Significance
Gene expression can be regulated at the level of transcription elongation. Coliphage HK022 Nun protein suppresses coliphage λ development by inhibiting transcription elongation. We have determined the mechanism of Nun action in a purified in vitro transcription system. Nun blocks RNA polymerase (RNAP) lateral movement on the DNA template, thereby preventing translocation. Nun requires a 9- to 10-bp RNA–DNA hybrid and recognizes RNA or DNA sequences at the 5′ end of the hybrid. Suppression of lateral movement of RNAP represents a newly described regulatory mechanism.
Keywords: Transcription Termination, Bacteriophage HK022, Bacteriophage Lambda, Transcription Elongation, Exclusion
Abstract
The Nun protein of coliphage HK022 arrests RNA polymerase (RNAP) in vivo and in vitro at pause sites distal to phage λ N-Utilization (nut) site RNA sequences. We tested the activity of Nun on ternary elongation complexes (TECs) assembled with templates lacking the λ nut sequence. We report that Nun stabilizes both translocation states of RNAP by restricting lateral movement of TEC along the DNA register. When Nun stabilized TEC in a pretranslocated register, immediately after NMP incorporation, it prevented binding of the next NTP and stimulated pyrophosphorolysis of the nascent transcript. In contrast, stabilization of TEC by Nun in a posttranslocated register allowed NTP binding and nucleotidyl transfer but inhibited pyrophosphorolysis and the next round of forward translocation. Nun binding to and action on the TEC requires a 9-bp RNA–DNA hybrid. We observed a Nun-dependent toe print upstream to the TEC. In addition, mutations in the RNAP β′ subunit near the upstream end of the transcription bubble suppress Nun binding and arrest. These results suggest that Nun interacts with RNAP near the 5′ edge of the RNA–DNA hybrid. By stabilizing translocation states through restriction of TEC lateral mobility, Nun represents a novel class of transcription arrest factors.
Transcription elongation is highly processive, yet the rate of nucleotide addition varies significantly for different ternary elongation complexes (TECs). In the normal elongation pathway, each nucleotide addition is followed by translocation of RNA polymerase (RNAP) along DNA in single-nucleotide increments. This process transfers the RNA 3′ end from the i + 1 site to the i site of the active center, thus allowing binding of the next NTP and subsequent phosphodiester bond formation. Translocation is thought to be a stochastic, rapid, and fully reversible process and not rate-limiting for elongation (1, 2). Instead, NTP sequestration, phosphodiester bond formation, or pyrophosphate release has been suggested to be rate-limiting for transcription elongation (3, 4). However, several reports strongly suggest that translocation may be at least partially rate-limiting for elongation (5–9).
Immediately following bond formation, a catalytically inactive and highly pyrophosphorolytic (pretranslocated) state is formed. In this state, the 3′ RNA end remains in the i + 1 site of the active center. The 3′ RNA thus prevents NTP binding and generates a substrate for pyrophosphorolysis. To bind the next NTP, RNAP must translocate 1-bp forward along the DNA register to form a catalytically active and pyrophosphate-resistant (posttranslocated) state. Stationary RNAP has been suggested to “ratchet” between the two states via Brownian motion (5). NTP bound to the transient posttranslocated state acts as a pawl that interferes with backward translocation, thereby favoring the formation of a phosphodiester bond. Retention of this NTP in the posttranslocated TEC is facilitated by isomerization of the active site that blocks substrate exit, and aligns it for catalysis (10, 11). According to a thermodynamic model for transcription elongation (12), annealing of a single RNA–DNA base pair at the newly formed 3′ end and melting of a single RNA–DNA bp at the 5′ end follow forward translocation (13, 14). In the pretranslocated state, the hybrid is 10 nt in length, whereas in the posttranslocated state, the hybrid retracts to 9 nt (10). Restriction of this transition in hybrid length may hinder forward translocation and induce subsequent pausing and/or arrest. Although a conventional view suggests that translocation is rapid and reversible (15), and occurs in both directions with a rate substantially faster than catalysis, recent data argue that, at least at some sequences, RNAP dwells primarily in the posttranslocated state with a slow return to the pretranslocated state (16, 17).
NusG is the only known elongation factor that promotes forward translocation; factors that directly restrict translocation in both directions have yet to be reported (6). It has been suggested that Nun may inhibit transcription elongation by blocking RNAP translocation (8). Using a combination of bulk biochemical and presteady-state approaches, we provide evidence that translocation inhibition is, in fact, the mode of Nun action. Furthermore, on the basis of physical and mutational analysis, we propose a mechanism by which Nun acts upon RNAP to restrict lateral movement of the RNAP along DNA.
Coliphage HK022-encoded Nun protein blocks elongation of λ early transcripts in vivo, suppressing phage λ growth in HK022 lysogens. Nun is a small (109-aa residue) protein with a functionally distinct N-terminal domain and C-terminal domain (CTD). The N-terminal arginine-rich motif of Nun binds the boxB RNA sequences within the N-Utilization (nut) regions of the λpL and λpR nascent transcripts (18, 19). Interactions between Nun, the RNA nut sites, and RNAP promote formation of stalled TECs at intrinsic pause sites downstream from nut both in vivo and in vitro. Escherichia coli Nus factors are required for Nun activity in vivo and stimulate Nun in vitro (20). The Nun CTD contacts DNA and is indispensable for Nun inhibition of transcription. Substitution of the penultimate tryptophan (W108) with a nonaromatic residue abrogates Nun arrest. Nun might intercalate W108 residue into dsDNA (21). Additionally, two neighboring CTD lysines are essential for Nun activity. These residues (K106/K107) are suggested to facilitate CTD binding to template by salt-bridging and hydrogen-bonding to DNA phosphate groups (22). Although Nun has been extensively analyzed functionally, the exact mechanism by which it acts upon elongating TEC has yet to be elucidated.
Inhibitors of translocation have been described that function by inducing global conformational changes in TEC, leading to physical rearrangements in the active center (7, 23–26). Here, we show that Nun affects translocation, and thereby the nucleotide addition cycle. By restricting physical movement of TEC along the DNA register, Nun freezes the translocation state. Such restriction can thereby result in both arrest (when RNAP is locked into a pretranslocated state) and activation (when it is frozen in a posttranslocated state). Nun therefore represents a newly described class of dual-function transcription factors that alter elongation rate by physically restricting translocation. We suggest that the action of Nun upon RNAP is due to a physical restraint imposed upon melting of the RNA–DNA hybrid.
Materials and Methods
Proteins for in Vitro Transcription.
Nun is toxic upon overproduction; thus, measures were taken to ensure tight control of induction during protein expression and purification. Note that the method of Nun purification used in this study was adapted from Kim and Gottesman (22). Improvements were implemented to increase the specific activity of the full-length protein. Nun protein was expressed from a pET-21d vector (22) and harvested from BL21 arabinose-inducible cells (Invitrogen). In these cells, expression of T7 RNAP is controlled by the stringent ara promoter. Overnight cultures were grown in noninducing media (MDAG-135 + 200 μg/mL carbenicillin) and inoculated 1:500 into autoinducing media (ZYM-5052 + 0.05% arabinose + 200 μg/mL carbenicillin). Media and methods of autoinduction were described by Studier (27). A total of 4 L of autoinducing media was used per purification, and culture volumes were kept to 70% of the flask capacity to ensure optimal aeration for protein production in the autoinducing media (27). Inoculated cultures were incubated for 12–16 h at 37 °C, shaking at 330 rpm. The final OD600 of autoinduced cultures were between 12 and 16, consistent with published methods (27). Cultures were then transferred on ice, and cells were pelleted at 5,000 rpm for 30 min in a Sorvall RC-5B centrifuge, using an SS-34 rotor, and resuspended in buffer A [50 mM Na-phosphate (pH 7.9), 200 mM NaCl, 1 mM DTT, 1.5 tablets of Complete Protease Inhibitor (Roche Applied Science)] in a final volume of 50 mL of buffer to ∼30–60 g of cells. Resuspended cells were then kept on ice before being passed three times through an EmulsiFlex C3 French press (Avestin) at 1,500 psi. Cell debris was then removed by centrifugation at 45,000 rpm for 20 min in an Optima L-80 XP ultracentrifuge (Beckman Coulter) using a type 45 Ti rotor. Clear supernatant was then loaded on a 5-mL Hi-trap SP-Sepharose column (Amersham/GE Healthcare) at a flow rate of 0.5 mL/min. The column was washed with 50 mL of 50 mM Na-phosphate (pH 7.9) and 200 mM NaCl (1 mL/min flow rate) before protein elution in 50 mM Na-phosphate (pH 7.9) and 1 M NaCl at a flow rate of 1 mL/min. Two-milliliter fractions were collected and analyzed via SDS/PAGE, and fractions containing Nun were pooled before the second step of purification with Mono-S column (Amersham/GE Healthcare), as described by Kim and Gottesman (22). Note that to maintain an extremely high standard of homogeneity in protein preparation, only ∼10–20% of fractions were retained. Therefore, although the final amount of protein harvested from a single purification was ∼20 mg, more protein could potentially have been acquired. After the final step of purification, resulting fractions were then analyzed via MALDI MS to ensure integrity and homogeneity of the preparation, and fractions of >95% purity were pooled. This step allowed us to eliminate fractions that contained trace amounts of nonfunctional CTD mutants. Buffer was then exchanged to 50 mM Na-phosphate (pH 7.9) and 100 mL of NaCl using a Slide-A-Lyzer dialysis cassette, with a molecular weight cutoff of 3,000 (Pierce). Glycerol was added to a final concentration of 10% (vol/vol), and aliquots were frozen in an ethanol-dry ice bath and stored at −80 °C.
The E. coli RNAP holoenzyme carrying the His6 tag at the C terminus of the β′ subunit was purified as described by Kashlev et al. (28) with minor modifications.
Assembly of TECs for Transcription Assays.
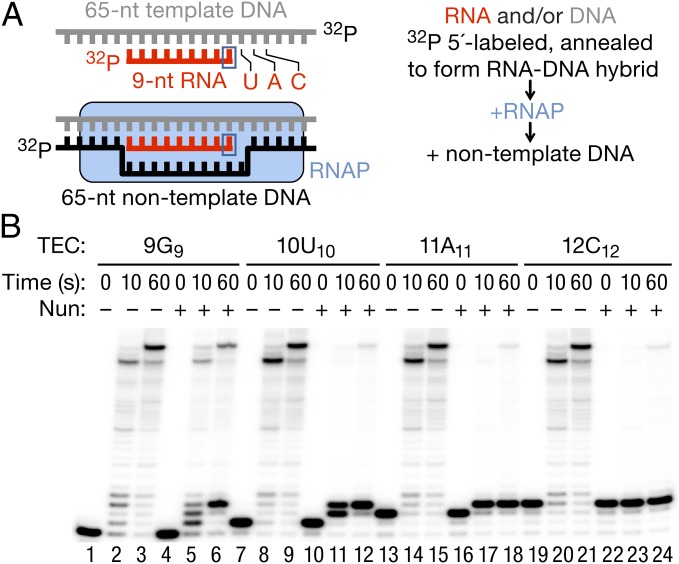
Synthetic RNA and DNA oligonucleotides used in this work (Table S1) were from Integrated DNA Technologies. The TECs were assembled in transcription buffer (TB) [20 mM Tris⋅HCl (pH 7.9), 5 mM MgCl2, 40 mM KCl, 2 mM 2-mercaptoethanol], with 10–20 pmol of purified RNAP, 5′-labeled RNA primer annealed to the template DNA strand (TDS) and nontemplate DNA strand (NDS) subsequently added (Fig. 1A) as previously described (14, 29). The nomenclature of the TECs is as follows: Taking, for example, the TEC 9G9, the RNA length is indicated by the first number (9), the letter (G) identifies the 3′ residue of the RNA primer/transcript, and the subscripted number (9) indicates the distance from the arbitrary +1A position in the nontemplate DNA to the 3′ end of the RNA (the transcription start site of the T7A1 promoter from which the oligonucleotide sequence was derived). When the RNA length and the distance from the arbitrary transcription start site are the same, the TECs may be referred to as G9 or A11, for example, omitting the specification of the RNA length. The assembled TECs were purified by immobilization on Ni2+-nitroloacetic acid (NTA) agarose, followed by TB washes (28) or by membrane filtration (30). When the 12C12 and 10C12 TECs were purified from excess CTP for pyrophosphorolysis experiments, MgCl2 in the buffer was substituted with 1 mM EDTA to prevent the RNA pyrophosphorolysis by the trace amounts of pyrophosphate present in the Nun storage buffer.
Fig. 1.
Assembly of Nun-sensitive TECs. (A) Cartoon representation of TEC assembly. (B) TEC G9 was assembled by addition of a 32P 5′-labeled RNA9 hybridized to a TDS65, followed by addition of NDS65. TEC A11 was obtained by addition of ATP and UTP (5 μM each) to the G9 assembly reaction. G9 and A11 were purified by membrane filtration. TECs U10 and C12 were obtained from G9 and A11 by 5 min of incubation with 10 μM UTP or CTP, respectively. TECs were then preincubated for 10 min at 25 °C with 5 μM Nun, or a comparable volume of Nun storage buffer before transcription was initiated by addition of 1 mM NTPs and stopped with an equal volume of 2× stop buffer. The 0-, 10-, and 60-s time points are shown.
To assess the effect of the Nun on transcription, 15 μM Nun or a corresponding volume of Nun buffer was added to the TEC immobilized on Ni2+-NTA agarose and incubated at room temperature for 10 min; Nun was then removed by two consecutive washes with TB. The TECs were eluted from Ni2+-NTA agarose by addition of 100 mM imidazole (pH 7.5) and 0.2 mg/mL acetylated BSA (Sigma–Aldrich). Eluted complexes were then incubated for 5 min at room temperature before 1 mM NTPs were added to begin transcription reactions. Alternatively, the TECs purified by membrane filtration were preincubated with 5 μM Nun or a comparable volume of Nun buffer at room temperature for 10 min and directly used in the transcription assays. The reactions were stopped manually by addition of an equal volume of gel-loading buffer [10 M urea, 50 mM EDTA (pH 7.9), 0.05% bromophenol blue, and xylene cyanol] or, for incubation times less than 5 s, by addition of HCl using an RQF-3 rapid quench flow instrument (Kintek) as described (31). Transcription products were resolved in 20% or 23% denaturing polyacrylamide gels (acrylamide/methylene-bis-acrylamide ratio was 19:1) containing 1× Tris-Borate-EDTA (TBE) and 7 M urea. Gel slabs (20 cm × 20 cm × 0.4 mm) were used, and electrophoresis was performed in 1× TBE at constant power of 50–55 W. The gels were exposed to phosphor screens, which were scanned with a Typhoon PhosphorImager (GE Healthcare Life Sciences). Transcription products were quantified using ImageQuant software (GE Healthcare Life Sciences).
Exonuclease III Toe Printing.
The TECs immobilized on Ni2+-NTA agarose were formed and purified as described for transcription assays, but one of the DNA strands, in addition to the RNA primer, was labeled at the 5′ end. For RNAP rear-end toe printing, the TDS was labeled, and the NDS contained two consecutive phosphothioate bonds at the 3′ end to ensure unidirectional degradation of the TDS by exonuclease III (ExoIII; Promega). Nun (5 μM) was added to the TECs for 10 min, and ExoIII was added to the TEC without washing off excess Nun protein. For the front-end toe printing, the TECs were assembled with TDS65 containing two consecutive phosphothioate bonds and NDS52 labeled at the 5′ end. The TECs were washed after incubation with Nun as described for the transcription assays. The final concentration of ExoIII was 3 units/μL. The reactions were stopped by addition of the gel-loading buffer as described above. ExoIII digestion products were resolved in 10% denaturing polyacrylamide gels (acrylamide/methylene-bis-acrylamide ratio was 19:1) containing 1× TBE and 7 M urea. Gel slabs (40 cm × 20 cm × 0.4 mm) were used, and electrophoresis was performed in 1× TBE at a constant power of 60–65 W.
Results
Nun Arrests TECs Assembled on Synthetic RNA–DNA Scaffolds.
To determine how Nun arrests elongating RNAP, we assembled a TEC in vitro from RNAP core enzyme carrying a His6 tag in the C terminus of the β′ subunit, as well as synthetic TDS65, RNA9 and NDS65 (14, 29) (Table S1). The template DNA sequence used in these studies was derived from the T7A1 promoter. Fig. 1A shows an overview of the assembly process. TECs U10, A11, and C12 were obtained from TEC G9 as described in the figure legend. The four TECs were chased with 1 mM NTPs for 10 s to detect transient pausing and for 1 min to establish the sites of transcription arrest after preincubation with Nun (Fig. 1B). All of the TECs were proficient in elongation despite some pausing at the +9G, +10U, and +12C sites in the absence of Nun (e.g., Fig. 1B, lane 2). Preincubation of 9G9 with Nun increased pausing at the +10U position, promoted pausing at +11A, and stably arrested ∼50% of RNAP after formation of three consecutive phosphodiester bonds (+12C position; Fig. 1B, compare lanes 3 and 6). Note that Nun did not induce a pause until at least one nucleotide was added. Thus, upon NTP addition, the G9 TEC paused at +10U and +11A (Fig. 1B, lane 5) and U10 paused at +11A (Fig. 1B, lane 11). When A11 was preincubated with Nun and chased with NTPs, a single CMP was incorporated and about 90% of the TECs were arrested at +12C (Fig. 1B, lane 18). Nun arrested the preformed TEC C12 with the same high efficiency (Fig. 1B, lane 24). Thus, A11 and C12 TECs represent a clear minimal system for Nun-induced transcription arrest that recapitulates nut-independent arrest in the regular promoter-driven transcription. We used these TECs to address the question of why some TECs incorporate one or more NMPs before being arrested, whereas others are instantly arrested by Nun.
Translocation States of Nun-Arrested TEC.
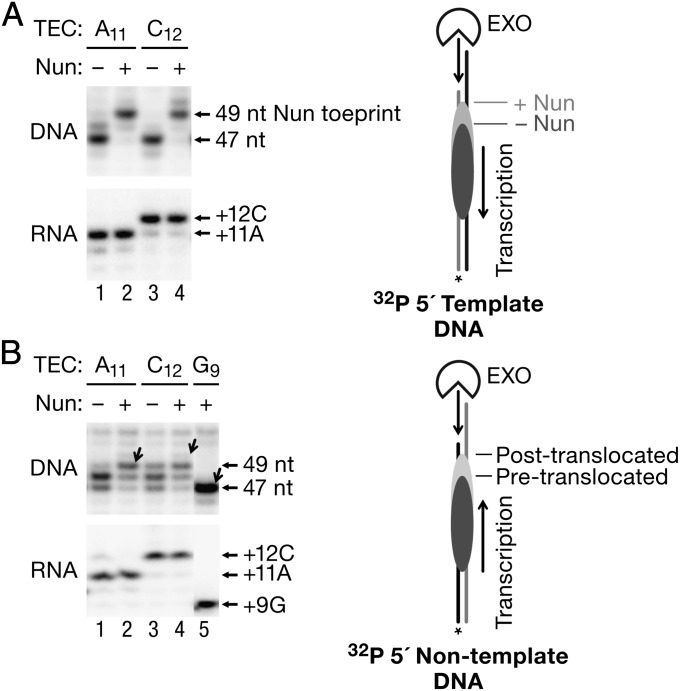
To test if Nun affects RNAP interaction with the DNA template, and to identify the site of Nun interaction with elongating RNAP, we used ExoIII, a 3′→5′ processive dsDNA exonuclease, to probe the upstream and downstream boundaries of the TECs with and without Nun. ExoIII has been widely used to determine translocation equilibria in TECs by different RNAPs and to locate DNA binding sites of proteins (32–34). Such experiments were performed as time courses, and representative time points were chosen to determine the translocation state. In TEC A11, RNAP protects 13 nt of the TDS upstream from the RNA 3′ end from degradation by ExoIII, yielding a 47-nt product (Fig. 2A, Upper, lane 1). Interestingly, incubation of TEC A11 with CTP generated TEC C12 (Fig. 2A, Lower, compare lanes 1 and 3 showing the RNA) but did not result in a significant shift of the rear-end RNAP boundary (Fig. 2A, lane 3). In TEC C12, RNAP protects 14 nt of the TDS from the RNA 3′ end. The invariant toe prints indicate that TEC A11 is primarily posttranslocated and C12 is primarily pretranslocated. In this case, the faint band above the major toe print of A11 (Fig. 2A, lane 1) corresponds to the pretranslocated boundary of TEC A11, and the band detected below the major toe print of TEC C12 (Fig. 2A, lane 3) is its posttranslocated boundary.
Fig. 2.
Nun effects on RNAP toe prints. (A) Rear-end toe printing of TECs A11 and C12 in the presence and absence of Nun. Upstream DNA was unidirectionally digested with ExoIII to the rear-end boundary of each TEC (the TDS was 5′-labeled). Reactions were stopped after 2 min of incubation. (Upper) Toe prints (including the length of the DNA fragments) are indicated by arrows on the immediate right side of the blot. (Right) Cartoon indicates how ExoIII digestion defines the pre- and posttranslocated states of TEC as determined by the rear-end toe print. (Lower) RNA synthesized by each TEC is shown. (B) Front-end toe printing of TECs A11 and C12. Downstream DNA was unidirectionally digested with ExoIII. Posttranslocated boundaries of A11, C12, and G9 (a translocation marker) are indicated by diagonal arrows next to the corresponding gel lanes. (Upper) Lengths of the ExoIII digestion products are indicated on the immediate right side of the blot. (Lower) RNA products are again shown. (Right) Cartoon illustrates front-end ExoIII digestion.
Addition of Nun extended the RNAP toe prints in both TECs. A clear Nun-dependent toe print located 2 bp upstream from the most prominent rear-end boundary of posttranslocated TEC A11 and pretranslocated TEC C12 (Fig. 2A, lanes 2 and 4) was observed. Importantly, the addition of CMP to TEC A11 did not change the site of the Nun-dependent toe print. The Nun-dependent toe print might reflect (i) a direct physical barrier imposed by Nun to ExoIII approaching the rear end of RNAP, (ii) Nun-induced DNA “scrunching” (retraction of upstream DNA into RNAP) protecting the DNA from nuclease digestion, or (iii) backtracking (i.e., reverse 1- to 2-bp translocation) of Nun-bound RNAP. A major fraction of TEC A11 efficiently and very rapidly incorporated CMP in the presence of Nun and was resistant to pyrophosphorolysis (Fig. 2A, RNA and the presteady-state experiment described below), further indicating that Nun did not induce backtracking of this TEC. A very high sensitivity of TEC C12 to pyrophosphorolysis in the presence of Nun also eliminated involvement of backtracking in arrest of this complex (see below).
When the front-end toe prints of TEC A11 and TEC C12 were determined, ExoIII revealed at least three RNAP boundaries (Fig. 2B, lanes 1 and 3). The 47-nt ExoIII digestion product is the same as the single-band front-end toe print of TEC G9 (Fig. 2B, lane 5), suggesting that ExoIII can induce RNAP backtracking by pushing the TEC backward (16, 35, 36). The longest 49-nt digestion product, barely detectable in TEC A11 (Fig. 2B, lane 1) and well pronounced in C12 (Fig. 2B, lane 3), corresponds to the posttranslocated boundary of TEC A11 and the pretranslocated boundary of C12. Nun stabilized the 49-nt posttranslocated boundary, indicating that Nun blocks the movement of this complex from a posttranslocated to pretranslocated state (Fig. 2B, compare lanes 1 and 2). Notably, we find no Nun toe print at the downstream border of TEC A11. In TEC C12, Nun reduced the abundance of reverse translocated bands and stabilized the complex in a pretranslocated state. In the presence and absence of Nun, incorporation of CMP to TEC A11 did not induce forward translocation of RNAP on DNA. Strikingly, the Nun-induced toe print also remained stationary upon RNA synthesis. Thus, we conclude that Nun restricts lateral movement of either pre- or posttranslocated TEC. TEC A11 is clearly not backtracked because it is catalytically active in the presence of Nun (see below). TEC C12 also did not show any evidence of backtracking in this assay; thus, we conclude that backtracking is not implicated in Nun arrest of this complex.
Effect of Nun-Resistant RNAP Mutants and a Nonfunctional Nun Mutant on Transcription Arrest and RNAP Toe Print.
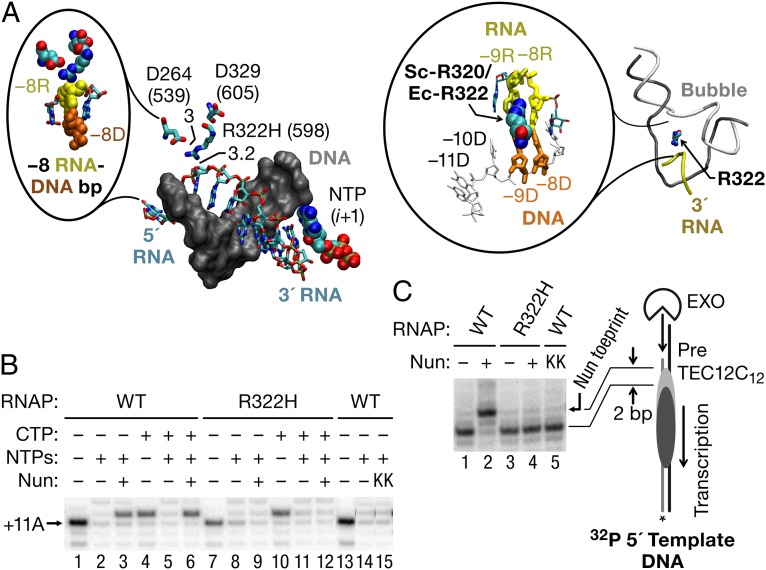
Three RNAP mutations that specifically inhibit Nun activity in vivo (β′ D264G, β′ D329G, and β′ R322H) have been previously identified (37). Other than inhibiting Nun arrest, all three mutant polymerases are fully functional in elongation and support λ N antitermination (37). β′ D329 and D264 are located in the base of the rudder and lid elements, respectively, and β′ R322 is located in the rudder. They are positioned within 3–9 Å of one another, measured from a structure of TEC determined by Vassylyev et al. (10) (Protein Data Bank ID code 2PPB) and the model for the transcription bubble and upstream DNA duplex in TEC from Andrecka et al. (38) (Fig. 3A, Right). All three residues localize near the −8 RNA base of the RNA–DNA hybrid in elongating Thermus thermophilus RNAP (Fig. 3A, Left).
Fig. 3.
RNAP β′ mutations near the upstream edge of the RNA–DNA hybrid inhibit Nun. (A, Left) Structure of 9-bp RNA–DNA hybrid in a TthRNAP TEC is shown (PDB ID code 2O5J). All three RNAP mutants that inhibit arrest by Nun in vivo locate in the β′ subunit of EcRNAP (β′ D264G, β′ D329G, and β′ R322H) between the lid and rudder domains in the zipper region. The mutations are clustered within 3–6 Å of the 5′ end of the transcription bubble in the vicinity of base pair −9 of the 9-bp RNA–DNA hybrid. Only the DNA (gray, space-filled model) and the RNA (colored elements, sticks) strands of the 9-bp RNA–DNA hybrid and the NTP in the active center (i + 1 site) are shown. Numbers in brackets indicate the corresponding amino acid residues in the β′ subunit of TtRNAP. (Inset) Zoomed area surrounding the −8/−9 bp of the hybrid. The −8 RNA and DNA paired bases (−8R and −8D) are shown in yellow and brown colors, respectively. The R322 (TtRNAP R598) residue is located 3 Å from the α-phosphate of the −8 residue of the nascent RNA. (A, Right) Model of the structure of the “complete” TEC by yeast RNAPII containing the intact transcription bubble (44). The colors are as in the left panel. (Inset) The −8 and −9 positions of the RNA–DNA hybrid are highlighted; the position of Rpb1-R320 (corresponds to EcRNAP β′ R322) is shown. (B) TECs A11 were assembled with WT and β′ R322H mutant RNAPs immobilized on Ni2+-NTA agarose for the TEC purification with RNA11, TDS65, and NDS65. C12 TECs were obtained by 90 s of incubation with 5 μM CTP. A nonfunctional mutant of Nun (K106/107A) was tested with WT TEC (lane 9). The +11A RNA is indicated by an arrow. (C) Upstream DNA was unidirectionally digested with ExoIII to the rear-end boundary of A11 or C12 TECs. Before ExoIII treatment, each TEC was incubated for 5 min with Nun or a comparable volume of Nun storage buffer. The Nun-specific toe print is indicated on the immediate right side of the blot by an arrow. (Right) Cartoon indicates the location of the Nun-specific toe print.
To confirm that the minimal in vitro transcription system accurately reflected the biological functions of Nun, we tested the β′ R322H RNAP mutant for arrest in vitro. TEC 11A11 was assembled with either mutant or WT RNAP and incubated with Nun either before or after addition of CTP (thus comparing TEC A11 with TEC C12), followed by incubation with all four NTPs to test for arrest. The β′ R322H was 10-fold less efficient than WT RNAP in supporting Nun arrest at the +12C site regardless of whether transcription was started from A11 or C12. (Fig. 3B, compare lanes 2 and 4 and lanes 6 and 8). Conversely, when WT TEC A11 was incubated with the nonfunctional Nun mutant (K106A/K107A) (22), arrest was inhibited to the same extent as with the mutant RNAP (Fig. 3B, compare lanes 14 and 15).
Consistent with the transcription data, the Nun-dependent toe print was absent whether mutant RNAP (Fig. 3C, lanes 3 and 4) or mutant Nun (Fig. 3C, lane 5) was used. Collectively, the positions of the β′ mutations suggest that Nun may physically interact with the upstream boundary of RNAP, dsDNA, the transcription bubble, or the RNA–DNA separation site.
Presteady-State Kinetic Analysis of Nun Activity on NMP Incorporation and Pyrophosphorolysis.
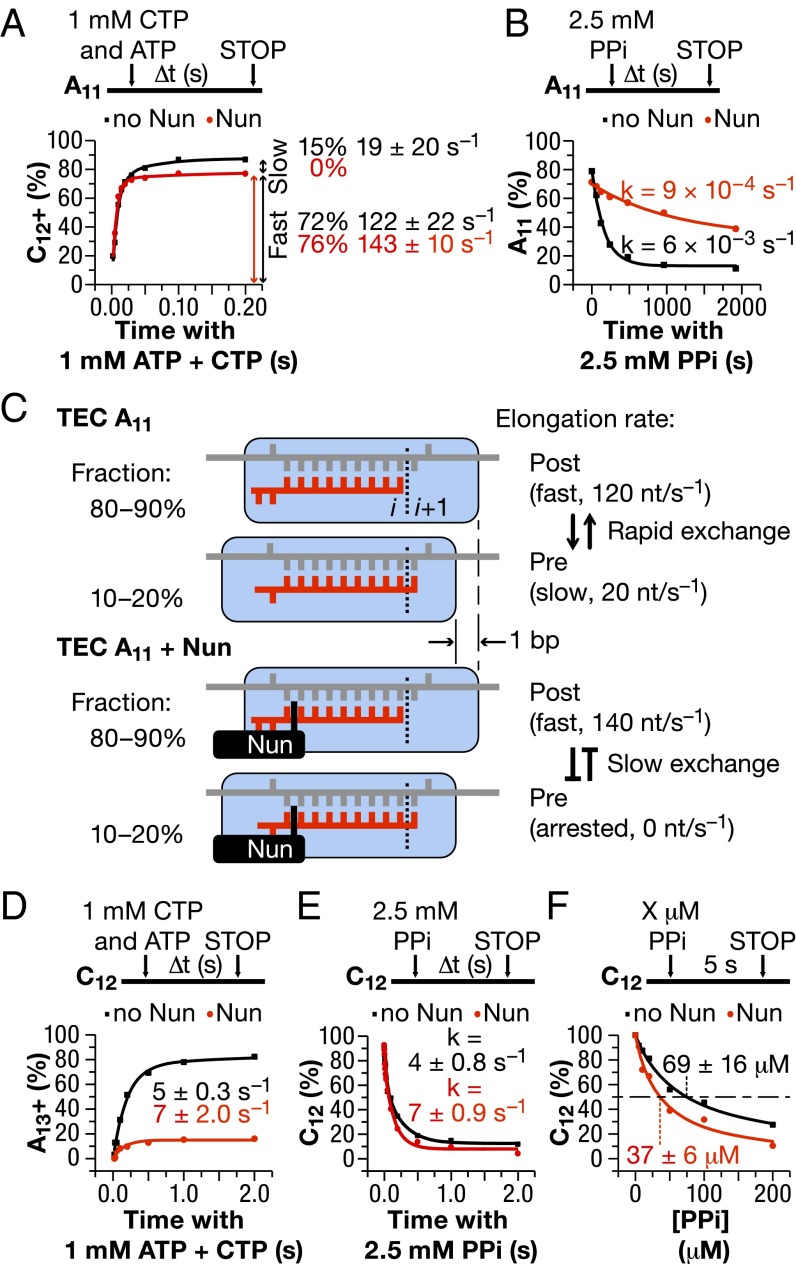
Presteady-state kinetic analysis can quantify the typically very rapid NMP addition rate at physiological concentrations of substrate NTP. It can identify alternative functional states of TEC during the translocation/catalysis cycle (5) at a resolution impossible to achieve from bulk biochemical experiments. We performed a presteady-state analysis of CMP addition to TEC A11 in the presence and absence of Nun. In the absence of Nun, the major (∼70%) fraction of TEC A11 showed a rapid incorporation rate (>100 s−1), whereas the rate of the minor (15%) fraction was substantially slower. The experimental data fitted a double-exponential function rather than a single-exponential function (i.e., two incorporation rates are observed, as indicated by the double-headed arrows in the graph in Fig. 4A.) We attributed these two fractions to posttranslocated TEC A11, which incorporates CMP12 rapidly, and pretranslocated TEC A11, which must translocate before nucleotide addition. The extra translocation step is responsible for the slow catalytic rate of the minor fraction of TEC A11. It has been shown recently that forward RNAP translocation is a rate-limiting step at some DNA sequences (7, 17, 39). Nun converted the double-exponential curve to a single-exponential curve, presumably by inactivating the minor pretranslocated fraction (compare the black and red curves in Fig. 4A, Left). Accordingly, TEC A11 treated with Nun incorporated ∼15% less CMP than untreated TEC A11 after a 0.2-s incubation with CTP (the red curve lies below the black curve in Fig. 4A). These results eliminated the possibility that TEC A11 backtracked on DNA in the presence of Nun. The robustness of the Nun effect was confirmed with a complex carrying a 3′ A11→T(U)11 substitution (Fig. S1). We conclude that in a 0.2- to 0.5-s time frame, Nun kinetically trapped or reduced the conversion of pretranslocated TEC to posttranslocated TEC at position +11A.
Fig. 4.
Presteady-state analyses of Nun effect on NMP addition and pyrophosphorolysis. (A) CMP addition time course by TEC A11. TEC A11 was obtained as described in Fig. 1B. Transcription was assayed by mixing the TEC with ATP and CTP (1 mM final concentration) using an RQF-3 rapid quench flow instrument (Kintek). The fraction of the 12-nt or longer products was plotted vs. time, and the data were fitted with a double-exponential function. The apparent rates and fractions of the TECs incorporating the CMP with these apparent rates are shown in the plot. (B) Pyrophosphorolysis of TEC A11. The TEC was incubated with 2.5 mM potassium pyrophosphate for the indicated times. The reaction was stopped manually by addition of gel-loading buffer. The fraction of the 11-nt RNA product was plotted vs. time, and the data were fitted with a single exponential function. (C) Proposed mechanism of Nun interaction with TEC A11. (D) AMP addition time course by TEC C12. The experiment was performed and analyzed as described in A, except that TEC C12 was obtained from TEC A11 before Nun addition. (E) Pyrophosphorolysis of TEC C12. The experiment was performed and analyzed as in B, but the reaction was carried out with an RQF-3 rapid quench flow instrument. (F) Nun effect on susceptibility of TEC C12 to pyrophosphate. The fraction of TEC C12 remaining intact after 5 s of incubation of TEC C12 with various pyrophosphate concentrations is plotted. The data were fitted to a Michaelis–Menten equation to determine the concentration of pyrophosphate promoting pyrophosphorolysis of 50% of the elongation complex after 5 s of incubation with pyrophosphate. Δ t (s) (time in seconds), threshold cycle.
As a second approach to determine the translocation state of TEC A11 and the effect of Nun on translocation, we measured pyrophosphorolysis rates and sensitivities (Fig. 4B). Pyrophosphorolysis in TEC depends on the identity of the 3′ RNA base and the translocation state of RNAP (7, 40). The 3′ RNA must reside in the i + 1 site to be susceptible to nucleolytic attack by exogenous pyrophosphate, leading to pyrophosphorolysis. Therefore, pretranslocated RNAP is more sensitive to pyrophosphorolysis than its posttranslocated counterpart. The latter requires backward translocation (i.e., reversion to the pretranslocated state) to pyrophosphorolize the 3′ RNA. Sensitivity to pyrophosphate also depends on the nature of 3′ RNA residue in TEC (40). As expected, TEC A11, which is primarily posttranslocated (Figs. 2A and 4A), was relatively resistant to pyrophosphorolysis. This result is not surprising, given the high catalytic rate of this complex, which indicates the posttranslocated state (Fig. 4B; note the difference in the time scale for nucleotide addition in Fig. 4A and pyrophosphorolysis in Fig. 4B). Nun further increased the resistance of A11 to pyrophosphate (Fig. 4B, red curve). This finding suggests that Nun blocked transient reversion of the posttranslocated TEC A11 to the pretranslocated TEC A11, just as it blocked forward translocation of the pretranslocated state to the posttranslocated state of TEC A11 in Fig. 4A. These results strongly argue that Nun blocks or slows down lateral movement of TEC independently of the translocation state (Fig. 4C). In contrast to TEC A11, Nun strongly prevented addition of the next cognate AMP to TEC C12 (Fig. 4D), supporting the idea that the Nun-arrested complex is primarily pretranslocated (Fig. 4C). TEC C12 was also hypersensitive to pyrophosphorolysis compared with TEC A11 (Fig. 4E, compare time scale with Fig. 4B). Nun stimulated pyrophosphorolysis approximately twofold in TEC C12 (Fig. 4 E and F), presumably by inhibiting conversion of the pretranslocated complex to the pyrophosphate-resistant posttranslocated state. The Nun-dependent increase in pyrophosphate sensitivity of TEC C12 further argues against backtracking of RNAP induced by Nun. In the backtracked TEC, the RNA 3′ end is extruded into the secondary channel, and is thus resistant to cleavage by exogenous pyrophosphate.
We conclude that Nun prevents lateral movement of TEC, blocking conversion of pretranslocated TEC to posttranslocated TEC, and vice versa (Fig. 4C). This idea is consistent with the observation that Nun stabilized TEC against dissociation by high (1–2 M) salt (20), indicating a tighter clamping of the nucleic acid scaffold by RNAP.
RNA–DNA Hybrid Length Influences Nun Activity.
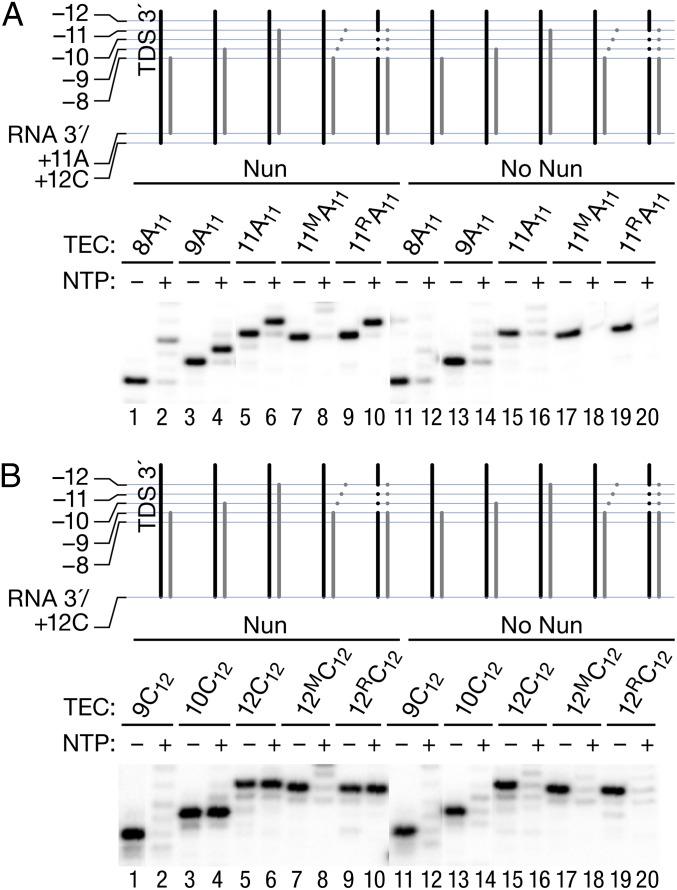
We next searched for regions in the RNA–DNA scaffold critical for Nun activity. We initiated transcription from a set of TECs with the same RNA 3′ end (+11A) but with different RNA lengths at the 5′ end, and compared elongation in the presence and absence of Nun. Cartoon representations of the complexes and their respective RNA lengths are depicted above each gel in Fig. 5. A complex formed with an 8-mer RNA (8A11) showed no Nun arrest at +12C, although a weak arrest site (27%) could be identified 2 nt downstream, at +14C (Fig. 5A, lane 2). In contrast, TEC 9A11 was strongly arrested by Nun at +12C [Fig. 5A, lane 4 (75%)]. The efficiency of arrest was comparable to that seen when transcription was initiated from a complex formed with an 11-mer RNA (TEC 11A11; Fig. 5A, lane 6). TEC 11MA11, which carries a 3-nt mismatch at the 5′ end of the 11-nt RNA, and thus an 8-bp RNA–DNA hybrid, behaved similar to TEC 8A11. Arrest at +12C was abolished, although weak arrest at +14C could be seen (Fig. 5A, lane 8). The sequence of the noncomplementary RNA did not account for this result, because changing the DNA sequence to restore complementarity (TEC 11RA11) fully restored Nun arrest (Fig. 5A, lane 10).
Fig. 5.
Hybrid length determines the ability of Nun to inhibit transcription. TECs were assembled as described in Fig. 1, but with RNA8A, RNA9A, and RNA11A (Table S1). All of the RNA primers shared the 3′ sequence, ending at +11A. RNA11A was fully complementary to TDS65, and RNA11AM carried a 3-nt 5′-end mismatch. TEC 11AR11 was assembled with RNA11AM, TDS65M, and NDS65M, restoring the full complementarity of the RNA to the template. The assembled TECs were preincubated with Nun or Nun storage buffer before (A) or after (B) addition of 5 μM CTP for 5 min. NTPs (1 mM) were added in even-numbered lanes for 60 s.
We then tested the effect of Nun on complexes assembled directly at the site of arrest (+12C) (Fig. 5B). In this context, a minimum of 10-mer RNA, rather than 9-mer RNA, was necessary for Nun arrest at +12C (Fig. 5B, compare lanes 2 and 4). Furthermore, a TEC formed with a 12-mer RNA containing a 3-nt mismatch at its 5′ end (TEC 12M C12), carrying 9 nt of 3′ cRNA, was also refractory to Nun arrest at +12C (Fig. 5B, lanes 7 and 8). Again, restoring complementarity by changing the template sequence restored Nun arrest (Fig. 5B, lanes 9 and 10). These results suggest that the minimal 10-bp length of the RNA–DNA hybrid, rather than the RNA length per se, determines Nun arrest efficacy at this site. A 9-bp cRNA corresponds exactly to the length of the RNA–DNA hybrid observed in the X-ray crystals of posttranslocated T. thermophilus TEC (10). The hybrid length should increase to 10 bp after conversion of this complex to the pretranslocated state upon a single bond formation. In conclusion, Nun arrested TEC with a 9-bp hybrid only after addition of a single NTP, whereas Nun-modified TEC with a 10-bp hybrid cannot incorporate another NMP.
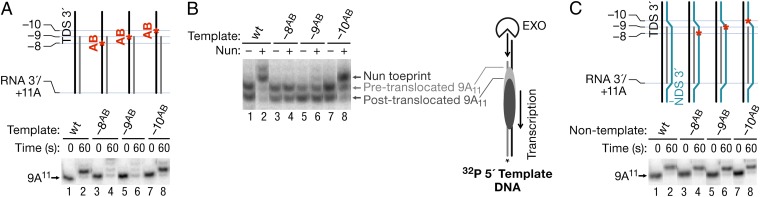
We next determined that a 9-bp RNA–DNA hybrid is the minimum required for Nun activity. We substituted single abasic residues in the template or NDSs of TEC 9A11 (Fig. 6). Note that only the DNA was modified in these scaffolds. Importantly, the ability to support Nun-mediated arrest was eliminated with templates that contained an abasic site opposite the −8 or −9 base position in the RNA, generating continuous hybrids of 7 bp or 8 bp, respectively (Fig. 6A, lanes 4 and 6). In contrast, an abasic substitution at the −10 position, which allows formation of a 9-bp RNA–DNA hybrid, did not inhibit Nun activity (Fig. 6A, lane 8). We next assayed Nun binding to TEC 9A11 carrying template-strand abasic substitutions (Fig. 6B). Nun generated a strong upstream toe print on this template (Fig. 6B, lanes 1 and 2). In contrast, we observed no Nun-specific toe prints on the Nun-resistant −8 and −9 abasic templates (Fig. 6B, lanes 3–6). Like WT TEC 9A11, these variants were roughly equivalent in the distribution between their pre- and posttranslocated states. In contrast, Nun generated the extended toe print in TEC 9A11 carrying a −10 abasic template (Fig. 6B, lanes 7 and 8). The toe print was slightly weaker on this template compared with its nonmodified counterpart, indicating that the −10 abasic site slightly reduced or altered Nun binding to RNAP (Fig. 6B, compare lanes 2 and 8). These results confirm the positive correlation between Nun-dependent toe print formation and Nun arrest efficiency. In support of the hypothesis that Nun recognizes template DNA and/or RNA–DNA hybrid, the abasic substitutions on the nontemplate strand had no impact on Nun-mediated arrest (Fig. 6C). We conclude that posttranslocated TEC, which includes a 9-bp RNA–DNA hybrid, is recognized by Nun, although arrest only occurs when the hybrid is extended to 10 bp, that is, at a pretranslocated state (Fig. 5A, lanes 3 and 4, and B, lanes 3 and 4).
Fig. 6.
Abasic sites in the TDS that disrupt the RNA–DNA hybrid block Nun-mediated transcription arrest and Nun binding to TEC. (A) TEC 9A11 was assembled with RNA9A; original TDS65; or TDS65 containing abasic substitutions −8, −9, and −10 relative to the location of the RNA 3′ end (TDS65 abasic series; Table S1) and NDS65. AB, abasic. (B) Rear-end boundary of TEC 9A11 on the original, −8 abasic, −9 abasic, and −10 abasic templates was determined by ExoIII toe printing. A short arrow indicates the position of the Nun-specific toe print. Bars and cartoon indicate the positions of pre- and post-translocated TEC 9A11. (C) TEC 9A11 was assembled with RNA9A; TDS65; original NDS65; or NDS65 containing abasic substitutions −8, −9, and −10 relative to the location of the RNA 3′ end (NDS65 abasic series; Table S1). Transcription assays in A and C were performed as described in Fig. 3B.
Discussion
The Nun protein of temperate coliphage HK022 is a unique transcription elongation factor that specifically excludes superinfecting phage λ. Nun arrests E. coli RNAP at intrinsic pause sites promoter-distal to phage λ RNA nut sequences. Nun arrest results in the abrogation of λ transcription (8, 19, 20). E. coli Nus proteins facilitate Nun-mediated arrest both in vivo and in vitro. We assayed Nun activity in vitro on templates that lack both λ nut RNA and ancillary protein factors. Thus, our system allowed us to study the direct effect of Nun on transcription elongation.
We demonstrate that Nun impedes lateral mobility of RNAP on the DNA template in both directions, immobilizing RNAP in either a pre- or posttranslocated register. Tethering of posttranslocated TEC inhibits RNA pyrophosphorolysis (as exemplified by TEC A11), whereas tethering of pretranslocated TEC enhances pyrophosphorolysis and blocks phosphodiester bond formation (e.g., TEC C12). Nun requires at least a 9-bp RNA–DNA hybrid for binding, and it arrests transcription once the hybrid is extended to a length of 10 bp. Nun binds to both pretranslocated and posttranslocated TEC, thus implying that the hybrid fluctuates between a length of 9 bp and 10 bp depending on the translocation state of TEC. This conclusion is consistent with structural data (10).
In addition to transcription arrest, Nun can modulate TEC pausing (Fig. 1). Nun effects on pausing are complex. When transcription was initiated from TEC G9, Nun stimulated pausing at +10U and +11A yet arrested transcription at +12C. The +10 and +12 positions are intrinsic pause sites that appear to be sensitive to Nun. This finding is in accordance with previous observations that Nun arrest corresponds to intrinsic pause sites on natural DNA templates (20). Nun may induce pausing of TEC at nonintrinsic pause sites, as illustrated by pausing at +11A of transcription initiated from TEC 10U10 in the presence of Nun (Fig. 1B, lanes 5 and 11).
Nun produced a unique toe print 1 to 2 nt upstream to the TEC edge of TECs A11 and C12 (Fig. 2B). This toe print did not change upon limited RNA synthesis (from +11A to +12C). Nun allows TEC A11 to add a single nucleotide (CMP) but blocks further nucleotide addition by freezing the pretranslocated TEC C12 (Fig. 1B). Note that TEC C12 has an intrinsically slow rate of forward translocation after bond formation. Even in the absence of Nun, the toe prints of TEC A11 and TEC C12 remain invariant. We propose that TEC C12 is a substrate for arrest by Nun because it does not undergo rapid equilibrium between the pre- and posttranslocated states, and remains primarily pretranslocated.
It has recently been shown that the translocation dynamics of DNA polymerases may significantly vary between different DNA sequences (41). Two distinct patterns have been identified: (i) prolonged dwelling in either the pretranslocated or posttranslocated state (e.g., TEC C12 and TEC A11, respectively) or (ii) rapid exchange between these states, accompanied by a short duration in each state (41). Comparatively Nun-resistant TECs, such as G9, may belong to the latter category of rapidly translocating complexes, which have very short-lived and poorly defined translocation states, presenting Nun with a challenge to interact with either of these states. An alternative model would be that Nun requires access to the hybrid fork junction for a suitable length of time to establish a tight interaction. Thus, frequent oscillation of the hybrid length due to rapid melting/reannealing nucleotides at the 5′ end (because of either a rapid exchange of translocation states or accelerated transcription) occlude the pocket into which Nun must establish tight contact with the upstream hybrid edge.
Our work indicates that the 5′ edge of the hybrid region plays a critical role in Nun action. This finding is supported by the fact that the Nun-specific rear-end toe print is correlated with arrest. Whether this toe print represents a barrier to ExoIII by Nun protein or is the consequence of a secondary effect of Nun binding (e.g., DNA distortion) is not known. Nevertheless, this effect is specific to the upstream region; we have not observed a Nun-induced front-end toe print. Furthermore, the three RNAP mutants that ablate Nun activity all localize near the 5′ edge of the RNA–DNA hybrid. One such representative mutant, β′ R322H, abolished both Nun-induced arrest and the Nun-specific rear-end toe print. Because forward translocation requires melting one base pair at the upstream end of the hybrid in TEC (12, 42), Nun may interfere with translocation by stabilizing the end of the hybrid.
Previous work using a C-terminal cross-linker showed contacts between the Nun C terminus and template DNA ∼9 bp downstream to the active center (43). The work presented here does not refute that data, because it did not monitor cross-linking of Nun to template. It is possible that Nun makes multiple simultaneous contacts at both the upstream and downstream boundaries of TEC. A downstream Nun toe print may not have been detected because the site of cross-linking, 9 bp downstream from the 3′ end of transcript, lies within the RNAP dsDNA-binding groove, and therefore is not accessible to ExoIII digestion. We cannot rule out the possibility that more than one Nun molecule is bound by TEC and that each one makes distinct contacts. Furthermore, different templates were used in each study. Those used in the cross-linking experiments, for example, were based upon λ sequences; the templates used here were not. Note that neither template contained BoxB.
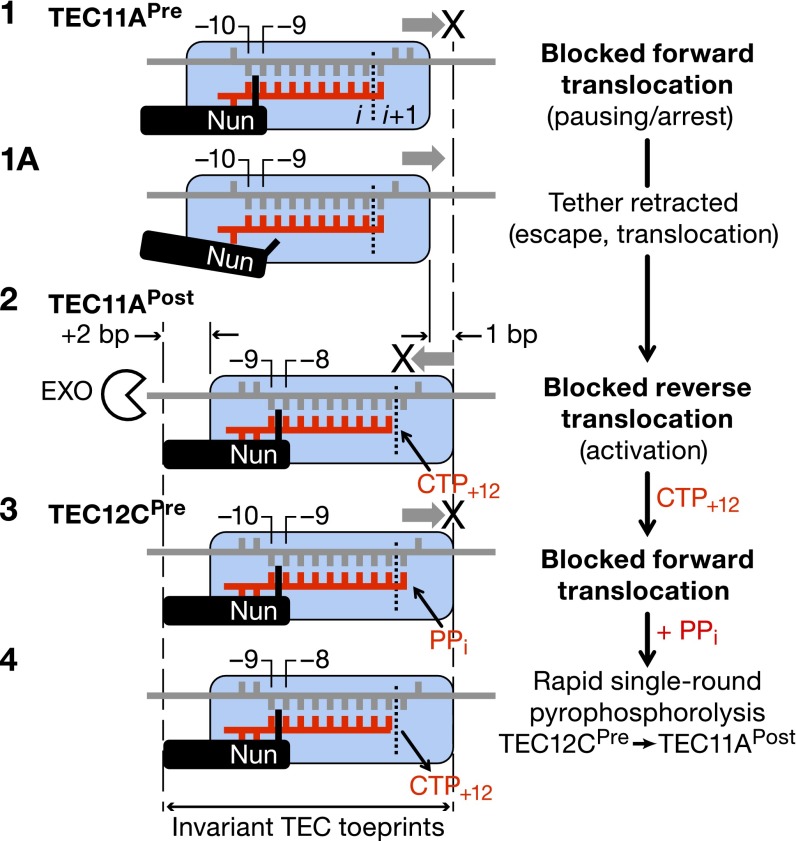
Fig. 7 shows how Nun might interfere with the transcription cycle by restricting translocation of RNAP. TEC A11, which is mostly posttranslocated, is used to illustrate the model. Nun does not block CMP incorporation by this TEC population. For the minor pretranslocated population of TEC A11, Nun immobilizes the upstream edge of the RNA–DNA hybrid (between −9 and −10 positions of the hybrid), thus blocking forward translocation and incorporation of CMP (step 1). Nun also blocks backward translocation of the posttranslocated fraction, thus increasing its resistance to pyrophosphate (step 2). Immobilization of posttranslocated TEC A11 allows CMP incorporation but not AMP, the next nucleotide (steps 2 and 3; Fig. 4 A and B). Nun-immobilized pretranslocated TEC C12 is sensitive to pyrophosphorolysis (steps 2 and 4). Note that the Nun-induced toe print does not change position in steps 2–4. Nun pauses, rather than arrests, pretranslocated TEC A11 (Fig. 1B). Nun releases its hold on the RNA–DNA hybrid, which melts, allowing the complex to move to the posttranslocated state and add CMP (step 1a). We suggest that the forward translocation potential of pretranslocated TEC A11 may be greater than that of pretranslocated TEC C12 due to the lower stability of this state at +11A compared with the +12C position. This difference may allow the former to break from Nun tethering and to escape from the pause.
Fig. 7.
Molecular mechanism of Nun action. Details are provided in the main text.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319740111/-/DCSupplemental.
References
- 1.Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421(6921):423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 2.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446(7137):820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 3.Larson MH, et al. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci USA. 2012;109(17):6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17(1):99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010;1799(5-6):389–401. doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Herbert KM, et al. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399(1):17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imashimizu M, et al. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol. 2013;425(4):697–712. doi: 10.1016/j.jmb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung SC, Gottesman ME. The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities. Genes Dev. 1997;11(20):2670–2678. doi: 10.1101/gad.11.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dangkulwanich M, et al. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. Elife. 2013;2:e00971. doi: 10.7554/eLife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448(7150):157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127(5):941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yager TD, von Hippel PH. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991;30(4):1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 13.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10(5):1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 14.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2(1):55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 15.Xiong Y, Burton ZF. A tunable ratchet driving human RNA polymerase II translocation adjusted by accurately templated nucleoside triphosphates loaded at downstream sites and by elongation factors. J Biol Chem. 2007;282(50):36582–36592. doi: 10.1074/jbc.M707014200. [DOI] [PubMed] [Google Scholar]
- 16.Nedialkov YA, Nudler E, Burton ZF. RNA polymerase stalls in a post-translocated register and can hyper-translocate. Transcription. 2012;3(5):260–269. doi: 10.4161/trns.22307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malinen AM, et al. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 2012;40(15):7442–7451. doi: 10.1093/nar/gks383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burmann BM, Uc-Mass A, Schweimer K, Gottesman ME, Rösch P. The Y39A mutation of HK022 Nun disrupts a boxB interaction but preserves termination activity. Biochemistry. 2008;47(28):7335–7341. doi: 10.1021/bi8004347. [DOI] [PubMed] [Google Scholar]
- 19.Chattopadhyay S, et al. Interaction between the phage HK022 Nun protein and the nut RNA of phage lambda. Proc Natl Acad Sci USA. 1995;92(26):12131–12135. doi: 10.1073/pnas.92.26.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung SC, Gottesman ME. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol. 1995;247(3):428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- 21.Watnick RS, Herring SC, Palmer AG, 3rd, Gottesman ME. The carboxyl terminus of phage HK022 Nun includes a novel zinc-binding motif and a tryptophan required for transcription termination. Genes Dev. 2000;14(6):731–739. [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HC, Gottesman ME. Transcription termination by phage HK022 Nun is facilitated by COOH-terminal lysine residues. J Biol Chem. 2004;279(14):13412–13417. doi: 10.1074/jbc.M313206200. [DOI] [PubMed] [Google Scholar]
- 23.Tagami S, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468(7326):978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 24.Tuske S, et al. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122(4):541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong XQ, Nedialkov YA, Burton ZF. Alpha-amanitin blocks translocation by human RNA polymerase II. J Biol Chem. 2004;279(26):27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- 26.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15(8):811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 27.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Kashlev M, et al. Histidine-tagged RNA polymerase: Dissection of the transcription cycle using immobilized enzyme. Gene. 1993;130(1):9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 29.Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 2003;371:233–251. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- 30.Kireeva ML, Domecq C, Coulombe B, Burton ZF, Kashlev M. Interaction of RNA polymerase II fork loop 2 with downstream non-template DNA regulates transcription elongation. J Biol Chem. 2011;286(35):30898–30910. doi: 10.1074/jbc.M111.260844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kireeva M, et al. Millisecond phase kinetic analysis of elongation catalyzed by human, yeast, and Escherichia coli RNA polymerase. Methods. 2009;48(4):333–345. doi: 10.1016/j.ymeth.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2009;106(22):8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nedialkov YA, et al. The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim Biophys Acta. 2013;1829(2):187–198. doi: 10.1016/j.bbagrm.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mymryk JS, Archer TK. Detection of transcription factor binding in vivo using lambda exonuclease. Nucleic Acids Res. 1994;22(20):4344–4345. doi: 10.1093/nar/22.20.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Q, Sousa R. Weakening of the T7 promoter-polymerase interaction facilitates promoter release. J Biol Chem. 2005;280(15):14956–14961. doi: 10.1074/jbc.M500518200. [DOI] [PubMed] [Google Scholar]
- 36.Kireeva ML, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30(5):557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robledo R, Atkinson BL, Gottesman ME. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991;220(3):613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 38.Andrecka J, et al. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci USA. 2008;105(1):135–140. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuzenkova Y, Roghanian M, Bochkareva A, Zenkin N. Tagetitoxin inhibits transcription by stabilizing pre-translocated state of the elongation complex. Nucleic Acids Res. 2013;41(20):9257–9265. doi: 10.1093/nar/gkt708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hein PP, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011;50(32):7002–7014. doi: 10.1021/bi200437q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahl JM, et al. Direct observation of translocation in individual DNA polymerase complexes. J Biol Chem. 2012;287(16):13407–13421. doi: 10.1074/jbc.M111.338418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 2012;31(3):630–639. doi: 10.1038/emboj.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watnick RS, Gottesman ME. Binding of transcription termination protein nun to nascent RNA and template DNA. Science. 1999;286(5448):2337–2339. doi: 10.1126/science.286.5448.2337. [DOI] [PubMed] [Google Scholar]
- 44.Andrecka J, et al. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res. 2009;37(17):5803–5809. doi: 10.1093/nar/gkp601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.