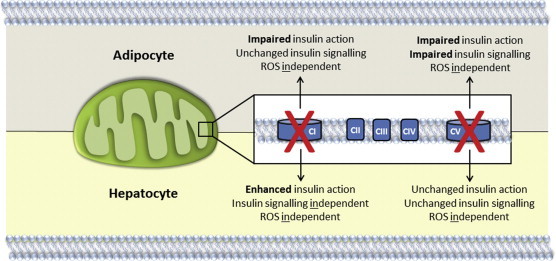
Complexity linking impaired mitochondrial function to insulin action. Cell type- and impairment site-specific relationships between mitochondrial dysfunction and insulin action in adipocytes and hepatocytes.
Our findings also revealed that mitochondrial ROS is not a universal driver of insulin resistance in the cellular systems examined. This theory on the aetiology of insulin resistance has gained momentum following a number of recent studies showing that skeletal muscle insulin resistance can be prevented through buffering of mitochondrial ROS [19,20]. Furthermore, buffering total cellular ROS in a number of adipocyte models of insulin resistance was sufficient to reverse insulin resistance [32]. However, the compartmentalisation of ROS production and the cell type-dependent responses to ROS require further consideration when interpreting these data, as ROS is also required for insulin action [33]. Nonetheless, mitochondrial ROS-mediated insulin resistance appears to be a valid mechanism in skeletal muscle. It is unclear why mitochondrial ROS production was not associated with insulin resistance in our models, however this is an area that warrants further investigation and could involve differences in anti-oxidant capacities. Such experiments could be important in determining the utility of anti-oxidant therapies in heterogeneous insulin resistant states. The mechanisms that link mitochondrial dysfunction and insulin action in adipocytes and hepatocytes are also unclear. In adipocytes, mitochondrial dysfunction-induced alterations in calcium handling could be important, as insulin-stimulated GLUT4 translocation and glucose uptake are calcium sensitive [34]. Protein signalling from the mitochondria under states of stress could also play a role. Notably, deletion of apoptosis-inducing factor (AIF), a protein typically released from the mitochondria by pro-apoptotic stimuli, protects against insulin resistance despite a reduction in mitochondrial respiration [22]. This could suggest that protein-mediated signalling from mitochondria might impact on insulin action. In hepatocytes, a recent study has found that reduced cellular energy status can impede glucagon signalling, which opposes insulin signalling in the control of glucogeogenesis [35]. Increased AMP might also allosterically inhibit gluconeogenic enzymes directly to reduce gluconeogenic flux [36]. However, we found no association between reduced ATP, or any involvement for the canonical insulin signalling pathway in our mitochondrial dysfunction models that reduced insulin suppression of glucose production. The mechanisms by which mitochondrial dysfunction impacts on insulin action in hepatocytes remain to be determined.
We also observed that the anti-diabetic agents phenformin and rosiglitazone showed cell type-dependent impairments in mitochondrial function and insulin action. This included enhanced suppression of glucose production and enhanced insulin action in hepatocytes, but also included induction of insulin resistance in adipocytes. The acute effects of these drugs in adipocytes might be offset by longer term remodelling of adipocyte metabolism, particularly in the case of rosiglitazone, which enhances the expression of PPAR dependent genes to increase glucose disposal and lipogenesis, thereby reducing hyperglycemia in vivo [37]. Indeed, the reduced insulin action we observed in rosiglitazone treated adipocytes was primarily due to an increase in basal glucose uptake, rather than reduced insulin-stimulated glucose uptake. Nonetheless, both anti-diabetic drugs altered mitochondrial function in adipocytes, which could have longer term implications in patients treated with these drugs for extended periods. Indeed, these findings could explain inconsistencies in efficacy associated with these drugs and their reduced efficacy over time. The finding that both drugs exacerbated insulin resistance in the context of existing mitochondrial dysfunction also highlights that personalised medicine approaches that identify any mitochondrial defects could be used to develop improved treatment strategies for patients.
In conclusion, our findings show that acute, physiological impairments in mitochondrial function are sufficient, but not necessary, for the development of cell type-dependent insulin resistance and this is independent of mitochondrial ROS production. The translational implications from these findings are that therapies for insulin resistance that involve modulation of mitochondrial function should be tissue specific and their mechanism of action on mitochondria well characterised to ensure optimal therapeutic outcomes. The complexity in the relationship between mitochondrial function and insulin action revealed by these studies provides insights into opposing conclusions on this issue and questions reductionist theories that attempt to describe this relationship.
Acknowledgements
The authors wish to thank Prof. Mark Febbraio, Prof. Ken Walder, Dr. Lyndal Kerr-Bayles and Dr. Kirsten Howlett for helpful comments and critical evaluation of this manuscript. This study was supported by grants from the National Health and Medical Research Council of Australia (APP1027726 and APP1027727), the Deakin University Molecular and Medical Research Strategic Research Centre to SLM and the Deakin University Central Research Grant Scheme to SDM and SLM. SLM is supported by a National Health and Medical Research Council of Australia Career Development Fellowship (APP1030474).
Disclosure statement
The authors have nothing to disclose.
Conflict of interest
None declared.
Appendix A. Supporting information
The following is the Supplementary data related to this article:
References
- 1.Johnson D.A., Olefsky J.M. Origins and drivers of insulin resistance. Cell. 2013;152:673–684. doi: 10.1016/j.cell.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 2.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochemical Journal. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley D.E., He J., Menshikova E.V., Ritov V.B. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 4.Phielix E., Schrauwen-Hinderling V.B., Mensink M., Lenaers E., Meex R., Hoeks J. Lower intrinsic ADP-stimulated mitochondrial respiration underlies in vivo mitochondrial dysfunction in muscle of male type 2 diabetic patients. Diabetes. 2005;57:2943–2949. doi: 10.2337/db08-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ritov V.B., Menshikova E.V., He J., Ferrell R.E., Goodpaster B.H., Kelley D.E. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 6.van Tienen F.H., Praet S.F., de Feyter M.H., van den Broek N.M., Lindsey P.J., Schoonderwoerd K.G. Physical activity is the key determinant of skeletal muscle mitochondrial function in type 2 diabetes. Journal of Clinical Endocrinology and Metabolism. 2012;97:3261–3269. doi: 10.1210/jc.2011-3454. [DOI] [PubMed] [Google Scholar]
- 7.Szendroedi J., Chmelik M., Schmid A.I., Nowotny P., Brehm A., Krssak M. Abnormal hepatic energy homeostasis in type 2 diabetes. Hepatology. 2009;50:1079–1086. doi: 10.1002/hep.23093. [DOI] [PubMed] [Google Scholar]
- 8.Schmid A.I., Szendroedi J., Chmelik M., Krššak M., Moser E., Roden M. Liver ATP synthesis is lower and relates to insulin sensitivity in patients with type 2 diabetes. Diabetes Care. 2011;34:448–453. doi: 10.2337/dc10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouderba S., Sanz M.N., Sánchez-Martín C., El-Mir M.Y., Villanueva G.R., Detaille D. Hepatic mitochondrial alterations and increased oxidative stress in nutritional diabetes-prone Psammomys obesus model. Experimental Diabetes Research. 2012;2012:430176. doi: 10.1155/2012/430176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogacka I., Xie H., Bray G.A., Smith S.R. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 2005;54:1392–1399. doi: 10.2337/diabetes.54.5.1392. [DOI] [PubMed] [Google Scholar]
- 11.Choo H.J., Kim J.H., Kwon O.B., Lee C.S., Mun J.Y., Han S.S. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–791. doi: 10.1007/s00125-006-0170-2. [DOI] [PubMed] [Google Scholar]
- 12.Mitrou P., Boutati E., Lambadiari V., Maratou E., Papakonstantinou A., Komesidou V. Rates of glucose uptake in adipose tissue and muscle in vivo after a mixed meal in women with morbid obesity. Journal of Clinical Endocrinology and Metabolism. 2009;94:2958–2961. doi: 10.1210/jc.2008-2297. [DOI] [PubMed] [Google Scholar]
- 13.Petersen K.,F., Dufour S., Befroy D.E., Garcia R., Shulman G.I. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. New England Journal of Medicine. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reardon W., Ross R.J., Sweeney M.G., Luxon L.M., Pembrey M.E., Harding A.E. Diabetes mellitus associated with a pathogenic point mutation in mitochondrial DNA. Lancet. 1992;340:1376–1379. doi: 10.1016/0140-6736(92)92560-3. [DOI] [PubMed] [Google Scholar]
- 15.Becker R., Laube H., Linn T., Damian M.S. Insulin resistance in patients with the mitochondrial tRNA(Leu(UUR)) gene mutation at position 3243. Experimental and Clinical Endocrinology and Diabetes. 2002;110:291–297. doi: 10.1055/s-2002-34592. [DOI] [PubMed] [Google Scholar]
- 16.Szendroedi J., Schmid A.I., Meyerspeer M., Cervin C., Kacerovsky M., Smekal G. Impaired mitochondrial function and insulin resistance of skeletal muscle in mitochondrial diabetes. Diabetes Care. 2009;32:677–679. doi: 10.2337/dc08-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowell B.B., Shulman G.I. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 18.Morino K., Petersen K.F., Dufour S., Befroy D., Frattini J., Shatzkes N. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. Journal of Clinical Investigation. 2005;115:3587–3593. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson E.J., Lustig M.E., Boyle K.E., Woodlief T.L., Kane D.A., Lin C.T. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. Journal of Clinical Investigation. 2009;119:573–581. doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoehn K.L., Salmon A.B., Hohnen-Behrens C., Turner N., Hoy A.J., Maghzal G.J. Insulin resistance is a cellular antioxidant defence mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17787–17792. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han D.H., Hancock C.R., Jung S.R., Higashida K., Kim S.H., Holloszy J.O. Deficiency of the mitochondrial electron transport chain in muscle does not cause insulin resistance. PLoS One. 2011;6:e19739. doi: 10.1371/journal.pone.0019739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pospisilik J.A., Knauf C., Joza N., Benit P., Orthofer M., Cani P.D. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 23.Quintens R., Singh S., Lemaire K., De Bock K., Granvik M., Schraenen A. Mice deficient in the respiratory chain gene Cox6a2 are protected against high-fat diet-induced obesity and insulin resistance. PLoS One. 2013;8:e56719. doi: 10.1371/journal.pone.0056719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen M.R., Doran E., Halestrap A.P. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochemical Journal. 2000;348:607–614. [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz M.N., Sánchez-Martín C., Detaille D., Vial G., Rigoulet M., El-Mir M.Y. Acute mitochondrial actions of glitazones on the liver: a crucial parameter for their antidiabetic properties. Cellular Physiology and Biochemistry. 2011;28:899–910. doi: 10.1159/000335804. [DOI] [PubMed] [Google Scholar]
- 26.Divakaruni A.S., Wiley S.E., Rogers G.W., Andreyev A.Y., Petrosyan S., Loviscach M. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5422–5427. doi: 10.1073/pnas.1303360110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane D.A., Anderson E.J., Price J.W., 3rd, Woodlief T.L., Lin C.T., Bikman B.T. Metformin selectively attenuates mitochondrial H2O2 emission without affecting respiratory capacity in skeletal muscle of obese rats. Free Radical Biology and Medicine. 2010;149:1082–1087. doi: 10.1016/j.freeradbiomed.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deschatrette J., Weiss M.C. Characterisation of differentiated and dedifferentiated clones from a rat hepatoma. Biochimie. 1974;56:1603–1611. doi: 10.1016/s0300-9084(75)80286-0. [DOI] [PubMed] [Google Scholar]
- 29.McGee S.L., Sadli N., Morrison S., Swinton C., Suphioglu C. DHA protects against zinc mediated alterations in neuronal cellular bioenergetics. Cellular Physiology and Biochemistry. 2011;28:57–162. doi: 10.1159/000331724. [DOI] [PubMed] [Google Scholar]
- 30.Brehm A., Krssak M., Schmid A.I., Nowotny P., Waldhausl W., Roden M. Increased lipid availability impairs insulin-stimulated ATP synthesis in human skeletal muscle. Diabetes. 2006;55:136–140. [PubMed] [Google Scholar]
- 31.Hoeks J., van Herpen N.A., Mensink M., Moonen-Kornips E., van Beurden D., Hesselink M.K. Prolonged fasting identifies skeletal muscle mitochondrial dysfunction as consequence rather than cause of human insulin resistance. Diabetes. 2010;59:2117–2125. doi: 10.2337/db10-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houstis N., Rosen E.D., Lander E.S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 33.Loh K., Deng H., Fukushima A., Cai X., Boivin B., Galic S. Reactive oxygen species enhance insulin sensitivity. Cell Metabolism. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yip M.F., Ramm G., Larance M., Hoehn K.L., Wagner M.C., Guilhaus M. CaMKII-mediated phosphorylation of the myosin motor Myo1c is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metabolism. 2008;8:384–398. doi: 10.1016/j.cmet.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Miller R.A., Chu Q., Xie J., Foretz M., Viollet B., Birnbaum M.J. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foretz M., Hébrard S., Leclerc J., Zarrinpashneh E., Soty M., Mithieux G. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. Journal of Clinical Investigation. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reginato M.J., Lazar M.A. Mechanisms by which thiazolidinediones enhance insulin action. Trends in Endocrinology and Metabolism. 1999;10:9–13. doi: 10.1016/s1043-2760(98)00110-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


