Significance
Much attention has been drawn to research, which suggests that environmental factors, including stress and maternal care, alter the way the genetic code is executed. These epigenetic changes in gene regulation are thought to be stable enough to be heritable and thus may influence subsequent generations. Such prospects are as intriguing as they are troubling, since it is possible to imagine that accumulation of stress memories over several generations could make life decisions difficult. Therefore, we have questioned whether mechanisms exist that could prevent the inheritance of stress-induced epigenetic changes and have discovered such mechanisms using forward genetics in Arabidopsis. Interestingly, one of the critical activities erasing stress memories is conserved between plants and mammals.
Keywords: chromatin regulation, DNA methylation
Abstract
Examples of transgenerational transmission of environmentally induced epigenetic traits remain rare and disputed. Abiotic stress can release the transcription of epigenetically suppressed transposons and, noticeably, this activation is only transient. Therefore, it is likely that mechanisms countering the mitotic and meiotic inheritance of stress-triggered chromatin changes must exist but are undefined. To reveal these mechanisms, we screened for Arabidopsis mutants impaired in the resetting of stress-induced loss of epigenetic silencing and found that two chromatin regulators, Decrease in DNA methylation1 (DDM1) and Morpheus' Molecule1 (MOM1), act redundantly to restore prestress state and thus erase “epigenetic stress memory”. In ddm1 mutants, stress hyperactivates heterochromatic transcription and transcription persists longer than in the wild type. However, this newly acquired state is not transmitted to the progeny. Strikingly, although stress-induced transcription in mom1 mutants is as rapidly silenced as in wild type, in ddm1 mom1 double mutants, transcriptional signatures of stress are able to persist and are found in the progeny of plants stressed as small seedlings. Our results reveal an important, previously unidentified function of DDM1 and MOM1 in rapid resetting of stress induced epigenetic states, and therefore also in preventing their mitotic propagation and transgenerational inheritance.
Although environmentally induced traits and their transgenerational transmission in plants have been described repeatedly, trait stability and the involvement of epigenetic mechanisms in their generation remain controversial (1–3). In contrast, it has been well documented that environmental challenges such as elevated temperature can transcriptionally activate chromosomal loci normally silenced by repressive chromatin (4–7). However, this release of epigenetic silencing, unaccompanied by changes in DNA methylation or histone modifications, is only transient (4, 5). Such noncanonical release of transcriptional gene silencing (TGS) is similar to alterations in epigenetic regulation observed in the mom1 mutant, where release of TGS occurs without major changes in epigenetic marks (8–13). Although, molecular mechanisms used by MOM1 in TGS regulation are not well understood, genetic studies have linked MOM1 activity to small interfering RNAs (14) and RNA processing (15). In addition, structure/function studies have suggested that a conserved domain of MOM1 forms a homodimer, which is possibly required as a binding platform for additional silencing factors (13, 16).
The rapid resilencing of heterochromatic transcription induced by heat stress seems to involve changes in nucleosome occupancy and resilencing is delayed in mutants with impaired chromatin assembly (5). These observations suggest that suppressive chromatin has certain plasticity in response to stress, but also a robust buffering system that resets its prestress state. This counters the persistence of stress-induced epigenetic alterations during subsequent development and thus their transmission to the progeny.
Results and Discussion
To identify factors involved in the erasure of “epigenetic stress memory,” we performed a genetic screen using Arabidopsis line LUC25 carrying a transcriptionally silenced transgene encoding firefly luciferase (LUC) (14), which as an endogenous chromosomal TGS target loci can be transiently activated after heat stress. First, we introduced the mom1 mutation into LUC25 (mom1 LUC25). The mom1 mutation partially releases silencing of LUC25, producing weak luciferase signals in roots but not aerial parts, where the LUC transgene remains silenced. Importantly, the LUC transgene in mom1 LUC25 is strongly activated by heat stress, similar to LUC25 (Fig. S1A). We presumed that the introduction of the mom1 mutation would enhance stress-induced luciferase signals, increasing clarity and thus the efficiency of the mutant screen. Moreover, although the mom1 mutation does not directly influence the kinetics of stress-induced TGS release, MOM1 is involved in buffering epigenetic states of chromatin (11). We hypothesized, therefore, that any deficiency in such buffering would facilitate phenotypic detection of additional epigenetic regulators involved in the rapid restoration of TGS after stress and, thus, in the erasure of epigenetic stress memory.
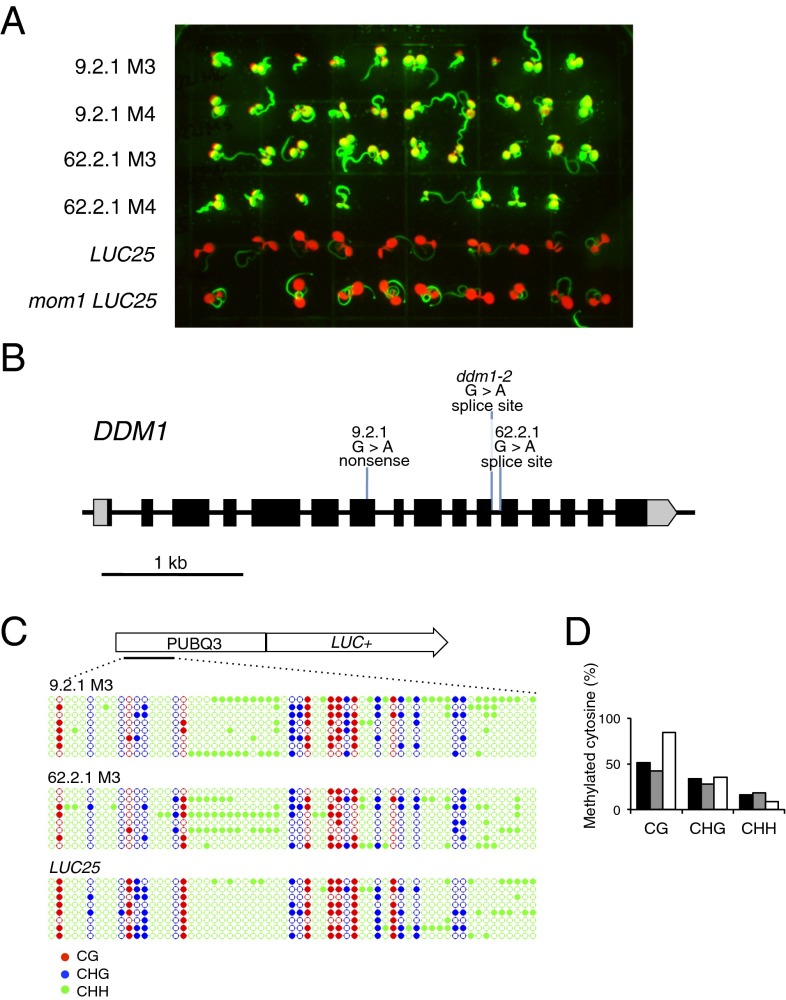
M2 seedlings of mutagenized mom1 LUC25 were germinated for 5 d and individuals showing enhanced luciferase signals before stress treatment were removed, because these plants release TGS constitutively. The remaining seedlings were subjected to heat stress and plants showing significantly stronger and/or longer-lasting luciferase signals were selected and grown to maturity (Fig. S1B). We examined their M3 progeny to determine whether selected phenotypes were heritable. Interestingly, several plants selected in the M2 produced progeny uniformly showing high luciferase signals prior to heat stress in the M3. Because such “constitutive” phenotypes had been discarded in the previous M2 generation, we concluded that their appearance in the M3 reflects transgenerational transmission of heat stress-induced TGS release that occurred in the previous plant generation (Fig. 1A). In other words, we may have recovered mutant plants severely impaired in the erasure of epigenetic stress memory. Focusing on four independent lines with these characteristics, we identified causal mutations by a combination of genetic mapping and whole genome sequencing. Two independent mutations resided in a gene encoding nucleosome remodeler DDM1 (17–20) (Fig. 1B). The DDM1 protein, conserved between plants and mammals, is required for maintenance of DNA methylation, thus TGS (21, 22). It has been suggested that DDM1 alters accessibility of H1-containing heterochromatin to DNA methyltransferases (23).
Fig. 1.
Identification and characterization of mutants showing transgenerational transmission of the heat-stress release of TGS. (A) Bioluminescence images of the progeny (M3 and M4) of two mutant candidates (9.2.1 and 62.2.1) and of the controls (LUC25, and mom1 LUC25), all grown under control conditions at 21 °C. The two mutant lines are M3 progeny of heat stressed M2 parents recovered from the mutant screen. The green and red signals are luciferase luminescence and auto-fluorescence of chlorophyll, respectively. (B) Two new mutant alleles of the DDM1 gene (9.2.1 and 62.2.1) were identified in the screen. The ddm1-2 allele was used for the experiments presented in Fig. 2. (C) DNA methylation distribution at the ubiqutin3 promoter of the LUC transgene in 9.2.1 (M3), 62.2.1 (M3), and LUC 25 (WT). Colored and open circles represent methylated and unmethylated cytosines respectively, with red representing CG sites, blue CHG and green CHH (H can be A, T, or C). (D) Percentage of cytosine methylation in the ubiqutin3 promoter. Black bars, 9.2.1; gray bars, 62.2.1; white bars, LUC25.
Recovery of ddm1 mutants in our screen was both surprising and disturbing. Surprising, because ddm1 mutants are known to release epigenetic silencing independently of stress and, therefore, should have been eliminated from the screen in the M2. Disturbing, because ddm1 mutants have a transgenerationally progressive effect on the loss of DNA methylation and silencing release (18, 19). Thus, luciferase activity observed in the M3 could simply reflect this ddm1 property rather than transgenerational memory of heat stress. To address these reservations, we first analyzed DNA methylation of the LUC promoter. Cytosine methylation patterns were only slightly altered, however, surprisingly high levels of methylation remained in the M3 of both mutant lines (Fig. 1 C and D), which is unusual for ddm1 mutants strictly associated with hypomethylation-mediated TGS release (17, 20, 24, 25). This result supported the possibility that the heat stress-activated state of the LUC transgene, which is independent of DNA methylation (4, 5), can in fact be maintained and transmitted to the next generation in the ddm1 background, and therefore, would define a previously unrecognized and potentially crucial activity of DDM1 in reversing TGS after it has been destabilized by environmental changes/stress.
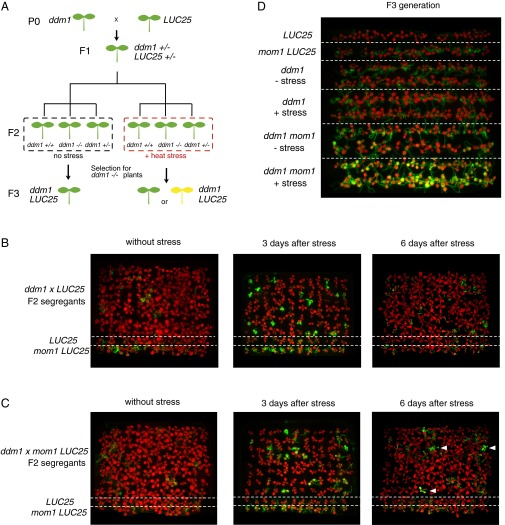
The only way to test this hypothesis was by the recreation and analysis of the ddm1 mutant line with a naïve and, thus, still-silenced LUC transgene (Fig. 2A). To obtain such a line, we crossed the commonly used allele of ddm1 (ddm1-2) (18) into LUC25 and subjected the F2-segregating progeny to temperature stress. Importantly, under control growth conditions, luciferase signals remained at the LUC25 level and no differences between segregating F2 individuals were recorded (Fig. 2B). This suggested that the LUC transgene remained silent in the first generation of ddm1 mutants and explains the initially unexpected presence of ddm1 among plants subjected to heat stress during the mutant screen. Notably, heat stress applied to segregating F2 seedlings revealed individuals with very strong luciferase signals (Fig. 2B) in proportion close to expected segregation ratio for plants homozygous ddm1 mutation containing LUC plants (18.75%). The genotyping of the segregating population at DDM1 and LUC loci confirmed that, all of these plants were found to be homozygous for the ddm1-2 mutation and contained LUC transgene in homozygous or hemizygous state, which had no influence on the intensity of LUC signals. Furthermore, we performed an analogous genetic experiment introducing the ddm1-2 allele into another line (L5) (26) carrying a silent transgenic locus for the glucuronidase marker gene (GUS). As with the LUC transgene, we observed heat-stress-dependent hyperactivation of GUS expression in ddm1-2 mutants (Fig. S2). These results demonstrated that DDM1 down-regulates stress-induced heterochromatic transcription. Moreover, this novel DDM1 activity appears to be independent of changes in DNA methylation, with which so far DDM1 was very tightly associated.
Fig. 2.
Inheritance of stress-induced transcriptional activation of LUC. (A) Crossing scheme for the recreation of the ddm1-2 mutant line with the naïve LUC transgene. P0: ddm1-2 was crossed with WT LUC25, F1: heterozygous for ddm1 and carrying the hemizygous naïve LUC transgene, F2: ddm1 homozygous mutants are segregated in the progeny. F2 seedlings were separated into two subpopulations, one of which was subjected to heat stress. Bioluminescence images were captured (B), and each plant was genotyped at the DDM1 and LUC loci. (B) Bioluminescence images of segregating progeny of a hybrid between ddm1-2 and LUC25 (A). The F2 seedlings were expected to include 18.75% of individuals homozygous for the ddm1-2 mutation and carrying the LUC transgene, these were predicted to display enhanced luminescence. Rows of LUC25 and mom1 LUC25 plants are shown as a control. Note, LUC signals in mom1 after heat stress are restricted to roots. (C) Bioluminescence images of segregating progeny of a cross between ddm1-2 mom1 LUC25 (for details, see Fig. S3). The F2 progeny is expected to include 4.69% of individuals homozygous for both ddm1-2 and mom1 and carrying the LUC transgene. White arrowheads point toward ddm1-2 mom1 double mutant plants, as revealed by the genotyping of the population at DDM1 and MOM1 loci. Quantification of LUC signals is displayed in Fig. S4. (D) Bioluminescence images of ddm1 LUC25 and ddm1 mom1 LUC25 F3 progenies (as depicted in A) of F2 heat stressed parents at the seedlings stage (+stress) or nonstressed controls (−stress). Two independent F3 populations (upper or lower row) derived from two ddm1 LUC25 or ddm1 mom1 LUC25 F2 plants (B and C).
However, we found that in ddm1-2 mutants the stress-hyperactivated LUC transgene was resilenced within a few days (Fig. 2B) and there was no difference in LUC signals in the progeny (F3 generation) between stressed and nonstressed ddm1-2 LUC25 plants (Fig. 2D).
Considering the genetic screen was performed in the mom1 LUC25 background, we repeated the genetic reconstruction experiment including the mom1 mutation. ddm1-2 was crossed with mom1 LUC25 and stress-induced LUC activation in the segregating F2 populations was examined as before, as well as its inheritance in the F3 (Fig. S3). LUC phenotyping and subsequent genotyping of F2 plants showed that the mom1 mutation alone did not affect stress-induced expression of LUC, confirming the previous observations with the mom1 LUC25 line. Although, the stress-induced LUC activation levels in the ddm1-2 mom1 double mutants were similar to those in the ddm1-2 single mutants (Fig. S4A), however, the LUC signals remained high only in ddm1-2 mom1 double mutants (Fig. 2C). Furthermore, in the next (F3) generation, progeny from stressed ddm1-2 mom1 LUC25 plants showed significantly higher LUC signals than nonstressed ddm1-2 mom1 LUC25 controls (Fig. 2D and Fig. S4B), indicating that the stress-induced active state of the LUC transgene initiated at the small seedling stage could persist throughout plant development and be transgenerationally transmitted, however only in ddm1 mom1 double mutants.
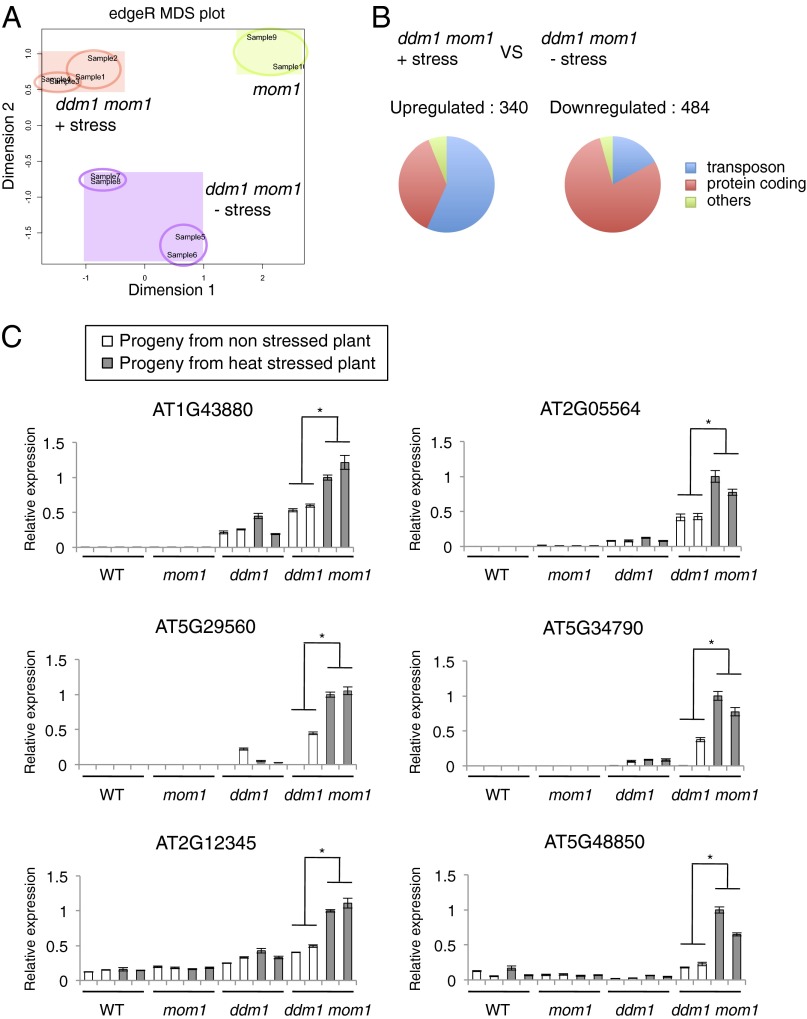
To further investigate stability and possible transgenerational inheritance in ddm1 mom1 double mutants of stress-triggered TGS release, we examined transcriptional changes at endogenous chromosomal loci. For this, we first performed RNA-seq analyses on four independent populations of the progeny of stressed and control-treated ddm1-2 mom1 double mutant plants. This genome-wide approach should uncover chromosomal loci stably activated following the stress subjected to the parental plants. The MDS plot analysis of the RNA-seq data revealed well-clustered biological repetitions for each individual line (Fig. 3A), indicating the robustness of the RNA-seq data. Noticeably, the largest difference between biological repetitions was seen for ddm1 mom1 double mutant seedlings derived from ancestors not subjected to stress. This may simply reflect an intrinsic property of ddm1 mom1 double mutants, which exhibit variation in heterochromatin silencing among individual plants (27). Most importantly, the samples were clearly clustered according to whether seedlings of the previous generation were stressed or not (Fig. 3A). Therefore, the genome-wide transcriptional profiles supported and extended our previous conclusion based on transgenic loci that temperature stress-activated transcription occurs genome wide, and that newly acquired transcriptional signatures can be transgenerationally inherited in ddm1 mom1 double mutants.
Fig. 3.
Genome-wide analysis of transcriptional changes in the progeny of heat-stressed ddm1 mom1. (A) Multidimensional scaling (MDS) plot (R/Bioconductor) showing the overall similarity of RNA expression patterns between samples using RNA-seq data of two biological repetitions. These were performed with two independent populations (circled) of ddm1 mom1 progeny plants obtained from stressed or nonstressed parents at the seedling stage (mom1 transcriptome was used as an additional control). (B) Pie charts showing the difference in functional distribution of up- and down-regulated loci between heat-stressed or nonstressed double mutants. (C) Relative levels of mRNA of selected targets as determined by quantitative RT-PCR. Values were normalized to 18s ribosomal RNA. One sample of stressed ddm1 mom1 was set to 1. White and gray bars indicate the progeny of control plants and heat-stressed plants, respectively. Pooled F3 plants (∼20 individuals), progeny of two independent F2 plants for each category were used for these analyses. Gene annotations of each target: AT1G43880 - ATLANTYS1 (TE), AT2G05564 - VANDAL2 (TE), AT5G29560 - caleosin-related family protein, AT5G34790 - VANDAL20 (TE), AT2G12345 - ATHILA3 (TE), AT5G48850 - ATSDI1, SULFUR DEFICIENCY-INDUCED 1. Error bars indicate SD of results from three repeated experiments. *P < 0.01, Student t test.
Of the loci affected in the progeny of stressed ddm1-2 mom1, compared with the progeny of control-treated ddm1-2 mom1, 340 loci were up-regulated more than twofold (P < 0.01) and 484 down-regulated less than twofold (P < 0.01) (Fig. 3B). Approximately 60% of the up-regulated transcripts and 20% of the down-regulated transcripts are derived from transposable elements (TEs) (Fig. 3B). These results are consistent with our previous demonstration that heterochromatic regions enriched in TEs are predominantly transcriptionally activated by temperature stress, and that euchromatic regions are either activated or repressed by this treatment (4). Such a transcriptional signature of the genome-wide stress-induced alteration of transcription appears to be inherited by the progeny of ddm1 mom1 double mutant plants.
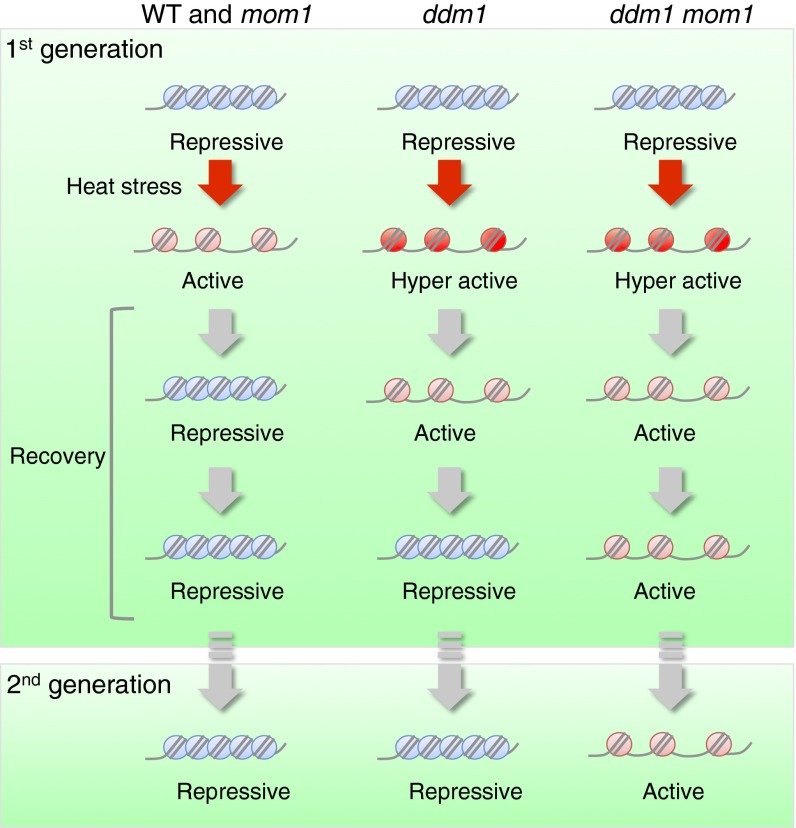
Due to economic constraints, we refrained from genome-wide analyses of plant populations constituting various experimental controls. Instead, using the transcriptional profiling data described above we selected several genomic loci displaying heritable stress-induced alteration of transcription in ddm1 mom1 double mutants, and examined by quantitative RT-PCR persistence of their transcriptional activation in WT and single mom1 or ddm1 mutant plants relative to stress treatments in the preceding generation. No significant differences in transcript levels were found between the progenies of stressed and control WT plants or single mutants, which is in contrast to ddm1 mom1 double mutants (Fig. 3C). This result supports our previous conclusion, derived from the properties of transgenic loci, that DDM1 and MOM1 both act redundantly in resetting chromatin status destabilized by heat stress, which prevents transgenerational propagation of transcriptional stress memory (Fig. 4).
Fig. 4.
Model summarizing a putative epigenetic mechanism preventing the transmission of “stress memory” to progeny. Schematic illustration on a possible chromatin states upon heat stress. Although heterochromatic loci are transcriptionally activated by temperature stress in WT and mom1 mutants, they are rapidly resilenced after stress is removed. Stress-induced transcription is hyper-activated and persists longer in ddm1 mutants than in WT and mom1; however, the altered transcriptional status is not transmitted to the progeny. In contrast, stress-induced transcriptional activation in ddm1 mom1 double mutants is transgenerationally inherited. The transcriptional activation in ddm1 mom1 could persist due to altered positioning of nucleosome or other modifications of chromatin properties.
A closer look at the results of the genome-wide transcriptional analyses reveals that DDM1 and MOM1 are not the only factors reverting the properties of chromatin affected by stress. We found previously that ∼3,000 loci are activated under stress conditions analogous to those used here (4) and, thus, only a small fraction (340) remain transgenerationally active in the ddm1 mom1 double mutants. This result suggests that the prevention of transgenerational transmission of stress memory extends far beyond the activities of DDM1 and MOM1 and, thus, that the unequivocal demonstration of transgenerational transmission of environmentally-induced epigenetic traits remains a significant challenge.
Materials and Methods
Plant Materials.
The LUC25 line was described (14). mom1 LUC25 was obtained by crossing mom1-6 and LUC25. mom1-6 seeds were obtained from INRA-Versailles, Genomic Resource Center (FLAG_340E12) and ddm1-2 seeds were provided by E. Richards (Boyce Thompson Institute for Plant Research, Cornell University, Ithaca, NY). LUC25 and mom1-6 are in the Wassilewskija (WS) background and ddm1-2 in the Columbia (Col-0) background.
Mutagenesis and Screening.
mom1 LUC25 seeds (20,000) were mutagenized in 0.3% EMS for 15 h. After washing with water, seeds were germinated on soil to give 78 M2 pools, each derived from ∼150 M1 plants. For each pool, 1,000 seeds were plated on 1/2 MS medium (0.8% agar, 1% sucrose) and screened for mutants by spraying with a luciferin (Biosynth) solution (31.5 mg per 100 mL of water) and examining treated seedlings using an Aequoria dark box with a mounted ORCAII CCD camera (Hamamatsu). Luciferase luminescence and chlorophyll auto-fluorescence image overlays were created using the Wasabi software package (Hamamatsu). Isolated mutants were crossed with WT in the Col-0 background to generate mapping populations.
DNA Methylation Analysis.
Bisulphite sequencing was performed as described (14). Sequencing data were analyzed with Kismeth (http://katahdin.mssm.edu/kismeth) (28). Primers used for bisulphite sequencing are listed in Table S1.
Histochemical GUS Staining.
Staining was performed on cotyledons of 12-d-old plants as described (13).
RAN-seq Analyses.
Total RNA samples were isolated from 7-d-old seedlings using TRI reagent (Sigma). The libraries were prepared using a TruSeq RNA Sample Prep Kit (Illumina) and sequenced using HiSEq. 2500 (Illumina) with single-end 50 base reads. The trimmed reads were mapped with the TopHat v1.3.3 software to the TAIR10 annotations. The normalization and differential expression analysis were performed with R/Bioconductor package edgeR v.2.6.12 (29).
Real-Time Quantitative PCR Analysis.
Total RNA was isolated from 7-d-old seedlings using TRI reagent (Sigma). After RQ1 DNase treatment (Promega), cDNA was synthesized with the SuperScript VILO cDNA synthesis kit (Life Technologies). Real-time quantitative PCR analyses were performed using Power SYBR Green PCR Master Mix (Life Technologies) in ABI7900FT (Life Technologies). PCR conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Primers are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Taisuke Nishimura for preparing EMS-mutagenized mom1 LUC25 seeds, Christian Megies for technical assistance, Mylène Docquier and Céline Delucinge for help with RNA-seq analysis, Detlef Weigel and Beth Rowan for help with whole genome sequencing analysis, and Patrick King for editing of the manuscript. We also thank all members of the J.P. laboratory for constructive discussions. This work was supported by grants from the Swiss National Science Foundation (31003A-125005), the European Commission through the AENEAS collaborative project (FP7 226477), the Gatsby Charitable Foundation, and the European Research Council.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402275111/-/DCSupplemental.
References
- 1.Paszkowski J, Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr Opin Plant Biol. 2011;14(2):195–203. doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Boyko A, Kovalchuk I. Genome instability and epigenetic modification—heritable responses to environmental stress? Curr Opin Plant Biol. 2011;14(3):260–266. doi: 10.1016/j.pbi.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14(3):267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Tittel-Elmer M, et al. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010;6(10):e1001175. doi: 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pecinka A, et al. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22(9):3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang-Mladek C, et al. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant. 2010;3(3):594–602. doi: 10.1093/mp/ssq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472(7341):115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 8.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405(6783):203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 9.Steimer A, et al. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell. 2000;12(7):1165–1178. doi: 10.1105/tpc.12.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33(4):743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 11.Habu Y, et al. Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep. 2006;7(12):1279–1284. doi: 10.1038/sj.embor.7400835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaillant I, Schubert I, Tourmente S, Mathieu O. MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 2006;7(12):1273–1278. doi: 10.1038/sj.embor.7400791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura T, et al. Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1. PLoS Genet. 2012;8(2):e1002484. doi: 10.1371/journal.pgen.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokthongwattana C, et al. MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 2010;29(2):340–351. doi: 10.1038/emboj.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Zhang J, Lin H, Guo G, Guo Y. MORPHEUS’ MOLECULE1 is required to prevent aberrant RNA transcriptional read-through in Arabidopsis. Plant Physiol. 2010;154(3):1272–1280. doi: 10.1104/pp.110.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caikovski M, et al. Divergent evolution of CHD3 proteins resulted in MOM1 refining epigenetic control in vascular plants. PLoS Genet. 2008;4(8):e1000165. doi: 10.1371/journal.pgen.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 18.Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA. 1996;93(22):12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeddeloh JA, Bender J, Richards EJ. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 1998;12(11):1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22(1):94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 21.Bourc’his D, Bestor TH. Helicase homologues maintain cytosine methylation in plants and mammals. Bioessays. 2002;24(4):297–299. doi: 10.1002/bies.10078. [DOI] [PubMed] [Google Scholar]
- 22.Tao Y, et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci USA. 2011;108(14):5626–5631. doi: 10.1073/pnas.1017000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakutani T, Munakata K, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151(2):831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430(6998):471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 26.Morel JB, Mourrain P, Béclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10(24):1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 27.Mittelsten Scheid O, Probst AV, Afsar K, Paszkowski J. Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(21):13659–13662. doi: 10.1073/pnas.202380499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruntman E, et al. Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics. 2008;9:371. doi: 10.1186/1471-2105-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.