Abstract
Introduction
The aim of this study was to characterize the mutations types present in the 23S rRNA gene related to H. pylori clarithromycin-resistance strains in Spain and evaluate a novel PCR-RFLP method for detection of the most frequent point mutation in our population.
Methods
Gastric biopsies were obtained by endoscopy from patients with gastric symptoms. H. pylori was cultured according to standard microbiological procedures and clarithromycin resistance was determined by E-test. DNA extraction was performed by NucliSens platform with the NucliSens magnetic extraction reagents (bioMérieux) according to the manufacturer instructions. Analyses for point mutations in 23S rRNA gene strains were performed by sequence analysis of amplified polymerase chain reaction products. Restriction fragment length polymorphism was performed using BsaI enzyme to detect restriction sites that correspond to the mutation (A2143G).
Result
We found 42 out of 118 (35.6%) strains resistant to clarithromycin by E-test. E-test results were confirmed for the presence of point mutation in 34 (88.1%) of these strains. Mutation A2143G was found in 85.3% of the strains. Analyses with the restriction enzyme BsaI was able to confirm the presence of A2143G mutation. There were 8 H. pylori strains resistant to clarithromycin by E-test but without any point mutation in the 23 rRNA gene.
Conclusion
We conclude that PCR-RFLP is a reliable method to detect clarithromycin-resistance H. pylori strains in countries with a high prevalence of clarithromycin-resistance as Spain It may be useful before choosing regimens of H. pylori eradication.
Keywords: clarithromycin, treatment failure and resistance
INTRODUCTION
Helicobacter pylori is a gram-negative bacterium that colonizes the human stomach and infects more than half of the world’s population(1). H. pylori is associated with chronic gastritis and peptic ulcer disease and may eventually result in the development of atrophic gastritis, mucosa-associated lymphoid tissue lymphoma, or gastric cancer(1, 2). Eradication therapy is recommended for patients with peptic ulcer disease, and mucosa-associated lymphoid tissue lymphoma, atrophic gastritis, first-degree relatives of gastric cancer patients, unexplained iron deficiency anaemia, and chronic idiopathic thrombocytopenic purpura(3). The Maastricht III consensus report recommended proton pump inhibitor (PPI) or ranitidine bismuth citrate based triple regimen with clarithromycin and amoxicillin or metronidazole as first-line therapy(4, 5). However, this therapy has been investigated because of the increased erradication failure(3).
Clarithromycin is recognised as key antibiotic for H. pylori treatment, since it has the most powerful bactericidal effect in vitro compared to other available molecules. The major cause of macrolide resistance in H. pylori is the lack of binding of the macrolides to the 23S rRNA components of the bacterial ribosome due to modification of the target site by methylation or point mutations in the peptidyltransferase region of domain V of the 23S rRNA(6, 7). H. pylori contains two copies of the 23S rRNA gene(8). Several points mutations have been reported that are associated with macrolide resistance, but the most common is A-G transitions at position 2143 (A2143G)(7, 8). Unfortunately, primary clarithromycin resistance is increasing worldwide, and it is regarded as the main factor reducing the efficacy of eradication therapy(9, 10). Clarithromycin resistance assessment is currently performed by Epsilometer(11), but it requires bacterial culture. Different polymerase chain reaction (PCR)-based approaches have been developed(12, 13). The aim of this study was to detect point mutations in the peptyltransferase region of 23S rRNA gen and to evaluate a molecular method for the detection of clarithromycin resistant H. pylori isolates from Spanish patients.
METHODS
Clinical isolates
H. pylori clinical isolates (n=118), obtained from 61 children (27 male, 34 female, aged 18 years old and under) and 57 adults (13 male and 44 female, more than 18 years old) were isolated from gastric biopsies obtained during upper gastroduodenal endoscopy from two children’s hospitals (Hospital Infantil Universitario Niño Jesús and Hospital Universitario Doce de Octubre, Madrid) and an adult hospital (Hospital Universitario de la Princesa, Madrid) from May 2008 to December 2008. 76.3% of patients were born in Spain and 20.3% have been previously treated, but patients had not received PPIs or antibiotics for at least two weeks. Hospitals were H. pylori-reference centers and parents and/or patients signed an informed consent form for the endoscopy.
Biopsies were received at the Department of Microbiology (Hospital Universitario de la Princesa, Madrid) and processed within the next 3 hours. Samples for culture were placed in sterile saline solution for transport. Tissue was streaked onto both non-selective (Columbia agar, with 5% sheep blood; BioMérieux, Marcy l’Etoile, France) and selective media (Pylori agar; BioMérieux, Marcy l’Etoile, France) incubated 10 days at 37°C in a microaerophilic atmosphere (5% O2, 10% CO2, 85% N2). Isolates were identified as H. pylori-based on colony morphology (small, grey and translucent), positive biochemical reactions for urease, catalase and oxidase test, and negative Gram staining. For the definition of a positive H. pylori infection status, a patient was considered positive if microbiological culture was positive.
Determination of clarithromycin resistance by phenotypic methods
Colonies of 48-hour H. pylori cultures were suspended in sterile saline and adjusted to a density equal to McFarland turbidity standard 3. The suspensions were spread onto the plates with sterile cotton swabs. The minimal inhibitory concentrations (MICs) of isolates were determined by the Epsilometer test (E-test; AB Biodisk, Solna, Sweden) on Mueller Hinton sheep blood agar (BBL, Becton Dickinson Microbiology Systems, Cockeysville, MD, USA). Plates with strip containing clarithromycin were incubated for 72 hours under microaerophilic conditions. MIC was considered the lowest concentration of drug which inhibited visible growth and read as the intercept of the elliptical zone of inhibition with the graded strip for the E-test. Based on CLSI(14) recommendations strains were resistant if MIC≥1mg/L and intermediate if MIC=0.5mg/L. Strains were susceptible below these thresholds.
DNA Extraction and PCR Amplification
The total bacterial genomic DNA was extracted by using NucliSens easyMAG instrument (BioMérieux, Inc., Durham, NC), according to manufacturer’s instructions. Briefly, sample was pre treated with lysis buffer and proteinase K overnight at 37°C. The lysed sample was then transferred to the well of a plastic vessel with 550 μl of silica. This was followed by automatic magnetic separation. Nucleic acid was recovered in 30 μl elution buffer.
To detect mutations related to clarithromycin resistance in the 23S rRNA gene, PCR amplification methods and oligonucleotide primers derived from a known sequence of the 23S rRNA gene were used (sense, 5′CCACAGCGATGTGGTCTCAG positions 1891 to 1911; antisense, 5′CTCCATAAGAGCCAAAGCCC positions 2200 to 2220). Amplification was carried out in a thermal cycler (Thermo Hybaid, MBS 0.5 S). PCR cycling conditions consisted of 35 cycles of 30 sec denaturation at 94°C, 30 sec annealing at 60°C and 30 sec extension at 72°C.
An extra PCR amplification was performed on eight strains that had discrepancies between phenotypic and genotypic methods. In order to identify new or unusual mutations that those previously published, we designed primers derived from a reference sequence of the 23S rRNA gene of H. pylori used (GenBank accession number U27270, sense, 5′TCCTCTCGTACTAGGGA positions 2057 to 2074; antisense 5′CTGCATGAATGGCGTAACGAG positions 2731 to 2752) and the PCR products were sequenced.
Sequence Analysis
PCR products were purified using a QIAquick PCR purification kit (QUIAGEN, Valencia, CA, USA). Sequencing reactions were performed by MACROGEN USA Company (Baltimore, MA, USA), and carried out in the BigDye TM terminator cycling. Computer sequence alignments were performed with CLUSTAL program (European Bioinformatics Institute [http://www.ebi.ac.uk/clustalw/]) and sequence comparisons were carried with GENEDOC program. [www.nrbsc.org/gfx/genedoc/index.htlm].
RFLP Analysis
To detect mutations at the position 2143 a PCR- restriction fragment length polymorphism (RFLP) technique was performed(15). To amplify the fragment of the peptidyltransferase region of the 23S rRNA, the primers described by Kyung et al. were used Primers K1 (5′ CCA CAG CGA TGT GGT CTC AG corresponding to position 2191 to 2615) and K2 (5′ CTC CAT AAG AGC CAA AGC CC corresponding to position 2596 to 2615) were used to amplify a fragment of 425 bp. PCR conditions were 1 cycle at 95°C for 5 min; 35 cycles of 95°C for 30 sec, 54°C for 30 sec, 72°C for 30 sec and a final step at 72°C for 10 min.
The amplified products were digested with BsaI (New England Biolabs, Inc., Beverly Mass., U.S.A) which allow discrimination between the wild type and the A2143G mutant. Two microliters of the PCR products with 8 microliters of destilated water was incubated for 24 hours with 2 microliters of enzyme BsaI at 50°C in order to detect the restriction site when the mutation was A to G at 2143, as shown in figure 1. The restriction products were analyzed by electrophoresis on 1.5% agarose gel.
Figure 1.

BsaI enzyme
RESULTS
Clarithromycin resistance
Clarithromycin resistance assessment showed that 75 patients (64.1%) were infected with susceptible strains and 42 (35.9%) with resistant bacteria, by E-test. In order to confirm the resistance to clarithromycin the correlation between E-test and sequence analyses of 23 sRNA in H. pylori strains based on clarithromycin status was studied. Overall, 34 of the 42 clarithromycin-resistant strains contained mutant 23S rRNA sequences. There was discrepancy between both methods in 8 strains: resistant by E-test, but no point mutation in the sequence. 1 strain does not have the E-test result, which was excluded from further statistical analysis. The predominant mutation among the 34 mutant strains was A2143G in 26 cases (76.58%), the A2142G in 2 cases (5.8%) and T2182C in 2 cases (5.8%), too, whilst double point mutations A2143G plus T2182C was found in 3 cases and A2142G plus T2182C was found in the remaining case, as shown in the table 1.
Table 1.
Frequency of point mutation found in 34 H. pylori clarithromycin resistant strains.
| MUTATION | N | % |
|---|---|---|
| A 2142 G | 2 | 5.8 |
| A 2143 G | 26 | 76.58 |
| T 2182 C | 2 | 5.8 |
| A 2143 G + T 2182 C | 3 | 8.8 |
| A 2142 G + T 2182 C | 1 | 2.9 |
Among the 29 resistant strains that had the point of mutation A2143G, BsaI restriction enzyme cut the PCR-products of the all strains to 304 bp and 101 bp bands, confirming that the 29 strains had an A to G mutation at position 2143.
In order to assess the specificity, this observation was confirmed by RFLP-PCR in 10 random clarithromycin-sensitive strains that do not have any point mutation and 2 strains with point mutation in position 2142. No BsaI digestion was observed in these 12 strains.
Relationship of clarithromycin resistance with host factors
The results for H. pylori isolates with different clarithromycin susceptibility patterns according to age of patient showed that H. pylori strains recovered from children were more often resistant to clarithromycin than strains isolated for adults (39.3% vs 17.5% p=0.003).
We found an association between clarithromycin susceptibility in H. pylori isolates and place of birth of the patients from whom the bacteria were isolated. H. pylori strains isolated from patients born in Spain were more often resistant to clarithromycin than strains isolated from patients born outside of Spain, (33.3% vs 14.3% p=0.05).
Analysis of a possible association between H. pylori clarithromycin resistance and gender of the patient showed that females and males were infected in almost identical proportions by resistant isolates (30.8% and 25% respectively).
DISCUSSION
Among Spanish subjects enrolled in this study, we found that the proportion of the clarithromycin resistant strains related to A2143G mutation among 23S rRNA gene was 85%, which was similar to the average reported in other studies in Spain(16).
Due to the high percentage of this mutation in our population we use a RFLP based method for detection of the most common point mutation occurring in the 23 rRNA gene of H. pylori that confer resistance to clarithromycin. This method consists in amplification of a fragment of the 23 S rRNA gene of H. pylori and the detection of the mutation in position A-G 2143 by restriction enzyme digestion.
Resistance of H. pylori to antimicrobial agents is the main cause of treatment failure(17,18), even a triple regimen, with two of the following antibiotics (clarithromycin, amoxicillin and metronidazole) plus a proton-pump inhibitors (PPI) during two weeks. These drugs are considered to be the most effective against H. pylori, but drug resistance is a growing problem. Clarithromycin is a key component of many combination therapies which were used to eradicate H. pylori(17). However, resistance to clarithromycin in H. pylori is becoming more and more common; such increase in resistant is the main reason of the failure to H. pylori eradication therapy(19).
The prevalence of mutant strains among the clarithromycin-resistant H. pylori varies in different parts of the world. In our study H. pylori resistance to clarithromycin is high, nearly 30 %. Furthermore, Spanish patients have one of the highest levels of clarithromycin resistance reported in Europe(20). In Spain, similar to other European countries, including France, Portugal, Poland, Turkey and Bulgaria, increase of resistance to clarithormycin has been observed. In Northern European countries, this increase was not observed(21). This difference probably depends on macrolide consumption. New macrolides were marketed in Spain at the beginning of the 1990, and clarithromycin in 1991. In this study, we confirmed that patients born in Spain were colonized more often with resistant strains than patients, born outside of Spain. The majority of these immigrant patients are from South America where the level of resistance to clarithromycin is lower than in the South of Europe(22, 23). Also, children were colonized more often with resistant strains. Children are exposed to macrolides constantly, and it is very frequent nowadays to treat respiratory infections in young children with this group of antibiotics(20, 24, 25).
Resistance to macrolides is caused by a decrease in binding of macrolides to the ribosome, which is associated with methylation of A2058 or A2059 (Escherichia coli gene reference numbering)(6). In 1996, Versalovic et al first reported the association of clarithromycin resistance of H. pylori with a single point mutation within the domain V of 23S rRNA(7).
Clarithromycin resistance in H. pylori is mostly due to point mutations A2142G/C, A2143G in the peptidyltransferase-encoding region of 23S rRNA gene, mutations at positions cognate with E. coli 23S rRNA positions 2058 and 2059. The prevalence of the A2143G mutation has been reported as 44% to 67%, however studies in Japan showed that more than 80% of the mutant strains are in the position A2143G, as in our study(26).
Other mutations in the 23S rRNA such as A2115G, G2141A, C2147G, T2190C, C2195T, A2223G and C2694A have been described and associated with clarithromycin resistance(27, 28). In our study, the strong association between resistance to macrolides and specific mutations in the 23S rRNA was confirmed in 81% of our cases. However, there were discrepancies between sequence analysis of 23S rRNA and clarithromycin resistance tested by E-test in 8 strains. All 8 strains were resistant by E-test but did not present any of the point mutations described, in the 23S rRNA gene sequence. PCR amplification was performed in order to analyze the conserved 695 bp region of H. pylori 23S rRNA to look for new point of mutations which have been described. We did not find any point mutation in the strains with phenotype of resistant to clarithromycin. Based on those results we suggested that other mechanisms which are not related to 23S rRNA could be involved in the resistance to clarithromycin in those strain, such as efflux pump has been described by other authors(29).
The detection of point mutations in the 23S rRNA gene associated with resistance to clarithromycin has significantly improved macrolide susceptibility testing H. pylori isolates. Knowledge about the molecular mechanism of resistance is important as it can be used to facilitate the development of other molecular methods to detect resistance(7, 12, 13, 15). It has been shown that PCR-RFLP can be used to detect mutations in clarithromycin-resistant isolates. Using this technique we have demonstrated that only strains with the A2143G mutations produced restriction fragments when digested or BsaI. This alleviated the need to use the more expensive and time-consuming processes.
In conclusion, A2143G was the main point mutation in the 23S RNAr gene. PCR-RFLP assay permits accurate, fast, and cost-effective detection of susceptibility of the strain to clarithromycin.
Figure 2.
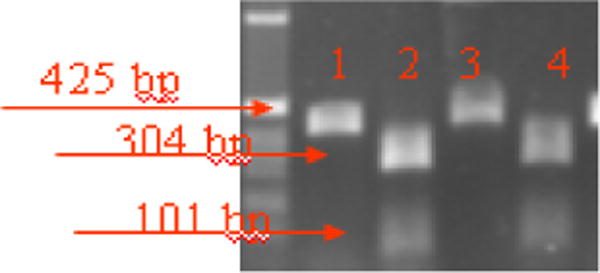
RFLP analysis of 23S rRNA to detect the A2143G using BsaI restriction enzyme.
Lanes 1 and 3: Digestion products of two clarithromycin-resistant isolates without enzyme
Lanes 2 and 4: Digestion products of two clarithromycin resistant strains with BsaI
Acknowledgments
This study was supported in part by Fondo de Investigaciones Sanitatias FIS 05/2442 and FIS 05/2452 and NYU Center for the Study of Asian American Health, NIH grant support (p60MD000538-05).
Bibliography
- 1.Shanks AM, El-Omar EM. Helicobacter pylori infection, host genetics and gastric cancer. J Dig Dis. 2000;10:157–164. doi: 10.1111/j.1751-2980.2009.00380.x. [DOI] [PubMed] [Google Scholar]
- 2.Chey WD, Wong BC. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol. 2007;102:1808–25. doi: 10.1111/j.1572-0241.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 3.O’Connor A, Gisbert J, O’Morain C. Treatment of Helicobacter pylori infection. Helicobacter. 2009;1:46–51. doi: 10.1111/j.1523-5378.2009.00704.x. [DOI] [PubMed] [Google Scholar]
- 4.Mana F. The Maastricht III consensus: summary and comments. Man Acta Gastroenterol Belg. 2009;72:344–49. [PubMed] [Google Scholar]
- 5.Malfertheiner P, Megraud F, O’Morain C, Bazzoli F, El-Omar E, Graham D, Hunt R, Rokkas T, Vakil N, Kuipers E. Concepts in the management of Helicobacter pylori infection. The Maastricht III Consensus Report. Gut. 2007;56:772–81. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman RC, Zakula R, Flamm J, Beyer, Capobianco J. Tight binding of clarithromycin, its 14-(R)-hydroxy metabolite and erythromycin to Helicobacter pylori ribosome. Antimicrob Agents Chemother. 1994;38:1496–500. doi: 10.1128/aac.38.7.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verlasovic J, Osato M, Spakovsky, Dore M, Reddy R, Stone G, Shortridge D, Flamm R, Tanaka D, Graham D. Point of mutations in the 23S rRNA gen of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–86. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 8.Taylor D, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and Sequence Analysis of Two copies of a 23 S rRNA Gene from Helicobacter pylori and Association of Clarithromycin Resistance with 23S RNAr Mutations. Antimicrob Agents Chemother. 2008;41:2621–28. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francesco V, Zullo A, Lerardi E, Vaira D. Minimal inhibitory concentration (MIC) values and different point mutations in the 23S rRNA gene for clarithromycin resistance in Helicobacter pylori. Digestive and Liver Disease. 2009;41:610–611. doi: 10.1016/j.dld.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Megraud F. H. pylori antibiotic resistance: prevalence, importance and advantages in testing. Gut. 2004;53:1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agudo S, Alarcón T, Urruzuno P, Martíez MJ, López-Brea M. Detection of Helicobacter pylori and clarithromycin resistance in gastric biopsies of paediatric patients by using a commercially available real-time polymerase chain reaction after NucliSens semiautomated DNA extraction. Diagn Microbiol Infect Dis. 2010;67:213–9. doi: 10.1016/j.diagmicrobio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L, Soussy CJ, Delchier JC, Megraud F. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;7:3600–07. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement M100-S17. CLSI; Wayne, PA, USA: 2009. [Google Scholar]
- 15.Kim KS, Kang JO, Eun CS, Han DS, Choi TY. Mutations in the 23S rRNA Gene of Helicobacter pylori associated with clarithromycin resistance. J Korean Med Sci. 2002;17:599–633. doi: 10.3346/jkms.2002.17.5.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alarcón T, Vega E, Domingo D, et al. Clarithromycin resistance among Helicobacter pylori strains isolated from children: Prevalence and Study of Mechanism of Resistance by PCR-Restriction Fragment Length Polymorphism Analysis. J Clin Microbiol. 2003;41:486–88. doi: 10.1128/JCM.41.1.486-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francesco V, Margiotta M, Zullo A, et al. Prevalence of primary clarithromycin resistance in Helicobacer pylori strains over a 15 year period in Italy. J Antimicrobial Chemother. 2007;59:783–5. doi: 10.1093/jac/dkm005. [DOI] [PubMed] [Google Scholar]
- 18.Graham D, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat Clin Pract Gastroenterol Hepatol. 2008;5:321–31. doi: 10.1038/ncpgasthep1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanches B, Coelho L, Moretzsohn L, Vieira G. Failure of Helicobacter pylori treatment after regimes containing clarithromycin: new practical therapeutic options. Helicobacter. 2008;13:572–6. doi: 10.1111/j.1523-5378.2008.00649.x. [DOI] [PubMed] [Google Scholar]
- 20.Agudo S, Alarcón T, Cibrelus L, Urruzuno P, Martínez M, López-Brea M. High percentage of clarithromycin and metronidazole resistance in Helicobacter pylori clinical isolates obtained from Spanish children. Rev Esp Quimioter. 2009;22:88–92. [PubMed] [Google Scholar]
- 21.Koletzko S, Richy F, Bontems P, Crone J, Kalach N, Monteiro ML, Gottrand F, Celinska-Cedro D, Roma-Giannikou E, Orderda G, Kolacek S, Urruzuno P, Martínez-Gómez MJ, Casswall T, Ashorn M, Bodanszky H, Mégraud F. Prospective multicentre study on antibiotic resistance of Helicobacter pylori strains obtained from children living in Europe. Gut. 2006;55:1711–16. doi: 10.1136/gut.2006.091272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Álvarez A, Moncayo J, Santacruz J, Santacoloma M, Corredor L, Reinosa E. Antimicrobial Susceptibility and Mutations involved in Claritrhromycin Resistance In Helicobacter pylori Isolates from Patients in the Western Central Region of Colombia. Antimicrobl Agent Chemother. 2009;53:4022–4024. doi: 10.1128/AAC.00145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fariña N, Kasamatsu E, Samudio M, Morán M, Sanabria R, Laspina Antimicrobial susceptibility of H pylori strains obtained from Paraguayan patients. Rev Med Chil. 2007;135:1009–1014. doi: 10.4067/s0034-98872007000800008. [DOI] [PubMed] [Google Scholar]
- 24.López-Brea, Martínez MJ, Domingo D, Alarcón T. A 9 year study of clarithromycin and metronidazole resistance in Helicobacter pylori from Spanish children. J Antimicrob Chemother. 2001;48:295–297. doi: 10.1093/jac/48.2.295. [DOI] [PubMed] [Google Scholar]
- 25.Prieto J, Calvo A, Gómez-Lus ML. Antimicrobial resistance: a class effect? J Antimicrob Chemother. 2002;50:7–12. doi: 10.1093/jac/dkf508. [DOI] [PubMed] [Google Scholar]
- 26.Omata F, Uemura M, Suzuki S, Ishii N, Iizuka Y, Fukuda K, Fujita Y, Katsurahara M, Ito T, Cesar GE, Imoto I, Takei Y. Annual change of primary resistance to clarithromycin among Helicobacter pylori isolates from 1996 through 2008 in Japan. Helicobacter. 2009;14:86–90. doi: 10.1111/j.1523-5378.2009.00714.x. [DOI] [PubMed] [Google Scholar]
- 27.Kim, Mogg J, Kim J, Kim N, Kim Y, Kim IY, Chee Y, Lee CH, Jung HC. New mutations of 23S RNAr associated with Clarithromycin resistance in Helicobacter pylori Strains Isolated from Korean Patients. J Microbiol Biotechnol. 2008;18:1584–1589. [PubMed] [Google Scholar]
- 28.Rimbara E, Kawai T, Sasatsu M. Novel mutation in 23S RNAr that confers Low-Level Resistance to clarithromycin in Helicobacter pylori. Antimicrob Agents Chemother. 2008;52:3465–3466. doi: 10.1128/AAC.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Zheng P, Yang P. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastronterol. 2008;14:5217–5222. doi: 10.3748/wjg.14.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]


