Significance
The ability of trimethylamine N-oxide (TMAO) to counteract osmotic stress along with the deleterious effects of denaturants such as urea is fascinating and has inspired many studies. To further our understanding of how TMAO acts to stabilize folded proteins, we carry out an infrared experiment designed to probe the microscopic details of this action explicitly from the perspective of the solute molecule. Our results reveal that the protein-stabilizing effect of TMAO originates from two contributions: One is entropic and the other is enthalpic in nature. Thus, this study provides not only microscopic details underlying the stabilizing action of TMAO, but also a method that can be used to study the stability-perturbing effect of other cosolvents.
Keywords: infrared, crowding, 2D IR, linear response function
Abstract
Although it is widely known that trimethylamine N-oxide (TMAO), an osmolyte used by nature, stabilizes the folded state of proteins, the underlying mechanism of action is not entirely understood. To gain further insight into this important biological phenomenon, we use the C≡N stretching vibration of an unnatural amino acid, p-cyano-phenylalanine, to directly probe how TMAO affects the hydration and conformational dynamics of a model peptide and a small protein. By assessing how the lineshape and spectral diffusion properties of this vibration change with cosolvent conditions, we are able to show that TMAO achieves its protein-stabilizing ability through the combination of (at least) two mechanisms: (i) It decreases the hydrogen bonding ability of water and hence the stability of the unfolded state, and (ii) it acts as a molecular crowder, as suggested by a recent computational study, that can increase the stability of the folded state via the excluded volume effect.
Nature employs a variety of small organic molecules to cope with osmotic stress. Trimethylamine N-oxide (TMAO) is one such naturally occurring osmolyte that protects intracellular components against disruptive stress conditions (1). In particular, previous studies have shown that TMAO is able to enhance protein stability and to counteract the denaturing effect of urea (2, 3). TMAO (Fig. 1, Inset) adopts a skewed tetrahedral structure with a charged oxygen capable of accepting hydrogen bonds (H bonds) and three hydrophobic (methyl) groups. This amphiphilic structural arrangement makes TMAO a rather special cosolvent, because it can form H bonds with water, self-associate in a manner similar to surfactants, and show preferential interactions with or exclusion (4–12) from certain protein functional groups (13–23). Indeed, these molecular properties of TMAO have been used, either individually or in combination, to rationalize its biological activities. For example, a prevailing view about TMAO is that its osmotic and protective role is caused by the molecule’s tendency to be preferentially depleted from protein surfaces, as suggested by physicochemical measurements (24–28). This thermodynamic picture, as described by Bolen and coworkers (29), implies that the protein is preferentially hydrated. Although this notion is consistent with TMAO’s being a protecting osmolyte, to the best of our knowledge no experimental studies have been carried out to directly examine the effect of TMAO on protein hydration dynamics, an aspect that is also important to protein function. Herein, we use two-dimensional infrared (2D IR) spectroscopy and a site-specific IR probe to explore this critical issue and gain insight toward achieving a microscopic understanding of the molecular mechanism of the protecting action of TMAO.
Fig. 1.
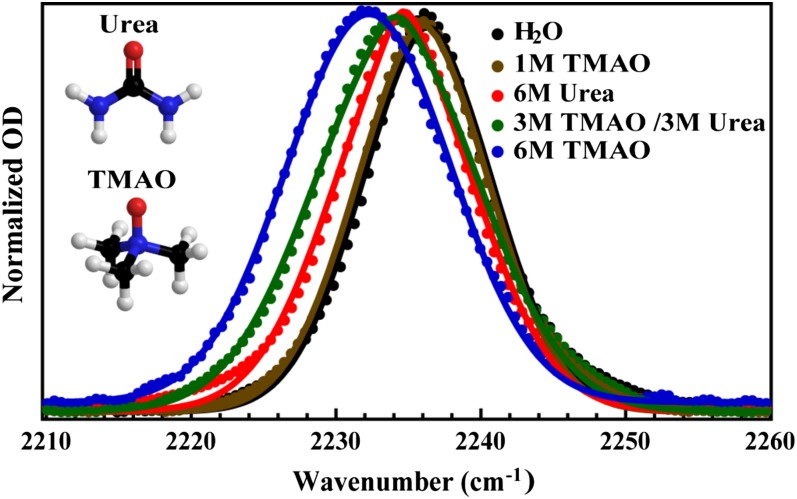
The C≡N stretch bands of GFCNG obtained under different solvent conditions, as indicated. The solid lines are fits of these spectra to the linear response function using Eqs. 2–4 in the text and the resulting fitting parameters are listed in Table 1. The structures of urea and TMAO are shown as insets.
2D IR spectroscopy is capable of assessing the frequency–frequency correlation function (FFCF) of a given IR probe, which reports on the underlying dynamics of events that lead to fluctuations of its vibrational frequency (30). For an IR probe that is able to interact with water via H bonding, measurement of its FFCF can provide, sometimes in a site-specific manner, detailed information about the hydration dynamics of the protein molecule of interest. For example, this approach has been used to identify the existence of mobile water molecules inside Aβ40 amyloid fibrils (31, 32) and to interrogate the water-assisted drug-binding mechanism of HIV-1 reverse transcriptase (33), among many other applications (34–39). In the current study, we capitalize on the established sensitivity of the nitrile stretching vibration (C≡N) to local hydration and electrostatic environment (40) and use the unnatural amino acid p-cyano-phenyalanine (PheCN) as a local IR reporter. The advantages of using the C≡N stretching vibration of PheCN are that (i) it is located in a spectrally uncongested region (2,000−2,400 cm−1), where water has a relatively low absorbance; (ii) it has a reasonably large extinction coefficient; and (iii) it is, in most cases, a simple transition that is decoupled from other vibrational modes of the molecule (41).
Recently, we have shown that upon addition of TMAO, the C≡N stretching vibration of PheCN in a short peptide shifts to a lower frequency compared with that obtained in pure water (42). As shown (Fig. 1), the peptide studied in the current case, Gly-PheCN-Gly (hereafter referred to as GFCNG), exhibits the same trend. Taken together, these results suggest that TMAO weakens the strength of the H bond formed between the nitrile group and water molecules. Whereas this finding is consistent with the ability of TMAO to increase protein stability, a microscopic picture regarding how TMAO alters protein–water interactions has yet to be established. To provide new insights into the molecular mechanism of the protecting action of TMAO, herein we measure the FFCF of the nitrile probe in two model systems, the tripeptide GFCNG and the villin headpiece subdomain (HP35) with PheCN mutations under different cosolvent conditions. In particular, we are able to extract both the homogeneous and the inhomogeneous contributions to the total lineshape of the C≡N stretching band and, perhaps more importantly, to provide a dynamic view, from the perspective of the PheCN sidechain, of how a given solution condition (i.e., in the presence of TMAO, urea, or both) affects the hydration dynamics of the peptide in question (43). Our results suggest that TMAO molecules produce a local protein environment that strengthens backbone–backbone H bonds and also reduces the conformational entropy of the system. In other words, TMAO increases protein stability by acting as a nano-crowder, as proposed by Thirumalai, Straub and coworkers based on computer simulations (44).
Results
Consistent with our previous study, TMAO affects the C≡N stretching band of GFCNG by shifting its peak to a lower frequency and broadening its width (42). Although these changes in lineshape indicate a change in the local electrostatic and H-bonding environment of the C≡N probe, and thus contain information about the molecular effect of TMAO, a quantitative assessment of the factors underlying these changes, based on linear IR measurements alone, is not feasible. Thus, herein we combine data from linear and nonlinear IR measurements and use response function theory to fully characterize the lineshape of the C≡N stretching vibrations obtained under different solvent conditions. This analysis allows us to dissect the homogeneous and inhomogeneous contributions to the linear IR spectrum of interest, which provides insights into the mechanism of action of TMAO.
2D IR Photon Echo Spectra of GFCNG.
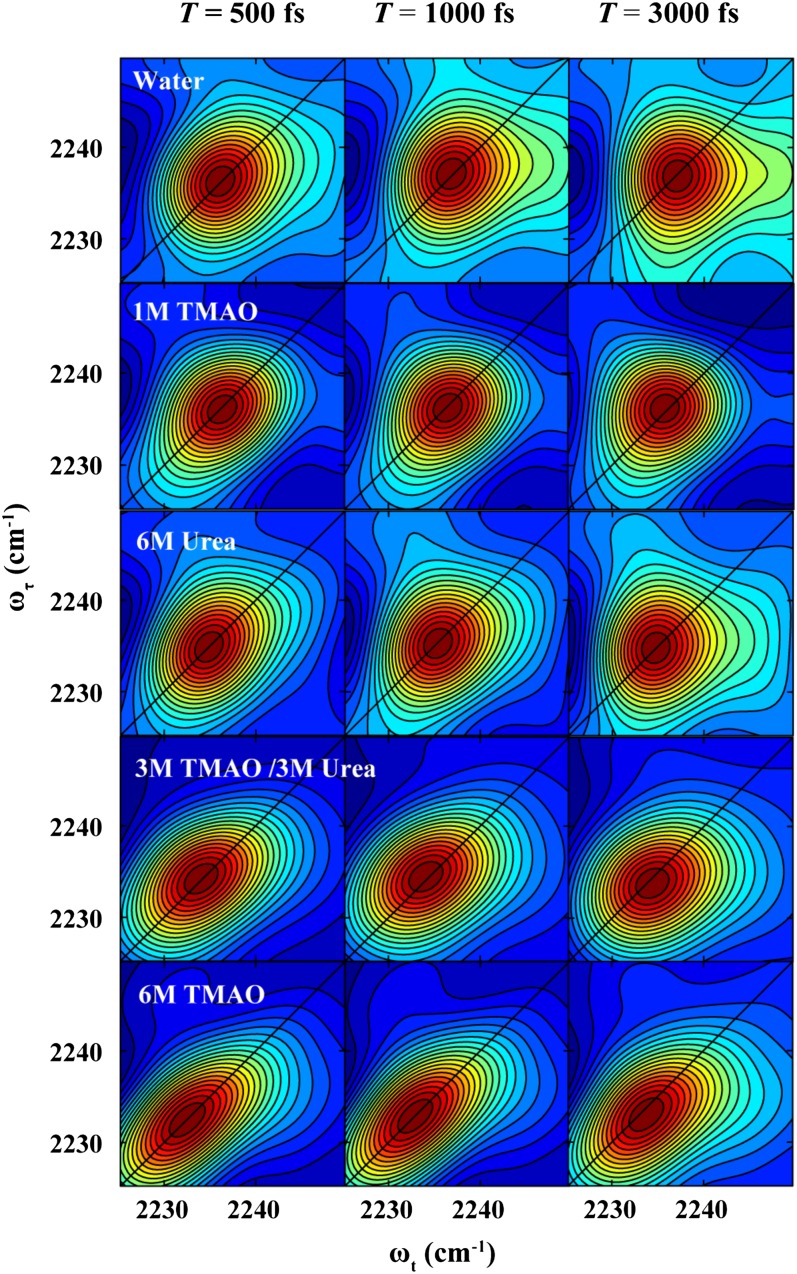
As indicated (Fig. 2), the absorptive 2D IR spectra of the nitrile group of GFCNG, collected at different waiting times (T) and solution conditions, clearly show that the 2D lineshapes and the spectral diffusion dynamics of the C≡N stretching vibration depend on the cosolvent. For example, at T = 500 fs, the 2D IR lineshape of this vibration in water becomes more circular than that in 6 M TMAO, suggesting that adding TMAO significantly changes contributions from the homogenous part, as well as the inhomogenous part, to the total lineshape of the vibrational transition. To provide a more quantitative assessment of how a particular cosolvent affects the microscopic environment of the nitrile probe and hence its vibrational transition, below we examine how the key spectral and dynamic properties of this vibrator change with solution condition.
Fig. 2.
Representative absorptive 2D IR photon echo spectra of GFCNG obtained under different solvent conditions and at different waiting times (T), as indicated.
First, we compare the vibrational lifetimes (T1) of the nitrile probe obtained under different solution conditions. As shown (Table 1), the T1 of the C≡N stretching vibration, consistent with previous studies (45, 46), is insensitive to solvent and, thus, is not a good reporter of its local environment. This finding is consistent with the study of Cho and coworkers (47), which showed that the vibrational relaxation of a nitrile moiety attached to a large molecule in water is dominated by intramolecular pathways. In addition, the measured T1 value (∼4 ps) indicates that in this case the lifetime-broadening contribution (∼1.3 cm−1) to the spectral width (10−15 cm−1) is small.
Table 1.
Spectroscopic parameters and lifetimes (T1) of the C≡N stretching vibration of GFCNG in different solvent conditions
| Cosolvent condition | ω0, cm−1 | FWHM, cm−1 | T1, ps | Γ, cm−1 | Δ, cm−1 | Δs, cm−1 |
| PheCN* | 2,236.3 | 12.2 | 4.5 | 5.8 ± 0.1 | 2.0 ± 0.1 | — |
| H2O | 2,236.3 | 10.4 | 4.2 ± 0.1 | 5.8 ± 0.1 | 2.1 ± 0.2 | 0 |
| 6 M urea | 2,234.7 | 11.2 | 4.0 ± 0.2 | 5.2 ± 0.2 | 1.8 ± 0.1 | 1.0 ± 0.2 |
| 1 M TMAO | 2,236.0 | 10.6 | 4.1 ± 0.2 | 5.1 ± 0.2 | 1.7 ± 0.1 | 1.2 ± 0.2 |
| TMAO/urea (3 M/3 M) | 2,234.2 | 13.3 | 3.8 ± 0.2 | 5.0 ± 0.2 | 2.0 ± 0.2 | 2.2 ± 0.1 |
| 6 M TMAO | 2,232.3 | 13.4 | 3.9 ± 0.2 | 4.8 ± 0.2 | 2.0 ± 0.2 | 2.9 ± 0.3 |
The PheCN parameters are taken from ref. 37, which were measured in water. The center frequency (ω0) and width (FWHM) were determined by fitting the linear IR spectra to a Gaussian function. The T1 values were determined by fitting the isotropic transient grating signals to an exponential function. Other lineshape parameters were obtained by fitting the C≡N stretching bands in Fig. 1 to Eqs. 2–4.
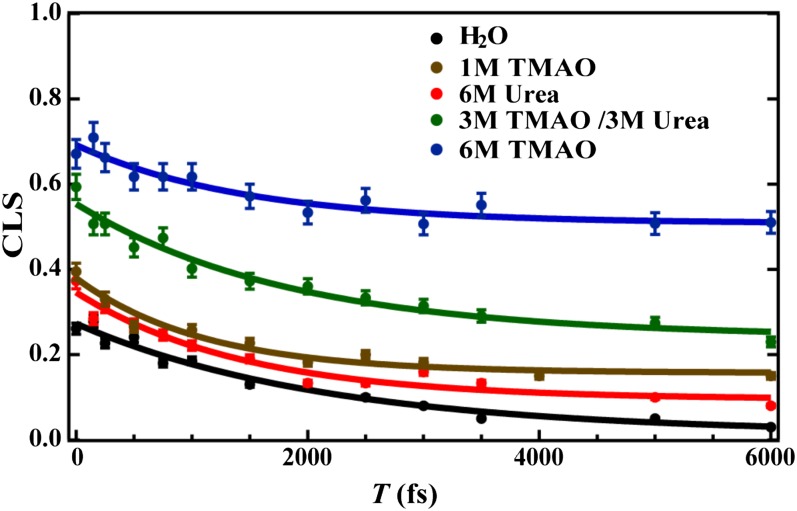
Second, we compare the effects of cosolvent on the spectral diffusion of this probe, using the center line slope (CLS) method developed by Fayer and coworkers (48). A center line is defined as the line that connects the maxima of the peaks of a series of cuts through a particular 2D IR spectrum that are parallel to the ωτ frequency axis. It has been shown that, within the short time approximation, the waiting-time (i.e., T) dependence of the CLS directly reports on the normalized FFCF of the vibrational probe in question (48). As indicated (Fig. 3), the CLS (T) of the nitrile probe, obtained under all solution conditions, can be described by the following equation:
| [1] |
where A, B, and τ are constants. The exponential term in Eq. 1 is often referred to as the Kubo term, which, in the current case, most likely reports on the dynamics of H-bond formation and breaking between the nitrile group and water, which cause the vibrational frequencies to fluctuate (49–51). As shown (Table 2), the decay time constant of this Kubo term determined for GFCNG in water is about 2.2 ps, which is in good agreement with previously reported values for the nitrile stretching vibration in aqueous solutions (30, 37, 46, 52). In addition, the offset term B obtained in water is practically zero. However, adding a cosolvent, either TMAO or urea, results in a measurable decrease in τ and an appreciable increase in B and, interestingly, TMAO is more effective than urea at increasing this offset. Fayer and coworkers (48) have shown, assuming that the system follows Bloch dynamics, that this offset term is related to the total inhomogeneous component of the lineshape. Thus, these results suggest that TMAO can effectively slow down or even suppress some motions that lead to local environmental changes of the nitrile probe and, hence, its spectral diffusion.
Fig. 3.
CLS versus T plots of GFCNG obtained under different solvent conditions, as indicated. The solid lines are fits of these data to Eq. 1 with those fitting parameters listed in Table 2.
Table 2.
| Cosolvent condition | A | τ, ps | B |
| H2O | 0.26 ± 0.02 | 2.17 ± 0.43 | 0.01 ± 0.02 |
| 6 M urea | 0.20 ± 0.01 | 1.44 ± 0.30 | 0.15 ± 0.30 |
| 1 M TMAO | 0.22 ± 0.02 | 1.30 ± 0.43 | 0.18 ± 0.02 |
| TMAO/urea (3 M/3 M) | 0.31 ± 0.02 | 1.85 ± 0.39 | 0.24 ± 0.02 |
| 6 M TMAO | 0.18 ± 0.02 | 1.46 ± 0.43 | 0.51 ± 0.02 |
To provide a more quantitative assessment of the cosolvent-induced changes in the spectral and dynamic properties of the nitrile stretching vibration, we calculate the homogeneous and inhomogeneous contributions to the total lineshape, by analyzing the linear IR spectrum using the linear response function (30). Specifically, we fit the IR spectrum, SIR(ω), to the following function (49):
| [2] |
where T1 is the vibrational lifetime, ω0 is the center frequency, and g(t) is the vibrational lineshape function, defined as
| [3] |
where C(t) represents the full FFCF of the IR probe and is described by the following equation (53):
| [4] |
where the first term represents the motional narrowing component determined by the pure dephasing time (T2*), the second term represents a static or slowly varying component, and the third term is the aforementioned Kubo term. As shown (Fig. 1), all of the linear IR spectra can be reasonably well described by Eq. 2 and the resultant fitting parameters (i.e., T2*, Δs, and Δ) are listed in Table 1. In particular, the parameters obtained for water are in good agreement with those previously reported for this probe (37). With the knowledge of T2*, we further calculated the pure dephasing linewidth (Γ) for each case using the relationship of Γ = 1/πT2*.
In the current case, we have neglected the contribution from Trot, which represents the rotational relaxation time of the probe (54) and is expected to be much longer than T1 or T2*. As indicated (Table 1), the spectral parameters produced from the above analysis clearly show that the pure dephasing linewidth (Γ) of the C≡N stretching vibration decreases in the following order: H2O > 6 M urea > 1 M TMAO > TMAO/urea (3 M/3 M) > 6 M TMAO, whereas the total inhomogeneous component (i.e., ) follows the exact opposite trend [i.e., H2O < 6 M urea < 1 M TMAO < TMAO/urea (3 M/3 M) < 6 M TMAO]. Moreover, the contribution from the Kubo term (i.e., Δ2) decreases upon adding cosolvents to the system, which, consistent with the above CLS analysis, indicates that cosolvents weaken the H bonding between the C≡N probe and water molecules.
As shown in Table 1, the most striking differences for the C≡N stretching vibration under different solution conditions are in the values of the static term . The origin of this static term, for large molecules such as proteins, is generally attributed to motions that can affect the local electrostatic environment of the IR probe but occur on a timescale that is much slower than the vibrational lifetime or the effective time window of the experiment, such as backbone conformational changes (55–57). Interestingly, in the current case, the static component obtained in water is nearly zero, indicating that during the time window of the experiment the nitrile probe can exhaustively sample, statistically speaking, all possible environments. This is consistent with the notion that the spectral diffusion of the nitrile probe in pure water is dominated by water H-bonding dynamics, which occur on the timescale of 1−3 ps (58). Furthermore, and perhaps more interestingly, adding TMAO to the peptide solution leads to a significant increase in the static component, which indicates that TMAO not only broadens the frequency distribution of the C≡N stretching vibration (Fig. 1), but also “freezes” a certain number of motions leading to vibrational frequency fluctuations. As shown in Table 1, although urea exhibits a similar effect, the net influence, in comparison with that of TMAO at the same concentration, is much smaller. Garcia and coworkers (59) have shown that the osmotic pressure of TMAO exhibits a positive deviation from its ideal value, whereas that of urea shows a negative deviation. Thus, a more rigorous comparison between the effects of TMAO and urea would require measurements on TMAO and urea samples that have identical osmotic pressures.
Linear IR and 2D IR Spectra of HP35 Mutants.
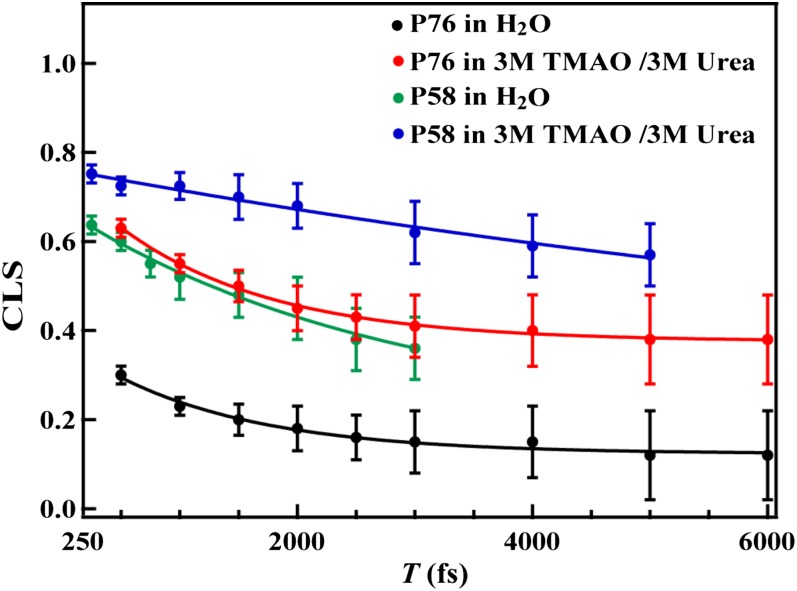
To confirm that the conclusions reached above are not limited to short peptides, we also performed similar experiments on two PheCN mutants of a well-studied helical protein, HP35 (60, 61). The first variant corresponds to a Phe76/PheCN mutation at the C terminus of the protein (referred to as HP35-P76), wherein the PheCN residue is exposed to solvent. Because several studies have suggested that protecting osmolytes, such as TMAO, could directly interact with aromatic sidechains (14), the second mutant we chose to study contains a Phe58/PheCN mutation at the core of the protein (referred to as HP35-P58). Because the PheCN residue in HP35-P58 is, to a large extent, buried in a hydrophobic environment (37, 53, 61), it is unlikely that TMAO will show any direct interactions with the nitrile probe. Thus, results obtained from both mutants will allow us to gain a more complete assessment of the effect of TMAO on protein stability. Unfortunately, we found that HP35 readily aggregates even in 1 M TMAO, which prevented us from making a direct evaluation of the effect of TMAO on the C≡N stretching vibration of both HP35 variants. To circumvent this problem, we evaluated the effect of TMAO in the presence of urea, which is known to help solubilize the protein. Specifically, we used a solution condition that was used in the study of GFCNG [i.e., TMAO/urea (3 M/3 M)]. As indicated, both the linear and 2D IR spectra of HP35-P76 (Figs. S1 and S2 and Table S1) show that the cosolvent-induced changes in the spectral and dynamic properties of the C≡N stretching vibration of HP35-P76 are very similar to those observed for the GFCNG peptide under the same solution conditions. For example, the 2D lineshape, at T = 500 fs, obtained in pure water is distinctly more circular than that measured in the presence of cosolvent. A more quantitative analysis using the method discussed above reveals that the cosolvent-induced spectral property changes in this case are similar to those obtained with GFCNG (Table 3), indicating that the findings for GFCNG, in terms of the effect of TMAO, are not unique to short peptides, but rather are generally applicable to proteins. Similar to that observed for HP35-P76, addition of the same cosolvent mixture results in a shift of the C≡N stretching frequency of HP35-P58 to lower wavenumbers (Fig. S3 and Table S2). However, the CLS of HP35-P58 obtained from its 2D IR spectra (Fig. S4) in water shows a slower decay and also a significant offset (Fig. 4 and Table 3). This is consistent with the study of Fayer and coworkers (37, 53, 61), which showed that the frequency fluctuation dynamics of the C≡N probe inside the hydrophobic core of HP35 are slowed down. However, addition of TMAO/urea (3 M/3 M) leads to a further increase in the amplitude of this offset (Fig. 4), indicating that the nitrile probe samples a more slowly varying or rigid environment (53). Because, in this case, the PheCN residue is unlikely to directly interact with cosolvent molecules, this increase in protein rigidity thus most likely arises from the TMAO-induced crowding effect, as discussed above.
Table 3.
| HP35-P76 (HP35-P58) | Γ, cm−1 | Δ, cm−1 | Δs, cm−1 |
| H2O | 5.6 ± 0.4 (4.8 ± 0.6) | 1.9 ± 0.3 (4.0 ± 0.4) | 0.4 ± 0.3 (2.0 ± 0.4) |
| TMAO/urea (3 M/3 M) | 5.1 ± 0.4 (4.2 ± 0.4) | 2.2 ± 0.3 (2.3 ± 0.4) | 2.3 ± 0.4 (4.1 ± 0.6) |
Fig. 4.
CLS versus T plots of HP35-P76 and HP35-P58 obtained under different solvent conditions, as indicated. The solid lines are fits of these data to Eq. 1 with those fitting parameters listed in Table 3. The relatively large uncertainties in the CLS data and the apparent asymmetric 2D lineshape in the spectra (Figs. S2 and S4) are due to distortions from background noise, because the absorbance of the nitrile group in these experiments was relatively low (∼2 mOD).
Discussion
The current view on how TMAO and urea affect protein stability has been recently reviewed by Canchi and García (62). Owing to various quantitative thermodynamic measurements, such as vapor pressure osmometry (63–65), volumetric measurements (66), and equilibrium dialysis (67), we now know a great deal about why urea and TMAO act differently. Urea’s protein denaturation effect is generally believed to arise from the molecule’s tendency to accumulate around both the protein backbone and sidechains, owing, for example, to a stronger dispersion interaction between urea and the protein backbone (68), which preferentially stabilizes the unfolded state. However, TMAO molecules are found to be excluded from various protein backbone and sidechain units, thus destabilizing the unfolded state (59). Moreover, many studies have shown that both TMAO and urea can alter the structure and dynamics of the H-bonding network of water, thus resulting in a change in protein stability (69). Although previous studies have significantly enhanced our understanding of the mechanisms by which TMAO/urea increase/decrease protein stability, especially from a thermodynamic point of view, many important microscopic details have yet to be revealed or verified by experiments. Herein, we use linear and nonlinear IR measurements to directly probe, from a sidechain’s perspective, how TMAO and urea affect the H-bonding dynamics of water at the protein–solvent interface.
Two key findings that emerged from the current study are worthy of further discussion. First, the spectral diffusion data of the C≡N stretching vibration clearly show that addition of either urea or TMAO makes the local H-bonding environment fluctuate faster (i.e., the smaller time constant of the Kubo term in Table 2). This picture is consistent with our early observation that TMAO and urea can weaken the H-bond strength between the nitrile probe and water molecules (42). It is obvious that a weakened H-bonding interaction will result in a shallower potential energy surface along the H-bond coordinate, thus making the diffusion dynamics along this coordinate faster (at the same temperature). In addition, weakening the H-bonding interactions will lead to a smaller vibrational energy splitting, which, in turn, would lead to a smaller contribution to the total FFCF amplitude by the corresponding H-bonding dynamics, as observed (A in Table 2 and Δ2 in Table 1). Furthermore, we can rule out the possibility that the observed changes in the H-bonding dynamics arise from an increase in viscosity (e.g., 6 M urea increases the solution viscosity by only 40%), because Kubarych and coworkers have recently shown that increasing bulk viscosity slows down spectral diffusion (70). Because water always competes, whenever possible, with a protein’s intrinsic H-bonding donors/acceptors to form additional H bonds with, for example, the backbone amides, the decreased strength of H bonds involving water would preferentially destabilize the unfolded state of proteins, where more amide units are exposed to the solvent. Thus, this is an important finding because it provides, to the best of our knowledge, the first experimental evidence indicating that TMAO can enhance protein stability by attenuating the strength of H bonds formed between protein polar groups and water. Interestingly, our IR measurements suggest, although to a lesser degree in comparison with TMAO, that urea can also weaken the H-bonding ability of water with proteins (42). At first glance, this finding seems to be inconsistent with the denaturing role of urea. However, upon consideration of the fact that urea, unlike TMAO, shows preferential accumulation in the vicinity of protein backbones and sidechains (14), this result provides strong evidence in support of the notion that urea’s ability to denature proteins does not arise from its effect on the structure of water, but is instead achieved through a more direct mechanism. This conclusion is in agreement with that reached by Pielak and coworkers (71), who showed that there is no correlation between a solute’s impact on water structure and its effect on protein stability.
The second major finding is that the static component in the FFCF, which is absent (or insignificant) in pure water, increases significantly upon addition of TMAO. This static component, together with the aforementioned Kubo term, determines the inhomogeneous broadening of the C≡N stretching vibration. Thus, the fact that TMAO increases the amplitude of provides strong evidence indicating that it either slows down the peptide’s conformational motions, creates new and slowly moving conformational states that are not populated in pure water, or both. This picture is entirely consistent with the NMR study of Loria and coworker (72), which showed that TMAO can significantly rigidify the protein backbone, and with the simulations of Thirumalai and coworkers (44), which demonstrated that TMAO can act as a nano-crowder, thus increasing protein stability via the excluded volume effect. Despite this consistency, we note that a study by Qu and Bolen (73) indicated that the efficacy of the macromolecular crowding in forcing proteins to fold may be modest and may also depend on the crowding agents. Thus, it would be very useful if new experiments can be designed to directly quantify the crowding efficacy of TMAO.
Taken together, the effect of TMAO on the Kubo and static terms of the FFCF of the nitrile probe provides insights into the mechanism of action of TMAO in increasing the stability of proteins. Primarily, our results indicate that the protein stabilizing ability of TMAO arises from both enthalpic and entropic contributions. The enthalpic factor results from a decreased H-bonding ability of water, whereas the entropic factor stems from the crowding effect of TMAO; both act to destabilize the unfolded state ensemble. Because the tripeptide used in the current study is relatively small, it is possible to use molecular dynamics simulations (68, 74, 75), in conjunction with vibrational frequency calculations (76), to directly calculate the FFCFs of the nitrile probe under different cosolvent conditions and to provide further molecular insights. We hope that the current study will inspire new computational efforts in this regard.
Materials and Methods
The details of sample preparation are given in SI Text. The linear FTIR spectra are collected at a resolution of 1 cm−1 on a Magna-IR spectrometer. The details of the 2D IR measurements are also given in SI Text.
Supplementary Material
Acknowledgments
We thank Dr. Arnaldo Serrano for synthesizing the HP35 variants. This work was supported by National Institutes of Health (NIH) Grant GM012592. The 2D IR instrumentation was developed with NIH Research Resource Grant P41 GM104605. I.M.P. was supported by National Science Foundation Graduate Research Fellowship DGE-0822.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403224111/-/DCSupplemental.
References
- 1.Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208(Pt 15):2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- 2.Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: Mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36(30):9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 3.Zou Q, Bennion BJ, Daggett V, Murphy KP. The molecular mechanism of stabilization of proteins by TMAO and its ability to counteract the effects of urea. J Am Chem Soc. 2002;124(7):1192–1202. doi: 10.1021/ja004206b. [DOI] [PubMed] [Google Scholar]
- 4.Sharp KA, Madan B, Manas E, Vanderkooi JM. Water structure changes induced by hydrophobic and polar solutes revealed by simulations and infrared spectroscopy. J Chem Phys. 2001;114(4):1791–1796. [Google Scholar]
- 5.Di Michele A, et al. Modulation of hydrophobic effect by cosolutes. J Phys Chem B. 2006;110(42):21077–21085. doi: 10.1021/jp068055w. [DOI] [PubMed] [Google Scholar]
- 6.Rezus YLA, Bakker HJ. Observation of immobilized water molecules around hydrophobic groups. Phys Rev Lett. 2007;99(14):148301. doi: 10.1103/PhysRevLett.99.148301. [DOI] [PubMed] [Google Scholar]
- 7.Rezus YLA, Bakker HJ. Destabilization of the hydrogen-bond structure of water by the osmolyte trimethylamine N-oxide. J Phys Chem B. 2009;113(13):4038–4044. doi: 10.1021/jp805458p. [DOI] [PubMed] [Google Scholar]
- 8.Koga Y, Westh P, Nishikawa K, Subramanian S. Is a methyl group always hydrophobic? Hydrophilicity of trimethylamine-N-oxide, tetramethyl urea and tetramethylammonium ion. J Phys Chem B. 2011;115(12):2995–3002. doi: 10.1021/jp108347b. [DOI] [PubMed] [Google Scholar]
- 9.Munroe KL, Magers DH, Hammer NI. Raman spectroscopic signatures of noncovalent interactions between trimethylamine N-oxide (TMAO) and water. J Phys Chem B. 2011;115(23):7699–7707. doi: 10.1021/jp203840w. [DOI] [PubMed] [Google Scholar]
- 10.Sagle LB, et al. Methyl groups of trimethylamine N-oxide orient away from hydrophobic interfaces. J Am Chem Soc. 2011;133(46):18707–18712. doi: 10.1021/ja205106e. [DOI] [PubMed] [Google Scholar]
- 11.Bakulin AA, Pshenichnikov MS, Bakker HJ, Petersen C. Hydrophobic molecules slow down the hydrogen-bond dynamics of water. J Phys Chem A. 2011;115(10):1821–1829. doi: 10.1021/jp107881j. [DOI] [PubMed] [Google Scholar]
- 12.Larini L, Shea J-E. Double resolution model for studying TMAO/water effective interactions. J Phys Chem B. 2013;117(42):13268–13277. doi: 10.1021/jp403635g. [DOI] [PubMed] [Google Scholar]
- 13.Athawale MV, Dordick JS, Garde S. Osmolyte trimethylamine-N-oxide does not affect the strength of hydrophobic interactions: Origin of osmolyte compatibility. Biophys J. 2005;89(2):858–866. doi: 10.1529/biophysj.104.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton M, Bolen DW. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci USA. 2005;102(42):15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul S, Patey GN. Structure and interaction in aqueous urea-trimethylamine-N-oxide solutions. J Am Chem Soc. 2007;129(14):4476–4482. doi: 10.1021/ja0685506. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien EP, Ziv G, Haran G, Brooks BR, Thirumalai D. Effects of denaturants and osmolytes on proteins are accurately predicted by the molecular transfer model. Proc Natl Acad Sci USA. 2008;105(36):13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meersman F, Bowron D, Soper AK, Koch MHJ. Counteraction of urea by trimethylamine N-oxide is due to direct interaction. Biophys J. 2009;97(9):2559–2566. doi: 10.1016/j.bpj.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu CY, Lynch GC, Kokubo H, Pettitt BM. Trimethylamine N-oxide influence on the backbone of proteins: An oligoglycine model. Proteins. 2010;78(3):695–704. doi: 10.1002/prot.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei H, Fan Y, Gao YQ. Effects of urea, tetramethyl urea, and trimethylamine N-oxide on aqueous solution structure and solvation of protein backbones: A molecular dynamics simulation study. J Phys Chem B. 2010;114(1):557–568. doi: 10.1021/jp9084926. [DOI] [PubMed] [Google Scholar]
- 20.Rösgen J, Jackson-Atogi R. Volume exclusion and H-bonding dominate the thermodynamics and solvation of trimethylamine-N-oxide in aqueous urea. J Am Chem Soc. 2012;134(7):3590–3597. doi: 10.1021/ja211530n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiore A, Venkateshwaran V, Garde S. Trimethylamine N-oxide (TMAO) and tert-butyl alcohol (TBA) at hydrophobic interfaces: Insights from molecular dynamics simulations. Langmuir. 2013;29(25):8017–8024. doi: 10.1021/la401203r. [DOI] [PubMed] [Google Scholar]
- 22.Macdonald RD, Khajehpour M. Effects of the osmolyte TMAO (Trimethylamine-N-oxide) on aqueous hydrophobic contact-pair interactions. Biophys Chem. 2013;184:101–107. doi: 10.1016/j.bpc.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Bruździak P, Panuszko A, Stangret J. Influence of osmolytes on protein and water structure: A step to understanding the mechanism of protein stabilization. J Phys Chem B. 2013;117(39):11502–11508. doi: 10.1021/jp404780c. [DOI] [PubMed] [Google Scholar]
- 24.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217(4566):1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 25.Uversky VN, Li J, Fink AL. Trimethylamine-N-oxide-induced folding of alpha-synuclein. FEBS Lett. 2001;509(1):31–35. doi: 10.1016/s0014-5793(01)03121-0. [DOI] [PubMed] [Google Scholar]
- 26.Mello CC, Barrick D. Measuring the stability of partly folded proteins using TMAO. Protein Sci. 2003;12(7):1522–1529. doi: 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu YX, Bolen DW. Hydrogen exchange kinetics of RNase A and the urea:TMAO paradigm. Biochemistry. 2003;42(19):5837–5849. doi: 10.1021/bi0206457. [DOI] [PubMed] [Google Scholar]
- 28.Panuszko A, Bruździak P, Zielkiewicz J, Wyrzykowski D, Stangret J. Effects of urea and trimethylamine-N-oxide on the properties of water and the secondary structure of hen egg white lysozyme. J Phys Chem B. 2009;113(44):14797–14809. doi: 10.1021/jp904001m. [DOI] [PubMed] [Google Scholar]
- 29.Auton M, Bolen DW, Rosgen J. Structural thermodynamics of protein preferential solvation: Osmolyte solvation of proteins, aminoacids, and peptides. Proteins. 2008;73(4):802–813. doi: 10.1002/prot.22103. [DOI] [PubMed] [Google Scholar]
- 30.Hamm P, Zanni MT. Concepts and Methods of 2D Infrared Spectroscopy. New York: Cambridge Univ Pres; 2011. p. ix. [Google Scholar]
- 31.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in β-amyloid fibrils. Proc Natl Acad Sci USA. 2009;106(42):17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J, et al. Intrinsic structural heterogeneity and long-term maturation of amyloid β peptide fibrils. ACS Chem Neurosci. 2013;4(8):1236–1243. doi: 10.1021/cn400092v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda DG, et al. Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase. Nat Chem. 2013;5(3):174–181. doi: 10.1038/nchem.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strasfeld DB, Ling YL, Shim S-H, Zanni MT. Tracking fiber formation in human islet amyloid polypeptide with automated 2D-IR spectroscopy. J Am Chem Soc. 2008;130(21):6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. Strategies for extracting structural information from 2D IR spectroscopy of amyloid: Application to islet amyloid polypeptide. J Phys Chem B. 2009;113(47):15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc Natl Acad Sci USA. 2011;108(15):6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung JK, Thielges MC, Fayer MD. Dynamics of the folded and unfolded villin headpiece (HP35) measured with ultrafast 2D IR vibrational echo spectroscopy. Proc Natl Acad Sci USA. 2011;108(9):3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bagchi S, Boxer SG, Fayer MD. Ribonuclease S dynamics measured using a nitrile label with 2D IR vibrational echo spectroscopy. J Phys Chem B. 2012;116(13):4034–4042. doi: 10.1021/jp2122856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Middleton CT, et al. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat Chem. 2012;4(5):355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Getahun Z, et al. Using nitrile-derivatized amino acids as infrared probes of local environment. J Am Chem Soc. 2003;125(2):405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 41.Waegele MM, Culik RM, Gai F. Site-specific spectroscopic reporters of the local electric field, hydration, structure, and dynamics of biomolecules. J Phys Chem Lett. 2011;2(20):2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pazos IM, Gai F. Solute’s perspective on how trimethylamine oxide, urea, and guanidine hydrochloride affect water’s hydrogen bonding ability. J Phys Chem B. 2012;116(41):12473–12478. doi: 10.1021/jp307414s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennion BJ, Daggett V. Counteraction of urea-induced protein denaturation by trimethylamine N-oxide: A chemical chaperone at atomic resolution. Proc Natl Acad Sci USA. 2004;101(17):6433–6438. doi: 10.1073/pnas.0308633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SS, Reddy G, Straub JE, Thirumalai D. Entropic stabilization of proteins by TMAO. J Phys Chem B. 2011;115(45):13401–13407. doi: 10.1021/jp207289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh A, Remorino A, Tucker MJ, Hochstrasser RM. 2D IR photon echo spectroscopy reveals hydrogen bond dynamics of aromatic nitriles. Chem Phys Lett. 2009;469(4-6):325–330. doi: 10.1016/j.cplett.2008.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbanek DC, Vorobyev DY, Serrano AL, Gai F, Hochstrasser RM. The two dimensional vibrational echo of a nitrile probe of the villin HP35 protein. J Phys Chem Lett. 2010;1(23):3311–3315. doi: 10.1021/jz101367d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park K-H, et al. Infrared probes based on nitrile-derivatized prolines: Thermal insulation effect and enhanced dynamic range. J. Phys. Chem. Lett. 2013;4(13):2105–2110. [Google Scholar]
- 48.Kwak K, Park S, Finkelstein IJ, Fayer MD. Frequency-frequency correlation functions and apodization in two-dimensional infrared vibrational echo spectroscopy: A new approach. J Chem Phys. 2007;127(12):124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 49.Mukamel S. Principles of Nonlinear Optical Spectroscopy. New York: Oxford Univ Press; 1995. p. xviii. [Google Scholar]
- 50.Cho M. 2009. Two-Dimensional Optical Spectroscopy (CRC, Boca Raton, FL), p 378.
- 51.Volkov VV, Palmer DJ, Righini R. Distinct water species confined at the interface of a phospholipid membrane. Phys Rev Lett. 2007;99(7):078302. doi: 10.1103/PhysRevLett.99.078302. [DOI] [PubMed] [Google Scholar]
- 52.Ghosh A, Hochstrasser RM. A peptide’s perspective of water dynamics. Chem Phys. 2011;390(1):1–13. doi: 10.1016/j.chemphys.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung JK, Thielges MC, Fayer MD. Conformational dynamics and stability of HP35 studied with 2D IR vibrational echoes. J Am Chem Soc. 2012;134(29):12118–12124. doi: 10.1021/ja303017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hochstrasser RM. Two-dimensional IR-spectroscopy: Polarization anisotropy effects. Chem Phys. 2001;266(2-3):273–284. [Google Scholar]
- 55.Huang CY, Getahun Z, Wang T, DeGrado WF, Gai F. Time-resolved infrared study of the helix-coil transition using (13)C-labeled helical peptides. J Am Chem Soc. 2001;123(48):12111–12112. doi: 10.1021/ja016631q. [DOI] [PubMed] [Google Scholar]
- 56.Wang T, Du DG, Gai F. Helix-coil kinetics of two 14-residue peptides. Chem Phys Lett. 2003;370(5-6):842–848. [Google Scholar]
- 57.Lin YS, Shorb JM, Mukherjee P, Zanni MT, Skinner JL. Empirical amide I vibrational frequency map: Application to 2D-IR line shapes for isotope-edited membrane peptide bundles. J Phys Chem B. 2008;113(3):592–602. doi: 10.1021/jp807528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asbury JB, et al. Water dynamics: Vibrational echo correlation spectroscopy and comparison to molecular dynamics simulations. J Phys Chem A. 2004;108(7):1107–1119. [Google Scholar]
- 59.Canchi DR, Jayasimha P, Rau DC, Makhatadze GI, Garcia AE. Molecular mechanism for the preferential exclusion of TMAO from protein surfaces. J Phys Chem B. 2012;116(40):12095–12104. doi: 10.1021/jp304298c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu KN, Havlin RH, Yau WM, Tycko R. Quantitative determination of site-specific conformational distributions in an unfolded protein by solid-state nuclear magnetic resonance. J Mol Biol. 2009;392(4):1055–1073. doi: 10.1016/j.jmb.2009.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung JK, Thielges MC, Lynch SR, Fayer MD. Fast dynamics of HP35 for folded and urea-unfolded conditions. J Phys Chem B. 2012;116(36):11024–11031. doi: 10.1021/jp304058x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Canchi DR, García AE. Cosolvent effects on protein stability. Annu Rev Phys Chem. 2013;64(1):273–293. doi: 10.1146/annurev-physchem-040412-110156. [DOI] [PubMed] [Google Scholar]
- 63.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT., Jr Quantifying why urea is a protein denaturant, whereas glycine betaine is a protein stabilizer. Proc Natl Acad Sci USA. 2011;108(41):16932–16937. doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cannon JG, Anderson CF, Record MT., Jr Urea-amide preferential interactions in water: Quantitative comparison of model compound data with biopolymer results using water accessible surface areas. J Phys Chem B. 2007;111(32):9675–9685. doi: 10.1021/jp072037c. [DOI] [PubMed] [Google Scholar]
- 65.Zhang W, Capp MW, Bond JP, Anderson CF, Record MT., Jr Thermodynamic characterization of interactions of native bovine serum albumin with highly excluded (glycine betaine) and moderately accumulated (urea) solutes by a novel application of vapor pressure osmometry. Biochemistry. 1996;35(32):10506–10516. doi: 10.1021/bi960795f. [DOI] [PubMed] [Google Scholar]
- 66.Lee S, Shek YL, Chalikian TV. Urea interactions with protein groups: A volumetric study. Biopolymers. 2010;93(10):866–879. doi: 10.1002/bip.21478. [DOI] [PubMed] [Google Scholar]
- 67.Arakawa T, Timasheff SN. Preferential interactions of proteins with salts in concentrated solutions. Biochemistry. 1982;21(25):6545–6552. doi: 10.1021/bi00268a034. [DOI] [PubMed] [Google Scholar]
- 68.Hua L, Zhou R, Thirumalai D, Berne BJ. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci USA. 2008;105(44):16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuffel A, Zielkiewicz J. The hydrogen bond network structure within the hydration shell around simple osmolytes: Urea, tetramethylurea, and trimethylamine-N-oxide, investigated using both a fixed charge and a polarizable water model. J Chem Phys. 2010;133(3):035102. doi: 10.1063/1.3464768. [DOI] [PubMed] [Google Scholar]
- 70.King JT, Baiz CR, Kubarych KJ. Solvent-dependent spectral diffusion in a hydrogen bonded “vibrational aggregate”. J Phys Chem A. 2010;114(39):10590–10604. doi: 10.1021/jp106142u. [DOI] [PubMed] [Google Scholar]
- 71.Batchelor JD, Olteanu A, Tripathy A, Pielak GJ. Impact of protein denaturants and stabilizers on water structure. J Am Chem Soc. 2004;126(7):1958–1961. doi: 10.1021/ja039335h. [DOI] [PubMed] [Google Scholar]
- 72.Doan-Nguyen V, Loria JP. The effects of cosolutes on protein dynamics: The reversal of denaturant-induced protein fluctuations by trimethylamine N-oxide. Protein Sci. 2007;16(1):20–29. doi: 10.1110/ps.062393707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qu Y, Bolen DW. Efficacy of macromolecular crowding in forcing proteins to fold. Biophys Chem. 2002;101–102:155–165. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 74.Zangi R, Zhou R, Berne BJ. Urea’s action on hydrophobic interactions. J Am Chem Soc. 2009;131(4):1535–1541. doi: 10.1021/ja807887g. [DOI] [PubMed] [Google Scholar]
- 75.Kohlhoff KJ, et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat Chem. 2014;6(1):15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi J-H, Cho M. Vibrational solvatochromism and electrochromism of infrared probe molecules containing C≡O, C≡N, C=O, or C-F vibrational chromophore. J Chem Phys. 2011;134(15):154513. doi: 10.1063/1.3580776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.