Abstract
Purpose of review
Diffuse large B cell lymphoma (DLBCL) is an aggressive disease featuring heterogeneous genetic, phenotypic and clinical characteristics. Understanding the basis for this heterogeneity represents a critical step toward further progress in the management of this disease, which remains a clinical challenge in approximately one third of patients. This review summarizes current knowledge about the molecular pathogenesis of DLBCL, and describes how recent advances in the genomic characterization of this cancer have provided new insights into its biology, revealing several potential targets for improved diagnosis and therapy.
Recent findings
In the past few years, the development of high-resolution technologies has provided significant help in identifying genetic lesions and/or disrupted signaling pathways that are required for DLBCL initiation and progression. These studies uncovered the involvement of cellular programs that had not been previously appreciated, including histone/chromatin remodeling and immune recognition. Alterations in these pathways could favor epigenetic reprogramming and escape from cellular immunity.
Summary
The identification of genetic alterations that contribute to the malignant transformation of a B cell into a DLBCL is helping to better understand the biology of this disease and to identify critical nodes driving tumor progression or resistance to therapy. The rapid pace at which these discoveries are taking place is poised to have significant impact for patients stratification based on molecular predictors and for the development of rational targeted therapies.
Keywords: DLBCL, genomic analysis, epigenetic modifiers, germinal center, immune escape
Introduction
Diffuse large B cell lymphoma (DLBCL) represents the most prevalent B cell non-Hodgkin lymphoma (B-NHL) in adulthood, accounting for 30–40% of diagnoses [1]. Although durable remissions can be achieved in a substantial proportion of cases by using combination immuno-chemotherapy, outcome is often unpredictable and over 30% of patients do not respond to available therapeutic regimens, underscoring the need for an improved understanding of its biology. It is now clear that one of the obstacles to therapeutic success in DLBCL is the heterogeneous nature of this disease, which can be appreciated at the morphologic, genetic, phenotypic, and clinical level. Indeed, genome-wide expression profile (GEP) studies over the past decade revealed that this single diagnostic entity comprises several distinct molecular subgroups, which differ in the expression of specific gene signatures and also in the oncogenic pathways that appear to be involved, often corresponding to discrete prognostic categories [2, 3]. Thus, the recognition of genes that are aberrantly regulated by structural alterations provides an important molecular framework for the development of more specific markers for risk stratification and rationally targeted therapeutic approaches.
This review summarizes current knowledge about the molecular pathogenesis of DLBCL, with emphasis on the recent identification of epigenetic remodeling and immune regulatory genes as common targets of genetic alterations in this disease.
Molecular heterogeneity of DLBCL
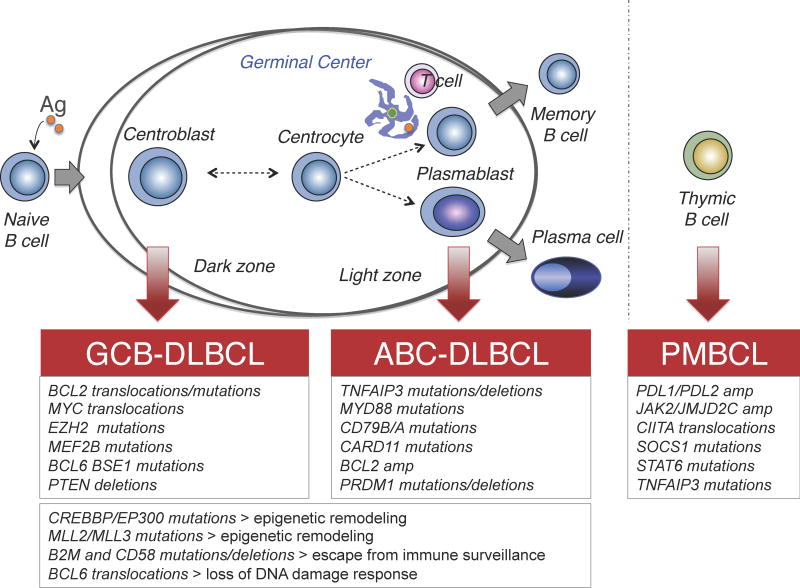
The advent of GEP technologies a decade ago allowed the recognition of multiple molecularly distinct DLBCL subgroups, reflecting the derivation from cells with discrete biological programs during normal B cell differentiation or the coordinated expression of comprehensive consensus clusters (CCC). Based on similarities with their putative cell of origin (COO), DLBCL have been categorized into at least three subgroups: i) germinal center B cell-like (GCB) DLBCL, presumably derived from a GC centroblast expressing high levels of the master regulator BCL6 and harboring highly mutated immunoglobulin genes with ongoing somatic hypermutation (SHM) [2, 3]; ii) activated B cell-like (ABC) DLBCL, whose cell of origin is related to a BCR-activated B cell or a plasmablastic B cell in the light zone of the GC [4]; and iii) primary mediastinal large B-cell lymphoma (PMBCL), postulated to arise from a thymic post-GC B cell (Figure 1) [3]. Of note, multiple genetic lesions of pathogenic significance segregate with different DLBCL subtypes, suggesting that these tumors rely on distinct oncogenic pathways [3]. The central roles of the involved pathways during distinct phases of the GC response indicate that lymphoma cells exploit the unique properties of their normal counterpart to their own selective advantage. Importantly, stratification of DLBCL patients according to the COO classification was shown to be predictive of different overall survival rates, with ABC-DLBCL representing the less curable subgroup [2].
Figure 1. Postulated normal counterpart of major DLBCL subtypes.
Schematic cartoon of the GC reaction, illustrating its relationship with major DLBCL subtypes. GCB-DLBCLs display phenotypic similarities with proliferating centroblasts, while ABC-DLBCLs appear to be related to a plasmablastc B cell; PMBCL is postulated to arise from a post-GC B cells in the thymic medulla. The most common genetic lesions that are associated with individual molecular subtypes, or shared by multiple subtypes, are indicated below.
The resemblance of distinct DLBCL subtypes to their presumed cell of origin is not the only aspect of the disease that can be captured by GEP. A separate classification schema using multiple clustering algorithms identified three discrete subgroups defined by the expression of genes involved in oxidative phosphorylation (OXP), B-cell receptor/proliferation (BCR), and tumor microenvironment/host inflammatory response (HR)[5]. Interestingly, BCR-DLBCL cell lines were uniquely sensitive to inhibition of BCR survival signaling, as well as to blockade of BCL6 activity by a specific peptide inhibitor, suggesting that these patients may be amenable to BCL6-targeted therapy [6–8]. Conversely, OxPhos-DLBCLs were found to be susceptible to approaches that inhibit mitochondrial fatty acid oxidation and glutathione synthesis [9]. While highly reproducible, COO-defined and CCC-defined molecular subtypes do not overlap, underscoring the complexity of this disease and highlighting the need for targeted medicine approaches to treating these patients.
The Genome of DLBCL
Over the past few years, the rapid expansion of novel sequencing technologies has provided an unprecedented opportunity to examine the cancer genome in a comprehensive and unbiased manner. The integration of whole-transcriptome or whole-exome next-generation sequencing studies and high-density SNP array analysis, together with functional shRNA screens, allowed significant progress in our understanding of DLBCL pathogenesis. These efforts revealed that the coding genome of DLBCL has a high degree of complexity compared to other B cell malignancies, harboring on average between 30 and >100 lesions/case, with great variability across patients [10–12]. Sequencing of the DLBCL coding genome also uncovered several previously unrecognized genes/pathways that are involved by genetic lesions in this disease. The observed lesions, which include mostly single nucleotide variants and copy number losses, with fewer amplifications and translocations [10–13], likely represent an underestimate of the DLBCL mutation load, since the sequencing methods used do not interrogate non-coding portions of the genome; as such, these studies are not informative about the activity of aberrant SHM, a mechanism of genetic damage that is due to an aberrant function of the physiologic IgV-associated SHM mechanism, and results in the accumulation of multiple mutations in the 5′ sequences of genes that are normally unaffected in GC B cells [14]. Indeed, a targeted approach integrating bioinformatics tools with the sequencing of gene regulatory regions marked by H3K4me3 uncovered increased numbers of mutation hotspots within genes that had not been previously reported as mutated [15]. Next-generation sequencing studies also confirmed that mutations of TP53 are relatively less common in DLBCL than in other epithelial malignancies [10–12]; however, a combined analysis of copy number data and transcriptional profiles revealed a complementary set of alterations that may lead to decreased p53 levels and activity and cell cycle deregulation [16]. While, for some of the identified candidates, a detailed functional characterization is still missing, these studies provided a significant gain in our knowledge of DLBCL biology, leading to the discovery of potential targets for therapy. Complementing the genetic findings, shRNA screens identified addiction of the tumor cells to several oncogenically active circuits –e.g., NF-κB and BCR– that are currently being tested as actionable targets [3].
Genomic lesions disrupt key cellular pathways in DLBCL
The molecular heterogeneity of DLBCL is reflected in the broad catalogue of structural aberrations that are associated with its pathogenesis. These include abnormalities that are common to various phenotypic subtypes, and lesions that appear to be preferentially or even exclusively associated with individual COO-defined DLBCL categories, pointing to distinct oncogenic pathways. The following paragraphs will focus on recently identified targets of genetic lesion in DLBCL, as emerged from genomic analyses. Other recurrent and historically well-characterized structural alterations have been extensively reviewed, and will thus be mentioned only briefly.
Alterations in epigenetic modifiers
One of the most relevant findings from recent DLBCL genome studies is the high frequency of lesions affecting histone/chromatin modification enzymes, most commonly acetyl-transferases and methyltransferases.
Acetyltransferases
Mutations and/or deletions inactivating CREBBP and, less frequently, EP300 are detectable in up to 35% of DLBCL (with some preference for the GCB-subtype) and 40% of follicular lymphoma (FL) [11, 12]. These ubiquitously expressed nuclear phosphoproteins modify lysine residues on both histone and non-histone nuclear proteins, serving as transcriptional coactivators for a large number of DNA-binding transcription factors [17]. Consistently, CREBBP/EP300 enhance transcription through various mechanisms, including targeted acetylation of chromatin [18, 19], acetylation of transcriptional activators (e.g., the tumor suppressor p53) [20–22] and acetylation-mediated inactivation of transcriptional repressors (e.g. BCL6)[23].
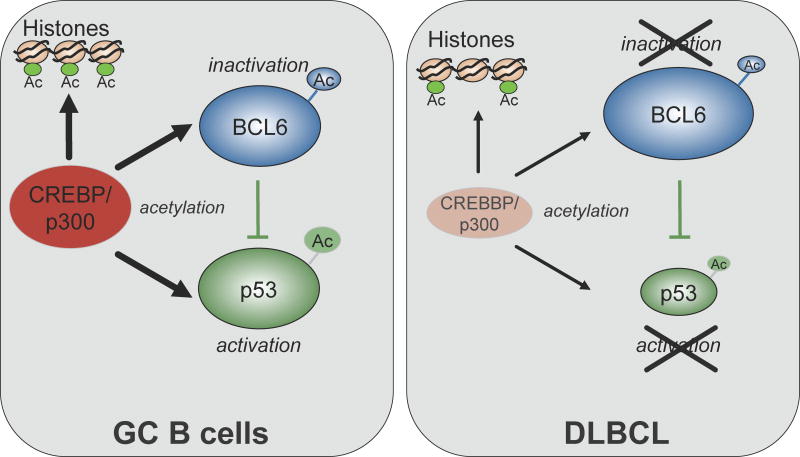
CREBBP mutations lead to truncations of the C-terminal HAT domain or to amino acid changes that impair its ability to acetylate known substrates such as BCL6 and p53, leading to constitutive activation of the oncoprotein and to decreased p53 tumor suppressor function (Figure 2) [24]. Since the balance between the activities of these two genes is critical for the regulation of DNA damage responses during immunoglobulin genes remodeling in the GC [25], one consequence of BCL6 activity overriding p53 would be an increased tolerance for DNA damage in the context of impaired apoptotic and cell cycle arrest responses (Figure 2). However, the general involvement of histone acetyltransferases in gene transcriptional regulation suggests that the consequences of CREBBP/EP300 alterations are much broader, warranting additional studies aimed at dissecting the entire set of cellular targets that are critically affected by HAT dose reduction in lymphoma. With few exceptions, alterations of CREBBP and EP300 are observed in heterozygosis, and coexist with the expression of the residual wild-type allele, suggesting a haploinsufficient tumor suppressor role [24]. Indeed, the dose-dependent pathogenic effect of these two genes is demonstrated by the causative role of CREBBP and EP300 haploinsufficiency in a rare congenital disorder known as Rubinstein-Taybi syndrome [26]. The identification of mutations in CREBBP and EP300 may have direct therapeutic implications in view of current attempts to use histone deacetylase inhibitors (HDAC) as anti-cancer drugs. Interestingly, p300 and its cofactor HLA-B-associated transcript 3 are both direct targets of BCL6 mediated transcriptional repression, and could be re-induced in DLBCL cell lines by a retro-inverso BCL6 inhibitory peptide (RI-BPI), resulting in acetylation of its critical targets p53 and Hsp90 [27]. Consistently, combination of RI-BPI with either an HDAC inhibitor or an Hsp90 inhibitor suppressed or even eradicated human DLBCL xenografts in mice [27].
Figure 2. Deregulation of the BCL6-p53 axis by inactivating mutations of CREBBP/EP300.
In normal B cells, CBP-mediated acetylation of BCL6 leads to inactivation of its transrepression function, while the same post-translational modification represents an essential requirement for activation of the p53 tumor suppressor functions (left panel). TP53 is also a direct target of BCL6 transcriptional repression in GC B cells, a mechanism that allows the execution of DNA remodeling events such as CSR and SHM, without eliciting DNA damage responses. This fine balance is disrupted in nearly one third of DLBCL, owing to the presence of mutations and/or deletions in CREBBP and, less frequently, EP300 (right panel). Deregulation of the BCL6 oncoprotein and impairment of the p53 tumor suppressor function represent two of presumably many mechanisms by which CREBBP/EP300 genetic lesions contribute to DLBCL transformation.
Histone methyltransferases
Among the newly identified candidates, MLL2 (mixed-lineage leukemia 2, previously termed MLL4) emerged as the single most common target of mutations in B-NHL, accounting for over 30% of DLBCL and 89% of FL [11, 12]. MLL2 encodes a highly conserved and ubiquitously expressed histone methyltransferase that controls gene transcription by modifying the lysine-4 position of Histone 3 (H3K4) [28]. H3K4 trimethylation represents an evolutionarily conserved mark of transcriptionally active chromatin and is closely associated with promoters and early transcribed regions of active genes, where it counteracts the repressive chromatin environment imposed by H3K9 and H3K27 methylation [28]. More recently, the Drosophila homolog of the mammalian MLL2/3 complex –thritorax related (trr)– was shown to function as a major H3K4 monomethyltransferase on enhancers, suggesting a critical function in regulating the transition from inactive/poised to active enhancers [29]. DLBCL-associated MLL2 mutations generate truncated proteins that lack the catalytic SET domain required for its methyltransferase activity, implicating a tumor suppressor role. In support of this hypothesis, Drosophila trr mutant cells display a significant growth advantage over their wild-type neighbors as the result of changes in the expression of multiple proteins regulating cell division and growth [30]. Of note, its paralogue MLL3 was also found mutated or deleted in ~15% of DLBCLs [12, 31], pointing to a selective role for this complex during transformation.
The polycomb-group oncogene EZH2 encodes for a histone methyltransferase that represses gene expression by trimethylating H3K27. In ~22% of GCB-DLBCL patients, gain-of-function mutations result in the replacement of a single evolutionary conserved tyrosine residue (Tyr641) within the EZH2 SET domain [32], leading to increased H3K27me3 levels through altered substrate specificity in vitro [33, 34]. EZH2 may represent a promising new therapeutic target in DLBCL, as suggested by several recent studies reporting the development of selective small molecule EZH2 inhibitors capable of inducing cell cycle arrest and apoptosis specifically in EZH2-mutated DLBCL cells [35–37].
Together, alterations in chromatin remodeling genes may impose significant effects on transcriptional regulation, and contribute to lymphomagenesis by epigenetically reprogramming the lymphoma cell.
Pathways leading to immune escape
A second set of lesions recurrently observed in both molecular subtypes of DLBCL involve immune recognition and antigen presenting functions. In ~30% of patients, biallelic inactivating mutations and focal deletions disrupt the β2-microglobulin (B2M) gene, which encodes an invariant subunit of the major histocompatibility complex (MHC) class I on the surface of nearly all nucleated cells [38]. Furthermore, 75% of DLBCLs lack membrane B2M expression, which is required for the assembly of HLA class I molecules and the recognition by cytotoxic T lymphocytes (CTL) [38]. The observation that the fraction of cases lacking surface B2M and HLA-I expression is much higher than that predicted by genetic alterations suggests that additional epigenetic mechanisms are involved. Focal homozygous deletions and truncating mutations were also frequently detected in the CD58 locus (~20% of cases) [38]. CD58 is a member of the immunoglobulin superfamily and functions as ligand of the CD2 protein on T lymphocytes and natural killer cells, participating in their activation [39]. The concurrent absence of B2M/HLA-I and CD58, observed in ~60% of DLBCLs, strongly suggests that these alterations are co-selected during lymphomagenesis for their complementary roles in protecting from both CTL- and NK cell-mediated lysis.
Other lesions affecting regulators of immune responses include genomic breakpoints and mutations of the MHC class II transactivator gene CIITA, as well as amplifications and breaks of the genes encoding for the immunomodulatory proteins PDL2 and PDL1, all of which are preferentially observed in PMBCL [40–42]. By downregulating surface HLA class II expression, rearrangements of CIITA may reduce tumor cell immunogenicity, whereas amplifications of the PDL1 locus have been linked to impaired anti-tumor immune responses in several cancers.
The ability of the above lesions to interfere with the interaction between tumor cells and the microenvironment suggests that escape from immune-surveillance mechanisms plays a central role in DLBCL.
Genetic lesions leading to constitutive NF-κB activity and chronic active BCR signaling
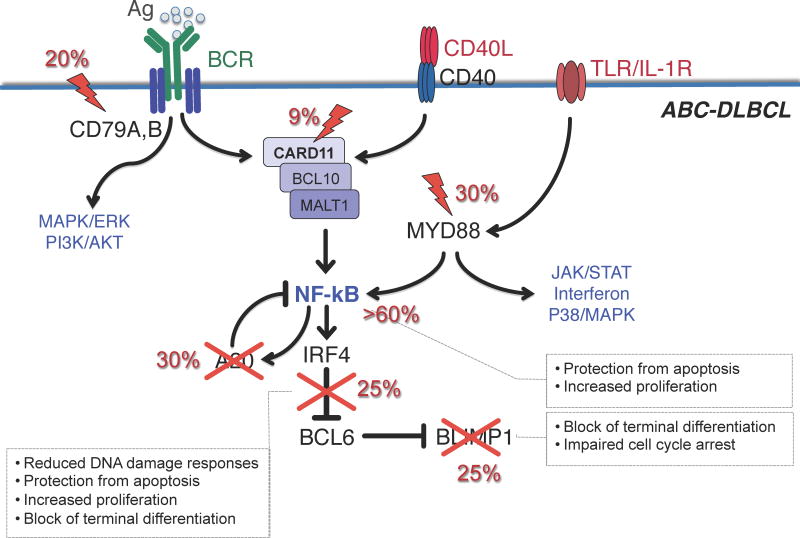
The critical role of NF-κB in the pathogenesis of ABC-DLBCL was first revealed by the observation that the gene expression signature of these tumors is significantly enriched in NF-κB target genes, and that NF-κB activity is required for their proliferation and survival [43]. Following on this lead, a number of studies have provided direct genetic evidence for the presence of structural alterations that affect a variety of pathways whose common denominator is the ability to induce constitutive activation of NF-κB, including those triggered by the B cell receptor (BCR), Toll-like receptor (TLR)/interleukin-1 receptor (IL-1R) and CD40 receptor (Figure 3).
Figure 3. Pathway lesions in ABC-DLBCL.
Schematic representation of a GC centrocyte, expressing a functional surface BCR, CD40 receptor and TLR/IL-1R. Engagement of these signaling cascades in normal B cells converge on the activation of the NF-κB transcription complex, and induces the expression of hundreds of targets genes, including IRF4 and the NF-κB negative regulator TNFAIP3/A20. IRF4, in turn, downregulates BCL6 expression, allowing the release of the plasma cell master regulator PRDM1 and the development into a differentiated plasma cell. In DLBCL, a variety of genetic lesions disrupt this circuit at multiple levels specifically in the ABC-subtype, and contribute to lymphomagenesis by favoring the anti-apoptotic function of NF-κB while blocking terminal B cell differentiation. Crosses indicate inactivating mutations/deletions; lightning bolts denote activating mutations. Modified with permission from [12]
CD79A/B mutations
Tonic BCR signaling is required for the manteinance of all B cells [44]; however, ABC-DLBCL depend on a chronically active form of BCR signaling, which is sustained in >20% of cases by hotspot mutations in the cytoplasmic immunoreceptor tyrosine-based activation motifs of the Ig superfamily members CD79B and CD79A [45]. Importantly, knockdown of several BCR proximal and distal subunits is specifically toxic to ABC-DLBCL, offering the rationale for the development of targeted therapies aimed at inhibiting BCR signaling in this lymphoma subtype [45]. Indeed, use of the BTK inhibitor ibrutinib is proving significantly effective in ABC-DLBCL patients carrying CD79A/B mutations, and synergized with lenalidomide to block IRF4 in most ABC-DLBCL cell lines [46]. Likewise, a small molecule inhibitor of MALT1 displayed selective activity against ABC-DLBCL cell lines both in vitro and in xenotransplanted tumors [47].
Oncogenic MYD88 mutations
Approximately 30% of ABC-DLBCL patients display a recurrent amino acid change (L265P) in the intracellular Toll/interleukin-1 receptor domain of this adaptor molecule, which has the potential to activate NF-κB as well as JAK/STAT3 transcriptional responses [3, 48]. Although the relationship between MYD88 mutations and TLR signaling has not been studied, the requirement of MYD88 for the survival of ABC-DLBCLs implicates a pathogenic role for TLR in this subtype.
Alterations in negative and positive regulators of NF-kB
In up to 30% of ABC-DLBCL cases (and a smaller fraction of GCB-DLBCL), disruptive mutations and/or focal deletions biallelically inactivate the gene encoding for TNFAIP3/A20 [49, 50], a dual function ubiquitin modification enzyme involved in the termination of NF-κB, TLR and BCR responses, the latter through its ability to interact with MALT1. Loss of A20 could thus promote lymphomagenesis at least in part by inducing inappropriately prolonged NF-κB signaling [49, 50]. Another ~9% of ABC-DLBCLs harbor oncogenically active mutations of CARD11, a major component of the “signalosome” complex, which is required for proper transduction of BCR signaling to NF-κB [51]. These events cluster within the protein coiled-coil domain and augmented its ability to mediate NF-κB activity [51].
In conclusion, distinct signaling pathways are disrupted by genetic lesions in most ABC-DLBCLs. While a common outcome of these lesions is the activation of NF-κB responses, the same signals can trigger a number of other downstream signaling cascades, including PI3K, ERK/MAP kinase and NF-AT (Figure 3). Thus, future studies will have to address the relative or coordinate contribution of these lesions to DLBCL development.
Deregulation of BCL6 activity
BCL6 is a transcriptional repressor that binds to specific sequences in the promoter of its target genes and modulates their expression via interaction with distinct co-repressor complexes [52]. BCL6 controls the biological program of GC B cells by suppressing a broad set of genes involved in multiple signaling pathways, including BCR and CD40 signaling, T-cell mediated B-cell activation, the sensing and response to DNA damage (via direct repression of TP53 and ATR), the induction of apoptosis (via repression of BCL2), and the differentiation to plasma cells (via suppression of PRDM1/BLIMP1)[52]. These activities sustain the proliferative status of GC B cells while allowing them to tolerate the DNA breaks that are associated with SHM and class switch recombination, without eliciting DNA damage responses. Moreover, BCL6 prevents the premature B cell activation and exit from the GC prior to the selection for the survival of clones producing high affinity antibodies [52].
Chromosomal rearrangements of the BCL6 locus are present in ~35% of DLBCL patients, with higher frequencies in the ABC-DLBCL subtype [3, 52]. These recombination events juxtapose the intact coding domain of BCL6 downstream to heterologous sequences derived from over 20 alternative chromosomal partners. Because of the broader window of activity of these alternative promoters throughout B-cell development, BCL6 translocations prevent the downregulation of BCL6 expression that is normally associated with differentiation into post-GC B cells [4, 52].
In addition to translocations, the BCL6 5′ sequences are targeted by hypermutation in up to 75% of DLBCLs [53, 54]. This phenomenon largely reflects the activity of the physiologic SHM mechanism operating in GC B cells [54, 55]. However, a subset of mutations found specifically in lymphoma affect two BCL6 binding sites located within the first noncoding exon of the gene, and deregulate its expression by disrupting a negative autoregulatory circuit by which the BCL6 protein controls its own transcription [56, 57]. Less frequently, mutations prevent the IRF4-mediated BCL6 repression induced upon CD40:CD40L interaction in the GC light zone (Figure 3) [52].
A separate set of genetic lesions deregulate BCL6 activity by interfering with post-transcriptional regulatory mechanisms: this is the case of CREBBP/EP300 mutations, which impair acetylation-mediated inactivation of BCL6 (see above)[24], and FBXO11 mutations (5% of cases), which inactivate this ubiquitin ligase involved in the control of BCL6 proteasomal degradation via the SKP1-CUL1-SCF complex [58]. Finally, 10–18% of patients, mostly of the GCB-subtype, harbor gain-of-function mutations in MEF2B [10–12], a transcription factor expressed specifically in the GC and recently shown to act as a positive regulator of BCL6 transcription [59].
The critical role of BCL6 in initiating lymphomagenesis has been confirmed in a mouse model in which deregulated BCL6 expression promotes the development of human-like DLBCL [60]. As such, BCL6 represents an attractive target for therapeutic intervention; indeed, BCL6 inhibitory molecules have shown potent anti-lymphoma activity and strong synergism in DLBCL-directed combinatorial therapies [6, 27, 61].
Disruption of the terminal differentiation pathway
In GC B cells undergoing terminal differentiation, BCL6 must be downregulated in order to relieve the expression of its target gene and plasma cell master regulator PRDM1/BLIMP1 [62]. This regulatory axis is disrupted in the majority of ABC-DLBCLs due to mutually exclusive genetic lesions that inactivate BLIMP1 directly –by disruptive mutations and deletions (25% of cases)– or indirectly –through its constitutive repression by deregulated BCL6 (25% of cases) (Figure 3)[63, 64]. BCL6 translocations and PRDM1 inactivation may thus represent alternative oncogenic mechanisms converging on the same pathway to promote lymphomagenesis by blocking terminal differentiation. Consistent with this, conditional deletion of Blimp1 in the GC leads to lymphoproliferative diseases akin to the human ABC-DLBCL[63].
Conclusion
Over the past few years, significant efforts have been focusing on the identification of molecular signatures and genetic alterations that might help in stratifying DLBCL patients with different prognosis and response to therapy. Targeted resequencing and genomic profiling have led to the discovery of important new genetic lesions, revealing the involvement of several previously unrecognized genes and pathways that may play critical roles in DLBCL pathogenesis. Some of these (e.g. NF-κB, BCR and BCL6) are already being exploited as potential targets for clinical application, while others represent highly promising candidates (e.g., EZH2). While it is likely that a broader picture of the DLBCL genome will become available soon, major efforts will be needed in the future to characterize the functional significance of the identified lesions, and their specific contribution to lymphoma development. These findings are expected to have major implications for the development of new diagnostic tests, and to inspire the design of rationale treatment approaches that could improve the survival of lymphoma patients with minimal toxicity.
KEY POINTS.
Genomic analysis of DLBCL reveals a high degree of complexity compared to other hematologic malignancies, with great variability across individual patients
Recurrent genetic lesions in histone/chromatin modifiers suggest a critical role for epigenetic remodeling during malignant transformation
Inactivating mutations and deletions of the acetyltransferases CREBBP and EP300 promote lymphomagenesis by tipping the balance between the activities of the BCL6 oncoprotein and the p53 tumor suppressor
The disruption of pathways leading to escape from immune-surveillance represents a major player in lymphomagenesis
Newly identified lesions may guide the development of improved biomarkers and targeted therapeutic approaches
Acknowledgments
Funding: LP is supported by NCI RO1 CA172492 and CA164152, the Lymphoma Research Foundation and the Leukemia and Lymphoma Society.
Footnotes
The author declares no conflicts of interest
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer (IARC); 2008. [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- *3.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. A comprehensive review of the molecular pathogenesis of B-NHL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 5.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 6.Polo JM, Dell’Oso T, Ranuncolo SM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 7.Polo JM, Juszczynski P, Monti S, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Monti S, Juszczynski P, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111:2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Caro P, Kishan AU, Norberg E, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. This elegant work characterizes a metabolic signature in the previously defined Ox/Phos subgroup of DLBCL, which may have therapeutic implications as perturbation of the identified metabolic program proved selectively toxic to this DLBCL subset. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. This study provides an unbiased view of the landscape of mutations in the DLBCL exome, extending the findings of two previous studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **11.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. The first analysis of the DLBCL coding genome using whole-exome/whole-transcriptome sequencing technologies; this study also revealed MLL2 as the most common target of somatic mutations in DLBCL (40% of cases) and FL (89% of cases) [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature genetics. 2011;43:830–837. doi: 10.1038/ng.892. This work integrates whole-exome sequencing and high resolution SNP array approaches to provide a comprehensive genomic analysis of DLBCL. It also identifies recurrent genetic lesions in multiple histone/chromatin modifiers and immune regulators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Scott DW, Mungall KL, Ben-Neriah S, et al. TBL1XR1/TP63: a novel recurrent gene fusion in B-cell non-Hodgkin lymphoma. Blood. 2012;119:4949–4952. doi: 10.1182/blood-2012-02-414441. The first novel gene fusion identified to date in DLBCL through RNA-sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- *15.Jiang Y, Soong TD, Wang L, et al. Genome-wide detection of genes targeted by non-Ig somatic hypermutation in lymphoma. PLoS One. 2012;7:e40332. doi: 10.1371/journal.pone.0040332. This is the only study to interrogate DLBCL patients and cell lines for the presence of mutations in regulatory sequences genome-wide, leading to the identification of non-Ig aberrant SHM hotspots and providing initial evidence for their functional effect in altering gene transcription of the target gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monti S, Chapuy B, Takeyama K, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012;22:359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 18.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 19.Ogryzko VV, Schiltz RL, Russanova V, et al. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 21.Lill NL, Grossman SR, Ginsberg D, et al. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 22.Avantaggiati ML, Ogryzko V, Gardner K, et al. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 23.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- **24.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. The first demonstration that CREBBP is targeted by loss-of-function genetic lesions in nearly 30% of DLBCL patients; this study also documents that CREBBP mutations impair the acetylation of BCL6 and p53, providing one mechanism by which these lesions may contribute to lymphomagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 26.Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 27.Cerchietti LC, Hatzi K, Caldas-Lopes E, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010 doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. Comprehensive overview discussing the role of histone H3K4 methyltransferases as a mark of active transcription, enhancer signatures, and developmentally poised genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **29.Herz HM, Mohan M, Garruss AS, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. Reports a previously unrecognized function for Trr, the Drosophila homolog of the mammalian Mll3/4 COMPASS-like complexes as a major H3K4 monomethyltransferase on enhancers in vivo, implying a central role in the transition from inactive/poised to active enhancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Kanda H, Nguyen A, Chen L, et al. The Drosophila ortholog of MLL 3/MLL 4, trithorax related, functions as a negative regulator of tissue growth. Molecular and cellular biology. 2013 doi: 10.1128/MCB.01585-12. Demonstrates that mutations in the Drosophila homolog of MLL3/MLL4 confer a growth advantage, supporting a tumor suppressor role for MLL2, with relevant implications for the mechanism by which loss-of-function mutations could contribute to lymphomagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Grubor V, Love CL, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yap DB, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2010 doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **35.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. This reference, along with Ref 35 and 36, represents the original report on the development of a potent, selective inhibitor of EZH2, demonstrating its efficacy in animal models and paving the way to Phase I clinical in DLBCL patients with EZH2 mutations. [DOI] [PubMed] [Google Scholar]
- **36.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- *37.Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of beta2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011;20:728–740. doi: 10.1016/j.ccr.2011.11.006. This study provides a comprehensive genomic analysis of B2M and CD58 in DLBCL, reveals that expression of these proteins, as well as of HLAI, is lost in the majority of patients, and provides functional evidence for the ability of these lesions to impair NK-cell mediated lysis, as a mechanism for evasion of immune surveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moingeon P, Chang HC, Wallner BP, et al. CD2-mediated adhesion facilitates T lymphocyte antigen recognition function. Nature. 1989;339:312–314. doi: 10.1038/339312a0. [DOI] [PubMed] [Google Scholar]
- 40.Steidl C, Shah SP, Woolcock BW, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rui L, Emre NC, Kruhlak MJ, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green MR, Monti S, Rodig SJ, et al. Integrative analysis reveals selective 9p24. 1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 45.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *46.Yang Y, Shaffer AL, 3rd, Emre NC, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21:723–737. doi: 10.1016/j.ccr.2012.05.024. Offers a mechanistic basis for clinical trials combining drugs that modulate NF-κB signaling, particularly the BCR inhibitor ibrutinib, and lenalidomide in ABC-DLBCL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Fontan L, Yang C, Kabaleeswaran V, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22:812–824. doi: 10.1016/j.ccr.2012.11.003. Through the development of a method for generating active paracaspases in vitro, this study identifies a potent and irreversible inhibitor of MALT1 that suppresses NF-κB activity in vitro and in xenotransplanted ABC-DLBCL tumors with minimal toxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2010 doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 51.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 52.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 53.Migliazza A, Martinotti S, Chen W, et al. Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci U S A. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasqualucci L, Migliazza A, Fracchiolla N, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen HM, Peters A, Baron B, et al. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Li Z, Naganuma A, Ye BH. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proc Natl Acad Sci U S A. 2002;99:15018–15023. doi: 10.1073/pnas.232581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pasqualucci L, Migliazza A, Basso K, et al. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- *58.Duan S, Cermak L, Pagan JK, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. Two important findings for normal GC B cell biology and lymphomagenesis: i) a novel mechanism for the regulation of BCL6 protein stability by the FBXO11 ubiquitin ligase, and ii) the presence of mutations targeting FBXO11 in ~5% of DLBCL patients, adding to the list of genetic lesions perturbing BCL6 activity in lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *59.Ying CY, Dominguez-Sola D, Fabi M, et al. MEF2B Mutations Lead to DeRegulated Expression of the BCL6 Oncogene in Diffuse Large B-Cell Lymphoma and Follicular Lymphoma. Blood. 2012:120. doi: 10.1038/ni.2688. This abstract, reported at the annual meeting of the American Society of Hematology, provides the first characterization of the functional consequences and biological role of MEF2B mutations in DLBCL; it also identifies BCL6 as a direct target of MEF2B, with implications for lymphomagenesis and its potential as a therapeutic target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 61.Cerchietti LC, Ghetu AF, Zhu X, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 63.Mandelbaum J, Bhagat G, Tang H, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasqualucci L, Compagno M, Houldsworth J, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]