Abstract
5-Methylcytosine (5mC) can be converted to 5-hydroxymethylcytosine (5hmC) in mammalian cells by the ten-eleven translocation (Tet) family of dioxygenases. While 5mC has been extensively studied, we have just started to understand the distribution and function of 5hmC in mammalian genomes. Despite the fact that this new epigenetic mark has only been discovered three years ago, exciting progress has been made in understanding its generation, fate, and genomic distribution. In this review we discuss these progresses as well as the recent advance in the single-base resolution mapping of 5hmC.
Introduction
DNA methylation at the 5-position of cytosine (5mC) in mammals is essential for normal development and plays important roles in a variety of biological processes, including transcriptional regulation and maintenance of genome stability. It is the only known epigenetic mark of DNA until 2009, when 5-hydroxymethylcytosine (5hmC) was discovered as another relatively abundant cytosine modification in mouse Purkinje neurons and embryonic stem cells (ESCs) [1,2]. The ten-eleven translocation (Tet) family proteins are responsible for the conversion of 5mC to 5hmC [2,3]. Follow-up studies showed that Tet proteins can further oxidize 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which can then be removed from the genome by thymine-DNA glycosylase (TDG) [4••,5••,6•]. This suggests that 5hmC may act as a DNA demethylation intermediate. In addition, because 5hmC not only impairs the binding of 5mC binding proteins [7], but also has its own unique binding protein [8] and shows unique distribution patterns in the genome [9–20,21•,22•], 5hmC may also serve as an epigenetic mark with unique regulatory functions.
In the following sections, we will briefly discuss recent progress in our understanding of 5hmC with an emphasis on its generation, fate, and distribution in mammalian cells.
TET family proteins oxidize 5mC to generate 5hmC in mammalian cells
Although it was not recognized as an epigenetic mark until recently, 5hmC has long been known to exist in natural DNA. About sixty years ago, it was found that all cytosines in the DNA of T-even bacteriophages (e.g. T-4 bacteriophage) are replaced by 5hmC [23], which can be further glucosylated to prevent the phage DNA from being degraded by bacterial restriction enzymes [24–26]. It is worth noting that 5hmC in phage DNA is not derived by in situ DNA modifications. Instead, premodified bases are incorporated into DNA by replacement of deoxycytidine triphosphate (dCTP) with hydroxymethyldeoxycytidine triphosphate (hmdCTP) during DNA synthesis [24].
While the generation and function of 5hmC in bacteriophages have been well-studied, we have only started to understand the function of 5hmC in mammalian genomes. Although 5hmC in mammalian DNA was first reported over forty years ago [27], it did not draw much attention as the experiments could not be reproduced by others and 5hmC itself was simply considered as a product of 5mC oxidative damage in mammalian genomes [28–30]. There were very few reports on 5hmC in mammals until 2009, when two groups provided compelling evidence for the existence of 5hmC in mouse Purkinje neurons and ESCs using both thin layer chromatography (TLC) and mass spectrometry analysis [1,2]. More importantly, through a delicate homology search for the trypanosome thymidine hydroxylases JBP1 and JBP2, human TET1 protein was identified to have the capacity to convert 5mC to 5hmC [2,31]. Using a mechanism-based approach coupled with sequence homology search, we independently identified and demonstrated that all members of the mouse Tet protein family (Tet1–3) have the 5mC hydroxylase activity both in vivo and in vitro [3].
Sequence comparisons revealed that Tet proteins are a distinct family of 2-oxoglutarate (2OG)-dependent and Fe(II)-dependent dioxygenases (2OGFeDOs). Similar to most 2OGFeDO superfamily members (e.g. JmjC-domain-containing histone demethylases), the catalytic domain of Tet proteins contains eight conserved strands, which constitute a putative double-stranded β-helix (DSBH) fold (Figure 1). Unique features are also found in Tet proteins. These features include the cysteine-rich domain adjacent to the N-terminal of the core DSBH fold and the large non-conserved low-complexity region between strands 4 and 5 [31,32]. While the functions of these distinct insertions are unknown, the hydroxylation of 5mC catalyzed by Tet proteins seems to be a canonical 2OGFeDO-catalyzing oxidative reaction that requires Fe(II) and 2OG as cofactors and uses oxygen to oxidize the 5-methyl group in 5mC to generate 5hmC [3].
Figure 1.
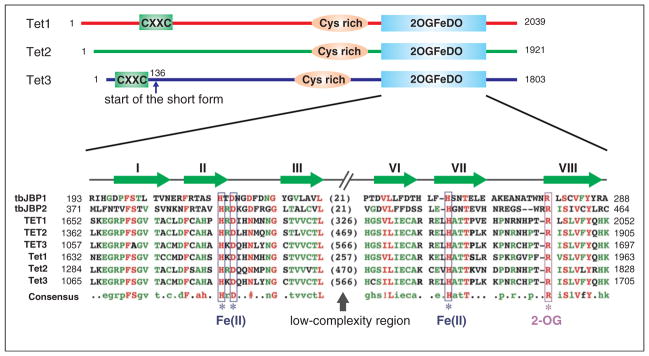
Schematic diagrams of the Tet proteins. Three conserved domains are indicated in mouse Tet proteins, including CXXC zinc finger, cysteine-rich region (Cys-rich), and the double-stranded β-helix (DSBH) fold of the 2OG-dependent and Fe(II)-dependent dioxygenase domain (2OGFeDO). Note that Tet3 has a shorter form, which starts at amino acid 136, that does not contain the CXXC domain. For the 2OGFeDO domain, a multiple sequence alignment of selected JBP/Tet family proteins is shown. Sequences used in the alignment include the Trypanosoma brucei JBP1 (Q9U6M3) and JBP2 (Q57X81); human TET1 (NP_085128), TET2 (NP_001120680), and TET3 (ADU77107); and mouse Tet1 (NP_001240786), Tet2 (ACY38292), and Tet3 (ADR57137). Predicted Fe(II) and 2OG-binding sites are indicated, and the conserved strands that constitute the DSBH fold are shown above the multiple sequence alignment. Numbers represent the amino acid numbers.
Although it is not rigorously confirmed, Tet-mediated oxidation of 5mC seems to be the only source of 5hmC in mammalian cells. First, the existence of 5hmC in the genome requires pre-existing 5mC, as 5hmC is eliminated in DNA methyltransferase (DNMT) triple knockout (Dnmt1−/−/Dnmt3a−/−/Dnmt3b−/−) ESCs in which 5mC is absent [9,18]. Second, depletion of Tet1 leads to a significant decrease of 5hmC in ESCs [2,3,9,33,34]. Third, paternal-genome conversion of 5mC into 5hmC fails to occur in Tet3-deficient mouse zygotes [35•].
5hmC-mediated DNA demethylation
Because 5hmC is converted from 5mC, it was naturally considered to have a direct role in DNA demethylation [36]. This notion has been supported by many recent studies [4••,5••,37,38•,39,40•,41]. Several 5hmC removal pathways have been reported (Figure 2). First, passive dilution of 5hmC during DNA replication is observed in preimplantation embryos [38•]. Consistently, the maintenance DNA methyltransferase DNMT1 methylates hemi-hydroxymethylated CpGs with a much lower efficiency to hemi-methylated CpGs in vitro [30,42].
Figure 2.
Proposed DNA demethylation pathways that involve 5hmC. DNA methylation (5mC) is established and maintained by DNA methyltransferases (DNMTs). In mammals, 5mC can be oxidized by the Tet proteins to generate 5hmC. 5hmC is recognized poorly by Dnmt1 and can be diluted during DNA replication. 5hmC can also be further oxidized by Tet proteins to produce 5fC and 5caC. Alternatively, 5hmC may be deaminated by AID/APOBECs to become 5hmU. 5fC, 5caC, and 5hmU can be excised from DNA by glycosylases. In addition, DNMT3A and DNMT3B may directly dehydroxymethylate 5hmC to generate unmodified C. Note that solid lines represent processes with strong evidence, while the dashed lines indicate processes which need to be further confirmed controversial process.
Second, Tet proteins can further oxidize 5hmC to 5fC and 5caC, which can be efficiently removed by TDG [4••,5••,6•]. Subsequent repair of the resulting abasic site by base excision repair (BER) can regenerate an unmethylated cytosine. This is a very plausible active demethylation pathway as all the involved enzymatic reactions have been demonstrated to be robust in vitro and in cultured cells [4••,5••,6•]. Interestingly, 5fC and 5caC can also be passively diluted in certain biological processes such as during mouse preimplantation development when Tdg is almost not detectable [43].
Third, deamination followed by the action of DNA glycosylases and the BER pathway is yet another pathway that may be involved in the removal of 5hmC. A previous study showed that co-transfection of various AID/APO-BEC deaminases with either a 5mC-containing or 5hmC-containing reporter into HEK293 cells promotes demethylation of the 5hmC-containing, but not 5mC-containing reporter, along with the generation of 5hmU in the cells [37]. In addition, DNA glycosylases SMUG1 and TDG exhibit robust activity on the 5hmU:G mismatch, the expected deamination product of 5hmC, in double-stranded DNA [40•]. However, a recent systematic in vitro biochemical study has revealed that AID/APOBEC deaminases have no detectable deamination activity on 5hmC and have reduced activity on 5mC relative to unmodified cytosine, which may be due to the increased steric bulk at C5 position [44]. Thus, the mechanism underlying AID-mediated DNA demethylation is likely to be due to 5mC deamination rather than 5hmC deamination [45,46]. Further studies are required to confirm this point in vivo.
Fourth, a recent in vitro study suggests an unexpected role of de novo DNA methyltransferases Dnmt3a and Dnmt3b in direct conversion of 5hmC to unmodified cytosine under oxidative conditions [47], indicating that Dnmt3 proteins may act as dehydroxymethylases. This attractive one-step removal of 5hmC is supported by an earlier in vitro study, which shows that the bacterial DNA methyltransferase M.HhaI has dehydroxymethylation activity toward 5hmC [48]. However, the reported in vitro activity needs further confirmation and more importantly the significance of this novel activity needs to be demonstrated in vivo. Finally, for all these 5hmC-mediated DNA demethylation pathways discussed above, their physiological context and relative importance need to be further addressed.
Maintenance of 5hmC during DNA replication
Accumulating evidence suggests that 5hmC not only can serve as an intermediate of DNA demethylation, but also functions as an epigenetic mark with unique regulatory functions. For instance, 5hmC has its own unique binding protein to read the epigenetic information it carries [8], and it also shows unique genomic distribution patterns that is related to transcriptional activities [9–20,21•,22•]. Most of the epigenetic marks are faithfully maintained to ensure that the epigenetic information they carry can propagate through mitotic divisions. However, whether/how 5hmC is maintained during DNA replication is still a complete mystery.
In mammalian cells, 5mC is maintained during DNA replication mainly by DNMT1 [49]. The SRA domain containing protein UHRF1 is also essential in maintenance methylation as it recruits DNMT1 to methylated DNA during DNA replication [50,51]. In vitro studies showed that UHRF1 binds to 5hmC-containing and 5mC-containing DNA with similar affinity [52]. However, DNMT1 methylates hemi-hydroxymethylated CpGs (5hmC-C) with a much lower efficiency than that of hemi-methylated CpGs (5mC-C) [30,42]. Thus, for 5hmC containing CpG sites, it is still difficult to predict the modification states of the cytosines in the newly synthesized strand. If fully hydroxymethylated CpGs are immediately generated during DNA replication, UHRF1, DNMT1, and Tet proteins may function in a complex to maintain 5hmC during DNA replication. Alternatively, if hemi-hydroxymethylated CpGs (5mC-5hmC or C-5hmC) are generated, Tet proteins or both DNMTs and TETs need to be targeted to these hemi-hydroxymethylated CpGs after DNA replication to maintain 5hmC. However, it is also possible that 5hmC is not maintained or maintained in a less faithful way during DNA replication. For instance, genomic DNA hydroxymethylation pattern may be re-established based on other epigenetic modifications after DNA replication is accomplished. More detailed studies using cell cycle synchronized cells are needed to address all these possibilities.
Genomic distributions of 5hmC
Since 5hmC is relatively abundant in ESCs and brain tissues, extensive studies have been carried out to determine the genomic distribution of 5hmC in both human and mouse ESCs and brain tissues [9–20,21•,22•]. Two types of approaches have been used in mapping the genomic distribution of 5hmC. The first uses affinity-based approaches, in which 5hmC-specific antibodies [9,14,18–20] or chemical/enzymatic labeling of 5hmC [10–13,15–17] are first used to enrich 5hmC-containing DNA from fragmented genomic DNA. Then, the 5hmC-enriched DNA is subjected to high-throughput sequencing. Through an evaluation with 5hmC-containing oligos, it has been suggested that chemical labeling-based methods can better enrich genomic DNA fragments with lower 5hmC density than antibody-based methods [11]. However, all the different affinity-based methods produced similar 5hmC distribution maps in mouse ESCs [53], demonstrating that these methods are largely equivalent in detecting 5hmC-enriched regions in the genome. This is likely explained by the fact that 5hmC are highly clustered in the genome [22•].
These affinity-based 5hmC-profiling studies have revealed several features regarding the genomic distribution of 5hmC in mouse ESCs (Figure 3). First, 5hmC is enriched in gene-rich euchromatic regions, particularly at transcription start sites (TSSs), promoters, and exons [9,11,18–20]. This agrees well with 5hmC immunostaining results of both ESCs and somatic cells, where 5hmC accumulates on euchromatin marked by H3K4me2/3, but not on heterochromatin marked by H3K9me3 [9,15,54]. Second, 5hmC is preferentially located at genomic regions with moderate CpG density [18,19]. Consistently, 5hmC is enriched at CpG islands (CGIs) with low to medium GC-content, as well as promoters with intermediate CpG density [9,18,20]. Third, 5hmC is specifically enriched at gene promoters associated with bivalent domains, which are marked with both the transcriptional permissive mark H3K4me3 and the Polycomb repressive complex 2 (PRC2) deposited repressive mark H3K27me3 [11,18,19]. Fourth, 5hmC is relatively enriched in the gene bodies of actively transcribed genes, especially at the 3′ end [19,20]. Finally, 5hmC is located at many intergenic cis-regulatory elements such as active enhancers, pluripotent transcription factor-binding sites, and insulator-binding sites [9,11,19]. The findings in human ESCs and in neuronal cells are largely consistent with the above observations in mouse ESCs, with only minor differences. 5hmC enrichment at cis-regulatory elements is more significant in human ESCs as compared to mouse ESCs [14,15]. Additionally, 5hmC is less enriched at TSSs and CGIs in both mouse and human cerebellums [13,17], suggesting that dynamic changes of 5hmC occur during differentiation.
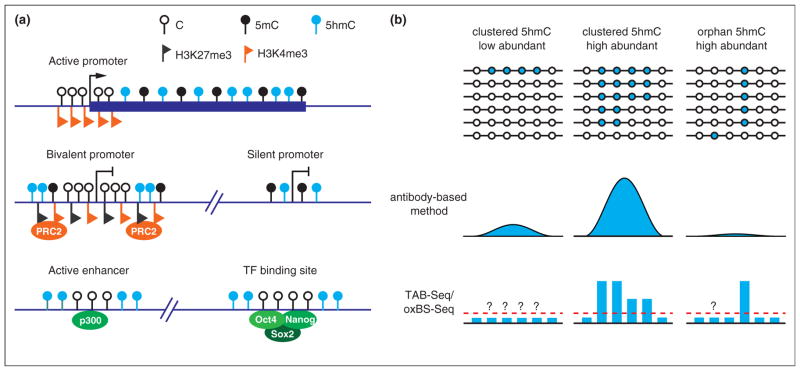
Figure 3.
Genomic distribution of 5hmC. 1. Schematic diagram illustrating the 5hmC distribution in the genome of mouse ESCs. 5hmC is preferentially enriched at gene bodies of actively transcribed genes, bivalent and silent promoters, as well as active enhancers and a cohort of pluripotency transcription factor binding sites. 2. Comparison between the antibody-based 5hmC profiling methods and the single-base resolution methods. Antibody-based methods are not sensitive enough to detect all orphan 5hmCs, and thus may not be able to detect low 5hmC-density regions. In contrast, the single-base resolution methods can quantitatively determine the levels of orphan 5hmCs. However, the single-base resolution methods still cannot achieve low detection limitation (red dashed line) with normal sequencing depth. Thus, 5hmCs with low abundance in a population of cells may not be confidently identified.
While these affinity-based methods have provided initial genome-wide profiles and biological insights of 5hmC, such approaches have relatively low resolution and cannot quantitatively determine 5hmC abundance at each modified site in a population of cells. The recently developed second category of approaches enables single-base resolution mapping of 5hmC, allowing for quantitative measurement of 5hmC levels. These approaches include oxidative bisulfite sequencing (oxBS-Seq) [21•] and Tet-assisted bisulfite sequencing (TAB-Seq) [22•]. In TAB-Seq, Yu et al. first used β-glucosyltransferase (βGT) to convert 5hmC to β-glucosyl-5-hydroxymethycytosine (5gmC), which protects 5hmC from further TET oxidation. They then treated the DNA from the first step with an excess amount of Tet1 protein to convert nearly all 5mCs to 5caCs. Finally, they performed bisulfite sequencing (BS-Seq) on the treated genomic DNA. In BS-Seq analysis, 5mC, 5hmC, and 5gmC all appear as Cs in the resulting sequence [22•,55]. On the other hand, 5fC and 5caC appear as thymines [4••,21•] (Table 1). Thus, all Cs from the TAB-Seq results are originally 5hmC.
Table 1.
Behavior of cytosine and its derivatives in bisulfite treatment and the following sequencing step. 5gmC, β-glucosyl-5-hydroxymethylcytosine; CMS, cytosine-5-methylenesulfonate.
| Base | After bisulfite treatment | Sequencing result |
|---|---|---|
| C | U | T |
| 5mC | 5mC | C |
| 5hmC | CMS | C |
| 5gmC | 5gmC | C |
| 5fC | U | T |
| 5caC | 5caU | T |
Similarly, in the oxBS-Seq method, Booth et al. used KRuO4 to chemically oxidize 5hmC to 5fC. Thus, all Cs from the oxBS-Seq result are originally 5mC. Subtracting oxBS-Seq hits (5mC) from conventional BS-Seq hits (both 5mC and 5hmC) gives base-resolution 5hmC distribution. Since there is no enrichment step before sequencing in either TAB-seq or oxBS-Seq, the final results from both methods reflect the absolute 5hmC level at each site.
The base-resolution 5hmC profiling methods are capable of detecting orphan 5hmCs (i.e. 5hmCs outside of the regions with clustered 5hmCs), which tend to escape the detection of affinity-based methods (Figure 3). Moreover, high-resolution profiles can also reveal some patterns that are difficult to discover from affinity-based method. For instance, while the affinity-based methods showed that 5hmC is enriched at pluripotent transcription factor binding sites [14,15], the TAB-Seq result showed that 5hmC is actually distributed around, but not within, transcription factor consensus motifs [22•].
It is worth noting, however, that these base-resolution approaches require very high sequencing depths to confidently identify 5hmC with low abundance. For instance, an average sequencing depth of 26.5× is required in TAB-Seq to resolve a single 5hmC site with 20% abundance of 5hmC at a false discovery rate < 5% [22•]. In addition, even higher sequencing depth is required when less abundant 5hmC needs to be identified or if the 5mC to 5caC conversion rate is not high enough. Similarly, high sequencing depth is also required for oxBS-Seq. A practical solution is to combine TAB-seq and oxBS-seq with reduced representation bisulfite sequencing (RRBS), which allows selective, but deep, sequencing of a fraction of the genome that is highly enriched for CGIs [56]. As illustrated by Booth et al., only ~25 million 40-bp short reads are required to achieve an average sequencing depth of ~120× by combining oxBS-Seq with RRBS (namely oxidative RRBS, or oxRRBS) [21•], making this method very cost-efficient and suitable for most applications. However, an important disadvantage of oxRRBS method is that the oxidation step leads to significant DNA degradation and requires relatively large amount of starting DNA, which may limit the application of oxRRBS to very rare samples such as preimplantation embryos and primordial germ cells (PGCs) [21•]. Since the enzymatic treatments in TAB-Seq are quite mild, examining 5hmC distribution through a combination of TAB-seq and RRBS may be promising. Such a method, namely Tet-assisted RRBS, is likely to provide the power of 5hmC base-resolution detection in CGIs for rare samples.
Concluding remarks
Ever since the discovery of 5hmC and Tet family proteins in 2009, there has been tremendous progress in understanding their distribution and function. Biochemical and genetic studies have demonstrated that the Tet family proteins play important roles in ESCs, hematopoiesis, PGC development, and embryonic development. It is also clear that 5hmC can serve as an intermediate in DNA demethylation. Despite these progresses, several important questions regarding the function of 5hmC still remain. First, what are the physiological context and relative importance of the various demethylation pathways that involve 5hmC? Second, what are the effects of 5hmC on transcriptional regulation and how is its role modulated by its location and other epigenetic modifications? Third, if 5hmC is a bona fide epigenetic mark, how is it maintained during DNA replication? Fourth, what is the dynamics of 5hmC in PGC and embryonic development and how is its dynamics linked to epigenetic reprogramming and function? The limitation of 5hmC detection is currently the major barrier to addressing these questions. With the Tet-assisted RRBS method we proposed above, and the fast development of single-molecule DNA sequencing technologies [57–59], we will soon be able to address these questions and have a greater understanding of 5hmC as well as other cytosine modifications in the genome.
Acknowledgments
We thank Diana Cai and Dr Hao Wu for critical reading of the manuscript. This work was supported by HHMI and NIH (U01DK089565). Y.Z. is an Investigator of the Howard Hughes Medical Institute.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. This paper demonstrates that 5mC and 5hmC can be oxidized to 5caC by Tet proteins. The authors also show that 5caC can be specifically recognized and excised from DNA by TDG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. This paper demonstrates that Tet dioxygenases can convert 5mC and 5hmC to both 5fC and 5caC in vitro and in vivo in an enzymatic activity-dependent manner. The authors also quantified the absolute amounts of all these cytosine derivatives in various mouse cell lines and organs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. This paper shows that TDG can rapidly excise 5fC and 5caC, but not 5hmC, from DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 10.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, Klungland A. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, Wu H, Warren ST, Jin P, Duan R, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. The authors of this paper utilize chemical oxidation of 5hmC to 5fC, and a bisulfite-sequencing-based method for quantitative mapping of 5hmC in genomic DNA at single-nucleotide resolution. [DOI] [PubMed] [Google Scholar]
- 22•.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. The authors of this paper describe a Tet-assisted bisulfite sequencing method for quantitative mapping of 5hmC in genomic DNA at single-nucleotide resolution in both human and mouse ESCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952;170:1072–1073. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg A, Zimmerman SB, Kornberg SR, Josse J. Enzymatic synthesis of deoxyribonucleic acid. Influence of bacteriophage T2 on the synthetic pathway in host cells. Proc Natl Acad Sci USA. 1959;45:772–785. doi: 10.1073/pnas.45.6.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shedlovsky A, Brenner S. A chemical basis for the host-induced modification of T-even bacteriophages. Proc Natl Acad Sci USA. 1963;50:300–305. doi: 10.1073/pnas.50.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hattman S, Fukasawa T. Host-induced modification of T-even phages due to defective glucosylation of their DNA. Proc Natl Acad Sci USA. 1963;50:297–300. doi: 10.1073/pnas.50.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kothari RM, Shankar V. 5-Methylcytosine content in the vertebrate deoxyribonucleic acids: species specificity. J Mol Evol. 1976;7:325–329. doi: 10.1007/BF01743628. [DOI] [PubMed] [Google Scholar]
- 29.Steinberg JJ, Cajigas A, Brownlee M. Enzymatic shot-gun 5′-phosphorylation and 3′-sister phosphate exchange: a two-dimensional thin-layer chromatographic technique to measure DNA deoxynucleotide modification. J Chromatogr. 1992;574:41–55. doi: 10.1016/0378-4347(92)80096-9. [DOI] [PubMed] [Google Scholar]
- 30.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 31.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 33.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. By using Tet3 conditional knockout mice, the authors of this study provided compelling evidence that Tet3 is a maternal effect gene that converts paternal 5mC to 5hmC in zygotes, and demonstrated that this process is essential in development. [DOI] [PubMed] [Google Scholar]
- 36.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. The authors of this paper showed mitotic chromosome spreads co-stained with 5hmC and 5mC antibodies at various stages of preimplantation embryos, which suggest that conversion of paternal 5mC to 5hmC, followed by replication-dependent dilution, contributes to paternal DNA demethylation in zygotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 40•.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. This study showed that Tdg depletion leads to mouse embryonic lethality, and demonstrated that TDG is necessary for recruiting p300 to retinoic acid-regulated promoters to protect CpG islands from hypermethylation, and active demethylation of tissue-specific developmentally and hormonally regulated promoters and enhancers. The authors also showed that TDG interacts with the deaminase AID and the damage response protein GADD45a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem. 2012;287:33116–33121. doi: 10.1074/jbc.C112.406975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 49.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 50.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 51.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 52.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10:2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kubiura M, Okano M, Kimura H, Kawamura F, Tada M. Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells. Chromosome Res. 2012 doi: 10.1007/s10577-012-9317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, He C, Korlach J. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2012;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2011;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb) 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]