Abstract
Ca2+ binding protein 1 (CaBP1) and caldendrin are alternatively spliced variants of a subfamily of Ca2+ binding proteins with high homology to calmodulin. Although CaBP1 and caldendrin regulate effectors including plasma membrane and intracellular Ca2+ channels in heterologous expression systems, little is known about their functions in vivo. Therefore, we generated mice deficient in CaBP1/ caldendrin expression (C-KO) and analyzed the expression and cellular localization of CaBP1 and caldendrin in the mouse brain. Immunoperoxidase labeling with antibodies recognizing both CaBP1 and caldendrin was absent in the brain of C-KO mice, but was intense in multiple brain regions of wild-type mice. By western blot, the antibodies detected two proteins that were absent in the C-KO mouse and consistent in size with caldendrin variants originating from alternative translation initiation sites. By quantitative PCR, caldendrin transcript levels were far greater than those for CaBP1 particularly in the cerebral cortex and hippocampus. In the frontal cortex but not in the hippocampus, caldendrin expression increased steadily from birth. By double-label immunofluorescence, CaBP1/caldendrin was localized in principal neurons and parvalbumin-positive interneurons. In the cerebellum, CaBP1/caldendrin antibodies labeled interneurons in the molecular layer and in basket cell terminals surrounding the soma and axon initial segment of Purkinje neurons, but immunolabeling was absent in Purkinje neurons. We conclude that CaBP1/ caldendrin is localized both pre- and postsynaptically where it may regulate Ca2+ signaling and excitability in select groups of excitatory and inhibitory neurons.
Keywords: calmodulin, Ca2+ channel, EF-hand, synaptic plasticity, neuron, Ca2+ sensor
In the nervous system, the generation and propagation of Ca2+ signals is crucial for the control of neuronal excitability, gene expression, and synaptic plasticity(Augustine et al., 2003). The ubiquitous Ca2+ sensing protein, calmodulin (CaM) is thought to be a major player in this process. CaM is the prototypical member of the superfamily of EF-hand Ca2+ binding proteins, so named because they contain one or more helix-loop-helix domains that coordinate a Ca2+ ion (Kretsinger, 1976). CaM modulates the activity of numerous effector proteins (Chin and Means, 2000) and is an essential regulator of neuronal development (Bolsover, 2005) and synaptic plasticity (Xia and Storm, 2005).
A subset of EF-hand proteins related to CaM is expressed mainly in neurons (Burgoyne et al., 2004). Most similar to CaM are the CaBPs (CaBP1-8), which are found primarily in the brain, retina, and inner ear (Haeseleer et al., 2000, Laube et al., 2002, Yang et al., 2006, Cui et al., 2007). A wealth of evidence supports a role for CaBPs as regulators of voltage-gated Cav Ca2+ channels. CaBP4 potentiates the activity of Cav1.4 (Haeseleer et al., 2004, Shaltiel et al., 2012) and Cav1.3 Ca2+ channels (Yang et al., 2006, Cui et al., 2007, Lee et al., 2007), and CaBP2 enhances the function of Cav1.3 channels (Schrauwen et al., 2012). Cav1.4 and Cav1.3 are the primary Cav channels that regulate neurotransmitter release from retinal photoreceptors and cochlear inner hair cells, respectively (Platzer et al., 2000, Mansergh et al., 2005). Human mutations in the genes encoding CaBP4 and CaBP2 impair Cav1 regulation and cause vision and hearing impairment, respectively (Zeitz et al., 2006, Littink et al., 2009, Schrauwen et al., 2012, Shaltiel et al., 2012).
Unlike CaBP4 and CaBP2, CaBP1 is highly expressed in the brain (Haeseleer et al., 2000). Alternative splicing of the N-terminal domain gives rise to multiple variants (CaBP1-S, CaBP1-L, and caldendrin) (Seidenbecher et al., 1998, Haeseleer et al., 2000). CaBP1 has powerful effects in opposing CaM regulation of Cav channels(Lee et al., 2002). For the major Cav1 channels in the brain (Cav1.2 and Cav1.3), CaBP1 competes with CaM for binding to the channel and facilitates channel opening by preventing Ca2+/CaM-dependent inactivation (Lee et al., 2002, Zhou et al., 2004, Zhou et al., 2005, Yang et al., 2006, Cui et al., 2007, Findeisen and Minor, 2010, Oz et al., 2011). CaBP1 also binds to and regulates inositol 1,4,5 – trisphosphate receptors (IP3Rs) (Yang et al., 2002, Haynes et al., 2004). The largest CaBP1 variant, caldendrin, was initially discovered as a component of the postsynaptic membrane of excitatory synapses in the brain (Seidenbecher et al., 1998), where it may interact with the cytoskeleton (Seidenbecher et al., 2004), A-kinase anchoring protein (AKAP)79/150 (Gorny et al., 2012), Cav1.2 (Tippens and Lee, 2007), and/or Jacob, a protein that couples N-methyl-D-aspartate (NMDA) receptor signaling to the nucleus (Dieterich et al., 2008).
The above findings suggest a role for CaBP1 and caldendrin in regulating neuronal Ca2+ signaling, but the neurophysiological significance of these proteins in vivo is unknown. To fill this gap in knowledge, we generated mice with targeted inactivation of the CaBP1/caldendrin gene (C-KO). While previous studies have characterized the distribution of CaBP1 and caldendrin in the nervous system of rat (Laube et al., 2002) and human (Bernstein et al., 2003), similar analyses in the mouse are necessary to understand phenotypes of the C-KO mice. Therefore, we used molecular, biochemical, and immunocytochemical strategies to characterize the expression and localization of CaBP1 and caldendrin in the mouse brain.
1. EXPERIMENTAL PROCEDURES
1.1 Generation of CaBP1/caldendrin knock-out mice
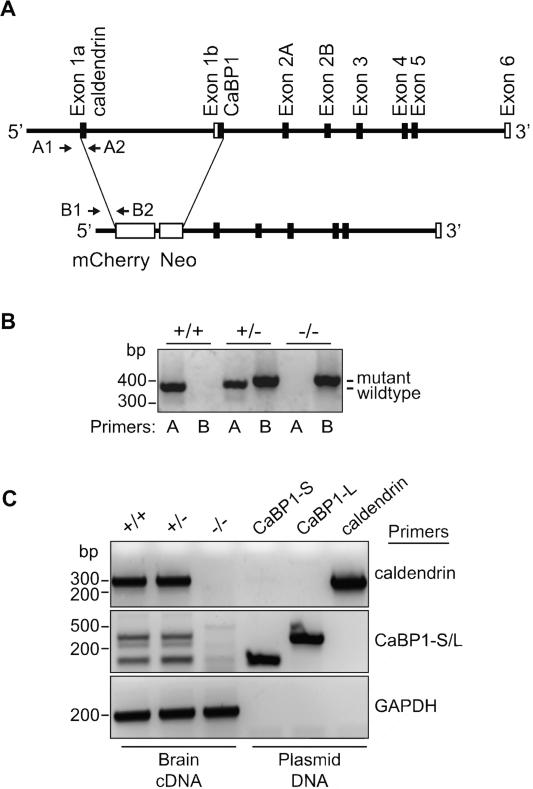
Genetic inactivation of CaBP1/caldendrin expression was accomplished with the assistance of the University of Iowa Gene Transfer Core. The strategy involved replacing exon 1 and exon 1b of the mouse CaBP1/caldendrin gene with sequences corresponding to mCherry and neomycin resistance gene. The targeting vector was constructed by amplifying a 2.4 kilobase (kb) DNA fragment upstream of exon 1 and a 5.8 kb fragment found downstream from exon 1b as the short and long arms, respectively. The 2.4 kb insert was ligated into NotI and NheI sites upstream of the mCherry start site and the 5.8 kb fragment was inserted into the SalI site downstream of the neomycin resistance cassette. The linearized construct was electroporated into 129/SvEv embryonic stem cells (ES). After selection in G418, surviving colonies were expanded, and PCR analysis was used to screen for homologous recombination with the following primers: oAL676 forward 5’-GTGTGCAAGATAACCAGCTTC-’3; oAL655 mCherry reverse 5’-CATGGTCTTCTTCTGCATTAC-3’. Seven ES cell lines were identified as positive for homologous recombination and each was microinjected into host C57BL6 blastocysts. The resulting chimeric mice were crossed with C57/Bl6 mice. Wild-type (WT) and mutant (KO) alleles were detected using the following primers: (oAL795 WT for. 5’-CTCGTGCTCACATTCAGTGC-3’; oAL796 WT rev. 5’-CAATGTGCGAGCTCATCG-3’; oAL797 KO rev. 5’-GATGATGGCCATGTTATCCTC-3’). PCR was performed using GoTaq Green Master Mix (Promega, Madison, WI) and 300 ng of DNA under the following conditions: 94°C × 1 min.; 94°C × 30 sec.; 50°C × 30 sec.; repeat cycles 2-4, 35 times; 72°C for 1 min). This strategy generated amplicons for WT and KO alleles that were 392 bp and 419 bp, respectively, which were electrophoretically resolved on a 2% agarose gel.
1.2 PCR analysis of CaBP1/caldendrin transcripts
For end-point PCR, total RNA was extracted from brain regions dissected from 3 mice that were 1 month old using TRIzol reagent (Life Technologies, Grand Island, NY) and cDNA synthesized using oligo d(T) primers from the Two-step Superscript III Kit (Life Technologies). CaBP1 and caldendrin transcripts were amplified with a common reverse primer (oAL703 rev. 5’-GTTGATCTGCTGAGACAGCTC-3’) and forward primers specific for CaBP1 (oAL701 for. 5’-CAAGTCGCCACTAAGAAACC-3’) or caldendrin (oAL702 CD for. 5’-CGGACCCGTTCCTCCAC-3’). GAPDH was amplified as a positive control (for. 5’-CCTCTGGAAAGCTGTGGCGTGATGG-3’; rev. 5’-AGATCCACGACGGACACATT-3’). PCR conditions were as follows: 94°C ×1 min.; 94°C × 15 sec.; 57°C × 30 sec.; 68°C × 30 sec.; repeat cycles 2-4, 30 times; 68°C × 30 sec.
For quantitative PCR (qRT-PCR), the cerebral cortex, cerebellum, and hippocampus were dissected from 3-4 mice that were one-month old. Total RNA was isolated using TRIzol Reagent (Life Technologies), and reverse transcription was performed with the SuperScript III First-Strand Synthesis System (Life Technologies). qRT-PCR was performed using the StepOnePlus Real-Time PCR system with TaqMan Gene Expression assays for CaBP1 (accession number Mm01203518_m1) and caldendrin. A custom design tool (Life Technologies) was used to create an assay for caldendrin, which recognizes coding regions from exon 1a to 2A (Fig. 1A). GAPDH was used as an endogenous normalizer. The assays were tested against CaBP1 and caldendrin plasmid DNAs to verify the specificity for the intended target. Assays were also tested on serial dilutions of cDNA prepared from total brain RNA to verify that caldendrin, CaBP1, and GAPDH assays demonstrated similar amplification efficiencies. ΔCT values obtained from cortex, cerebellum, and hippocampus samples were averaged over three separate PCR experiments, with 3 technical replicates from each sample per experiment. For developmental qRT-PCR (Fig.3D), frontal cortex, hippocampus, and cerebellum were dissected from 3-5 mice each at postnatal ages from 0 to 31 days. RNA isolation and qRT-PCR were performed as above. Data were averaged from 3 technical replicates from each sample in a single PCR experiment. The relative quantities (RQ) of caldendrin and CaBP1 were calculated by the ΔΔCT method using StepOne data analysis software (Life Technologies). Error bar ranges represent the minimum (RQmin) and maximum (RQmax) possible RQ values calculated from a 95% confidence interval of CT values.
Figure 1. Disruption of the gene encoding CaBP1 and caldendrin.
(A) Schematic of the targeting construct that interrupts alternatively spliced exons 1a and exon 1b with the sequence encoding mcherry and neomycin resistance cassette (Neo). Locations of genotyping primers that identify wild-type (WT; A1,A2) and mutant (B1,B2) alleles are indicated. (B) PCR amplification with WT (A) and mutant (B) primers indicated in (A). Reactions were performed on genomic DNA isolated from WT (+/+), heterozygous (+/-), and homozygous (-/-) mice. (C) PCR amplification of caldendrin and CaBP1 variants (CaBP1-S/L) from the corresponding plasmid DNA or brain cDNA generated from +/+, +/-, or -/- mice. Amplification of GAPDH was performed as a positive control.
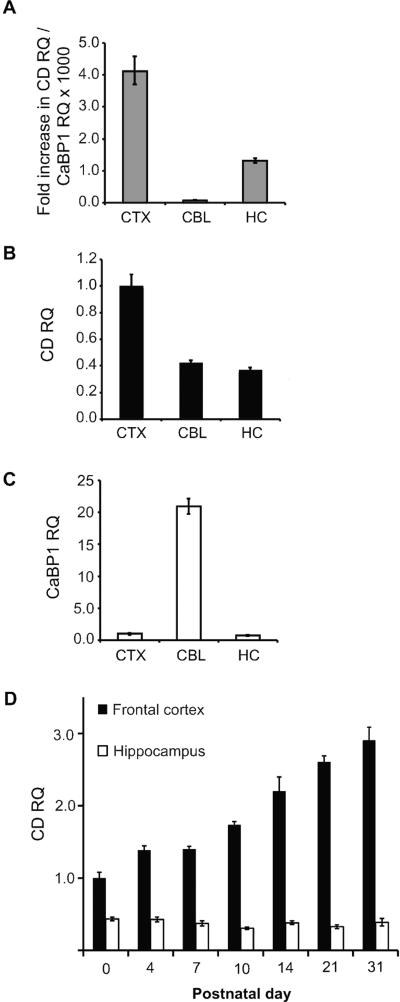
Figure 3. Quantitative PCR analysis of CaBP1 and caldendrin in the adult and developing mouse brain.
(A-C) Quantitative reverse-transcription PCR showing relative quantities (RQ) of caldendrin (CD) and calcium binding protein (CaBP1) RNA in brain regions isolated from 1-month-old mice. (A) Relative quantity (RQ) of caldendrin RNA expressed relative to CaBP1 RNA in the same region, indicating fold increase in caldendrin compared to CaBP1 RNA. (B,C) Quantities of caldendrin RNA (B) and CaBP1 RNA (C), expressed in cerebellum (CBL) and hippocampus (HC) relative to levels in the cerebral cortex. (D) Relative quantities of caldendrin in the frontal cortex (black bars) and hippocampus (white bars) at the indicated postnatal days, expressed relative to levels at birth (P0). N = 3-4 animals, data averaged from 3 separate PCR experiments with 3 technical replicates per sample per experiment (A-C) or 3-5 animals per age, data averaged from 3 technical replicates per sample (D). Error bars represent minimum and maximum possible RQ values calculated based on a 95% confidence interval of ΔCT values.
1.3 Western blot analysis of CaBP1/caldendrin
For western blot analysis of transfected HEK293T cells, mammalian expression constructs corresponding to mouse CaBP1-S and CaBP1-L cDNAs (Genbank #AF169153.1 and AF169152.1, respectively) cloned into the pcDNA3 vector (Life technologies) were used and described previously (Haeseleer et al., 2000). Caldendrin-S and –L variants were amplified from mouse brain cDNA by PCR using the following primers: oAL798 CD-S for. 5’-ATGAGCTCGCACATTGCC AAG-3’; oAL799 CD-L for. 5’- AGA GGCTGCGCTGTCACATG -3’; oAL800 common rev. 5’- TCAGCGAGACATCATCCGGAC-3’. PCR products were then ligated into pCR2.1TOPO vector (Life Technologies), then excised and inserted into CMV-promoter containing vector, pcDNA3.1+ (Life Technologies) using restriction endonucleases (New England Biolabs, Ipswich, MA) and sites EcoRI for caldendrin -L and BamHI/XhoI for caldendrin-S. The DNA constructs were sequenced at University of Iowa's DNA Sequencing Facility.
Human embryonic kidney, HEK293T (CRL-11268; ATCC, Manassas, VA), cells were grown and maintained in DMEM (14190-144, Life Technologies) containing 10% Fetal Bovine Serum (S11150; Atlanta Biologicals, Lawrenceville, GA), penicillin and streptomycin (Life Technologies), and held in a humidified atmosphere at 37°C and 5% CO2. Once cells have reached 70-80% confluence, cells were transfected with Gene Porter reagent (Genlantis, San Diego, CA) according to the manufacturer's protocol. Cells plated on 35 mm dishes were transfected with 2.0 μg of CaBP1/CD cDNAs. One day later, cells were rinsed and suspended with Versene (15040; Life Technologies), and pelleted at 800 × g for 5 min (4°C). Cells were homogenized using ice-cold RIPA buffer (150 mM sodium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 50 mM Tris, pH 8.0) and sterile pellet pestles (Kimble Chase Kontes, Vineland, NJ). Insoluble material was removed by centrifugation at 16,200 × g for 15 min (4°C), and the cell lysate used immediately or aliquoted and stored at -20°C.
For preparation of mouse brain lysates, 1-2 month old male or female WT or C-KO mice were anesthetized with isofluorane and brain regions were dissected and collected in 0.5 ml cold HB (HEPES Buffer: 0.32 M sucrose, 10 mM HEPES pH 7.4, 2 mM EDTA pH 8, EDTA-free protease inhibitor mix cocktail tablet; Roche, Indianapolis, IN). Tissue was homogenized using a plastic pestle and pellet pestle cordless motor (Kimble Chase Kontes) and the nuclei removed by centrifugation at 1000 × g for 15 min (4°C). The crude membrane fraction was isolated by centrifugation of the supernatant at 100,000 × g for 15 min (4°C). The membrane pellet was resuspended with 0.5 ml of HB, and centrifuged at 200,000 × g for 15 min (4°C). The membrane pellet was resuspended using 0.1 ml of HLB (50 mM HEPES, pH 7.4, 2 mM EDTA, EDTA-free protease inhibitor cocktail mix; Roche), aliquotted, and stored at -80°C. For phosphatase inhibitor assays (Fig.3C), lysates were incubated overnight at 4°C with PhosStop phosphatase inhibitor cocktail (Roche) prior to western blotting.
For western blotting, samples from transfected HEK293T cells or mouse brain were incubated with 4X LDS buffer (Life Technologies) in a total volume of 36 μl at 95-100°C for 15 min., and loaded into NuPage Novex 10% Bis-Tris gels (NP0304BOX, Life Technologies). Polyacrylamide gels were run at 100 volt × 1.5-2 h. Proteins isolated on polyacrylamide gels were transferred onto a nitrocellulose membrane (BioRad, Hercules, CA) at 80 V for 3 h. The membrane was blocked for 1 h at room temperature in blocking buffer (BB: 3% milk and TBST (0.1% Tween20, 1X TBS)). Blots were then incubated with UW72 antibodies (Haeseleer et al., 2000) diluted in BB (1:1000 for transfected HEK293T cell lysates and 1:4000 for brain fractions) for 1 h at room temperature. After rinsing 3 times in TBST, the blot was incubated with rabbit anti-horseradish peroxidase secondary antibody (Jackson immunoresearch) at 1:4000 in TBST for 45 min-1 h at room temperature. The blot was finally rinsed 3 times with TBST before applying the chemiluminiscent substrate (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific) and exposing to autoradiography film.
1.4 Immunohistochemistry
Mice (45-100 days old) were first anesthetized by inhalation with isofluorane and intraperitoneal injection of ketamine. Fixation of the brain was achieved by intracardial perfusion with 0.1 M sodium phosphate buffer (PB) followed by 4% paraformaldehyde in 0.1 M PB. The brain was removed and immersed in cryoprotectant solution (15% sucrose, 0.1 M PB) overnight at 4°C, followed by a second incubation in a solution containing 30% sucrose in 0.1 M PB for 24 h at 4°C. Brain sections (40 μm) were collected on a cryostat, and either used immediately or stored in cryoprotectant solution (500 ml: 0.88 M sucrose, 200 ml 0.1M PB, 30% ethylene glycol) at -20°C.
For immunoperoxidase staining, free-floating sections were rinsed in TBS 3 × 10 min at room temperature before blocking endogenous peroxides with hydrogen peroxide (0.3% H2O2 in TBS) and rinsed in TBS 2 × 5 min. Sections were then blocked in blocking buffer (BB: 10% normal goat serum (NGS; Jackson Immunoresearch) and 0.3 % Triton X-100) for 1 h at 4°C prior to incubation with the primary antibody (UW72, 1:15,000 in BB) overnight at 4°C. The sections were then incubated with secondary antibody (biotinylated donkey anti-rabbit IgG, Abcam, Cambridge, MA, 1:400 in BB) for 1 h at room temperature. After each antibody incubation step, sections were rinsed in TBS 3 × 10 min. The sections were then immersed in ABC-Elite (Vectastain) mix for 30 min. Signal was developed by incubation with DAB substrate solution (Vector Laboratories), prepared according to manufacturer's instructions, for 7 min. The reaction was stopped by rinsing sections in distilled water. The brain sections were then mounted onto gelatin-subbed slides (Southern Biotech, Birmingham, AL), and subject to dehydration and delipidation through incubation in a graded series of ethanols and xylenes. The tissue was then coverslipped with DPX mounting medium (Gallard Schlesinger Industries, Plainview, NY). The tissue was analyzed on an Olympus BX53 microscope using 2x, 20x nonoil and 60x oil immersion objectives with a numerical aperture of 0.08, 0.50 and 1.35. Bright field images were captured with an Olympus D72 camera driven by CellSens Standard software (Olympus).
For double-label immunofluorescence, tissue sections were rinsed with phosphate-buffered saline (PBS) 3 × 10 min before incubating in blocking buffer (BB: 10% normal goat serum (NGS; Jackson Immunoresearch) and 0.3 % Triton X-100) for 1 h at room temperature. All antibodies were diluted in BB at varying dilutions [anti-PV clone PARV-19 mouse, 1:500 (Sigma-Aldrich); anti-calbindin-D-28K clone CB-955 mouse (Sigma-Aldrich), 1:6000; anti-Kv1.2 mouse (Neuromab), 1:400; anti-TH clone 2/40/15 mouse (Millipore), 1:1000; UW72, 1:500; Alexa 488-conjugated goat anti-rabbit, IgG 1:2000; Alexa 546-conjugated goat anti-mouse IgG, 1:2000 (both from Life Technologies)]. Sections were rinsed 2-3 times for 15 min in PBS containing 1% NGS and 0.3 % Triton X-100 [washing buffer (WB)] after each antibody incubation step. Sections were incubated in primary antibody overnight at 4°C, and 1 hr at room temperature in secondary antibody solution. The sections were further washed with PBS 4 × 15 min, and 1 × 15 min in 0.05 M PB after the fourth wash in PBS. Some sections were incubated in Hoechst stain (Life Technologies, 10 min) prior to washing. Sections were mounted using Prolong Gold Antifade Reagent (Life Technologies), and viewed under a confocal laser scanning microscope (Olympus Fluoview FV1000, Melville, NY). Images were taken using 60x and 100x oil immersion objectives with numerical apertures of 1.45 and 1.30, respectively. Specific CaBP1/caldendrin immunolabeling was captured after setting fluorescent signals from C-KO sections as the threshold. In double labeling experiments, images were acquired independently, and scanned every 0.3-0.4 μm. Single optical slices (18-30 in total) were compiled into one image (Z-stacked) using Olympus Fluoview software. Colocalization was analyzed in single optical sections. Images were processed only for brightness and contrast adjustment using Adobe Photoshop software.
2. RESULTS
2.1 Generation of CaBP1/caldendrin knock-out mice
To generate C-KO mice, a targeting construct was designed in which the DNA sequences encoding mCherry fluorescent protein and the neomycin resistance cassette would be expressed in place of exon 1a of caldendrin and exon 1b of CaBP1 upon homologous recombination (Fig.1A). C-KO mice were viable and bred normally, with no differences in survival or apparent physiological deficits. Body weight measured between 3 and 21 weeks of age was similar to that of WT mice (data not shown). The presence of the mutant allele in heterozygous (+/-) and homozygous (-/-), but not wild-type (WT, +/+) mice was confirmed by PCR of genomic DNA (Fig.1B). The absence of caldendrin and CaBP1 expression in C-KO mice was confirmed by PCR of cDNA synthesized from brain RNA (Fig.1C). Primers specific for caldendrin or the short and long forms of CaBP1 (CaBP1-S, CaBP1-L;(Haeseleer et al., 2000)) yielded amplicons of the appropriate size from cDNA synthesized from the brain of +/+ and +/-, but not -/- mice (Fig.1C). These results confirmed that C-KO mice do not express CaBP1 or caldendrin transcripts.
2.2 Variants of caldendrin may result from alternative translation start sites
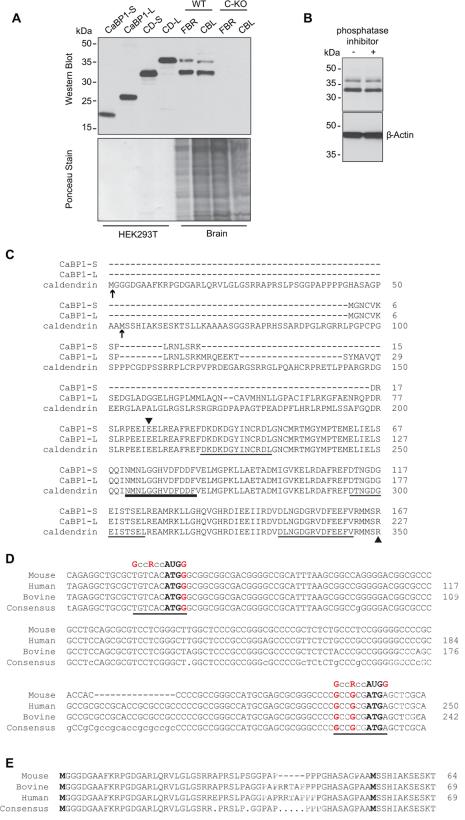
To verify that CaBP1 and caldendrin are eliminated at the protein level in C-KO mice, we performed western blotting with polyclonal rabbit antibodies, generated against amino acids 25-227 of human CaBP1 (96% homologous to mouse CaBP1), that recognize all CaBP1 variants, characterized previously (Haeseleer et al., 2000). While these antibodies detected bands corresponding to CaBP1-S, CaBP1-L, and caldendrin in lysates of transfected HEK293T cells, they only recognized two bands (33 and 38 kDa) in the brain of WT but not C-KO mice (Fig.2A). Similar results were reported using different CaBP1/caldendrin antibodies in rat brain, and were attributed to post-translational modification, possibly by phosphorylation (Laube et al., 2002). To test this, we incubated brain lysates from WT mice with phosphatase inhibitors prior to western blotting with CaBP1/caldendrin antibodies. If the smaller variant resulted from dephosphorylation of the larger variant, this treatment should result in a reduction in the intensity of the lower band. However, no differences were seen in the intensity of either 33- or 38-kDa band (Fig.2B), which argued against a role for phosphorylation.
Figure 2. Detection of two variants of caldendrin but not CaBP1 in WT but not C-KO mouse brain.
(A) Western blot with CaBP1/CD antibodies (top panel) and ponceau staining of the same blot (lower panel) to indicate amount of protein loaded. Lanes 1-4 contain lysates from HEK293T cells transfected with cDNAs for CaBP1-S, CaBP1-L or caldendrin starting from Met53 (CD-S) or from Met1 (CD-L) according to the sequence numbering in (C). Lanes 5-8 contain lysates from the forebrain (FBR) or cerebellum (CBL) of WT or C-KO mice. (B) Effect of phosphatase inhibitor on bands detected by CaBP1/CD antibodies. Brain lysates were incubated with (+) or without (-) PhosStop phosphatase inhibitor prior to SDS-PAGE and western blotting with CaBP1/CD antibodies. Detection with β-actin antibodies was used to confirm equal loading in the lanes. (C) Alignment of the amino acid sequence of CaBP1 variants (CaBP1-S, CaBP1-L) and caldendrin. Potential alternative translation start sites for caldendrin are indicated by arrows. Arrowheads mark region corresponding to antigen used to raise CaBP1/CD antibodies. EF-hand domains are underlined, with bold line indicating non-functional EF-hand 2. Numbers correspond to amino acids of the corresponding mouse sequences. (D) Alignment of caldendrin N-terminal mRNA sequences from different species with canonical Kozak sequence (GccRccAUGG), indicated. Potential translation start sites are indicated in bold type. Highly conserved purine (R) at the -3 position and guanine residues at -6 and +4 are indicated in red. (E) Alignment of caldendrin N-terminal amino acid sequences from different species. In C-E, numbers in correspond to nucleotide or amino acid sequences available in GenBank. Alignments were generated using the MultAlin multiple sequence alignment application.
Analysis of the 5’ region of the caldendrin sequence revealed the presence of an additional methionine residue (Met1, Fig.2C) upstream of the methionine (Met53) proposed to be the translation start site (Seidenbecher et al., 1998). While Met1 has a weaker Kozak consensus sequence for translation initiation compared to Met53, this sequence and that leading to Met53 are highly conserved between species at the nucleotide and amino acid level (Fig.2D,E). To determine if Met1 could serve as an alternative start site for translation, we generated a cDNA expression construct that contained both Met 1 and Met53. In HEK293T cells transfected with this variant (CD-L), only one band (38-kDa) was detected by CaBP1/caldendrin antibodies (Fig.2A). The fact that this band was the same size as the larger band detected in the brain (Fig.2A) suggested that alternative translation initiation may lead to expression of the two caldendrin variants in the brain but not in HEK293T cells.
Despite the fact that our gene targeting strategy should lead to expression of mcherry in place of caldendrin, we were unable to detect mcherry either by western blot or by immunofluorescence in the brain of C-KO mice (data not shown). Since the sequence encoding mcherry was inserted after that for Met53 in the targeting construct (Fig.1A), one might expect relatively weak mcherry expression if caldendrin translation is initiated partially at the upstream Met1. In addition, we cannot exclude that the caldendrin promoter might drive mcherry expression at levels too weak to be detected.
2.3 Caldendrin is expressed at higher levels than CaBP1 throughout the brain
While bands corresponding to the predicted molecular weight of caldendrin were detected in western blots of brain lysates, those corresponding to either CaBP1 variant were not (Fig.2A). To determine if this was due to reduced transcription of CaBP1 compared to caldendrin, we performed quantitative PCR using RNA isolated from different brain regions (Fig.3). Consistent with the western blotting results, caldendrin RNA levels far exceeded those of CaBP1. The ratio of caldendrin RNA to CaBP1 RNA was greatest (~4000-fold) in the cerebral cortex and least (~80-fold) in the cerebellum (Fig.3A). In the 3 brain regions analyzed, caldendrin was highest in the cerebral cortex, with less but equivalent levels in the cerebellum and hippocampus (Fig.3B). In contrast, CaBP1 was highest in the cerebellum and weakest in the cerebral cortex and hippocampus (Fig.3C). These results confirm previous findings that caldendrin is the major variant in the rodent brain (Laube et al., 2002), and show that CaBP1 and caldendrin exhibit variable expression levels in different brain regions. Caldendrin RNA levels increased in the frontal cortex from birth (postnatal day 0 (P0)) until about 1 month of age (P31). By contrast, relatively lower levels of caldendrin RNA in the hippocampus remained constant between P0 and P31 (Fig.3D).
2.4 CaBP1/caldendrin antibodies specifically label subsets of neurons in the brain
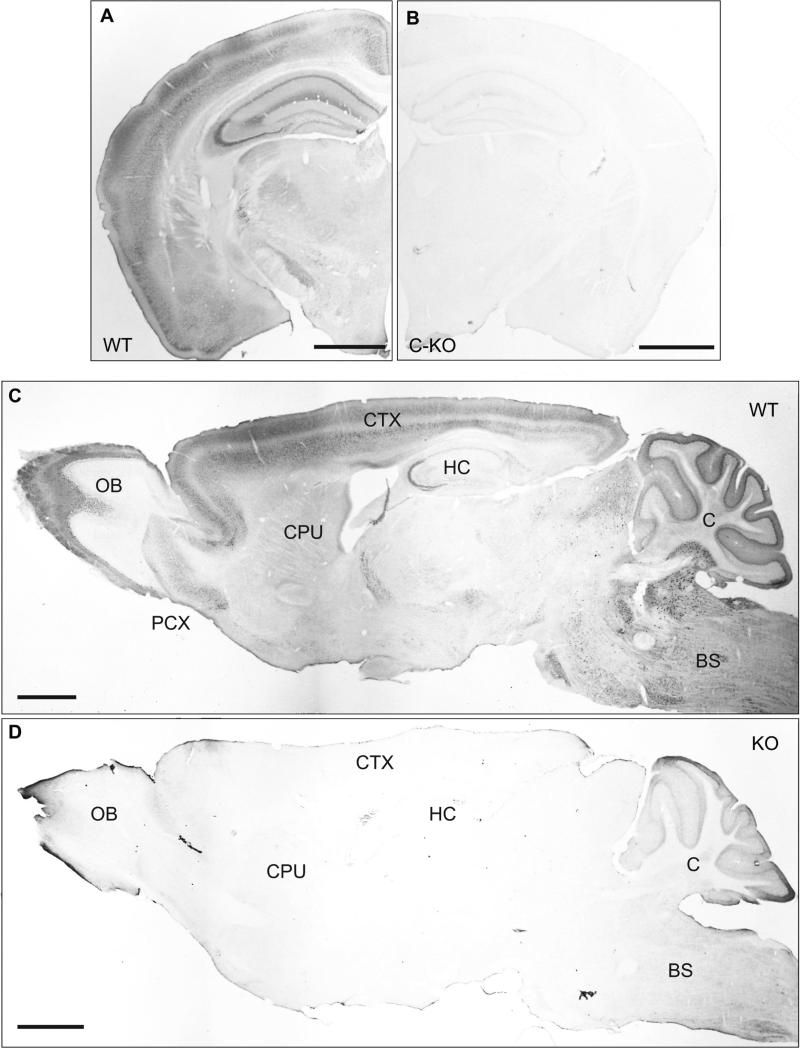
To confirm that our CaBP1/caldendrin antibodies specifically detect the corresponding proteins by immunohistochemistry, we compared immunoperoxidase labeling with these antibodies in brain sections obtained from WT and C-KO mice (Fig.4). Consistent with our western blotting results, the antibodies strongly labeled the cerebral cortex, hippocampus, and cerebellum in WT but not C-KO mice (Fig.4A-D). Similar patterns of labeling were observed in WT but not C-KO mice by immunofluorescence methods (data not shown). Specific labeling was also observed in the olfactory bulb and various brain stem nuclei (Fig.4C,D). Despite the relatively low levels of CaBP1 compared to caldendrin in our analyses (Fig.2,3), we cannot rule out that some of the labeling corresponds to CaBP1 rather than caldendrin since our antibodies recognize both. Therefore, we will refer to this labeling as CaBP1/caldendrin-immunoreactivity (CaBP1/CD-IR).
Figure 4. Specificity of CaBP1/CD antibodies for immunohistochemistry in mouse brain.
Immunoperoxidase labeling with CaBP1/CD antibodies in coronal (A,B) or sagittal (C,D) brain sections from WT (A,C) or C-KO (B,D) mice. OB, olfactory bulb; PCX, pyriform cortex; CPU, caudate putamen; CTX, cortex; HC, hippocampus; BS, brainstem; C, cerebellum. Scale bars, 1 mm.
2.5 Localization of CaBP1/CD-IR in principal neurons and interneurons in the forebrain
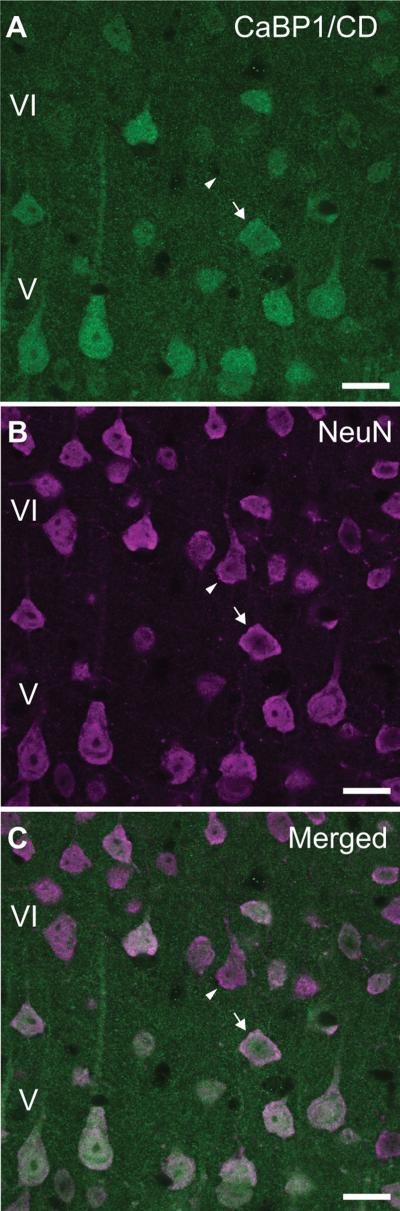
To characterize the cell-types exhibiting CaBP1/CD-IR, we performed double-labeling experiments. In the cerebral cortex, CaBP1/CD-IR was localized in the soma and dendrites of pyramidal cells (Fig.5A). Double-labeling revealed that all cells with CaBP1/CD-IR co-localized with the neuronal protein, NeuN (Fig.5B-C). However, some cells that were NeuN-positive were void of CaBP1/CD-IR (Fig.5B,C). Therefore, unlike NeuN, CaBP1/CD is not expressed in all neuronal cell-types.
Figure 5. Localization of CaBP1/CD-IR in principal neurons in the cerebral cortex.
Double immunofluorescence labeling of layer V-VI pyramidal neurons in the cerebral cortex with CaBP1/CD antibody (A,C) and the neuron-specific neuronal nuclei (NeuN) antibody (B,C). Arrow indicates co-localization of CaBP1/CD-IR with NeuN. Arrowhead indicates NeuN-positive neurons that do not show CaBP1/CD-IR. Scale bars, 20 μm.
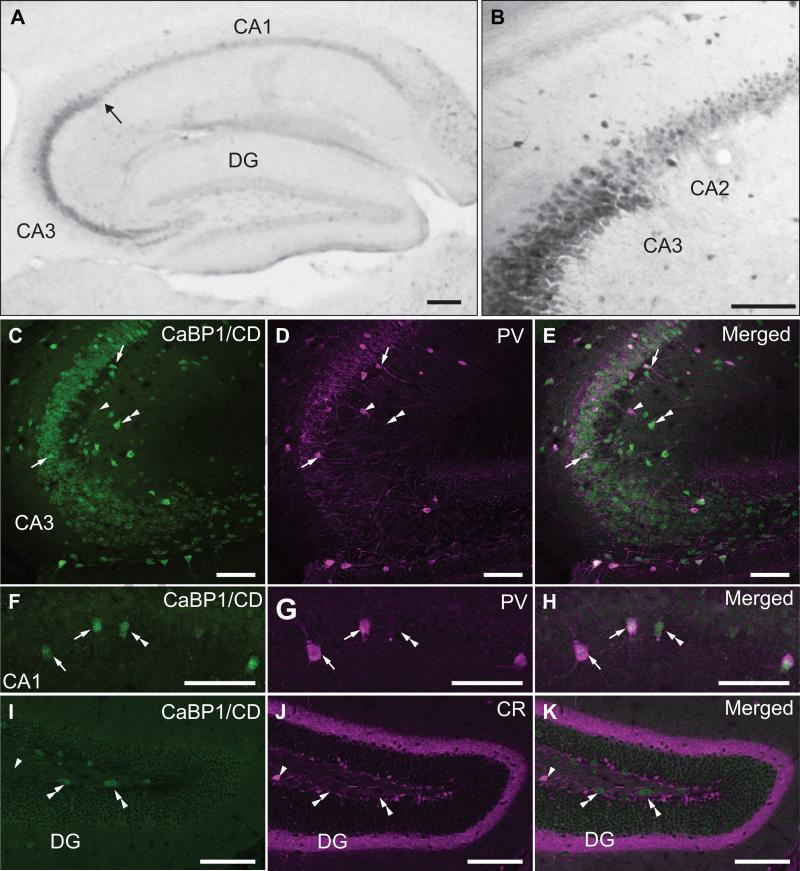
In the hippocampus, CaBP1/CD-IR was strongest in the stratum pyramidale in the CA3 region and relatively weak in CA1 and the dentate gyrus (Fig.6A). Consistent with previous results in rat brain (Laube et al., 2002), the intensity of CaBP1/CD-IR in pyramidal cell soma dropped markedly at the CA3/CA2 boundary (Fig.6B). Throughout CA3-CA1 regions, strong CaBP1/CD-IR was observed in cells resembling interneurons in the stratum pyrimidale and stratum oriens. Some of these cells were likely fast-spiking GABA-ergic interneurons since they were double-labeled with antibodies against parvalbumin, a Ca2+ binding protein highly expressed in these cells (Jones and Buhl, 1993). In double-labeled cells, parvalbumin labeling was concentrated in the soma as well as in fibers and puncta resembling synaptic terminals in the stratum pyramidale, while CaBP1/CD-IR was restricted to the soma (Fig.6C-E). Relatively large non-pyramidal neurons exhibiting single-labeling for parvalbumin or CaBP1/CD were often observed (Fig.6C-H), indicating some heterogeneity in the expression of these Ca2+ binding proteins in different classes of interneurons.
Figure 6. Localization of CaBP1/CD-IR in the hippocampal formation.
(A,B) Immunoperoxidase labeling with CaBP1/CD antibodies. Arrow in (A) indicates boundary between CA3 and CA2 regions, magnified in (B). (C-H) Double immunofluorescence labeling with CaBP1/CD antibodies (C,F) and parvalbumin antibody (PV; D,G) in CA3 (C-E) and CA1 regions (F-H). (I-K) Double immunofluorescence labeling with CaBP1/CD antibodies (I) and calretinin antibody (CR; J) in the dentate gyrus (DG). Arrows indicate neurons double-labeled for CaBP1/CD-IR and PV. Arrowheads indicate PV-positive (C-E) or CR-positive (I-K) neurons that do not show CaBP1/CD-IR. Double arrowheads indicate CaBP1/CD-positive neurons that are PV-negative (C-H) or CR (I-K). Scale bars, 100 μm.
In the dentate gyrus, we double-labeled with antibodies against calretinin, a Ca2+ binding protein highly concentrated in mossy cells in the ventral hilus and axon terminals in the superficial stratum granulosum (Gulyas et al., 1992). These experiments indicated no overlap of calretinin labeling and CaBP1/CD-IR (Fig.6I-K). However, CaBP1/CD-IR was observed in hilar cells with an elongated, fusiform morphology, which may represent a subset of calretinin-negative mossy cells identified previously in the dorsal hilus (Fujise et al., 1998).
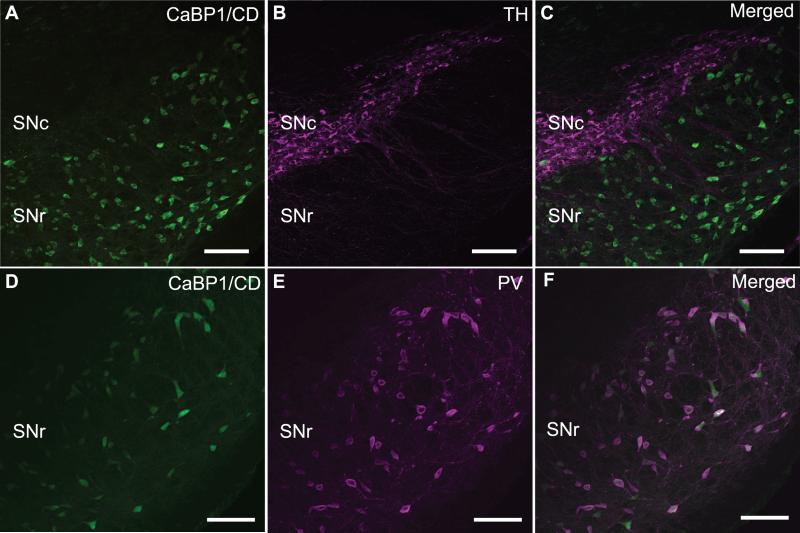
2.5 Localization of CaBP1/CD-IR in the substantia nigra
In dopaminergic neurons of the substantia nigra, Cav1.3 Ca2+ channels regulate spontaneous firing, which may predispose these neurons to pathogenic insults due to excessive activity-dependent Ca2+ influx (Chan et al., 2007). Since CaBP1 greatly potentiates the activity of Cav1.3 (Yang et al., 2006, Cui et al., 2007), we determined if CaBP1/CD is present in these neurons by double-labeling with antibodies against tyrosine hydroxylase, the rate-limiting enzyme in dopamine biosynthesis. These experiments showed no colocalization of tyrosine hydroxylase and CaBP1/CD-IR in the substantia nigra pars compacta (Fig.7A-C), which argued against the possibility that CaBP1/CD is a physiological partner of Cav1.3 channels in dopaminergic neurons in this region. However, double-labeling with antibodies against parvalbumin revealed strong concordance with CaBP1/CD-IR in the substantia nigra pars reticulata (Fig.7D-F). In this brain region, parvalbumin is present in most GABA-ergic interneurons (Gonzalez-Hernandez and Rodriguez, 2000). Together with the colocalization of CaBP1/CD with parvalbumin in the hippocampus (Fig.6C-H), these results demonstrate that CaBP1/CD is prominently expressed in some populations of GABA-ergic interneurons.
Figure 7. Localization of CaBP1/CD-IR in the substantia nigra.
Double immunofluorescence labeling with CaBP1/CD antibodies (A,D) and tyrosine hydroxylase antibody (TH; B) or parvalbumin antibody (PV; E). Co-localization of CaBP1/CD-IR and PV is indicated by white labeling in (F). SNc, substantia nigra pars compacta; SNr, substantia nigra pars reticulata. Scale bars, 50 μm.
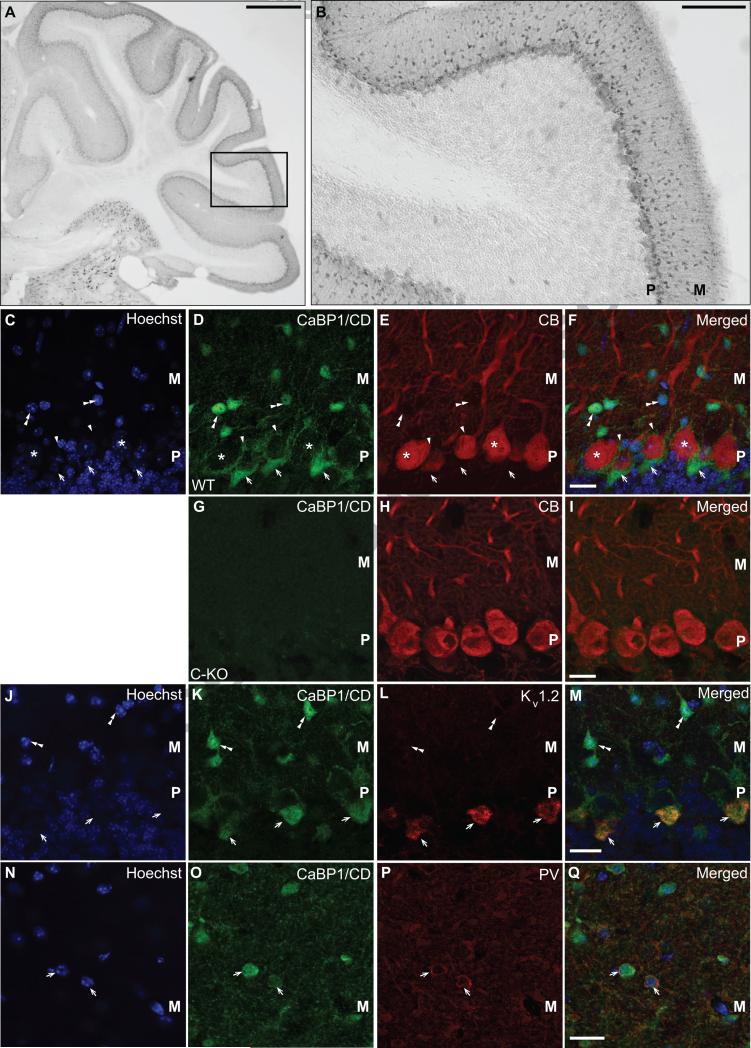
2.6 Localization of CaBP1/CD in cerebellar pinceaux
CaBP1 strongly enhances inactivation of Cav2.1 P/Q-type Ca2+ channels (Lee et al., 2002), which are the major Cav channels in cerebellar Purkinje neurons (Mintz et al., 1992). However, our previous studies indicated that CaBP1/CD-IR in the cerebellum was very weak in Purkinje neurons or granule cells (Zhou et al., 2004). To characterize the cellular and subcellular distribution of CaBP1/CD-IR in this brain region, we first performed single-labeling with CaBP1/CD antibodies with immunoperoxidase methods. CaBP1/CD-IR was most evident in cells in the molecular layer (Fig.8A-B). We next performed double labeling first with antibodies against calbindin, a Ca2+ binding protein enriched in Purkinje neurons (Schwaller et al., 2002). As expected, CaBP1/CD-IR did not overlap significantly with calbindin labeling, but was strongly localized in smaller cells in the molecular layer (Fig.8C-F). No CaBP1-CD-IR was observed in tissue samples from C-KO mice (Fig.8G-I). In addition, CaBP1/CD-IR was intense in perisomatic puncta surrounding the Purkinje cell soma and in structures resembling the specialized pinceaux formed by basket cell axon terminals on the axon initial segment of the Purkinje cell (Fig.8C-F; (Palay and Chan-Palay, 1974)). Double-labeling with antibodies against Kv1.2 K+ channels, which are localized in the pinceaux (Laube et al., 1996), revealed strong colocalization with CaBP1/CD-IR (Fig.8J-M). We also performed double-labeling with parvalbumin antibodies, which should identify basket cells and stellate cells in the molecular layer (Bastianelli, 2003). Consistent with its localization in the pinceaux, CaBP1/CD-IR was found in parvalbumin-positive cells in the molecular layer (Fig.8N-Q). Taken together, these results suggest that CaBP1/CD is present primarily in basket cell soma and terminals in the cerebellar cortex.
Figure 8. Localization of CaBP1/CD-IR in the cerebellum.
(A,B) Immunoperoxidase labeling with CaBP1/CD antibodies. Box in (A) indicates magnified region in (B). (C-Q) Double-immunofluorescence labeling in WT (C-F, J-Q) or CaBP1/CD-KO (C-KO, G-I) mice with CaBP1/CD antibodies (D,G,K,O) and: calbindin antibody (CB; C-I), Kv1.2 antibody (J-M), or parvalbumin antibody (PV; N-Q). (C,J,N) Hoechst nuclear labeling. Asterisks in (C-F) indicate CB-positive Purkinje neurons. Arrowheads in (C-F) indicate periosomatic CaBP1/CD-IR surrounding Purkinje neuron soma. Arrows in (C-M) indicate CaBP1-CD-positive pinceaux formations, which exhibit labeling for Kv1.2 (L-M). Arrows in (N-Q) indicate co-localization between CaBP1/CD-IR and PV labeling. Double arrowheads indicate CaBP1/CD-positive neurons that do not exhibit labeling for CB (C-F) or Kv1.2 (J-M). M, molecular layer. P, Purkinje cell layer. Scale bars, 500 μm (A), 100 μm (B), 20 μm (C-Q).
3. DISCUSSION
Our study provides the first report of the expression and localization of CaBP1 and caldendrin in the mouse brain. Our results largely agree with those of Laube and colleagues in the rat brain (Laube et al., 2002), but add the following new insights. First, we verify that the labeling pattern obtained with our CaBP1/CD antibodies is specific by its absence in the C-KO mouse brain. Second, we confirm quantitatively at the transcriptional level that caldendrin is the major CaBP1 variant expressed in the brain and undergoes a developmental increase in expression in the frontal cortex postnatally. Third, by double-label immunofluorescence we show that CaBP1/CD is not only expressed in principal neurons but also in inhibitory interneurons throughout the brain. Finally, we demonstrate that in addition to its localization in somatodendritic domains, CaBP1/CD is also present in axon terminals and so may have both pre- and postsynaptic functions. Our findings provide a framework for understanding the neurophysiological roles of CaBP1/CD, and will guide future phenotypic analyses of C-KO mice.
3.1 Greater prominence of caldendrin compared to CaBP1 variants in mouse brain
Our findings that caldendrin is expressed at higher levels than CaBP1 in the mouse brain (Fig.3A) support previous results in rat brain (Laube et al., 2002). However, CaBP1 was detected by PCR (Fig.1C) and coimmunoprecipitates with Cav1.2 channels from the postsynaptic density fractions of rat brain (Zhou et al., 2004). Therefore, we do not discount the possibility that CaBP1 has key regulatory functions in a select, but small, number of cell groups.
The developmental increase in caldendrin transcript levels (Fig.3D) in the frontal cortex parallels the time course of cortical synaptogenesis (Micheva and Beaulieu, 1996). Since it is a component of the postsynaptic density (Seidenbecher et al., 1998), caldendrin may regulate synapse formation. Caldendrin binds to the transcriptional regulatory protein, Jacob, which prevents reductions in synapse number upon activation of extrasynaptic NMDA receptors containing the NR2B subunit (Dieterich et al., 2008). Since a shift in NMDA receptor subunit composition from NR2B to NR2A occurs in the first two weeks postnatally (Yashiro and Philpot, 2008), and this enhances synapse formation and maintenance (Gambrill et al., 2011), caldendrin may support synaptogenesis by negatively regulating NR2B-dependent reductions in synapse number. Given the role of caldendrin in potentiating Cav1 channels (Zhou et al., 2004) and the importance of these channels for dendritic growth in cortical neurons (Redmond et al., 2002), it is also possible that caldendrin regulates activity-dependent processes involved in shaping dendritic arbors during development. Caldendrin may have different developmental roles in the frontal cortex and the hippocampus, given the lack of any postnatal changes in caldendrin levels in the latter brain region (Fig.3D).
3.2 Potential for two variants of caldendrin resulting from alternative translation initiation
By western blotting with antibodies that recognize CaBP1 and/or caldendrin, we and others have detected two bands in rat and mouse brain lysates (Seidenbecher et al., 1998, Laube et al., 2002, Tippens and Lee, 2007). Based on findings that caldendrin is subject to phosphorylation in rat hippocampal slices, it has been proposed that the two bands represent the phosphorylated and unphosphorylated forms. In addition, transfection of HEK293T cells with a cDNA containing only the downstream methionine (corresponding to Met53, Fig.2C) yielded both a 33- and 36-kDa band (Seidenbecher et al., 1998). This is in contrast to our results, where a similar cDNA containing only Met53 (CD-S) yielded only the smaller protein (~33 kDa), and a cDNA containing the upstream methionine (Met1, CD-L, Fig.2A) yielded only the larger protein. The larger protein (~38 kDa) corresponds well to the predicted increase in molecular weight due to translation initiating at Met1 and migrates at the same position on SDS PAGE gels with the larger variant in the mouse brain lysates (Fig.2A). Although caldendrin possesses 7 putative protein kinase phosphorylation sites (Seidenbecher et al., 1998), it is unlikely that phosphorylation at all these sites could account for the larger band as the addition of phosphate to a given residue should cause only an 80 Da increase in molecular weight. Considering also the high level of species conservation of the sequence between Met1 and Met53 (Fig.2D,E), we propose that the two variants more likely represent translation from alternate start sites, a process known to regulate protein expression in neurons (Thomas et al., 2008). It is also possible that there could be “leaky mRNA scanning” of the first initiation methionine and translation at the second methionine because the first initiation site is in a suboptimal context (pyrimidine in position -3 before the ATG (Kozak, 1991)). This is not observed in HEK293 cells after transfection of CD-L, but it is possible that Met53 is a tissue-specific translation initiation site. Finally, we do not discount that the smaller variant represents a proteolytic degradation product of the larger variant. Regardless of the underlying mechanism, it will be important to determine if these variants have different functional properties in terms of how they may interact with and modulate effectors.
3.3 CaBP1/CD is localized in inhibitory as well as excitatory neurons
Similar to many Ca2+ binding proteins, CaBP1/CD shows a widespread but discrete localization pattern, with expression in subsets of neurons throughout the brain (Braunewell and Gundelfinger, 1999). Consistent with previous studies (Seidenbecher et al., 1998, Laube et al., 2002), we observed intense CaBP1/CD-IR in the soma and dendrites of excitatory, principal neurons throughout the forebrain. However, we also found strong CaBP1/CD-IR in parvalbumin-positive interneurons in the CA3 region of the hippocampus (Fig.6-8). In the hippocampus, parvalbumin is expressed in fast-spiking GABA-ergic interneurons that regulate gamma frequency oscillations during cognitive processing or memory tasks through perisomatic or axoaxonic synapses with pyramidal neurons (Gulyas et al., 2010). Activation of Cav1 channels aids the growth and maturation of parvalbumin-positive fast-spiking interneurons (Jiang and Swann, 2005), so caldendrin-mediated enhancement of Cav1 channel function may be important for this process. The restriction of hippocampal CaBP1/CD-IR to the CA3 region is distinct from parvalbumin and other Ca2+ binding proteins such as calretinin and calbindin, which show broader distributions in the CA1-CA3 regions, dentate gyrus, and subiculum (Hof et al., 1996). The localization of CaBP1/CD in the hippocampus is, however, similar to that of chromogranin B, a Ca2+ buffering protein that regulates intracellular Ca2+ release through IP3Rs (Nicolay et al., 2007).
In the cerebellum, the discrete localization of CaBP1/CD-IR in the basket cell pinceaux and perisomatic synapses formed with Purkinje neurons diverges from the predominantly somatodendritic localization of CaBP1/CD-IR throughout the brain (Fig.8). Basket cells inhibit Purkinje cell firing with rapid kinetics and distinct Ca2+ dependent release properties compared to other synapses (Sakaba, 2008). Cav2.1 P/Q-type Ca2+ channels are localized in the perisomatic basket cell terminals, and likely mediate this inhibitory transmission (Stephens et al., 2001). Given the strong inhibitory modulation of Cav2.1 by CaBP1 (Lee et al., 2002), CaBP1/CD may be important for shaping presynaptic Ca2+ signals that regulate precise timing of Purkinje cell inhibition. Although morphological evidence argues against a role for the pinceaux in the release of GABA onto Purkinje neurons (Iwakura et al., 2012), it has been proposed that the pinceau mediates a “field effect” whereby electrical activity in the basket cell terminal influences the membrane potential of the ensheathed Purkinje neurons (Korn and Axelrad, 1980). Although we did not observe gross motor impairment in C-KO mice, further analyses will be required in these mice to define a potential role of CaBP1/CD in controlling cerebellar output. Other CaBPs may also play a role in shaping cerebellar Ca2+ signaling. While CaBP2 and CaBP5 show no expression in the brain (Haeseleer et al., 2000), a recent study demonstrates the presence of CaBP4 transcripts in the cerebellum (Yang et al., 2014). Further studies will be necessary to define the cellular localization and potential function of CaBP4 in the cerebellum.
3.4 Clinical implications
Both caldendrin and one of its known partners, the Cav1.2 Ca2+ channel, are implicated in neuropsychiatric disease. The number of caldendrin-positive neurons is reduced in the prefrontal cortex of postmortem brains from chronic schizophrenics (Bernstein et al., 2007) and caldendrin levels at the postsynaptic density are reduced in a rat model of psychosis (Smalla et al., 2009). In addition, variations in the gene encoding Cav1.2, CACNA1C, are strongly associated with major depression, autism spectrum, schizophrenia, bipolar disorder, and attention deficit hyperactivity disorder (Cross-Disorder Group of the Psychiatric Genomics and Genetic Risk Outcome of Psychosis, 2013). Both caldendrin and Cav1.2 are highly expressed in brain regions known to regulate cognition and mood (Hell et al., 1993, Laube et al., 2002, Bernstein et al., 2003, Tippens et al., 2008). Reductions in caldendrin would lead to greater Ca2+-dependent inactivation of Cav1.2 channels (Tippens and Lee, 2007), which could cause weaker Ca2+ signaling with subsequent alterations in neuronal excitability. Thus, the C-KO mouse may provide a valuable resource in understanding how dysregulation of caldendrin and Cav1.2 may derail neurophysiological processes underlying the pathogenesis of some forms of mental illness.
CaBP1 and caldendrin are alternate splice variants of Ca2+ binding proteins related to calmodulin
Caldendrin is the major variant and is developmentally upregulated in the cerebral cortex
Antibodies recognizing both variants label select groups of excitatory and inhibitory neurons
CaBP1/caldendrin immunostaining is absent in CaBP1/caldendrin knockout mice
CaBP1/caldendrin may regulate Ca2+ signaling from both pre- and post- synaptic sites of action
ACKNOWLEDGEMENTS
This work was supported by the NIH (DC009433, NS084190 to A.L.; DC010362 to the Iowa Center for Molecular Auditory Neuroscience; EY020850 to F.H.) and a Carver Research Program of Excellence Award to A.L. The authors thank Baoli Yang (U. Iowa Gene Targeting Core) for guidance on generating CaBP1/caldendrin KO mice, Daniel Seoh for care of mouse colonies and DP Mohapatra for support and the use of equipment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Augustine GJ, Santamaria F, Tanaka K. Local calcium signaling in neurons. Neuron. 2003;40:331–346. doi: 10.1016/s0896-6273(03)00639-1. [DOI] [PubMed] [Google Scholar]
- Bastianelli E. Distribution of calcium-binding proteins in the cerebellum. Cerebellum. 2003;2:242–262. doi: 10.1080/14734220310022289. [DOI] [PubMed] [Google Scholar]
- Bernstein HG, Sahin J, Smalla KH, Gundelfinger ED, Bogerts B, Kreutz MR. A reduced number of cortical neurons show increased Caldendrin protein levels in chronic schizophrenia. Schizophr Res. 2007;96:246–256. doi: 10.1016/j.schres.2007.05.038. [DOI] [PubMed] [Google Scholar]
- Bernstein HG, Seidenbecher CI, Smalla KH, Gundelfinger ED, Bogerts B, Kreutz MR. Distribution and cellular localization of caldendrin immunoreactivity in adult human forebrain. J Histochem Cytochem. 2003;51:1109–1112. doi: 10.1177/002215540305100816. [DOI] [PubMed] [Google Scholar]
- Bolsover SR. Calcium signalling in growth cone migration. Cell calcium. 2005;37:395–402. doi: 10.1016/j.ceca.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Braunewell KH, Gundelfinger ED. Intracellular neuronal calcium sensor proteins: A Family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 1999;295:1–12. doi: 10.1007/s004410051207. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD, O'Callaghan DW, Hasdemir B, Haynes LP, Tepikin AV. Neuronal Ca2+-sensor proteins: multitalented regulators of neuronal function. Trends Neurosci. 2004;27:203–209. doi: 10.1016/j.tins.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Chan CS, Guzman JN, Ilijic E, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. ‘Rejuvenation’ protects neurons in mouse models of Parkinson's disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics C. Genetic Risk Outcome of Psychosis C Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Meyer AC, Calin-Jageman I, Neef J, Haeseleer F, Moser T, Lee A. Ca2+-binding proteins tune Ca2+-feedback to Cav1.3 channels in auditory hair cells. J Physiol. 2007;585:791–803. doi: 10.1113/jphysiol.2007.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, Reissner C, Boeckers TM, Zuschratter W, Spilker C, Seidenbecher CI, Garner CC, Gundelfinger ED, Kreutz MR. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS biology. 2008;6:e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen F, Minor DL., Jr Structural basis for the differential effects of CaBP1 and calmodulin on Ca(V)1.2 calcium-dependent inactivation. Structure. 2010;18:1617–1631. doi: 10.1016/j.str.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise N, Liu Y, Hori N, Kosaka T. Distribution of calretinin immunoreactivity in the mouse dentate gyrus: II. Mossy cells, with special reference to their dorsoventral difference in calretinin immunoreactivity. Neuroscience. 1998;82:181–200. doi: 10.1016/s0306-4522(97)00261-3. [DOI] [PubMed] [Google Scholar]
- Gambrill AC, Storey GP, Barria A. Dynamic regulation of NMDA receptor transmission. J Neurophysiol. 2011;105:162–171. doi: 10.1152/jn.00457.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hernandez T, Rodriguez M. Compartmental organization and chemical profile of dopaminergic and GABAergic neurons in the substantia nigra of the rat. J Comp Neurol. 2000;421:107–135. doi: 10.1002/(sici)1096-9861(20000522)421:1<107::aid-cne7>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- Gorny X, Mikhaylova M, Seeger C, Reddy PP, Reissner C, Schott BH, Helena Danielson U, Kreutz MR, Seidenbecher C. AKAP79/150 interacts with the neuronal calcium-binding protein caldendrin. J Neurochem. 2012;122:714–726. doi: 10.1111/j.1471-4159.2012.07828.x. [DOI] [PubMed] [Google Scholar]
- Gulyas AI, Miettinen R, Jacobowitz DM, Freund TF. Calretinin is present in non-pyramidal cells of the rat hippocampus--I. A new type of neuron specifically associated with the mossy fibre system. Neuroscience. 1992;48:1–27. doi: 10.1016/0306-4522(92)90334-x. [DOI] [PubMed] [Google Scholar]
- Gulyas AI, Szabo GG, Ulbert I, Holderith N, Monyer H, Erdelyi F, Szabo G, Freund TF, Hajos N. Parvalbumin-containing fast-spiking basket cells generate the field potential oscillations induced by cholinergic receptor activation in the hippocampus. J Neurosci. 2010;30:15134–15145. doi: 10.1523/JNEUROSCI.4104-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, Lee A, Rieke F, Palczewski K. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci. 2004;7:1079–1087. doi: 10.1038/nn1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F, Sokal I, Verlinde CL, Erdjument-Bromage H, Tempst P, Pronin AN, Benovic JL, Fariss RN, Palczewski K. Five members of a novel Ca2+-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem. 2000;275:1247–1260. doi: 10.1074/jbc.275.2.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes LP, Tepikin AV, Burgoyne RD. Calcium-binding protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphosphate-mediated calcium signaling. J Biol Chem. 2004;279:547–555. doi: 10.1074/jbc.M309617200. [DOI] [PubMed] [Google Scholar]
- Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel a1 subunits. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Rosenthal RE, Fiskum G. Distribution of neurofilament protein and calcium-binding proteins parvalbumin, calbindin, and calretinin in the canine hippocampus. J Chem Neuroanat. 1996;11:1–12. doi: 10.1016/0891-0618(96)00117-2. [DOI] [PubMed] [Google Scholar]
- Iwakura A, Uchigashima M, Miyazaki T, Yamasaki M, Watanabe M. Lack of molecular-anatomical evidence for GABAergic influence on axon initial segment of cerebellar Purkinje cells by the pinceau formation. J Neurosci. 2012;32:9438–9448. doi: 10.1523/JNEUROSCI.1651-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Swann JW. A role for L-type calcium channels in the maturation of parvalbumin-containing hippocampal interneurons. Neuroscience. 2005;135:839–850. doi: 10.1016/j.neuroscience.2005.06.073. [DOI] [PubMed] [Google Scholar]
- Jones RS, Buhl EH. Basket-like interneurones in layer II of the entorhinal cortex exhibit a powerful NMDA-mediated synaptic excitation. Neuroscience letters. 1993;149:35–39. doi: 10.1016/0304-3940(93)90341-h. [DOI] [PubMed] [Google Scholar]
- Korn H, Axelrad H. Electrical inhibition of Purkinje cells in the cerebellum of the rat. Proc Natl Acad Sci U S A. 1980;77:6244–6247. doi: 10.1073/pnas.77.10.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. JBiolChem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kretsinger RH. Calcium-binding proteins. Annu Rev Biochem. 1976;45:239–266. doi: 10.1146/annurev.bi.45.070176.001323. [DOI] [PubMed] [Google Scholar]
- Laube G, Roper J, Pitt JC, Sewing S, Kistner U, Garner CC, Pongs O, Veh RW. Ultrastructural localization of Shaker-related potassium channel subunits and synapse-associated protein 90 to septate-like junctions in rat cerebellar Pinceaux. Brain Res Mol Brain Res. 1996;42:51–61. doi: 10.1016/s0169-328x(96)00120-9. [DOI] [PubMed] [Google Scholar]
- Laube G, Seidenbecher CI, Richter K, Dieterich DC, Hoffmann B, Landwehr M, Smalla KH, Winter C, Bockers TM, Wolf G, Gundelfinger ED, Kreutz MR. The neuron-specific Ca2+-binding protein caldendrin: gene structure, splice isoforms, and expression in the rat central nervous system. Mol Cell Neurosci. 2002;19:459–475. doi: 10.1006/mcne.2001.1078. [DOI] [PubMed] [Google Scholar]
- Lee A, Jimenez A, Cui G, Haeseleer F. Phosphorylation of Ca2+ binding protein CaBP4 by protein kinase C zeta in photoreceptors. J Neurosci. 2007;27:12743–12754. doi: 10.1523/JNEUROSCI.4264-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Westenbroek RE, Haeseleer F, Palczewski K, Scheuer T, Catterall WA. Differential modulation of Cav2.1 channels by calmodulin and Ca2+-binding protein 1. Nat Neurosci. 2002;5:210–217. doi: 10.1038/nn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littink KW, van Genderen MM, Collin RW, Roosing S, de Brouwer AP, Riemslag FC, Venselaar H, Thiadens AA, Hoyng CB, Rohrschneider K, den Hollander AI, Cremers FP, van den Born LI. A novel homozygous nonsense mutation in CABP4 causes congenital cone-rod synaptic disorder. Invest Ophthalmol Vis Sci. 2009;50:2344–2350. doi: 10.1167/iovs.08-2553. [DOI] [PubMed] [Google Scholar]
- Mansergh F, Orton NC, Vessey JP, Lalonde MR, Stell WK, Tremblay F, Barnes S, Rancourt DE, Bech-Hansen NT. Mutation of the calcium channel gene Cacna1f disrupts calcium signaling, synaptic transmission and cellular organization in mouse retina. Hum Mol Genet. 2005;14:3035–3046. doi: 10.1093/hmg/ddi336. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Beaulieu C. Quantitative aspects of synaptogenesis in the rat barrel field cortex with special reference to GABA circuitry. J Comp Neurol. 1996;373:340–354. doi: 10.1002/(SICI)1096-9861(19960923)373:3<340::AID-CNE3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Mintz IM, Adams ME, Bean BP. P-type calcium channels in rat central and peripheral neurons. Neuron. 1992;9:85–95. doi: 10.1016/0896-6273(92)90223-z. [DOI] [PubMed] [Google Scholar]
- Nicolay NH, Hertle D, Boehmerle W, Heidrich FM, Yeckel M, Ehrlich BE. Inositol 1,4,5 triphosphate receptor and chromogranin B are concentrated in different regions of hippocampus. J Neurosci Res. 2007;85:2026–2036. doi: 10.1002/jnr.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz S, Tsemakhovich V, Christel CJ, Lee A, Dascal N. CaBP1 regulates voltage-dependent inactivation and activation of Cav1.2 (L-type) calcium channels. The Journal of biological chemistry. 2011;286:13945–13953. doi: 10.1074/jbc.M110.198424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay V. Cerebellar Cortex, Cytology and Organization. Springer Verlag; New York: 1974. [Google Scholar]
- Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Sakaba T. Two Ca2+-dependent steps controlling synaptic vesicle fusion and replenishment at the cerebellar basket cell terminal. Neuron. 2008;57:406–419. doi: 10.1016/j.neuron.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Schrauwen I, Helfmann S, Inagaki A, Predoehl F, Tabatabaiefar MA, Picher MM, Sommen M, Seco CZ, Oostrik J, Kremer H, Dheedene A, Claes C, Fransen E, Chaleshtori MH, Coucke P, Lee A, Moser T, Van Camp G. A mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. American journal of human genetics. 2012;91:636–645. doi: 10.1016/j.ajhg.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller B, Meyer M, Schiffmann S. ‘New’ functions for ‘old’ proteins: the role of the calcium-binding proteins calbindin D-28k, calretinin and parvalbumin, in cerebellar physiology. Studies with knockout mice. Cerebellum. 2002;1:241–258. doi: 10.1080/147342202320883551. [DOI] [PubMed] [Google Scholar]
- Seidenbecher CI, Landwehr M, Smalla KH, Kreutz M, Dieterich DC, Zuschratter W, Reissner C, Hammarback JA, Bockers TM, Gundelfinger ED, Kreutz MR. Caldendrin but not calmodulin binds to light chain 3 of MAP1A/B: an association with the microtubule cytoskeleton highlighting exclusive binding partners for neuronal Ca2+-sensor proteins. J Mol Biol. 2004;336:957–970. doi: 10.1016/j.jmb.2003.12.054. [DOI] [PubMed] [Google Scholar]
- Seidenbecher CI, Langnaese K, Sanmarti-Vila L, Boeckers TM, Smalla KH, Sabel BA, Garner CC, Gundelfinger ED, Kreutz MR. Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J Biol Chem. 1998;273:21324–21331. doi: 10.1074/jbc.273.33.21324. [DOI] [PubMed] [Google Scholar]
- Shaltiel L, Paparizos C, Fenske S, Hassan S, Gruner C, Rotzer K, Biel M, Wahl-Schott CA. Complex regulation of voltage-dependent activation and inactivation properties of retinal voltage-gated Cav1.4 L-type Ca2+ channels by Ca2+-binding protein 4 (CaBP4). J Biol Chem. 2012;287:36312–36321. doi: 10.1074/jbc.M112.392811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla KH, Sahin J, Putzke J, Tischmeyer W, Gundelfinger ED, Kreutz MR. Altered postsynaptic-density-levels of caldendrin in the para-chloroamphetamine-induced serotonin syndrome but not in the rat ketamine model of psychosis. Neurochemical research. 2009;34:1405–1409. doi: 10.1007/s11064-009-9925-8. [DOI] [PubMed] [Google Scholar]
- Stephens GJ, Morris NP, Fyffe RE, Robertson B. The Cav2.1/alpha1A (P/Q-type) voltage-dependent calcium channel mediates inhibitory neurotransmission onto mouse cerebellar Purkinje cells. Eur J Neurosci. 2001;13:1902–1912. doi: 10.1046/j.0953-816x.2001.01566.x. [DOI] [PubMed] [Google Scholar]
- Thomas D, Plant LD, Wilkens CM, McCrossan ZA, Goldstein SA. Alternative translation initiation in rat brain yields K2P2.1 potassium channels permeable to sodium. Neuron. 2008;58:859–870. doi: 10.1016/j.neuron.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippens AL, Lee A. Caldendrin: a neuron-specific modulator of Cav1.2 (L-type) Ca2+ channels. J Biol Chem. 2007;282:8464–8473. doi: 10.1074/jbc.M611384200. [DOI] [PubMed] [Google Scholar]
- Tippens AL, Pare J-F, Langwieser N, Moosmang S, Milner TA, Smith Y, Lee A. Ultrastructural evidence for pre- and post-synaptic localization of Cav1.2 L-type Ca2+ channels in the rat hippocampus. J Comp Neurol. 2008;506:569–583. doi: 10.1002/cne.21567. [DOI] [PubMed] [Google Scholar]
- Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6:267–276. doi: 10.1038/nrn1647. [DOI] [PubMed] [Google Scholar]
- Yang J, McBride S, Mak DO, Vardi N, Palczewski K, Haeseleer F, Foskett JK. Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2+ release channels. Proc Natl Acad Sci U S A. 2002;99:7711–7716. doi: 10.1073/pnas.102006299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PS, Alseikhan BA, Hiel H, Grant L, Mori MX, Yang W, Fuchs PA, Yue DT. Switching of Ca2+-dependent inactivation of Cav1.3 channels by calcium binding proteins of auditory hair cells. J Neurosci. 2006;26:10677–10689. doi: 10.1523/JNEUROSCI.3236-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PS, Johny Yue DT. Allostery in Ca2+ channel modulation by calcium-binding proteins. Nat Chem Biol. 2014 doi: 10.1038/nchembio.1436. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C, Kloeckener-Gruissem B, Forster U, Kohl S, Magyar I, Wissinger B, Matyas G, Borruat FX, Schorderet DF, Zrenner E, Munier FL, Berger W. Mutations in CABP4, the Gene Encoding the Ca2+-Binding Protein 4, Cause Autosomal Recessive Night Blindness. American journal of human genetics. 2006;79:657–667. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Kim SA, Kirk EA, Tippens AL, Sun H, Haeseleer F, Lee A. Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Cav 1.2 (L-type) Ca2+ channels. J Neurosci. 2004;24:4698–4708. doi: 10.1523/JNEUROSCI.5523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yu K, McCoy KL, Lee A. Molecular mechanism for divergent regulation of Cav 1.2 Ca2+ channels by calmodulin and Ca2+-binding protein-1. J Biol Chem. 2005;280:29612–29619. doi: 10.1074/jbc.M504167200. [DOI] [PubMed] [Google Scholar]