Abstract
When the sta6 (starch-null) strain of the green microalga Chlamydomonas reinhardtii is nitrogen starved in acetate and then “boosted” after 2 days with additional acetate, the cells become “obese” after 8 days, with triacylglyceride (TAG)-filled lipid bodies filling their cytoplasm and chloroplasts. To assess the transcriptional correlates of this response, the sta6 strain and the starch-forming cw15 strain were subjected to RNA-Seq analysis during the 2 days prior and 2 days after the boost, and the data were compared with published reports using other strains and growth conditions. During the 2 h after the boost, ∼425 genes are upregulated ≥2-fold and ∼875 genes are downregulated ≥2-fold in each strain. Expression of a small subset of “sensitive” genes, encoding enzymes involved in the glyoxylate and Calvin-Benson cycles, gluconeogenesis, and the pentose phosphate pathway, is responsive to culture conditions and genetic background as well as to boosting. Four genes—encoding a diacylglycerol acyltransferase (DGTT2), a glycerol-3-P dehydrogenase (GPD3), and two candidate lipases (Cre03.g155250 and Cre17.g735600)—are selectively upregulated in the sta6 strain. Although the bulk rate of acetate depletion from the medium is not boost enhanced, three candidate acetate permease-encoding genes in the GPR1/FUN34/YaaH superfamily are boost upregulated, and 13 of the “sensitive” genes are strongly responsive to the cell's acetate status. A cohort of 64 autophagy-related genes is downregulated by the boost. Our results indicate that the boost serves both to avert an autophagy program and to prolong the operation of key pathways that shuttle carbon from acetate into storage lipid, the combined outcome being enhanced TAG accumulation, notably in the sta6 strain.
INTRODUCTION
Eukaryotic microalgae accumulate storage products—polysaccharides [starch and (chryso)laminarin] and lipids (triacylglycerides [TAG])—when subjected to growth-arresting conditions, such as transfer to nitrogen-free medium (1, 2). When conditions improve, the products are broken down and utilized as sources of carbon backbones, ATP, and reductant. Since TAG represents a potential feedstock for liquid transportation fuel (2–5), much recent research has explored the molecular and cellular parameters associated with TAG biosynthesis.
The green microalga Chlamydomonas reinhardtii has been the subject of many of these studies, since it has a rich history of genetic and biochemical analysis (6), a well-annotated genome (7), powerful molecular-genetic tools (6), and a strong starch/TAG response to N deprivation in wild-type strains (8–13). Of particular interest has been the mutant sta6 strain, which contains a deletion of the gene encoding an ADP-glucose pyrophosphorylase subunit (14, 15) and hence is incapable of starch formation. In most studies, the sta6 strain produces more TAG than starch-forming strains, such as the cw15 strain (10, 14, 16–18, 21, 23–25, 48, 72), apparently in large part because it assembles TAG-filled lipid bodies (LBs) in both the chloroplast and the cytoplasm, whereas starch-forming strains produce only cytoplasmic LBs (18). When provided with a “boost” of additional acetate, moreover, the sta6 strain proceeds to become obese, such that it floats when centrifuged (18). The boost also enhances LB formation in the cw15 strain, but the cells fail to achieve obesity and do not float (18).
Here we report studies on gene expression patterns during the path to obesity. The Merchant/Pellegrini and Los Alamos laboratories recently generated and analyzed RNA-Seq transcriptomes of the cw15 and sta6 mutants and several complemented sta6 strains during 2 days of N starvation (0→48 h NF) (14). In collaboration with these groups, the Goodenough lab generated a second pair of transcriptomes using the cw15 and sta6 strains, tracing 0→48 h NF gene expression patterns under a different set of culture conditions and taking the time course to 96 h NF, with an intervening acetate boost. Analysis of these data was deeply informed by cross-comparisons with the data obtained by Blaby et al. (14). Three additional RNA-Seq studies of wild-type strains (8, 11, 26) were also considered.
By consolidating these data, it has been possible to identify “robust” biochemical pathways, like starch, fatty acid, and TAG biosynthesis, wherein patterns of expression of the relevant genes are largely concordant regardless of genetic background or culture conditions, thereby calling attention to the few exceptional cases. Also identified are “sensitive” genes, encoding products operating in several pathways that are influenced by ongoing carbon flux; their expression is coordinated but varies within strains and conditions, suggesting that they play a role in monitoring and responding to N depletion in particular biosynthetic/metabolic contexts. We propose that the several enzymes that are differentially expressed in the sta6 strain, combined with a glucose-6-phosphate (glucose-6-P) “backflow,” participate in generating the chloroplast LBs of this starchless strain. We also propose that the acetate boost deters an autophagy pathway that compromises maximal TAG accumulation.
MATERIALS AND METHODS
Strains.
The sta6 strain (CC-4348; Chlamydomonas Center) is flagella-less and cell wall-less and carries an insertional deletion of the STA6 gene, encoding the small subunit of ADP-glucose pyrophosphorylase (14, 15) essential for starch biosynthesis. Blaby et al. (14) documented that the sta6 deletion extends into the neighboring RBO1 gene and that its contiguous orthologue, RBO2, is also attenuated in expression. The cw15 strain, CC-4349, was considered the clonal parent of the sta6 strain; however, recent genomic analyses (14) indicate that it is not the parent of the sta6 strain, since it is the opposite mating type and carries distinctive single nucleotide polymorphisms (SNPs). While its origin is unclear, its flagellar and wall phenotypes are morphologically indistinguishable from those of the sta6 strain.
Culture conditions in analyzed RNA-Seq data sets.
The current RNA-Seq study of the cw15 and sta6 strains, designated WUSTL (17 samples per strain), employed cultures grown to 4 × 106 cells/ml in phosphate-buffered high-salt medium (HSM) (28) containing 20 mM potassium acetate at 25 μE m−2 s−1 light intensity. Cells were harvested by centrifugation (1,153 × g for 3 min) and resuspended in acetate-containing HSM (HSM+acetate) lacking ammonium (nitrogen free [NF]). For the acetate boost, an appropriate aliquot of a 1.5 M potassium acetate stock was added to a culture to augment its acetate concentration by an additional 20 mM.
The two sta6/cw15 data sets from the Merchant/Pellegrini laboratories at the University of California Los Angeles (UCLA) (14), designated UCLA1 (8 samples per strain) and UCLA2 (3 samples per strain), were obtained with cultures grown to 4 × 106 cells/ml in Tris-buffered Tris-acetate-phosphate (TAP) medium (29) containing 17 mM acetate at 95 μE m−2 s−1 light intensity, harvested by centrifugation (1,006 × g for 5 min), and washed once in NF TAP before resuspension in NF TAP. In some cases, data were also assessed from two RNA-Seq studies of wild-type strains. The first (8), designated UCLA-WT (6 samples), reportedly employed strain CC-3269/2137, but the strain has since been ascertained to be CC-4532; cells were grown and N starved as in the other experiments at UCLA. The second (26), from the Snell laboratory at University of Texas Southwestern Medical school and designated UTSW-WT (3 samples per strain), employed strains CC-1690 mt+ and CC-1691 mt− grown in phosphate-buffered Sager and Granick medium (6) without acetate at 50 μE m−2 s−1 light intensity in either unsynchronized or synchronous cultures before transfer. The mt+ and mt− asynchronous cultures were pooled during log phase and prior to RNA extraction, as were the mt+ and mt− synchronous cultures, to yield the log reads in Table 8; samples from the synchronous culture were then transferred to N-free, acetate-free Sager and Granick medium for 18 h, and the reads are reported as separate mating types in Table 8. Primary data are found in Table S3 of reference 26.
TABLE 8.
Expression profiles (RPKM) of genes listed in Table 7 in the presence or absence of acetatea
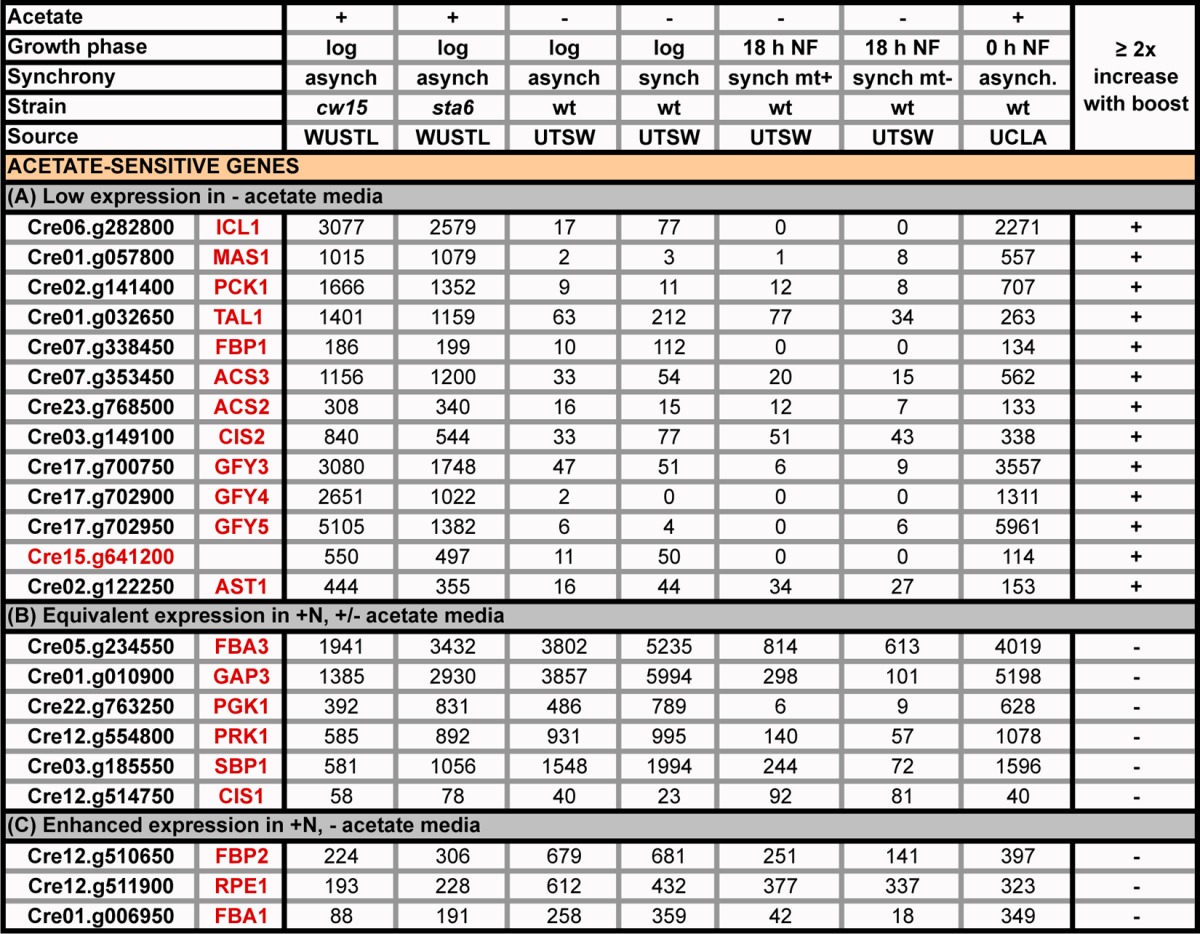
All genes with low expression without exogenous acetate have ≥2-fold increases in expression with acetate boost (Table 7).
The WUSTL and the UCLA culture conditions differ in the following respects: (i) medium (HSM+acetate versus TAP), (ii) trace elements (Hutner et al. [30] versus Kropat et al. [31]), (iii) light intensity (25 μE m−2 s−1 versus 95 μE m−2 s−1), (iv) culture configuration (500 ml in 1-liter Erlenmeyer flasks versus 1 liter in 2.8-liter Fernbach flasks); (v) flask rotation speed (125 rpm versus 180 rpm), and (vi) the protocol used for transfer to N-free medium (centrifugation duration and one versus two centrifugation steps). Blaby et al. (14) reported a transient stimulation of gene expression in the UCLA samples in conjunction with the centrifugation steps, whereas this was not observed in the WUSTL samples.
Microscopy.
Phase and bright-field light microscopy and quick-freeze deep-etch electron microscopy were performed as previously described (18).
Viability analyses.
Two methods of analyzing viability were used.
(i) Plating efficiency.
A log-phase culture was resuspended in N-free HSM+acetate; after 2 days, the culture was divided, and half was acetate boosted. At each time point, cells were counted, subjected to serial dilutions in TAP medium, mixed with top agar, overlaid on 1.5% TAP agar plates, and allowed to grow until colonies were visible. Plates with scorable colony numbers (50 to 150) were recorded, and the colony number/number of cells plated (plating efficiency) was calculated. The plating efficiency for log-phase cells was set as 100%, and values for N-starved cells, with or without the boost, were expressed proportionately.
(ii) Evans Blue exclusion.
A log-phase culture was resuspended in N-free HSM+acetate; after 2 days, the culture was divided, and half was acetate boosted. At each time point, cells were counted, mixed 1:1 with a 0.1% aqueous solution of Evans blue, and scored by light microscopy as viable if dye was excluded. Percent viability was calculated as (viable cell count/original cell count) × 100.
Acetate uptake.
The acetate uptake experiment was performed 3 times with equivalent results; data from one experiment are shown. Vegetative sta6 cells were grown to 4 × 106 cells/ml in HSM+acetate, pelleted, and resuspended at the same density in N-free HSM+acetate. At the time points indicated in Fig. 4, 7-ml samples were centrifuged at 10,000 × g for 5 min; the supernatant was collected by aspiration, passed through a 0.22-μm filter, snap-frozen in liquid N2, and stored at −20°C until analysis. The filtered medium samples were diluted 50% (vol/vol) with D2O containing a known amount of alanine, which served as an internal standard. Proton nuclear magnetic resonance (NMR) for these samples were collected on a 14.09-T NMR spectrometer (600 MHz 1H resonance) using a water suppression pulse sequence to suppress the 1H peak due to water in the medium. Each spectrum was collected for 4 scans with a recycle delay of 10 s. The CH3 protons of alanine are visible at 1.5 ppm, and the CH3 protons of acetate are visible at 1.9 ppm. The integrated proton peak intensities are directly proportional to the molar ratios of those protons. Hence, the acetate concentrations in the medium samples were determined by comparing the peak integrals of the CH3 protons of acetate to those of the CH3 protons of alanine of known concentration.
FIG 4.
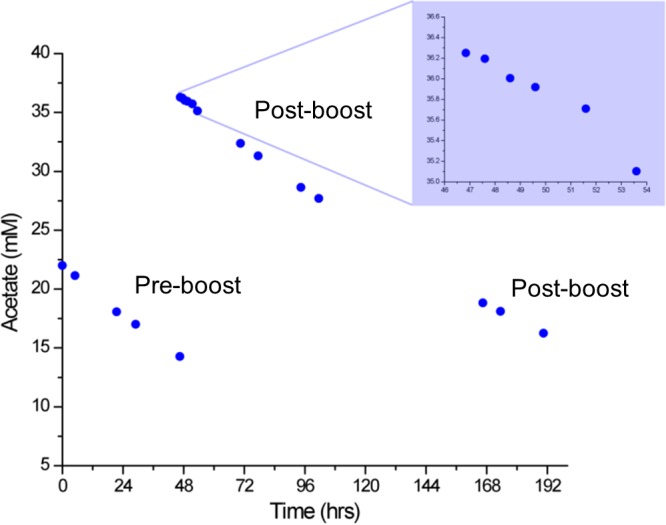
Medium acetate concentrations in an N-starved sta6 culture preboost and postboost.
RNA-Seq analysis.
For RNA extraction, cell density was determined with a hemacytometer at each time point. Twenty milliliters of culture was transferred at the time points indicated in Table 1 (maximum cell number per reaction = 2 × 108) to a 50-ml Falcon tube and centrifuged at 2,000 × g for 5 min at room temperature. The supernatant was immediately decanted, and the pellets were snap-frozen in liquid N2 and stored at −80°C.
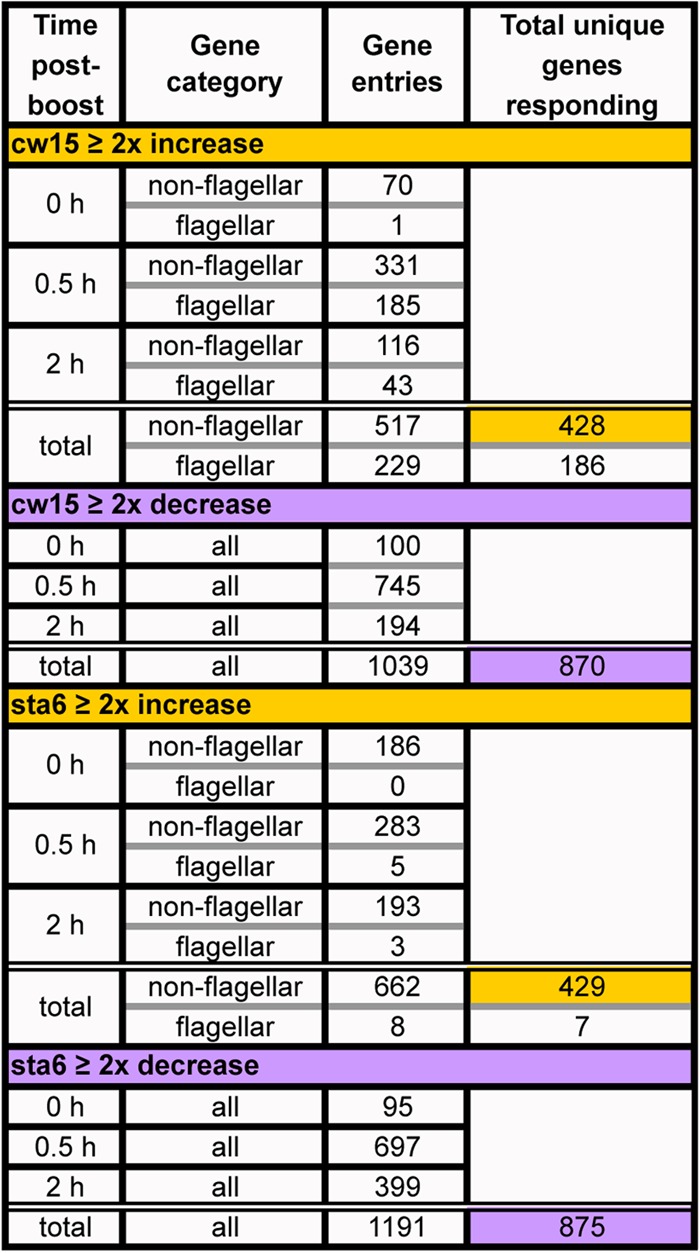
TABLE 1.
Summary of genes increasing or decreasing expression ≥2-fold relative to 48-h NF levels in response to the acetate boost
For processing, samples were brought to room temperature, and the pellets were resuspended in 1 ml freshly made lysis buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 20 mM EDTA [stock adjusted to pH 8.0], 2% sodium dodecyl sulfate). Ten milliliters of TRIzol (Invitrogen) was then added with thorough mixing, and the samples were incubated for 5 min at room temperature, after which the cw15 samples were centrifuged at 600 × g for 2 min to pellet starch. The TRIzol solution/lysate was mixed with 1/5 volume of chloroform-isoamyl alcohol (24:1) and shaken vigorously for 15 s. The mixture was incubated for 5 min at room temperature before being transferred to a MaXtract HD (Qiagen) tube. The nucleic acid-containing phase was subsequently separated according to the manufacturer's instructions. To extract RNA, samples were processed using the miRNeasy minikit (Qiagen) according to the manufacturer's instructions. To remove contaminating DNA, samples were on-column digested using the RNase-free DNase set (Qiagen) according to the manufacturer's instructions.
Prior to library preparation, each RNA sample was subjected to quality control evaluation as follows. The concentration and purity of RNA samples were assayed by a NanoDrop spectrophotometer (Thermo Scientific). Each sample was required to have an A260/A280 ratio between 2.0 and 2.2 and an A260/A230 ratio above 2.0. RNA quality was evaluated by Bioanalyzer (Agilent Technologies) on an Agilent RNA 6000 nanochip following the manufacturer's instructions. RNA integrity was quantified by Agilent 2100 Expert software. Each sample was required to have a RNA integrity number (RIN) above 7.0. The lowest RIN of the WUSTL samples was 7.4; the medians were 8.6 for the cw15 strain and 8.7 for the sta6 strain.
cDNA libraries were prepared as described by Boyle et al. (8), and alignments were performed as described by Blaby et al. (14), where reads were aligned to the Aug10.2 gene models (based on the v4 assembly [http://genome.jgi-psf.org/Chlre4/Chlre4.home.html]).
Protein localizations followed the predictions of Blaby et al. (14) and those determined by Predalgo (32) using their web interface (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main).
Phylogenetic analyses.
The PDG1 phylogeny (see Fig. S1 in the supplemental material) is a Bayesian consensus tree with bootstrap values from 4,000 iterative samplings using MrBayes (33). PGD1 homologs were collected from the genome assemblies of V10.2 C. reinhardtii (V10.2), Volvox carteri (V2), V4 Ostreococcus tauri (V4), Coccomyxa subellipsoidea C169 (V1), and Arabidopsis thaliana (V10). Protein sequences were aligned using MAFFT aligner, followed by manual refinement.
The GPD phylogeny (see Fig. S2 in the supplemental material) is a neighbor-joining tree with bootstrap values from 500 replicates. GPD homologs were collected from gene models for C. reinhardtii (V10.2), V. carteri (V1.0), and Arabidopsis thaliana (V10). Protein sequences were aligned using the MAFFT aligner, followed by manual refinement. Homology domain information was obtained at the Pfam site (http://pfam.sanger.ac.uk).
The FBA phylogeny (see Fig. S3 in the supplemental material) is a Bayesian consensus tree with bootstrap values from 1,000 iterative samplings using MrBayes (33). FBA homologs were collected from gene models for C. reinhardtii (V10.2), V. carteri (V1.0), and A. thaliana (V10). Protein sequences were aligned using the MAFFT aligner, followed by manual refinement. The FBA4 gene is truncated (apparently not due to a gene model error), deleting 100 amino acids at the C terminus, but retains homology to a full-length V. carteri member, forming a divergent clade. The topology of the tree was modified to generate coherent family groupings.
The GFY (GPR1/FUN30/YaaH family) phylogenies (see Fig. S5A, C, and D in the supplemental material) were constructed as follows. Multiple sequence alignments were performed using MUSCLE (34) in MEGA 5.2.2 (35). The unrooted neighbor-joining tree for chlorophycean pfam01184 proteins (see Fig. S5D) was generated in MEGA using 500 bootstrap replicates with the model JTT+G (1.4) and pairwise removal of gaps. The unrooted maximum likelihood tree (see Fig. S5C) was generated using PhyML (36) with the model LG+G (1.2) selected using ProtTest (37). Branch scores for the ML tree are derived from an approximate likelihood ratio test. Sequences for phylogenies were obtained as follows. C. reinhardtii sequences are from Phytozome gene models as listed. V. carteri gene models were based on Phytozome model numbers but manually curated and improved; the protein sequences of the improved V. carteri models are found in Data Set S5 in the supplemental material. The remaining sequences were obtained from the Phytozome, JGI UniProt, or NCBI database with the following accession numbers. Phytozome protein IDs are as follows: Coccomyxa subellipsoidea C-169, 44355 and 65361; Ostreococcus lucimarinus, gwEuk.3.605.1; Physcomitrella patens, Pp1s32_336V6.1, Pp1s40_45V6.1, and Pp1_s44_75V6.1. The JGI protein ID for Emiliania huxleyi is 240134. UniProt protein IDs were as follows: Vibrio vulnificus, Q8DF09; Escherichia coli, Q8FLC8; Leishmania major, Q9N686; Methanosarcina acetivorans, Q8TUG4; Pasteurella multocida, Q9CKZ8; Yarrowia lipolytica, Q96VC8; Saccharomyces cerevisiae, P32907; and Schizosaccharomyces pombe, P25613. NCBI protein IDs were as follows: Wickerhamomyces ciferrii, GI:406605912; Ustilago hordei, GI:388852517.
The GFY similarity network (see Fig. S5B in the supplemental material) was generated according to Atkinson et al. (38) and Blaby-Haas and Merchant (39). Briefly, protein sequences used to generate the network were obtained from the Uniprot90 database (40) using the GPR1/FUN34/YaaH domain of Cre17.g702900 (genome version 5.3) as a search query. Any duplicate sequences in the retrieved data were removed, as were sequences resulting from metagenome projects due to unknown eukaryote/prokaryote origin. The resulting 355 protein sequences are found in Data Set S6 in the supplemental material. The network was constructed using a local all-against-all BLASTP (v2.2.28+) search with an E value of 1e−29. Visualization of the BLASTP output was performed with Cytoscape v2.8.2 (41) using the BLAST2similarityGraph plugin (42).
Gene data accession number.
Raw and processed sequence files are available at the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE55253).
RESULTS
Acquisition of obesity by the sta6 strain.
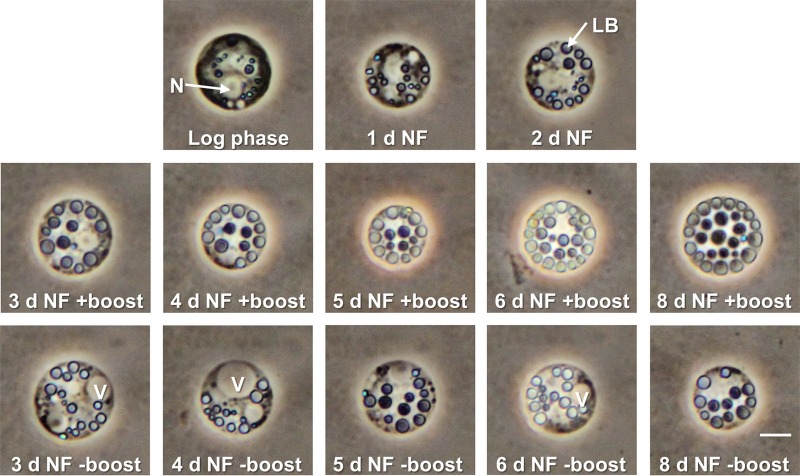
Figure 1 shows sta6 cultures that were acetate boosted 2 days after N starvation (48 h NF) and centrifuged (10,000 × g for 5 min) 2, 3, 4, 5, and 8 days after the boost. The insets show cells at 2 and 8 days postboost.
FIG 1.

Samples of a sta6 culture, boosted with 20 mM acetate after 48 h NF and centrifuged at 10,000 × g for 5 min at 2, 3, 4, 5, and 8 days postboost. Micrographs show phase and bright-field (lower right) microscopy. Asterisk, cellular debris. Bars, 10 μm.
Three features are evident. (i) As documented by Goodson et al. (18), the LBs greatly increase in size. (ii) The cells progressively degrade their chlorophyll and become bright yellow, perhaps reflecting the increase in carotenoid content reported for N-starved C. reinhardtii (9). (iii) The cells become sufficiently TAG filled that they float, even when centrifuged, the hallmark feature of obesity. Subsequently, the boosted cells die, turn white, and lyse; the released LBs float along with the cells, while the white cellular debris pellets (Fig. 1, asterisk).
Viability of boosted versus nonboosted cells.
Figure 2 compares living boosted and nonboosted sta6 cells using phase microscopy. As previously noted (18), the nonboosted cells display large vacuoles by 3 days in N-free medium, which we interpreted as an indication of morbidity. However, as ascertained by two different assays (see Table S1 in the supplemental material), viability is not compromised until 6 days in N-free medium, after which it slowly declines. Boosted cells display similar viability profiles (see Table S1), but they do not develop large vacuoles and contain more abundant LBs.
FIG 2.
Living sta6 cells after 1 to 8 days of N starvation with (+) or without (−) acetate boost (phase microscopy). N, nucleus; V, vacuole; LB, lipid body; refractile blue bodies, eyespots. Bar, 5 μm.
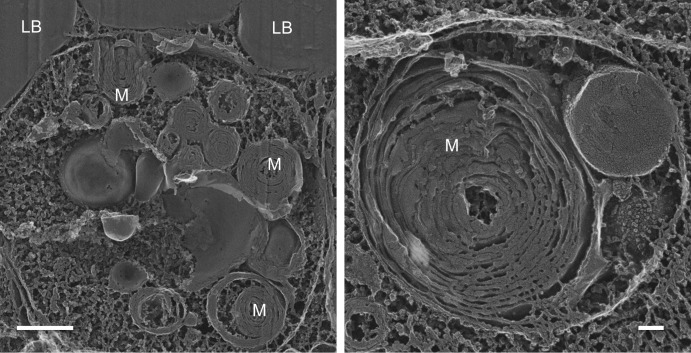
Quick-freeze deep-etch EM images of the vacuoles in 96-h NF nonboosted cells are shown in Fig. 3. Contents include profiles of membrane whorls (“myelin figures”), a hallmark of autophagy. No such autophagosomes are encountered in boosted cells. These observations indicate that the boost somehow averts the initiation of an autophagocytic response at 48 h NF, a response that is accompanied by diminished TAG accumulation.
FIG 3.
Autophagosomes in 96 h NF nonboosted sta6 cells. M, myelin figures; LB, cytoplasmic lipid bodies. Bars, 500 nm (left) and 100 nm (right).
Rates of acetate uptake.
An obvious explanation for the boost's ability to enhance TAG content is that after 0→48 h NF, acetate levels in the medium are exhausted and restored by the boost. Alternatively, the boost might enhance the rate of acetate uptake. Either scenario would provide the cells with more substrate for TAG synthesis.
To test these possibilities, NMR was used to determine acetate levels in the culture medium. As previously reported (25), log-phase sta6 cells take up acetate very rapidly, such that it is exhausted within 48 h of growth, whereas nongrowing N-starved cells utilize it much more slowly. Our results (Fig. 4) confirm these observations: more than half the medium acetate remains after 0→48 h NF, with the mean rate of depletion (193 μmol/h) being similar to the rate observed by Blaby et al. (14) (166 μmol/h). The rate of depletion following the acetate boost (157 μmol/h) is, if anything, lower than before the boost, and samples taken at short intervals following the boost (Fig. 4, inset) show no spike in acetate depletion rates with acetate addition. Hence, neither hypothesis is supported, although a small transient influx is not likely to be detectable by these measurements.
Evidence for a transient acetate influx has instead come from RNA-Seq analysis of boosted cw15 cells. As documented in Data Set S1B in the supplemental material and summarized in Table 1, an increase in expression of 229 flagellum-related genes is observed within the first 2 h after boost. It has been known for some time that when the pH of the medium is dropped to pH 4.5 with 0.5 N acetic acid for 1 min and then neutralized, C. reinhardtii cells first deflagellate and then upregulate expression of their flagellum-related genes and construct new flagella; a recent RNA-Seq profile of these genes (43) strongly overlaps the genes listed in Data Set S1B. Although the cw15 and sta6 strains are flagella-less, Cheshire et al. (44) reported that such gene upregulation also occurs in “bald” strains. Deflagellation is induced by several organic acids but not by inorganic acids (45), indicating that entry of organic acids, and not external pH, is the causative event. Even at near-neutral pH, increasing the concentration of exogenous acetate stimulates the deflagellation response (45).
Intriguingly, only 5 flagellum-related genes are upregulated with the boost in the sta6 strain (Table 1; also, see Data Set S3B in the supplemental material). Acetate (pKa = 4.76) is known to cross cell membranes in its protonated state and then release the proton into the cytoplasm (46, 47). Therefore, either the cytoplasm of sta6 cells is at a higher resting pH and/or better buffered than that of cw15 cells, or the sta6 strain is for some reason not responsive to some feature of the deflagellation/reflagellation signal, hypotheses we plan to test. Meanwhile, the boost-induced changes in expression of nonflagellar genes, described below, are apparently elicited by stimuli that can act independently of the pathway that elicits the flagellar-gene response, since the nonflagellar responses occur equivalently in both the cw15 and the sta6 strains (Table 1).
RNA-Seq experiments: general considerations.
Another way that the boost might enhance TAG levels is by influencing gene expression such that, for example, enzymes involved in TAG biosynthesis and/or polar-lipid recycling are more abundant. To test this thesis, RNA was sampled from cw15 and sta6 cultures at nine intervals during 0→48 h NF and at eight intervals during the 2 days following the acetate boost at 48 h NF and subjected to RNA-Seq analysis.
We first identified genes whose expression levels increased or decreased ≥2-fold within the 2 h following acetate boost compared with levels at 48 h NF, where in most cases the boost effect subsided several hours later. Data Sets S1 to S4 in the supplemental material provide complete lists of these genes and their RPKM (reads per kilobase of exon model per million mapped reads) values, where, notably, some half of the genes are not annotated, so we may well be missing some important or even key participants. Table 1 summarizes the outcome. Of the estimated 17,301 genes in v.4 of the C. reinhardtii genome, 875 nonflagellar genes displayed a ≥2-fold upshift in the sta6 strain and 870 displayed such an upshift in the cw15 strain with the acetate boost, while 429 genes displayed a ≥2-fold downshift in the sta6 strain and 428 displayed a ≥2-fold down-shift in the cw15 strain. These numbers might suggest that the same gene sets are responsive in both strains, but this is not the case: only 34% of the upregulated genes are shared, and only 43% of the downregulated genes are shared (identified in Data Sets S1 to S4 in the supplemental material, last column).
We then focused on genes that encode participants in biosynthetic and metabolic pathways related to C and N flux and stress-related processes. The genes considered in this report are highlighted in Data Sets S1 to S4 in the supplemental material. We also queried non-boost-responsive genes that participate in the same pathways as boost-responsive genes.
In analyzing these data, designated WUSTL, we ran comparative studies with the RNA-Seq data on 0→48 h NF in the cw15 and sta6 strains, designated UCLA1 (8 samples per strain) and, in some cases, UCLA2 (3 samples per strain) (14), derived from cells cultured under different conditions than the WUSTL conditions. On occasion, we also included data from N-starved wild-type cells maintained in the presence (UCLA-WT) (8) or absence (UTSW-WT) (26) of acetate. Details of strains and culture conditions are provided in Materials and Methods.
A comparison of expression patterns revealed that most of genes involved in the assessed metabolic and biosynthetic pathways were expressed concordantly in the WUSTL and UCLA experiments—e.g., “holding steady,” “increasing/decreasing,” or “rising and then falling”—even though culture conditions and input transcript levels were disparate, establishing these genes' response to N depletion as “robust” to environmental influence. This steady baseline allowed recognition of the few genes whose expression patterns were not concordant between experiments, where in many cases these genes also proved to be responsive to the acetate boost.
Absent from our data are no-boost controls, and such information would be of particular value with respect to the autophagy-related genes described below. In general, however, the thousands of genes in the data set that did not respond to the boost continued to follow their 0→48 h NF trajectory during the ensuing 48→96 h NF interval, most either holding steady or drifting downward. There is every reason to assume, therefore, that this would also be the case for the boost-responsive genes had a boost not been administered.
Genes related to carbon flux with robust expression patterns.
(i) Starch, fatty acid, and TAG biosynthesis.
Expression levels for genes involved with starch, fatty acid, and TAG biosynthesis are shown in Tables 2, 3, and 4.
TABLE 2.
Expression profiles (RPKM) of genes participating in starch biosynthesisa
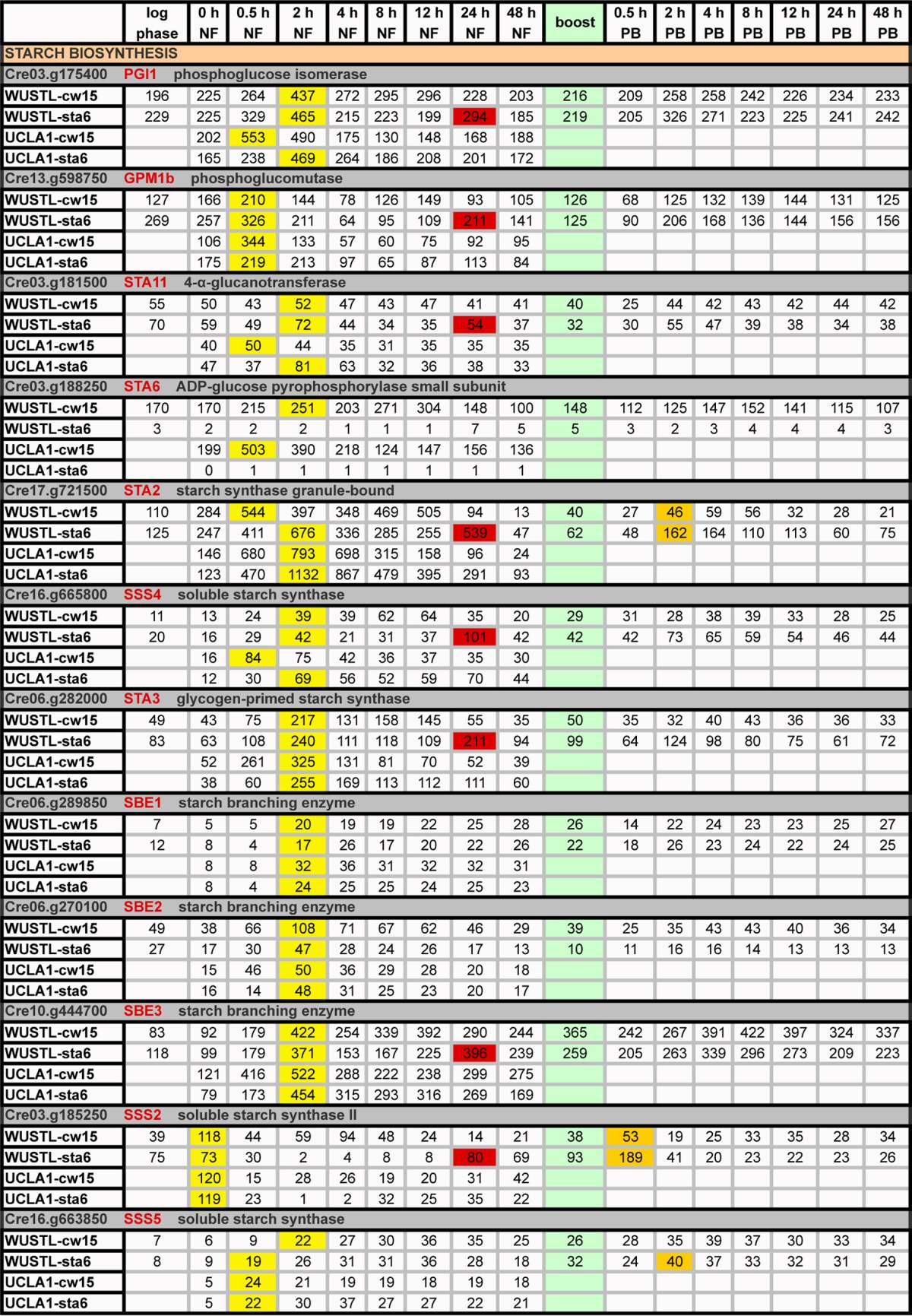
Green, boost addition; PB, postboost; yellow, time points of maximum transcripts during 0→48 h NF; orange, genes with ≥2-fold increases in expression relative to 48-h NF levels following acetate boost; red, increased gene expression in WUSTL-sta6 at 24 h NF.
TABLE 3.
Expression profiles (RPKM) of genes participating in fatty acid biosynthesisa
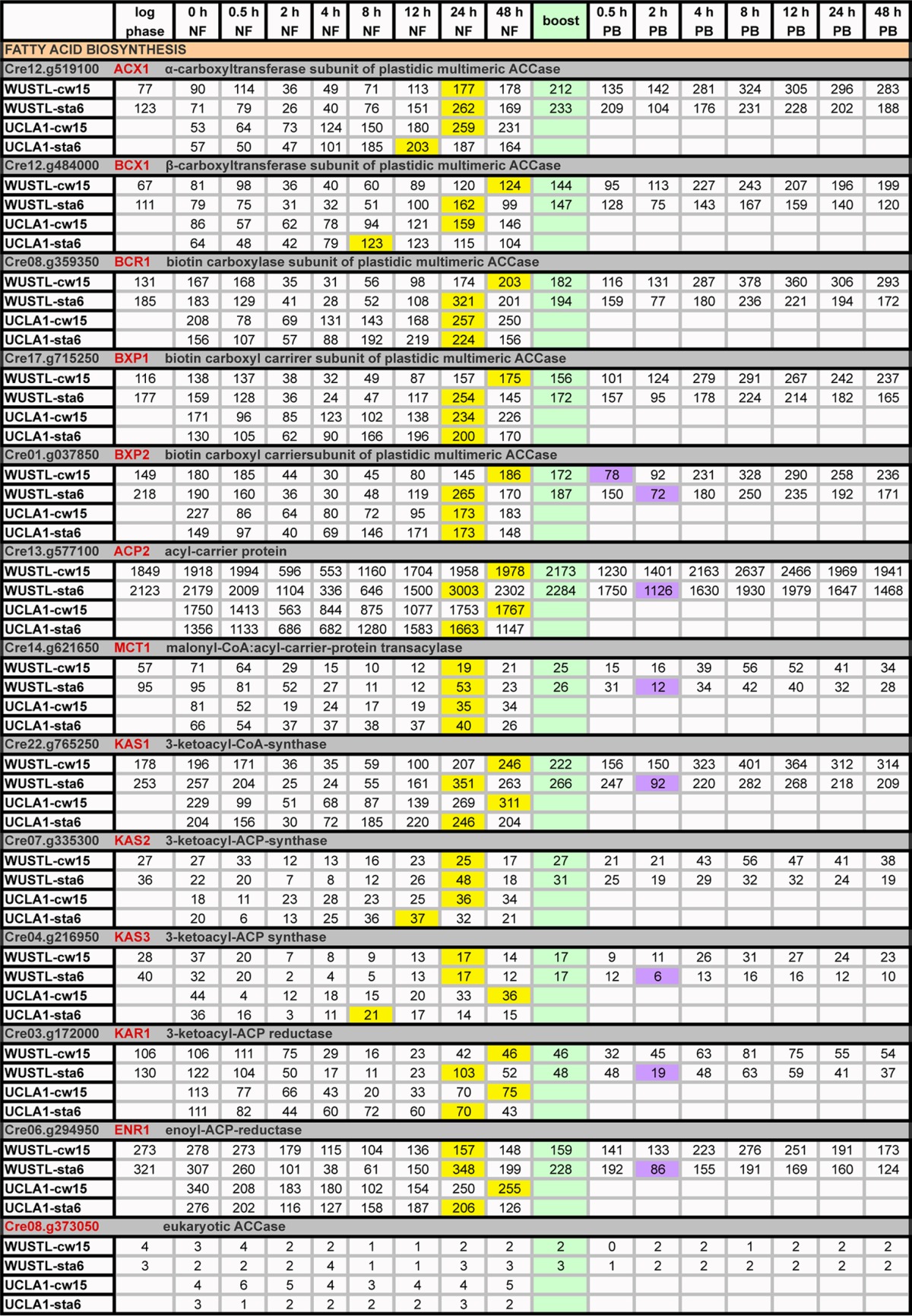
Green, boost addition; PB, postboost; yellow, time points of maximum transcripts during 0→48 h NF; purple, genes ≥2-fold decreases in expression relative to 48-h NF levels following acetate boost.
TABLE 4.
Expression profiles (RPKM) of genes participating in TAG biosynthesisa
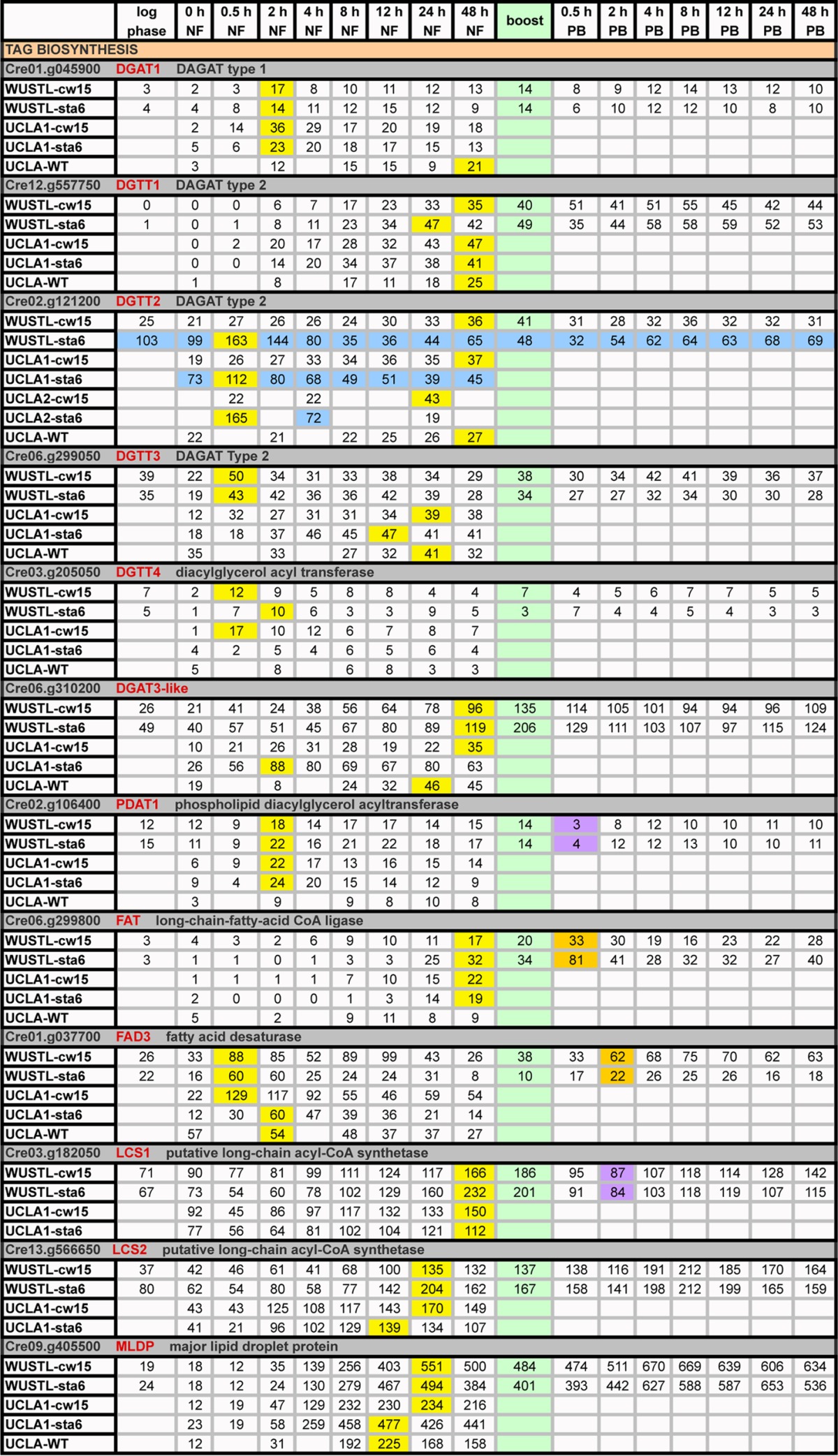
Green, boost addition; PB, postboost. Yellow, time points of maximum transcripts during 0→48 h NF; orange, genes with ≥2-fold increases in expression relative to 48-h NF levels following acetate boost; purple, genes decreasing expression ≥2-fold relative to 48-h NF levels following acetate boost; blue, genes showing strong differential expression between the sta6 and cw15 strains.
As initially noted by Blaby et al. (14) and confirmed here, the starch-related genes are uniformly upregulated soon after cells are subjected to N depletion (Table 2, yellow), while genes encoding enzymes involved in fatty acid biosynthesis are upregulated only at 24 h NF (Table 3, yellow), patterns that mirror the observed early increase in starch formation and the later increase in TAG (48). Genes encoding TAG-related enzymes show two patterns: some are upregulated early and then decline, while others show a steady increase, with maximal values at 48 h NF that persist to 96 h NF (Table 4, yellow). The strong concordance of these patterns between the WUSTL and UCLA data sets indicates that these are “robust” genetic programs, akin to those governing expression of chloroplast ribosomal proteins (see Table S2 in the supplemental material), persisting despite differences in strains and culture conditions and despite the fact that the sta6 strain fails to synthesize any starch and produces chloroplast LBs.
Starch-related genes are generally insensitive to boost; the three exceptions are highlighted in orange in Table 2. Most show a transient increase in expression at 24 h NF in the WUSTL-sta6 samples (Table 2, red).
Fatty acid synthesis-related genes are downregulated at 2 h NF and start recovering at 12 h NF, with maximal expression occurring at 24 h NF and continuing to 96 h NF (Table 3). Transcript levels are consistently higher in the sta6 strain than the cw15 strain at 48 h NF and, in the WUSTL data, consistently higher in the cw15 strain by 96 h NF. While transcription of these genes is not enhanced by the boost, it is briefly depressed in several cases, usually in the sta6 strain (Table 3, purple). Notably, the five genes encoding subunits of the multimeric “prokaryotic” acetyl coenzyme A (acetyl-CoA) carboxylase (ACCase)—ACX1, BCX1, BCR1, BXP1, and BXP2—are robustly expressed in both strains (Table 3), whereas the gene Cre08.g373050, encoding a monomeric “eukaryotic” ACCase, produces few (0 to 6 RPKMs) transcripts in both strains (Table 3).
DAG-to-TAG conversion is canonically catalyzed by type 1 (DGAT) and type 2 (DGTT) diacylglycerol acyltransferases. Confirming four reports (8, 11, 13, 14), expression of DGTT1 is strongly upregulated during 0→48 h NF, with further increases during the next 2 days without a boost response (Table 4), establishing this enzyme as a robust player in the TAG biosynthesis response in all tested strains. Confirming the results reported by Blaby et al. (14), expression of DGTT2 in the sta6 strain is initially 3- to 6-fold higher than in the cw15 strain; by 8 h NF, it tapers to the levels seen in the cw15 strain, but following the boost, levels are 2-fold higher in the sta6 strain to 96 h NF (Table 4, blue). This distinctive DGTT2 transcription pattern is the first of four differences between the sta6 and cw15 strains that are highlighted in this report. DGTT4 and DGAT1 peak early, and their enhanced levels are sustained to 96 h NF in both strains (Table 4). The WUSTL and UCLA data are dissimilar in two respects: (i) DGTT3 transcripts peak early and then decline in WUSTL, whereas they peak late in UCLA; and (ii) Cre06.g310200 (DGAT3-like) peaks late in the WUSTL sta6 sample and in the two cw15 samples, whereas it peaks early in the UCLA sta6 sample.
TAG synthesis can also be catalyzed by phospholipid diacylglycerol acyltransferase (PDAT), which uses fatty acids from polar lipids to acylate the DAG hydroxyl group. Recent studies variously reported that PDAT makes a 25% contribution (8) or only a minor contribution (27) to TAG synthesis under N-deprivation conditions. Transcription from PDAT1 peaks early and remains steady in all three strains, with a transient downregulation at the boost (Table 4).
Two other lipid-related genes augmented with the boost in both strains are Cre06.g299800 (long-chain-fatty acid CoA ligase) and FAD3 (fatty acid desaturase) (Table 4). A putative long-chain acyl-CoA synthetase (LCS1) decreases with the boost, while a second (LCS2) is not affected (Table 4).
Expression of the MLDP1 gene increases 28-fold in the cw15 strain and 21-fold in the sta6 strain during 0→48 h NF and is sustained at high levels for the next 2 days without a boost response; similar increases are seen in the UCLA and UCLA-WT data (Table 4). The gene product, originally posited to be associated with lipid bodies (major lipid droplet protein) (12), has recently been shown to instead be associated with endoplasmic reticulum (ER) membranes (49), where it possibly participates in the intimate ER/LB associations that are established during cytoplasmic LB formation in N-free medium (18).
(ii) Lipases.
In their pioneering RNA-Seq study, Miller et al. (11) identified 130 C. reinhardtii genes carrying the GXSXG motif expected of lipases, of which 46 were either upregulated (75%) or downregulated (25%) ≥2-fold in wild-type cells during 0→48 h NF, and they posited that a subset of these might be involved in releasing fatty acids from polar lipids for use in TAG formation in the fashion of the PDAT enzyme. In a subsequent paper (50), the lab reported that one of these genes, designated Cre03.g193500 and now named PGD1 (plastid galactoglycerolipid degradation), indeed contributes to TAG formation by breaking down pre-existing chloroplast monogalactosyldiacylglycerol (MGDG) into its lyso-lipid form for reacylation as TAG; knockdown of this gene results in reduced TAG accumulation with N starvation.
The WUSTL RNA-Seq data for these 46 candidate lipase genes are shown in Table S3 in the supplemental material; four genes annotated as TAG lipases and not included in the 46-gene set are also listed. Expression is generally equivalent for the cw15 and sta6 strains and generally equivalent in the WUSTL and UCLA data sets (data not shown). Except for a spike at 2 h NF for two of the genes (see Table S3, yellow), most display either steady or gradually increasing expression, with some showing small boost responses. The three genes in blue in Table S3 are displayed in Table 5 and given additional attention below.
TABLE 5.
Expression profiles (RPKM) of genes encoding candidate lipasesa
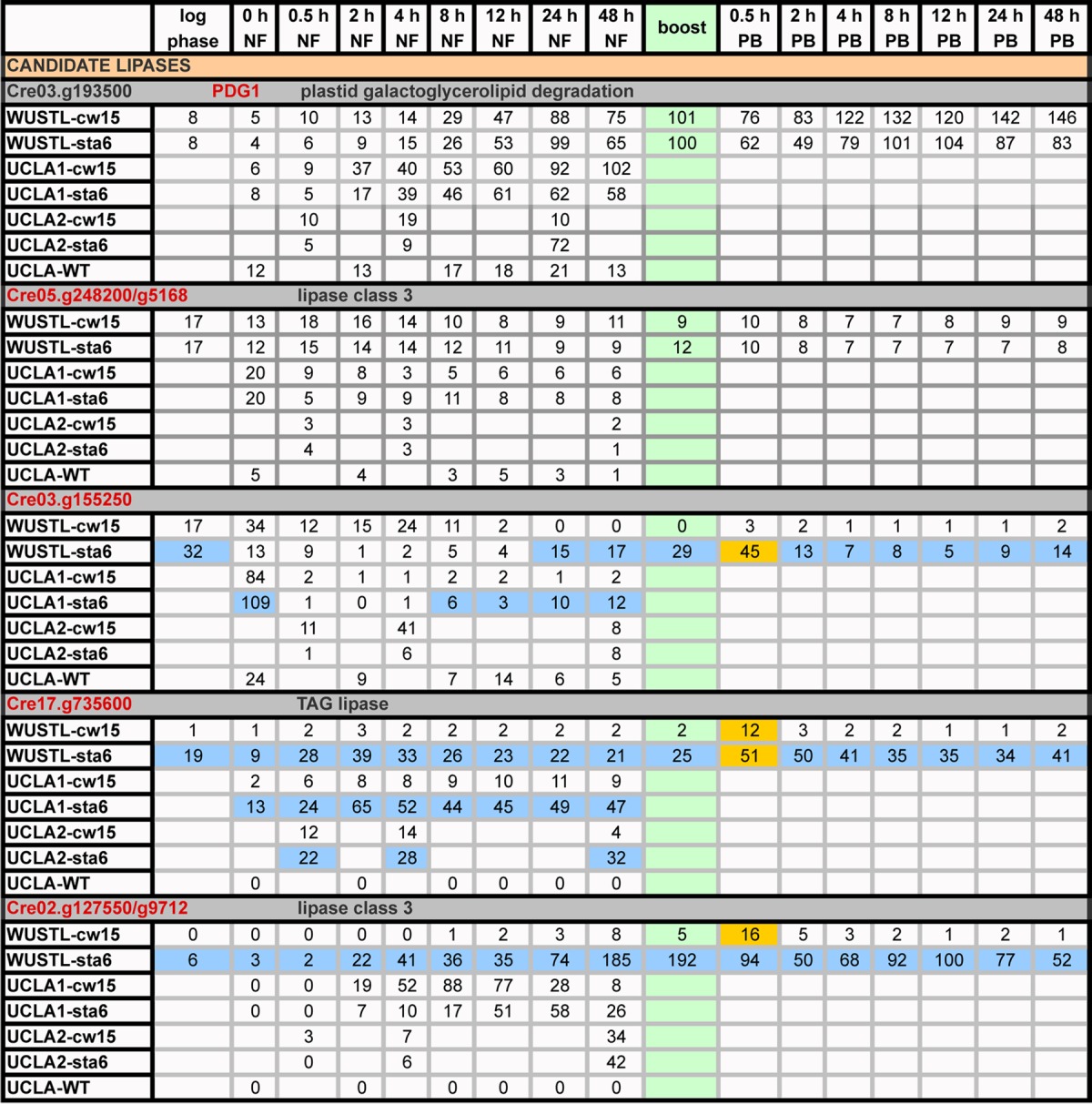
Green, boost addition; PB, postboost; orange, genes with ≥2-fold increases in expression relative to 48-h NF levels following acetate boost; blue, genes showing strong differential expression between the sta6 and cw15 strains.
As anticipated, PGD1 expression increases strongly throughout the N starvation time course in all strains (Table 5). The PDG1 protein lacks a predicted leader sequence, but since MGDG is restricted to thylakoids, a chloroplast location is considered likely.
Two homologues of PDG1 were identified (see Fig. S1 in the supplemental material). The first, Cre05.g248200/g5168, carries a high-scoring chloroplast transit sequence and is the only gene in the candidate-lipase cohort that decreases expression in all strains during the time course (Table 5).
The second PGD1 homologue, Cre03.g155250 (not included in the 46-gene set), is more strongly predicted to be mitochondrion localized (M score, 0.825) than chloroplast localized (C score, 0.444), and a mitochondrial TAG lipase was recently detected in yeast (51); that said, organelle transit sequences can be difficult to differentiate in C. reinhardtii, and direct localization experiments are needed. Cre03.g155250 is expressed in all strains at early time points, but transcript levels then plummet in the cw15 strain and the wild type, whereas they strongly increase in the sta6 strain, with a 2.7-fold boost (Table 5, blue). The distinctive transcription pattern of Cre03.g155250 is the second of four sta6/cw15 differences that are highlighted in this report.
Another candidate lipase-encoding gene also displays differential expression in the sta6 strain. Cre17.g735600 (not included in the 46-gene set) encodes a protein with a strongly predicted signal peptide, which usually indicates an ER→secretory destination but in some cases directs proteins to the chloroplast (20, 52). The gene is not expressed in UCLA-WT or in the cw15 strain, except for a brief spike at the boost, but is robustly expressed by sta6 in both the WUSTL and UCLA1/UCLA2 experiments (Table 5, blue), with a 2.5-fold increase with the boost (Table 5, orange). Cre17.g735600 is related to three other C. reinhardtii genes (data not shown): Cre09.g399400 and Cre14.g615550 yield no transcripts in either strain, while Cre02.g127300 shows a steady increase in both (see Table S3 in the supplemental material). The distinctive transcription pattern of Cre17.g735600 is the third of four sta6/cw15 differences that are highlighted in this report.
Cre02.g127550 (g9712 in the v.5.3 assembly), which lacks a predicted leader sequence, displays strong expression in the sta6 strain but weak expression in the cw15 strain, except for a brief spike at the boost, throughout the 96-h time course in the WUSTL experiment (Table 5). However, expression of this gene is anomalous. In the two UCLA experiments, transcripts are either somewhat more abundant in the cw15 strain than the sta6 strain (UCLA1) or equivalent (UCLA2); moreover, the gene shows no expression in UCLA-WT (Table 5), whereas it is the most strongly upregulated candidate lipase (5.6-fold) in the wild-type experiment described by Miller et al. (11). This example emphasizes the value of having several data sets when assigning expression patterns to particular strains: such anomalies, and this one is obviously of interest, presumably indicate sensitivity to particular culture conditions rather than a strain-specific trait.
(iii) Fatty acid β-oxidation enzymes.
TAG accumulation operates in opposition to TAG breakdown. The fatty acids released from the breakdown of both neutral and polar lipids are processed by the β-oxidation pathway. In their RNA-Seq comparison of wild-type cells at 0 h and 48 h NF, Miller et al. (11) noted >3-fold-decreased transcript levels for two enzymes in this pathway, acyl-CoA oxidase (Cre16.g689050) and 3-oxoacyl-CoA thiolase (Cre17.g723650, ATO1), and a 2-fold increase for a third, enoyl-CoA hydratase (Cre03.g190850, ECH1). Similar patterns were seen in the present experiments (see Table S4 in the supplemental material): expression levels of the oxidase gene declined slightly and those of the thiolase gene more substantially, while the hydratase levels increased. Interestingly, levels of thiolase and hydratase transcripts increased 3.9-fold and 1.6-fold, respectively, with the acetate boost for the sta6 strain, with no boost effect for the cw15 strain, and thiolase gene expression remained stronger in the sta6 strain to 96 h NF (see Table S4). Therefore, the boost appears to selectively enhance the machinery for fatty acid breakdown in the sta6 strain, perhaps in part in conjunction with the breakdown and remodeling of thylakoid membranes undertaken by sta6 at later time points in the N-starvation sequence (18).
(iv) Glycerol-3-P dehydrogenase.
Glycerol-3-P serves as the backbone for DAG and TAG biosynthesis and can thus be said to serve as a bridge between carbohydrate and lipid biosynthesis. It also drives a mitochondrial shuttle system engaged in the import of NADH (53). Glycerol-3-P is generated from dihydroxyacetone phosphate (DHAP), a sugar produced during both gluconeogenesis and the Calvin-Benson cycle, via NADH-dependent glycerol-3-P dehydrogenases (GPDH) (the mitochondrial enzymes are FADH2 dependent). NADH-dependent GPDHs are encoded by five genes in C. reinhardtii (Table 6), one of which (GPD5) was identified during this study.
TABLE 6.
Expression profiles (RPKM) of genes encoding glycerol-3-P dehydrogenasea
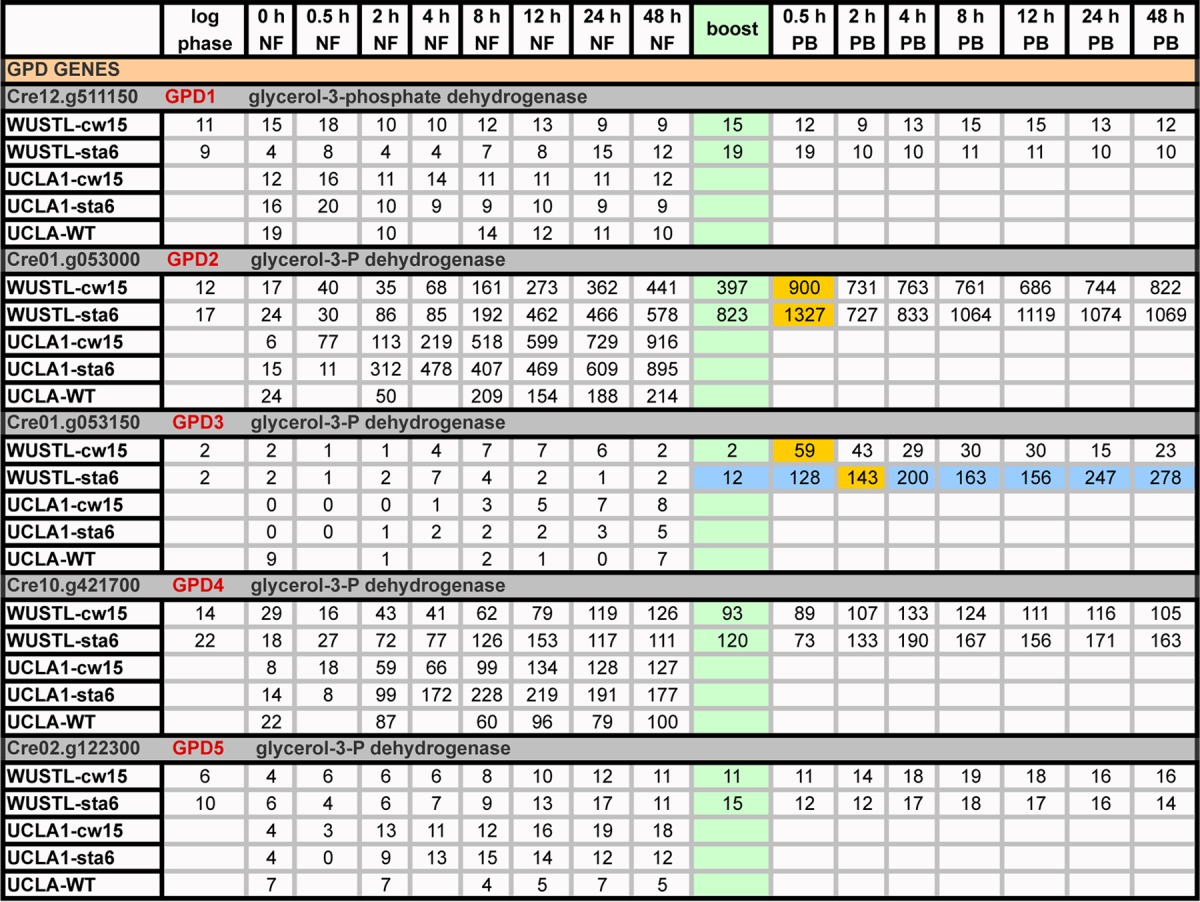
Green, boost addition; PB, postboost; orange, genes with ≥2-fold increases in expression relative to 48-h NF levels following acetate boost; blue, gene showing strong differential expression between the sta6 and cw15 strains.
Table 6 summarizes their expression patterns. GPD1 (Cre12.g511150) and GPD5 (Cre02.g122300/g9595), with no predicted targeting sequences, are expressed constitutively at low levels throughout the time course. GPD2 (Cre01.g053000) transcripts, predicted to be chloroplast targeted, increase 26-fold in the cw15 strain and 24-fold in the sta6 strain during 0→48 h NF; the boost increases transcript levels 2-fold more in the cw15 strain and 2.3-fold more in the sta6 strain, and high levels are sustained to 96 h NF. GPD4 (Cre10.g421700), also predicted to be chloroplast directed, increases expression during 0→48 h NF (4.3-fold for the cw15 strain and 6.2-fold for the sta6 strain) and sustains high levels to 96 h NF without responding to the boost, with levels being consistently higher in the sta6 strain than in the cw15 strain. Large increases in GPD2 and GPD4 transcripts during 0→48 h NF, and the sta6 bias for GPD4, are also seen in the UCLA and UCLA-WT data (Table 6).
Particularly striking is the pattern of GPD3 (Cre01.g053150), which carries no predicted leader sequence. Its expression level remained very low throughout the 0→48 h NF period in both strains in both experiments but then shot up 30-fold with the boost in the cw15 strain and 72-fold in the sta6 strain (Table 6). Moreover, while transcript levels then abated in the cw15 strain to 39% of their maximum boost levels by 96 h NF, they continued to strongly increase in the sta6 strain, reaching 217% of their maximum boost levels at 96 h NF (Table 6, blue), such that GPD3 expression levels at 96 h NF are 12-fold higher in the sta6 strain than in the cw15 strain. The distinctive GPD3 transcription pattern is the fourth of four sta6/cw15 differences that are highlighted this report.
The five GDP genes are members of three subfamilies (see Fig. S2 in the supplemental material). The genes in the third subfamily (GDP2 to GDP4) have an additional feature: they carry an N-terminal HAD domain encoding a hydrolase sequence with homology to the enzyme 3-phosphoserine phosphatase (PSP), which catalyzes the final and irreversible step of serine biosynthesis (54). The Arabidopsis member of the subfamily lacks this domain, but it is present in the V. carteri member. An independent PSP1 gene in C. reinhardtii maintains steady expression throughout 0→96 h NF (not shown). Assuming that the PSP domains of the GPDH proteins are operant, then two enzymatic activities would be stimulated with up-expression of GPD2-GPD4.
(v) TCA and glyoxylate cycles.
Genes encoding enzymes proposed (8) as components of the tricarboxylic acid (TCA) cycle show either steady or slowly declining 0→48 h NF expression in both strains, with concordant WUSTL and UCLA trajectories (see Table S5A in the supplemental material). Only three genes show upregulation with the boost (see Table S5A, orange). The CIS2 gene is given further consideration below.
The less familiar glyoxylate cycle, which is absent in animals, shares many enzymes with the TCA cycle and permits the net synthesis of a 4-C product (succinate) from two acetyl units; the succinate is then metabolized to malate/oxaloacetate, which feed into gluconeogenesis. Table S5B in the supplemental material shows the three genes encoding enzymes proposed (8) as components of the glyoxylate cycle that show robust expression, where only MDH2 shows a modest increase in the sta6 strain with the acetate boost. Two other genes unique to the glyoxylate cycle, ICL1 and MAS1, show a sensitive expression pattern and a strong stimulation with the boost (see below).
(vi) Acetyl-CoA.
Acetate enters both the TCA and glyoxylate cycles as acetyl-CoA, and acetyl-CoA is also the acetate donor for fatty acid biosynthesis. Acetyl-CoA synthetases are encoded by three genes. ACS1 expression, while steady, is far lower in the sta6 strain than the cw15 strain in the WUSTL data but equivalent in the UCLA data (see Table S5C in the supplemental material), again emphasizing the value of having multiple data sets when attempting to discern strain-specific expression patterns. Expression of both ACS2 and ACS3 increases with N deprivation in a sensitive fashion, and these genes are discussed below.
A second avenue to acetyl-CoA synthesis is catalyzed by multimeric pyruvate dehydrogenases that convert pyruvate to acetyl-CoA and CO2. The expression patterns of two genes, PDH2 (mitochondrial subunit) and PDC2 (chloroplast subunit) (see Table S5C in the supplemental material), exemplify those encoding the other subunits: transcription is strong and steady to 96 h NF without a boost effect.
A third avenue, the cleavage of citrate via ATP citrate lyase, is important in the generation of acetyl-CoA in the oleaginous yeast Yarrowia lipolytica (55), but ACLA1 and ACLB1 transcripts are steady during 0→96 h NF, and ACLA1 expression decreases with the boost (see Table S5C in the supplemental material).
Acetyl-CoA can also be generated via acetate kinase (ACK1 and ACK2) and acetylphosphotransferase (PAT1 and PAT2), and while PAT1 levels are steady, the other three decline >2-fold in expression with the boost (see Table S5C in the supplemental material), suggesting that this pathway is not a major participant.
(vii) Gluconeogenesis/glycolysis and the Calvin-Benson cycle.
Gluconeogenesis and the Calvin-Benson cycle both engage in generating hexose phosphates that have two alternative anabolic destinations: feeding into the oxidative pentose phosphate pathway with the concomitant generation of NADPH, or feeding into starch biosynthesis. They can also be catabolized via glycolysis. In a recent review, Johnson and Alric (56) noted that enzymes mediating the “upper half” of gluconeogenesis (from 3-phosphoglycerate to hexose phosphates) appear to be plastid localized in C. reinhardtii, where their activities may overlap Calvin-Benson cycle enzyme activities.
The robust members of the gluconeogenesis pathway and Calvin-Benson cycle are listed in Tables S5D and E in the supplemental material, respectively; sensitive members are discussed below. An obvious anomaly is the high initial level of expression of the RBCS2 gene, encoding the RuBisCO small subunit, in the sta6 strain compared with the cw15 strain; however, even higher levels are expressed in the UCLA-WT sample, so the significance of this difference is not clear [the large subunit is chloroplast encoded and not represented in these poly(A)-selected RNA samples]. A few gluconeogenesis/glycolysis genes are mildly up- or downregulated by the boost (see Table S5D in the supplemental material, orange and purple), but overall, expression of both gene sets in both strains is quite steady to 96 h NF except for a decrease in RBCS2 transcripts and increases in GAP1 (glyceraldehyde-3-P dehydrogenase), PYK1 (pyruvate kinase), and GND1 (6-phosphogluconate dehydrogenase) transcripts.
Fructose-1,6-bisphosphate aldolases (FBA) function in both gluconeogenesis/glycolysis and in the Calvin-Benson cycle. During gluconeogenesis, they catalyze the formation of 6-C fructose-bisphosphate from two 3-C sugars, glyceraldehyde-3-P and dihydroxyacetone phosphate (DHAP), the latter also being the substrate for the glycerol-3-P dehydrogenases mentioned above. The four FBA genes in the C. reinhardtii genome belong to two subfamilies (see Fig. S3 in the supplemental material). FBA2 (no predicted leader sequence) and FBA4 (predicted chloroplast leader), in one subfamily, are expressed at low levels (FBA4 apparently carries a C-terminal deletion), and only FBA2 shows a modest (1.7-fold) boost response in the sta6 strain. FBA1, also in this subfamily, and FBA3, in the second subfamily, have predicted chloroplast leaders; both show a sensitive expression pattern and a modest upregulation with the boost, as detailed below.
(viii) Pentose phosphate pathway.
The oxidative phase of the plastid-localized pentose phosphate pathway takes glucose-6-P to ribulose-5-P, generating 2 NADPHs needed for fatty acid synthesis; the nonoxidative phase generates fructose-6-P and glyceraldehyde-3-P to regenerate the hexose-P that keeps the cycle running. During 0→48 h NF, four of the genes encoding enzymes in the pathway—GND1, GLD1, GLD2, and FSA1—increase in expression and remain elevated during the next 2 days, while others—PGL2, RPI1, TRK1, and TAL2—decrease expression and remain low; PGL1 transcripts are steady (see Table S5F in the supplemental material). The one gene to display an acetate boost is TAL1, encoding transaldolase; it is considered with the other sensitive genes.
Nitrogen-related genes that increase in expression with boost.
As expected, and as previously reported (8, 11, 14), N starvation elicits upregulation of numerous genes involved with nitrogen uptake and scavenging. Table S6 in the supplemental material lists the genes encoding N transporters (Table S6A) and enzymes engaged in the transfer of amino groups (Table S6B) whose transcription is stimulated ≥2-fold with acetate boost. Expression patterns are generally concordant between WUSTL and UCLA experiments. Noteworthy are the relatively low levels of expression of LAO1 and LAO2, encoding periplasmic amino acid oxidases, in the sta6 strain but not the cw15 strain, the strong and enduring rescue of PROB1 (glutamate-5-kinase) transcription with the acetate boost in both strains, and the quirky expression patterns of DUR3A, a urea transporter. The AST1 gene, encoding aspartate aminotransferase, is discussed below as a member of the sensitive gene set.
Boyle et al. (8) presented molecular and genetic evidence that the gene NRR1 encodes a regulator for induction of the TAG accumulation pathway in N-free medium. Supporting this proposal, we found a strong increase in its expression starting at 2 h NF and a 2.3-fold increase with boost in the sta6 strain (see Table S6C in the supplemental material).
Stress-related genes whose expression increases with the boost.
Abrupt N starvation of log-phase cells is by definition stressful. An early response is the stimulated expression of members of the target of rapamycin (TOR)-related autophagy pathway (57). Of the seven annotated APG genes (called ATG in most organisms) that respond to TOR signaling in other organisms, all are strongly and coordinately upregulated starting at 2 h NF, and all except APG10 remain elevated for the next 96 h, with only APG4 showing a positive boost response and several showing a modest negative response (see Table S7A in the supplemental material).
Expression of most PEX, PRX, and MSD genes that participate in ROS scavenging remains steady or decreases in both experiments throughout the time course (data not shown), perhaps because chlorophyll levels (8, 9, 13, 25, 48) and photosynthetic electron transport activity (25, 58) decrease and such toxic products are not a major issue. Table S7B in the supplemental material shows the four genes in this category with a modest boost response. None of the genes encoding SRR scavenger receptors responds to the boost; SRR16 shows stronger expression in the sta6 strain than the cw15 strain in the WUSTL but not the UCLA data (see Table S7B), yet another example of the value of having 2 data sets.
Table S7C in the supplemental material includes several stress-related low-CO2-inducible (LCI) genes that are boost upregulated and a high-CO2-inducible (FEA1) gene to illustrate that members of this cohort have highly variable expression patterns both between strains and between experiments.
Respiratory burst oxidase.
The gene RBO1, encoding a homologue of the respiratory burst oxidase that responds to stress in land plants (59, 60), is contiguous to the STA6 gene and deleted in the sta6 mutant (14), suggesting that its absence might influence the sta6 phenotype (14). As shown in Table S7D in the supplemental material, RBO1 produces few transcripts in the cw15 strain and UCLA-WT during 0→48 h NF and is unresponsive to the boost.
A full copy of the contiguous orthologue RBO2 is present in the sta6 strain, but it yields few transcripts (see Table S7D in the supplemental material), suggesting that the deletion of RBO1 curtails RBO2 expression as well (14). RBO2 expression is also low in the wild type; in the cw15 strain, it decreases in the UCLA data and increases in the WUSTL data, where it stabilizes without the boost (see Table S7D).
Blaby et al. (14) noted that in the UCLA experiments, expression of LHCSR1 and LHCSR2, which are both involved in photoquenching, is higher in the cw15 strain than in the sta6 strain (see Table S7D in the supplemental material) and suggested that RBO1/RBO2 might play a role in their induction. However, in the WUSTL data, both genes are more strongly transcribed in the sta6 strain than in the cw15 strain (see Table S7D).
A key test of this hypothesis—the effect of an RBO1 transgene on the sta6 phenotype—is currently in progress in the UCLA labs.
Carbon-related genes with “sensitive” expression patterns.
(i) The blue/green cohort.
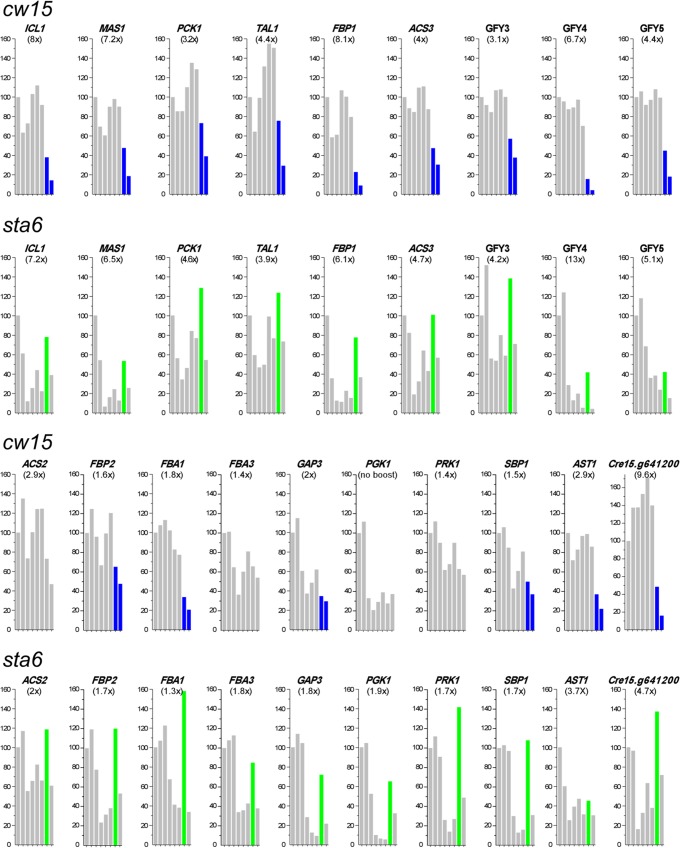
Five boost-enhanced genes—ICL1, MAS1, PCK1, TAL1, and FBP1—were of immediate interest because these genes were identified in the UCLA study (14) as having patterns of expression that were similar within a strain but markedly different when the cw15 and sta6 strains were compared. Specifically, expression of the five genes sank or stayed low during 0→48 h NF in the cw15 strain but increased, after an initial drop, in the sta6 strain (14).
When we analyzed the 0→48 h NF expression patterns of these five genes in the WUSTL data, they also proved to be similar within a strain and distinctive between strains. However, the patterns observed were quite different from those observed in the UCLA study. As shown in Fig. 5, transcription of these genes in the cw15 strain is high through 12 h NF, drops at 24 h NF, and is <40% of starting values by 48 h NF (blue bars), while transcription in the sta6 strain generally stays low throughout the time course except for a strong spike at 24 h NF (green bars), when the transcripts are >50% more abundant than at 48 h NF. The RPKM values for these gene sets are presented in Table 7.
FIG 5.
Sensitive gene set. RPKM values at 0 h NF (postcentrifugation) were set at 100; subsequent percentiles are at 0.5, 2, 4, 8, 12, 24, and 48 h NF. RPKM data for these and additional sensitive genes appear in Table 7. A drop in expression in the cw15 strain occurs at late time points (blue when the 48-h NF value is <40% of the initial value), and a drop in sta6 expression occurs early, with a spike at 24 h NF (green when the 24-h NF value is at least 50% greater than the 48-h NF value). Numbers in parentheses are fold increases in gene expression in response to the boost (maximum RPKM level during the 2 h postboost divided by RPKM level at 48 h NF).
TABLE 7.
Expression profiles (RPKM) of genes showing “sensitive” transcription patternsa
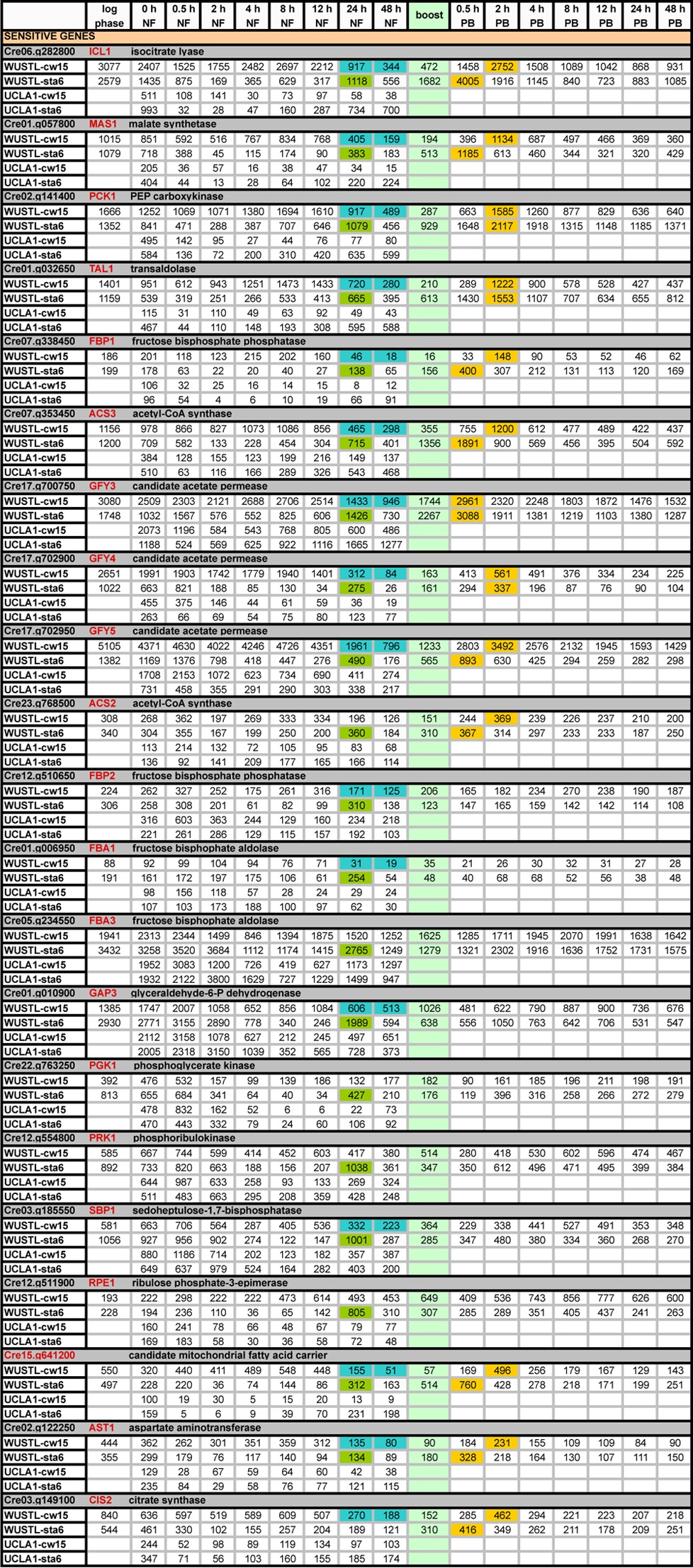
Light green, boost addition; PB, postboost; blue, drop in cw15 transcripts at 24 h NF and 48 h NF; dark green, increase in sta6 transcripts at 24 h NF; orange, genes with ≥2-fold increases in expression relative to 48 h NF levels following the acetate boost.
We went on to identify 16 additional genes that display the blue/green pattern in the WUSTL data set; all but ACS2, FBA3, PGK1, PRK1, and RPE1 show the late <40% drop in expression (blue) in the cw15 strain compared with starting values, and all but CIS2 show the >50% spike in expression (green) in the sta6 strain at 24 h NF compared with 48 h NF (Fig. 5 and Table 7). Figure S4 in the supplemental material shows WUSTL expression profiles for other genes whose products function in the same pathways as the genes shown in Figure 5, and none displays the blue/green pattern. Most of the 21 genes responded to the acetate boost to at least some extent (Fig. 5); those with a ≥2-fold increase are highlighted in orange in Table 7.
None of the additional 16 genes displays the blue/green pattern in the UCLA1 data, and only ACS3, GFY3, and Cre15.g641200 display the cw15 down/sta6 up pattern of the five founder UCLA genes (Table 7). Instead, in both UCLA experiments, their expression tends to decrease gradually in both strains (Table 7; also, see Table S8 in the supplemental material). Table S8 also displays data from the UCLA-WT experiment, where expression patterns are again different, with transcript levels generally being higher than those in UCLA-cw.
Unlike the genes grouped together in previous sections, the 21 genes listed in Table 7 encode proteins that operate in a number of different pathways. Isocitrate lyase (ICL1) and malate synthase (MAS1) are the linchpin enzymes of the glyoxylate cycle; PEP carboxykinase (PCK1) and fructose-1,6-bisphosphatase (FBP1 and FBP2), respectively, drive entry into and a late step in gluconeogenesis; acetyl-CoA synthetase (ACS2 and ACS3) feeds acetate into the TCA and glyoxylate cycles and into fatty acid synthesis; candidate acetate permeases (GFY3, GFY4, and GFY5), if verified, would mediate acetate uptake; transaldolase (TAL1) and ribulose-phosphate-3-epimerase (RPE1) serve in the nonoxidative pentose phosphate pathway; sedoheptulose-1,7-bisphosphatase (SBP1) and phosphoribulokinase (PRK1) are unique to the Calvin-Benson cycle; Cre15.g641200 is annotated as a candidate mitochondrial fatty acid carrier, although it lacks a mitochondrial targeting sequence; and aspartate aminotransferase (AST1) catalyzes the interconversion of aspartate and α-ketoglutarate to glutamate and oxaloacetate, which feeds into the pentose phosphate pathway. The gene products fructose-1,6-bisphosphate aldolase (FBA1 and FBA3), phosphoglycerate kinase (PGK1), and glyceraldehyde-6-phosphate dehydrogenase (GAP3) are predicted to be members of the Calvin-Benson cycle (14). It should be noted, however, that FBA, PGK, and GAPDH also function in gluconeogenesis, and given that both pathways operate in the chloroplast stroma (56), their activities may not be strictly segregated. An anomaly related to CIS2 is considered below.
Taken together, it appears that a group of 21 genes, whose products function in various pathways, respond to 0→48 h NF and the acetate boost as a cohort in the WUSTL experiment, whereas their expression patterns differ in both the UCLA and UCLA-WT experiments. We suggest that these differences relate to the fact that the three experiments were performed using different strains (wild type versus mutants) and laboratory conditions (e.g., medium and light intensity; see Materials and Methods) and propose that the listed genes are singularly sensitive to the cell's metabolic status, perhaps because their products serve as “gateway” members of their respective pathways. They might, for example, have short half-lives and/or govern rate-limiting reactions and hence serve as nodes that permit gene expression levels to influence the course of metabolism or biosynthesis. We therefore refer to these as “sensitive genes,” as contrasted with the “robust genes,” whose expression patterns are concordant, with minor variations, within the three experiments.
Support for this proposal comes from the recent study of an ICL1 deletion mutant (61), which is devoid of a glyoxylate cycle and displays many anomalies in central carbon metabolism. 14N/15N labeling experiments show that many of the proteins designated here as sensitive—specifically, those encoded by MAS1, CIS2, FBA1, PCK1, ACS3, TAL1, and AST1—are either strongly increased or decreased in the mutant relative to controls.
(ii) Genes whose expression is stimulated by acetate.
Thirteen of the 21 sensitive genes in Table 7 have ≥2-fold increases in expression with the acetate boost (orange highlighting), whereas the others show a weaker or no response. Expression of the strongly boost-responsive set is also highly sensitive to the presence of exogenous acetate in N-replete medium. Table 8 compares their transcript levels during N-replete log-phase growth in acetate-containing medium (WUSTL) versus acetate-free medium (UTSW) (26). The genes that show ≥2-fold responses to the acetate boost prove to be expressed at very low levels when the strain is grown on acetate-free medium (Table 8A, columns 3 and 4) compared with acetate-containing medium (Table 8A, columns 1, 2, and 7), with low expression persisting after 18 h NF in acetate-free medium (Table 8A, columns 5 and 6). In contrast, sensitive genes showing modest or no boost responses are expressed equivalently (Table 8B) or at higher levels (Table 8C) when the strains are grown on acetate-free medium compared with acetate-containing medium, with expression usually declining after 18 h NF in acetate-free medium. Hence sensitive genes displaying a strong acetate boost prove to also display a strong transcriptional sensitivity to the presence/absence of exogenous acetate. Notably, 7 of these 13 proteins, listed in the previous paragraph, are also expressed aberrantly in the ICL1 mutant (61).
In the course of this analysis, we encountered an anomaly pertaining to the CIS1 and CIS2 (citrate synthase) genes. The former has been predicted to encode a glyoxylate cycle enzyme and the latter to encode a TCA cycle enzyme (14), assignments that have been adopted in Table S5A and B in the supplemental material. However, CIS2 shows a strong acetate boost upregulation and low expression on acetate-free medium (Table 8A), akin to the dedicated ICL1 and MAS1 enzymes of the glyoxylate cycle (Table 8A), whereas CIS1 is non-boost-responsive (see Table S5A), like other TCA cycle genes (Table S5A), and equivalently expressed in both media (Table 8B). Plancke et al. (61) also presented arguments for CIS2 as a member of the glyoxylate cycle.
A second anomaly relates to the glyoxylate cycle itself. Both the UCLA1 and UCLA2 studies document increased levels of expression of ICL1, MAS1, and CIS2 in the sta6 strain and low expression levels in the cw15 strain and complemented sta6 strains during 0→48 h NF and suggest that enhanced glyoxylate activity plays a role in enhanced TAG production in the sta6 strain. However, this increase is not observed in the WUSTL data. Miller et al. (11), who also observed decreases in ICL1 and MAS1 expression in wild-type N-starved cells, pointed out that depressed glyoxylate cycle activity would result in more acetate being available for fatty acid biosynthesis. The phenotype of the ICL1 deletion mutant (57) supports their proposal: the mutant generates enhanced levels of fatty acids and TAGs compared with non-deletion-containing controls even under N-replete conditions. It has also been reported that hexose phosphates, which accumulate in the sta6 strain (14), inhibit expression of ICL and MAS genes in cucumber (62). Taken together, the enhanced sta6 glyoxylate cycle profiles in the UCLA studies during 0→48 h NF, confirmed with enzyme assays, presumably reflect particular features of the UCLA experimental conditions and may explain, at least in part, why TAG levels in the sta6 strain were found to be equivalent to those in the cw15 strain at 48 h NF and were not enhanced until 96 h NF, whereas they were enhanced by 48 h NF under WUSTL experimental conditions (18, 24).
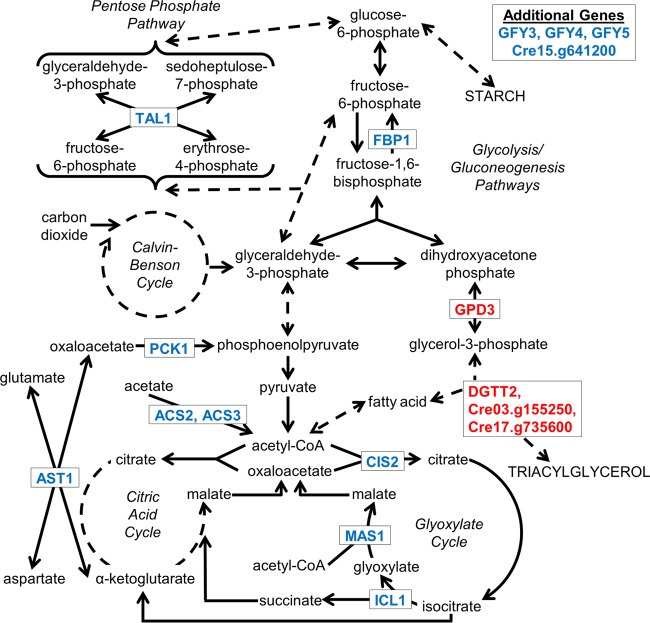
Figure 6 shows (in blue) the products of the 13 acetate-sensitive genes and their positions in various metabolic/biosynthetic pathways. We propose that of the 21 blue/green genes identified in this study (Tables 7 and 8), this cluster monitors a variety of activities related to acetate utilization, while the remaining 8 genes largely monitor activities related to the Calvin-Benson cycle.
FIG 6.
Proteins encoded by the acetate-sensitive gene set (blue font) and by genes selectively upregulated in the sta6 strain (red font). Abbreviations: ACS, acetyl-CoA synthetase; AST, aspartate transaminase; CIS, citrate synthase; Cre03.g155250 and Cre17.g735600, candidate TAG synthase enzymes; Cre15.g641200, candidate mitochondrial fatty acid carrier; DGTT, diacylglycerol acyltransferase type 2; FBP, fructose-1,6-bisphosphatase; GFY, GPR1/FUN30/YaaH family (candidate acetate transporters); GPD, glycerol phosphate dehydrogenase; ICL, isocitrate lyase; MAS, malate synthase; PCK, phosphoenolpyruvate carboxykinase; TAL, transaldolase.
(iii) Candidate acetate permeases.
The genes designated GFY3, GFY4, and GFY5 in Tables 7 and 8, as well as the genes GFY1 and GFY2, have been identified as members of the GPR1/FUN30/YaaH (pfam01184) gene family (63) (see Fig. S4A in the supplemental material). Several fungal proteins encoded by genes in this family have been shown or suggested to mediate acetate uptake (64, 65), and acetate permease activity has recently been demonstrated for the E. coli protein YaaH (66). In S. cerevisiae, both exogenous acetate and induction of the glyoxylate cycle are accompanied by strong upregulation of its GPR1/FUN30/YaaH gene (67, 68).
In C. reinhardtii, the five genes are closely linked on LG17, with GFY1 to GFY3 in one cluster and GFY4 and GFY5 apparently contiguous in a second cluster.
While GFY1 and GFY2 show constitutive expression and GFY1 transcripts actually decrease with the boost, GFY3 to GFY5 show the blue/green pattern during 0→48 h NF (Fig. 5) and an increase in expression of 3- to 13-fold with the acetate boost in both strains (Fig. 5 and Table 7). Expression of GFY3 to GFY5 is also strong in the UCLA and UCLA-WT samples (Table 7), all of which derive from cells maintained in acetate, whereas cells grown or maintained in the absence of acetate have very low reads (Table 8A).
We went on to analyze this family in more detail using two approaches. Figure S5B in the supplemental material shows a similarity network (38, 39) of 355 representative GPR1/FUN30/YaaH sequences (a key is provided in Data Set S6 in the supplemental material). The proteins are widely disseminated and clearly separate along the eukaryote/prokaryote divide, with members absent from animals and vascular land plants. The genomes of sequenced algal species encode orthologs (see Data Set S6) that bear greater similarity to the prokaryotic proteins than the other eukaryotic proteins (see Fig. S5B), suggesting that the nonalgal eukaryotes (mostly fungi) and algae independently acquired the genes from prokaryotes by horizontal gene transfer (HGT) in two distinct and early events. Maximum-likelihood analysis (Fig. S5C) shows that the green-algal genes form a loose clade, clade III, that is again more closely related to prokaryotic (clade I) than to eukaryotic (clade II) family members, and Fig. S5D in the supplemental material shows that the six genes in V. carteri form a subfamily distinct from the five genes in the closely related C. reinhardtii, highlighting the rapid evolution of these sequences in algal lineages. The phylogenetic distributions of GPR1/FUN30/YaaH genes can be highly unusual, with both moss and fungal genes being found in clade II, a Leishmania gene being found in the prokaryotic clade I (presumably a recent HGT), and volvocacean genes being more closely related to a haptophyte gene (E. huxleyi) than to other chlorophyte genes in clade III.
Schönknecht et al. (69) independently performed a phylogenetic analysis of this gene family and published a topology wherein eukaryotic algal and fungal/moss proteins share a direct common ancestry and together form a clade that is sister to the prokaryotic members. Subsequent analyses using their data (kindly provided by G. Schönknecht) revealed that the topology of the two major eukaryotic clusters is highly sensitive to evolutionary models and parameters used for phylogenetic reconstruction and can yield well-supported trees in either of two configurations: green algae and fungi/moss as separate HGT events from bacteria, as depicted in Fig. S5C in the supplemental material, or green algae (but not Galdieria) and fungi/moss as a single HGT event, as depicted in reference 69.
Our network analysis of the GPR1/FUN30/YaaH family (see Fig. S5B in the supplemental material) supports the tree topology in Fig. S5C in clearly distinguishing the fungal/moss members and the green-algal members as separate offshoots of the central prokaryotic cluster. In both of our analyses, the unusual phylogenetic relationships among the eukaryotic members of the family, noted above, suggest that subsequent HGT took place between disparate eukaryotic groups.
Stress-related genes that decrease in expression with the boost.
Whereas only a few stress-related genes are upregulated with the acetate boost (see Table S7 in the supplemental material), a far larger group of stress-related genes is down-regulated ≥2-fold with the boost, with the low point usually occurring at 30 min postboost (Table 9, purple). More genes are so affected in the sta6 strain (64 genes) than in the cw15 strain (40 genes); in Table 9, those not downregulated in the cw15 strain are italicized. The downregulated genes, some of which have been identified in a recent study of autophagy in C. reinhardtii (73), encode proteasome subunits, chaperones, heat shock proteins, and proteins that participate in degradative processes, the one apparent outlier being BCS1, which encodes a mitochondrial biogenesis factor. We went on to identify 20 additional nonannotated genes that also show this pattern in both strains (Table 9, purple); these may prove to be members of the same response cohort.
TABLE 9.
Expression profiles (RPKM) of sta6 genes whose expression decreases ≥2-fold relative to 48 h NF levels following acetate boost (purple)a
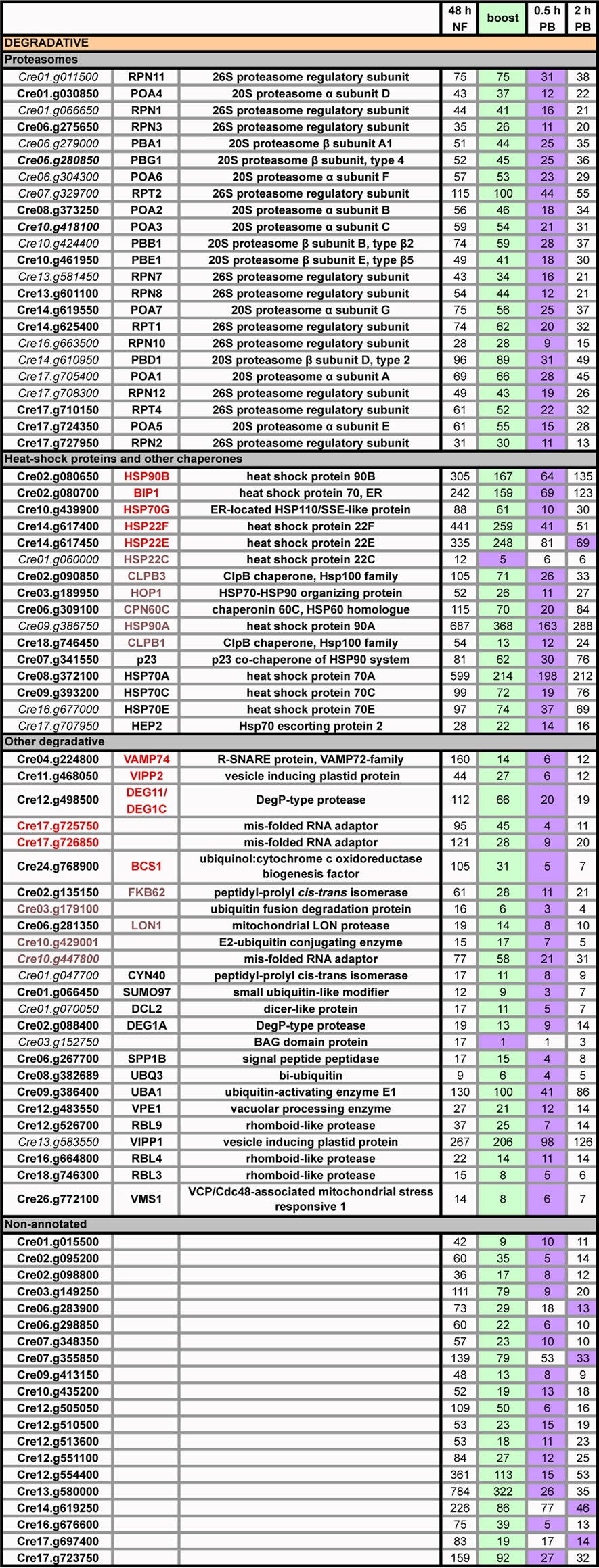
Genes that do not show this pattern in the cw15 strain are shown in italics. The genes in colored type also show an increase in expression at 48 h NF relative to 24 h NF (see Table S7 in the supplemental material): red indicates that the pattern is present in cw15 and sta6 strains, and blue indicates that it is present in the sta6 strain only. PB, postboost.
Of the 41 non-proteasome-encoding genes in Table 9, 22 share an additional pattern: their expression increases 2- to 6-fold (means of 3.3-fold ± 0.9-fold for the cw15 strain and 3.5-fold ± 1.4-fold for the sta6 strain) between the 24-h NF sample (see Table S9 in the supplemental material, blue) and the 48-h NF sample (see Table S9, yellow), followed by the sharp boost-induced decrease lifted up in Table 9 (see Table S9, purple). The 11 genes in red in Table 9 show this blue/yellow/purple pattern in both strains (see Table S9A); the 11 in blue show the pattern in the sta6 strain only (see Table S9B). The 20 nonannotated genes in Table 9 also show the blue/yellow/purple pattern (see Table S9C), except for the 5 italicized entries, where it is seen only in the sta6 strain, again suggesting that these genes are members of the same cohort as the annotated set. While not invariably the case, the genes in the sta6 strain usually show a larger increase at 48 h NF than the genes in the cw15 strain.
In the UCLA experiments, where the boost was not performed, none of the 22 annotated genes shows increased expression at 48 h NF in the cw15 strain (see Table S9A in the supplemental material). However, 12 of the 22 show a modest (mean of 2.1-fold ± 0.4-fold) increase in the sta6 strain (see Table S9A and B in the supplemental material), as do 8 of the 20 nonannotated genes (data not shown).
As detailed in the Discussion, we propose that this increase/decrease pattern in degradation-related genes may relate to our morphological observation (Fig. 2 and 3) that an acetate boost at 48 h NF appears to divert cells from pursuing an autophagocytic pathway.
DISCUSSION
General overview of the TAG-accumulation response.
This study, combined with those previously published, generates the following picture of the TAG accumulation response, which, while provisional and incomplete, can serve to provide context for a discussion of our results.
C. reinhardtii cells growing in acetate-supplemented N-replete medium actively run acetate-fed glyoxylate cycles and photosynthetic electron transport-fed Calvin-Benson cycles. When the cells are transferred into N-free acetate medium, these cycles presumably continue to operate in the short term, generating glucose-6-P via both the Calvin-Benson cycle and the glyoxylate cycle-derived oxaloacetate, which feeds into gluconeogenesis.
Early in the first day, genes encoding starch-related enzymes are upregulated, and much of the glucose-6-P is funneled into ADP-glucose and then starch biosynthesis in the cw15 strain but not in the sta6 strain. During the first day, genes encoding enzymes for glycerol-3-P, fatty acid, and TAG biosynthesis are also upregulated in specific patterns, while genes encoding enzymes for the glyoxylate cycle are in most cases downregulated, presumably allowing exogenous acetate to funnel into fatty acid biosynthesis. TAGs proceed to accumulate in cytoplasmic LBs (in both strains) and in chloroplast LBs (in the sta6 strain but not the cw15 strain).
By the end of the second day, when starch levels start to plateau (9, 17, 48), the cells ordinarily shift into an autophagy program that limits the extent of TAG accumulation, possibly because its execution is dependent on TAG breakdown. If, however, they are subjected to an acetate boost, this program is bypassed, for unknown reasons, and the cells instead continue to accumulate TAG until, in the sta6 strain, they reach full obesity. Along the way, chlorophyll levels and rates of photosynthetic electron transport diminish, meaning that cells are increasingly reliant on glucose-6-P entry into the pentose phosphate pathway to generate the NADPH needed for fatty acid synthesis. Key participants in these various transitions are 21 “sensitive” genes whose transcription levels, we suggest, are responsive to the overall operation of various metabolic pathways, with 13 being responsive to the cells' acetate status.
Given this context, we first discuss what has been learned about the differences between starch-forming strains, in particular the cw15 strain, and the starchless sta6 mutant. We then discuss what has been learned about the role of the acetate boost in long-term TAG accumulation.
Comparisons of the cw15 and sta6 strains.
The original premise of this study was that the cw15 strain was the parent of the sta6 strain, but the careful work of Blaby et al. (14) has established that this is not the case, leading them to focus much of their inquiry on comparing the sta6 mutant with complemented sta6 strains. However, the responses to N-free conditions in the cw15 strain and in complemented sta6 strains proved to be generally similar (14).
A major observation of the study by Blaby et al. (14), supported by assays of enzyme activity and metabolite profiles, is that sta6 cells increase expression of five key genes—ICL1, MAS1, PCK1, TAL1, and FBP1—during 0→48 h NF, whereas expression of these genes in starch-forming strains is low during this period, suggesting that the glyoxylate and gluconeogenesis pathways are more active in the sta6 strain than in starch-producing strains. In the WUSTL study, wherein different media and light conditions were used (see Materials and Methods), these five genes also share common patterns of expression, but the shared patterns are different from the UCLA patterns: in the cw15 strain, transcripts remain elevated until 24 h NF and then drop to lower levels, while in the sta6 strain, transcript levels are low except for a curious and unexplained spike in abundance at 24 h NF which abates by 48 h NF. We went on to identify 16 additional genes in the WUSTL data set whose patterns of expression match those of the first five, where most of these 21 genes also respond to acetate boost (Fig. 5 and Table 7). Several of the genes in this cohort are also anomalously expressed in a mutant strain that has a deletion of the isocitrate lyase gene (ICL1) and hence is blocked in its glyoxylate cycle (61). We suggest that the transcription patterns of this “sensitive” gene set are indicative of the biochemical pathways being pursued by cells under a given set of environmental/genetic conditions and that they may be less informative in indicating the defining differences between starch-forming and starch-null cells.
With the important caveat that half the C. reinhardtii genes have not yet been annotated, and some of these may play key roles in storage product biology, four genes, in red in Fig. 6, have been identified whose expression is markedly distinctive between the sta6 and starch-forming strains under various laboratory conditions: DGTT2, encoding one of several diacylglycerol acyltransferases (first noted in reference 14); Cre03.g155250 and Cre17.g735600, encoding candidate lipases; and GPD3, encoding one of five glycerol-3-P dehydrogenases.
DGTT2 and the candidate lipases are strongly overexpressed in the sta6 strain compared with starch-forming strains even in N-replete medium (Tables 4 and 5). While it is straightforward to posit correlations between enhanced DGTT2 levels and TAG accumulation, it is counterintuitive to posit such correlations for lipases. However, the recent work of Li et al. (50) documents that a gene annotated as encoding a TAG lipase, and now called PDG1, in fact participates in TAG biosynthesis under N-free conditions, and Cre03.g155250 is a homologue of PDG. A full characterization of the Cre03.g155250 and Cre17.g735600 gene products and gene knockdowns is clearly highly warranted.
Glycerol-3-P dehydrogenases (GPDHs) catalyze the formation of glycerol-3-P, the backbone of lipid molecules, from dihydroxyacetone phosphate (DHAP), which is, in turn, formed by the cleavage of fructose bisphosphate via fructose bisphosphate aldolase (FBA). Aldolases associate with both gluconeogenesis and the Calvin-Benson cycle, and two of the four FBA-encoding genes in C. reinhardtii are members of the sensitive gene cohort (Table 7).
The C. reinhardtii genome encodes five GPDH enzymes. Of these, GPDH2 and GPDH4 are predicted to be chloroplastic, and expression of their corresponding genes increases steadily during 48 h→N, with GPD4 transcripts generally being 1.5- to 2-fold higher in the sta6 strain than in the cw15 strain (Table 6). The GPD3 gene, with no predicted targeting sequence, is poorly expressed in all tested strains during 0→48 h NF. However, with the acetate boost, its transcription is strongly enhanced, far more so in the sta6 strain than in the cw15 strain, to 96 h NF (Table 6). Little attention has been given to a possible role for glycerol-3-P levels in influencing rates of DAG/TAG biosynthesis (19, 22), but these profiles suggest that an exploration of this possibility could be fruitful.
While these four gene expression differences between the cw15 and sta6 strains may well prove to participate in generating the sta6 phenotype, the phenotype is also likely to be influenced at the metabolic level by what we can term a glucose-6-P backflow. Blaby et al. (14) showed that levels of glucose-6-P are 2-fold higher in the sta6 strain at 96 h NF than in complemented starch-forming strains, similar to two starch-null mutants of Arabidopsis that accumulate hexose monomers (70, 71). Moreover, the WUSTL and UCLA data both indicate that sta6 cells are fully committed to forming starch, expressing the relevant enzymes at the same levels and for the same time periods as do starch-forming cells (Table 2). Hence glucose-6-P is presumably generated and sent to the starch-biosynthetic apparatus in a normal fashion. When, in the sta6 strain, it fails to be converted into glucose-ADP and undergo polymerization, it presumably has to go somewhere, the obvious possibility being that it is somehow involved in the formation of chloroplast LBs (18).
The widely accepted model for LB formation in land plants is that fatty acids are synthesized in the chloroplast and then shuttled to the ER, where they are conjugated to glycerol-3-P backbones by resident ER enzymes to generate DAGs and then TAGs. Recent studies, however, indicate that TAG biosynthesis in C. reinhardtii has a number of distinctive features (summarized in Fig. 9 of reference 50): (i) the DAG moieties are largely assembled in the plastid, and some are then shipped to the ER for the addition of a third acyl group (16); (ii) many of the TAG fatty acids derive from pre-existing chloroplast glycerolipids that are cleaved by dedicated lipases such as PDG1 (50); and (iii) the closely apposed chloroplast outer envelope membrane and ER membranes (18) are coparticipants in cytoplasmic LB assembly, perhaps assisted by alga-specific and ER-localized MLDP proteins (12, 49). Another relevant consideration is that C. reinhardtii likely possesses at least one chloroplast-localized diacylglycerol acyltransferase that mediates the constitutive formation of TAG-filled plastoglobules and eyespot granules in all strains (12, 18).
One sta6 scenario, then, would go as follows. (i) Some of the posited glucose-6-P backflow feeds into the plastid-localized pentose phosphate pathway to generate the NADPH required for additional fatty acid synthesis as photosynthetic electron transfer abates. (ii) Some of the backflow moves in the glycolysis direction until it forms fructose bisphosphate, some of which is then shunted into glycerol-3-P via enhanced levels of GPDH enzymes, where the sharply reduced level of fructose bisphosphate in N-starved sta6 cells (14) is consistent with this suggestion. (iii) The backflow may also inhibit operation of the glyoxylate cycle, as is observed in land plants (62), directing acetate into fatty acid biosynthesis. (iv) The augmented fatty acid and glycerol-3-P pools, supplemented by the products of thylakoid breakdown (18), generate augmented levels of chloroplast DAG, some of which is then converted into chloroplast TAG, events mediated by the enhanced levels of DGTT2 and Cre03.g155250/Cre17.g735600 enzymes. (v) The TAG is stored in chloroplast-localized LBs, perhaps via an expansion of pre-existing plastoglobules (18). Testable features of this model include the prediction that mutations in the four sta6-enhanced enzymes would compromise chloroplast LB formation in a sta6 background and that these enzymes are chloroplast localized.
Mutations like the sta6 deletion are expected to generate a loss-of-function phenotype, in this case an inability to form starch. Unexpected is a gain-of-function phenotype, in this case the formation of a novel class of LBs with the attendant upregulation of at least four lipid-related genes. Chloroplast LBs have a well-defined organization and architecture (18), which is also unexpected for a cellular trait generated by a biochemical defect. Hence, it is possible that the sta6 genotype elicits the expression of a chloroplast-LB biosynthetic program, encoded in the C. reinhardtii genome, which is not called upon in wild-type cells under normal laboratory conditions but is stimulated in wild-type cells under to-be-identified conditions.
Acetate boost.
The acetate boost was discovered by accident: an additional 20 mM acetate was inadvertently added to a culture at 48 h NF, and we noticed that the cells went on to accumulate far larger LBs than nonboosted cells. In our earlier study (18), the effects of the boost were monitored by light and electron microscopy, where both cytoplasmic and chloroplast LB size was more strongly enhanced in the sta6 strain than in the cw15 strain.
We document here that the rate of acetate depletion from medium boosted to 40 mM acetate is unchanged from the rate of depletion at 20 mM acetate (Fig. 4), countering hypotheses that the boosted cells simply take up additional acetate for fatty acid and hence TAG biosynthesis.
That said, there apparently occurs at least a pulse of acetate entry when the boost is administered, as documented by two events: (i) 229 flagellum-related genes are transiently upregulated in expression in the cw15 strain, the classic response to the acetic acid-mediated “pH shock” used to deflagellate C. reinhardtii cells (43); and (ii) ∼1,300 additional genes are either up- or downregulated in expression ≥2-fold following boost in both strains (Table 1), with most quickly returning to preboost levels. Only a few of the upregulated genes encode enzymes in pathways for starch or lipid biosynthesis (Tables 2 to 5), but many encode proteins involved in nitrogen uptake and scavenging (see Table S6 in the supplemental material) and in pathways of central carbon metabolism (Tables 6 and 7; also, see Table S5 in the supplemental material).
Of particular interest are 13 strongly boost-upregulated genes whose expression is low when cells are grown or N starved in medium lacking exogenous acetate (Table 8 and Fig. 6, blue font), suggesting that these genes carry upstream regulatory elements responsive to the cell's acetate status. Interestingly, a different set of genes, involved with spore formation, is coordinately upregulated by acetate in S. cerevisiae (68).
Hence, assuming that these transcripts are translated, one consequence of the boost is to endow cells with enhanced levels of key transporters and enzymes for the ensuing days of N starvation and TAG formation.
Two observations form the basis for an additional hypothesis on the influence of the boost. By microscopy, we noticed that starting at 48 h NF, nonboosted cells come to contain large cytoplasmic vacuoles (Fig. 2), filled with degrading cellular material (Fig. 3), a response that does not occur in boosted cells. The nonboosted cells are fully viable (see Table S1 in the supplemental material), indicating that this autophagy program is a “natural” and not a toxic response to N deprivation, but it is accompanied by a smaller accumulation of TAG in nonboosted cells.
We then noticed, in analyzing the 875 sta6 genes whose transcription is downregulated by the acetate boost, a set of 64 genes encoding proteins that are expected to participate in protein quality control and autophagocytic processes, including proteasome subunits, chaperones, heat shock proteins, cyclophilins, and proteases (Table 9). Moreover, 22 of these genes show a sharp increase in expression in the sta6 strain during the day immediately prior to the boost (see Table S9 in the supplemental material), where an additional 20 nonannotated genes also show this pattern as well and may represent additional members of the cohort. Hence, expression of a sizable autophagy-related gene subset is either simply downregulated by the boost or else first stimulated at 48 h NF and the stimulation then aborted by boost. In both sets of experiments, the cw15 strain is less responsive to the proposed “autophagy signal” than is the sta6 strain, possibly because it is less stressed due to its starch reserves. These data suggest that the boost is able, for unknown reasons, to signal to the sta6 strain that it is not necessary to pursue the autophagy program, thereby enabling the cells to follow the path to full obesity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bill Snell (UT Southwestern) for helpful discussions of his RNA-Seq results.
This work was supported by contract DE-EE0003046 (to U.G., S.J., S.S.M., and M.P.) via the National Alliance for Advanced Biofuels and Bioproducts (NAABB) from the U.S. Department of Energy (DOE) and by Cooperative Agreement DE-FC02-02ER63421 from the DOE Office of Science (BER) to the Institute of Genomics and Proteomics, UCLA. Support to U.G. from grant DE-SC0006873 from the DOE Office of Science (BER), to J.-H.L. from grant 2013M1A8A1056300 from the Korea CCS R&D Center (KCRC), Korean Ministry of Science, to I.B. from training grant T32 ES015457 from the National Institutes of Health, and to S.D.G. and S.S.M. from grant R24 GM092473 from the National Institutes of Health is acknowledged.
Footnotes
Published ahead of print 28 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00013-14.
REFERENCES
- 1.Liu B, Benning C. 2013. Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24:300–309. 10.1016/j.copbio.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 2.Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J. 2012. TAG, you're it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23:352–363. 10.1016/j.copbio.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 3.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54:621–639. 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- 4.Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR. 2009. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102:100–112. 10.1002/bit.22033 [DOI] [PubMed] [Google Scholar]
- 5.Wijffels RH, Kruse O, Hellingwerf KJ. 2013. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr. Opin. Biotechnol. 24:405–413. 10.1016/j.copbio.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 6.Harris EH. 2009. The Chlamydomonas sourcebook. I. Introduction to Chlamydomonas and its laboratory use. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 7.Merchant SS, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250. 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus S, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, Gallaher S, Johnson S, Benning C, Pellegrini M, Grossman A, Merchant SS. 2012. Three acyltransferases and a nitrogen responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287:15811–15825. 10.1074/jbc.M111.334052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cakmak T, Angun P, Demiray YE, Ozkan AD, Elibol Z, Tekinay T. 2012. Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol. Bioeng. 109:1947–1957. 10.1002/bit.24474 [DOI] [PubMed] [Google Scholar]
- 10.James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price GD, Djuordjevic MA. 2011. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour. Technol. 102:3343–3351. 10.1016/j.biortech.2010.11.051 [DOI] [PubMed] [Google Scholar]
- 11.Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B, Bullard B, Sears BB, Kuo M-H, Hegg EL, Shachar-Hill Y, Shiu S-H, Benning C. 2010. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154:1737–1752. 10.1104/pp.110.165159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affecting lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9:97–106. 10.1128/EC.00203-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Msanne J, Xu D, Konda AR, Casas-Mollano JA, Awada T, Cahoon EB, Cerutti H. 2012. Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75:50–59. 10.1016/j.phytochem.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 14.Blaby IK, Glaesener AG, Mettler T, Fitz-Gibbon ST, Gallaher SD, Liu B, Boyle NR, Kropat J, Stitt M, Johnson S, Benning C, Pellegrini M, Casero D, Merchant SS. 2013. Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25:4305–4323. 10.1105/tpc.113.117580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zabawinski C, Van Den Koornhuyse N, D'Hulst C, Schlichting R, Giersch C, Delrue B, Lacroix J-M, Preiss J, Ball S. 2001. Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183:1069–1077. 10.1128/JB.183.3.1069-1077.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan J, Andre C, Xu C. 2011. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585:1985–1991. 10.1016/j.febslet.2011.05.018 [DOI] [PubMed] [Google Scholar]
- 17.Fan J, Yan C, Andre C, Shanklin J, Schwender J, Xu C. 2012. Oil accumulation is controlled by carbon precursor supply for fatty acid synthesis in Chlamydomonas reinhardtii. Plant Cell Physiol. 53:1380–1390. 10.1093/pcp/pcs082 [DOI] [PubMed] [Google Scholar]
- 18.Goodson C, Roth R, Wang ZT, Goodenough U. 2011. Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot. Cell 10:1592–1606. 10.1128/EC.05242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YJ, Jeschke GR, Roelants FM, Thorner J, Turk BE. 2012. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 32:4705–4717. 10.1128/MCB.00897-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Chiu C. 2010. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61:157–180. 10.1146/annurev-arplant-042809-112222 [DOI] [PubMed] [Google Scholar]
- 21.Ramanan R, Kim B-H, Cho D-H, Ko S-R, Oh H-M, Kim H-S. 2013. Lipid droplet synthesis is limited by acetate availability in starchless mutant of Chlamydomonas reinhardtii. FEBS Lett. 587:370–377. 10.1074/jbc.M109.097758 [DOI] [PubMed] [Google Scholar]
- 22.Shen W, Li JQ, Dauk M, Huang Y, Periappuram C, Wei Y, Zou J. 2010. Metabolic and transcriptional responses of glycerolipid pathways to a perturbation of glycerol 3-phosphate metabolism in Arabidopsis. J. Biol. Chem. 285:22957–22965. 10.1074/jbc.M109.097758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velmurugan N, Sung M, Yim SS, Park MS, Yang JW, Jeong KJ. 2013. Evaluation of intracellular lipid bodies in Chlamydomonas reinhardtii strains by flow cytometry. Bioresour. Technol. 138:30–37. 10.1016/j.biortech.2013.03.078 [DOI] [PubMed] [Google Scholar]
- 24.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. 2009. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8:1856–1868. 10.1128/EC.00272-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LML, Dismukes GC, Posewitz MC. 2010. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot. Cell 9:1251–1261. 10.1128/EC.00075-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, Billker O, Snell WJ. 2013. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27:1198–1215. 10.1101/gad.212746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon K, Han D, Li Y, Sommerfeld M, Hu Q. 2012. Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24:3708–3724. 10.1105/tpc.112.100701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sueoka N. 1960. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi [sic]. Proc. Natl. Acad. Sci. U. S. A. 46:83–91. 10.1073/pnas.46.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorman DS, Levine RP. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 54:1665–1669. 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutner SH, Provasoli L, Schatz A, Haskins CP. 1950. Some approaches to the study of the role of metals in the metabolism of microorganisms. Proc. Am. Philos. Soc. 94:152–170 [Google Scholar]
- 31.Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. 2011. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66:770–780. 10.1111/j.1365-313X.2011.04537.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, Vallon O, Cournac L. 2012. Predalgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29:3625–3639. 10.1093/molbev/mss178 [DOI] [PubMed] [Google Scholar]
- 33.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 37.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]
- 38.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. 2009. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4:e4345. 10.1371/journal.pone.0004345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaby-Haas CE, Merchant SS. 2012. The ins and outs of algal metal transport. Biochim. Biophys. Acta 1823:1531–1552. 10.1016/j.bbamcr.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.UniProt Consortium. 2011. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39:D214–D219. 10.1093/nar/gkq1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432. 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittkop T, Emig D, Truss A, Albrecht M, Böcker S, Baumbach J. 2011. Comprehensive cluster analysis with transitivity clustering. Nat. Protoc. 6:285–295. 10.1038/nprot.2010.197 [DOI] [PubMed] [Google Scholar]
- 43.Albee AJ, Kwan AL, Lin H, Granas D, Stormo GD, Dutcher SK. 2013. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtii. G3 3:979–991. 10.1534/g3.113.006338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheshire JL, Evans JH, Keller LR. 1994. Ca2+ signaling in the Chlamydomonas flagellar regeneration system: cellular and molecular responses. J. Cell Sci. 107:2491–2498 [DOI] [PubMed] [Google Scholar]
- 45.Hartzell LB, Hartzell HC, Quarmby LM. 1993. Mechanisms of flagellar excision. I. The role of intracellular acidification. Exp. Cell Res. 208:148–153 [DOI] [PubMed] [Google Scholar]
- 46.Augstein A, Barth K, Gentsch M, Kohlwein SD, Barth G. 2003. Characterization, localization and functional analysis of Bpr1p, a protein affecting sensitivity to acetic acid in the yeast Yarrowia lipolytica. Microbiology 149:589–600. 10.1099/mic.0.25917-0 [DOI] [PubMed] [Google Scholar]
- 47.Gentsch M, Barth G. 2005. Carbon source dependent phosphorylation of the Gpr1 protein in the yeast Yarrowia lipolytica. FEMS Yeast Res. 5:909–917. 10.1016/j.femsyr.2005.04.009 [DOI] [PubMed] [Google Scholar]
- 48.Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, Peltier G. 2011. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. 10.1186/1472-6750-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang NL, Huang MD, Chen TL, Huang AH. 2013. Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 161:1862–1874. 10.1104/pp.112.212514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C. 2012. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24:4670–4686. 10.1105/tpc.112.105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ham HJ, Rho HJ, Shin SK, Yoon H-J. 2010. The TGL2 gene of Saccharomyces cerevisiae encodes and active acylglycerol lipase located in the mitochondria. J. Biol. Chem. 285:3005–3013. 10.1074/jbc.M109.046946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villarejo A., Buren S., Larsson S., Dejardin A., Monne M., Rudhe C., Karlsson J., Jansson S., Lerouge P., Rolland N., et al. 2005. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7:1224–1231. 10.1038/ncb1330 [DOI] [PubMed] [Google Scholar]
- 53.Larsson C, Pahlman I, Ansell R, Rigoulet M, Adler L, Gustafsson L. 1998. The importance of the glycerol 3-phosphate shuttle during aerobic growth of Saccharomyces cerevisiae. Yeast 14:347–357. [DOI] [PubMed] [Google Scholar]
- 54.Jung T-Y, Kim Y-S, Oh B-H, Woo E. 2013. Identification of a novel ligand binding site in phosphoserine phosphatase from the hyperthermophilic archaeon Thermococcus onnumineus. Proteins 81:819–829. 10.1002/prot.24238 [DOI] [PubMed] [Google Scholar]
- 55.Beopoulos A, Chardot T, Nicaud J-M. 2009. Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 91:692–696. 10.1016/j.biochi.2009.02.004 [DOI] [PubMed] [Google Scholar]
- 56.Johnson X, Alric J. 2013. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot. Cell 12:776–793. 10.1128/EC.00318-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pérez-Pérez ME, Florencio FJ, Crespo JL. 2010. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152:1874–1888. 10.1104/pp.109.152520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Han D, Hu G, Dauvillee D, Sommerfeld M, Ball S, Hu Q. 2010. Chlamydomonas starchless mutant defective in ADP glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab. Eng. 12:387–391. 10.1016/j.ymben.2010.02.002 [DOI] [PubMed] [Google Scholar]
- 59.Marino D, Dunand C, Puppo A, Pauly N. 2012. A burst of plant NADPH oxidases. Trends Plant Sci. 17:9–15. 10.1016/j.tplants.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 60.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. 2011. Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14:691–699. 10.1016/j.pbi.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 61.Plancke C, Vigeolas H, Höhner R, Roberty S, Emonds-Alt B, Larosa V, Willamme F, Duby F, Onga Dhali D, Thonart P, Hiligsmann S, Franck F, Eppe G, Cardol P, Hippler M, Remacle C. 2014. Lack of isocitrate lyase in Chlamydomonas leads to changes in carbon metabolism and in the response to oxidative stress under mixotrophic growth. Plant J. 77:404–417. 10.1111/tpj.12392 [DOI] [PubMed] [Google Scholar]
- 62.Graham IA, Denby KJ, Leaver CJ. 1994. Carbon catabolite repression regulates glyoxylate cycle gene expression in cucumber. Plant Cell 6:761–772. 10.1105/tpc.6.5.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casal M, Paiva S, Queirós O, Soares-Silva I. 2008. Transport of carboxylic acids in yeast. FEMS Microbiol. Rev. 32:974–994. 10.1111/j.1574-6976.2008.00128.x [DOI] [PubMed] [Google Scholar]
- 64.Gentsch M, Kuschel M, Schlegel S, Barth G. 2007. Mutations at different sites in members of the Fpr1/Fun34/YaaH protein family cause hypersensitivity to acetic acid in Saccharomyces cerevisiae as well as in Yarrowia lipolytica. FEMS Yeast Res. 7:380–390. 10.1111/j.1567-1364.2006.00191.x [DOI] [PubMed] [Google Scholar]
- 65.Paiva S, Devaux F, Barbosa S, Jacq C, Casal M. 2004. Ady2P is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast 21:201–210. 10.1002/yea.1056 [DOI] [PubMed] [Google Scholar]
- 66.Sá-Pessoa J, Paiva S, Ribas D, Silva IJ, Viegas SC, Arraiano CM, Casal M. 2013. SATP (YaaH), a succinate-acetate transporter protein in Escherichia coli. Biochem. J. 454:585–595. 10.1042/BJ20130412 [DOI] [PubMed] [Google Scholar]
- 67.Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86. 10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- 68.Taxis C, Keller P, Kavagiou Z, Juhl Jensen L, Colombelli J, Bork P, Stelzer EHK, Knop M. 2005. Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J. Cell Biol. 171:627–640. 10.1083/jcb.200507168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schönknecht G, Chen W-H, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, Carr K, Wilkerson C, Rensing SS, Gagneul D, Dicenson NE, Oesterhelt C, Lercher MJ, Weber APM. 2013. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339:1207–1210. 10.1126/science.1231707 [DOI] [PubMed] [Google Scholar]
- 70.Caspar T, Huber SC, Somerville C. 1985. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 79:11–17. 10.1104/pp.79.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ragel P, Streb S, Feil R, Sahrawy M, Grazia Annunziata M, Lunn JE, Zeeman S, Mérida Á. 2013. Loss of starch granule initiation has a deleterious effect on the growth of Arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol. 163:75–85. 10.1104/pp.113.223420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Ham D, Hu G, Sommerfeld M, Hu Q. 2010. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 107:258–268. 10.1002/bit.22807 [DOI] [PubMed] [Google Scholar]
- 73.Ramundo S, Casero D, Mühlhaus T, Hemme D, Sommer F, Crèvecoeur M, Rahire M, Schroda M, Rusch J, Goodenough U, Pellegrini M, Perez-Perez ME, Crespo JL, Merchant S, Schaad O, Civic N, Rochaix JD. Conditional depletion of the chloroplast ClpP1 protein activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.