Significance
The retrosplenial cortex (RSC) is a key part of a network of brain regions that processes and stores spatial information. However, it is unclear whether the RSC actually encodes or stores spatial information. With time-lapse two-photon in vivo imaging that allowed us to study the immediate early gene c-fos expression in tens of thousands of cells in the RSC, we uncovered repetitive activation of a fraction of cells in this structure during spatial learning. We also showed that RSC inactivation disrupts performance in a spatial task, whereas overexpressing of the transcription factor CREB results in enhanced spatial memory. Silencing the CREB-expressing neurons occluded these memory enhancements. These results demonstrate that the retrosplenial cortex is involved in the formation and storage of spatial information.
Abstract
The retrosplenial cortex (RSC) is part of a network of interconnected cortical, hippocampal, and thalamic structures harboring spatially modulated neurons. The RSC contains head direction cells and connects to the parahippocampal region and anterior thalamus. Manipulations of the RSC can affect spatial and contextual tasks. A considerable amount of evidence implicates the role of the RSC in spatial navigation, but it is unclear whether this structure actually encodes or stores spatial information. We used a transgenic mouse in which the expression of green fluorescent protein was under the control of the immediate early gene c-fos promoter as well as time-lapse two-photon in vivo imaging to monitor neuronal activation triggered by spatial learning in the Morris water maze. We uncovered a repetitive pattern of cell activation in the RSC consistent with the hypothesis that during spatial learning an experience-dependent memory trace is formed in this structure. In support of this hypothesis, we also report three other observations. First, temporary RSC inactivation disrupts performance in a spatial learning task. Second, we show that overexpressing the transcription factor CREB in the RSC with a viral vector, a manipulation known to enhance memory consolidation in other circuits, results in spatial memory enhancements. Third, silencing the viral CREB-expressing neurons with the allatostatin system occludes the spatial memory enhancement. Taken together, these results indicate that the retrosplenial cortex engages in the formation and storage of memory traces for spatial information.
Spatial navigation, learning, and memory depend on several interconnected brain regions. The hippocampus is known to have a central role in spatial learning and memory. Place cells in this structure form a spatial map of an animal’s environment (1). The main input to the hippocampus comes from the entorhinal cortex, an area that integrates cortical inputs before forwarding them into the hippocampal formation. The dorsal medial entorhinal cortex (MEC) contains spatially modulated cells (grid cells) that fire at the nodes of a hexagonal lattice as the animal traverses its environment (2). Beyond hippocampal place cells and MEC grid cells, there is also extensive evidence for another type of spatially modulated neurons that increase firing when the animal’s head points in a specific direction [head direction (HD) cells] (3). These cells reside in several subcortical and cortical regions, including the retrosplenial cortex (RSC). The RSC is one of the most important cortical afferents to MEC (4, 5). It is the most caudal part of the cingulate cortex, positioned between anterior cingulate, parietal/visual, and parahippocampal cortices. It is further subdivided into areas A29a–c (granular cortex) and area A30 (dysgranular cortex) (6). The RSC has been implicated in a number of cognitive tasks, including navigation based on external spatial (allothetic) information (7). Human functional MRI (fMRI) studies showed that the RSC is activated during route planning (8) and during recognition of a familiar environment (9). Damage to the RSC severely impairs ability to navigate within known locations (10). In rats, lesions of the RSC impair acquisition of behavioral tasks involving spatial navigation (11–16). These impairments are particularly large when the difficulty of the tasks is increased, for example during navigation in darkness (17, 18) or when there is a conflict between proximal and distal cues (19). Inactivation of this structure temporarily remaps place fields in the hippocampus (20), which suggests that hippocampal spatial processing is affected by the RSC. Therefore, this structure seems to be an important component of spatial circuits in both humans and rodents. Based on this evidence, it has been proposed that the RSC role in spatial navigation is due to either its ability to process the sequence of heading vectors required for navigating in space or its ability to support the transition between allocentric (world-centered) references, encoded by the hippocampus, and egocentric (self-centered) references, supported by the parietal cortex (21).
Recent studies suggested that, in addition to the hippocampus, other connected cortical regions can support encoding and storage of spatial memory (22–25). Elevated expression of immediate early genes (IEGs) is thought to reflect this involvement (22, 25, 26). Induction of IEGs, including c-fos, has been linked to behaviorally relevant transcriptional activity in multiple brain regions (27). The formation of a stable, nonrandom network of cells that repeatedly expresses Fos during acquisition of a specific behavioral task would suggest that a memory trace is formed within that structure. To address this hypothesis, we used a FosGFP reporter mouse (28) to chronically monitor in vivo patterns of neuronal activation in the RSC following a spatial task. We then tested whether the mouse RSC is necessary for precise retrieval of recently acquired spatial information. Importantly, we also determined the impact on spatial memory of increasing cAMP response-element binding protein (CREB) levels specifically within the RSC, a manipulation known to enhance memory consolidation in several other brain structures and in other species. Finally, we used the allatostatin neuronal inactivation system to specifically silence RSC cells with increased CREB levels before a spatial memory test. Our findings indicate that, as a part of its role in spatial navigation, the RSC encodes and stores memory traces for spatial information.
Results
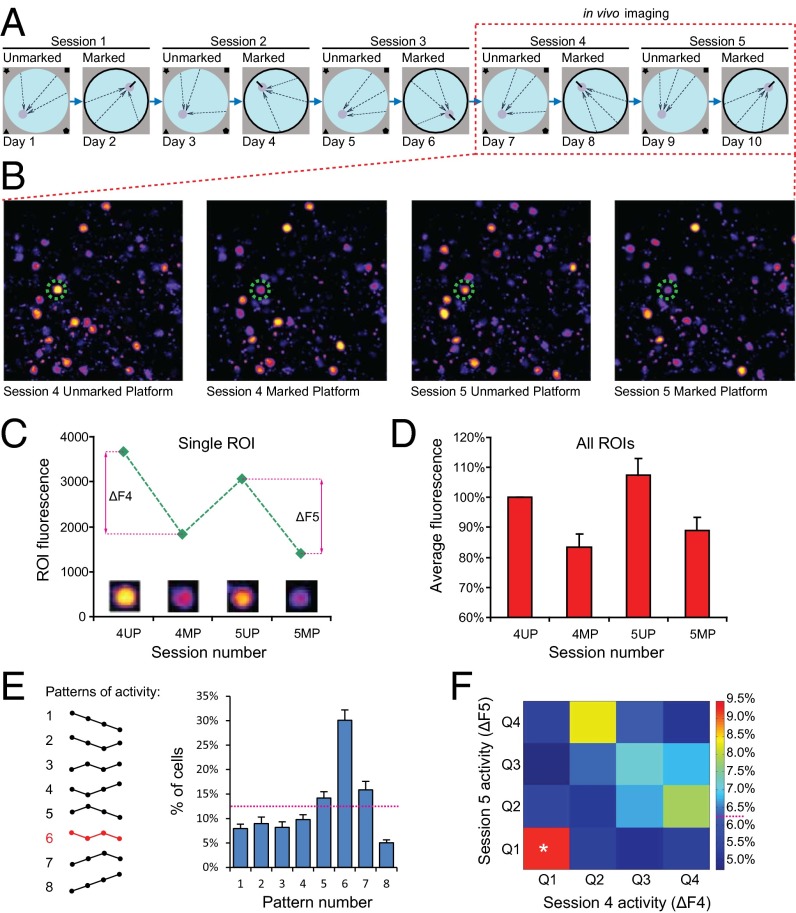
To test whether the RSC encodes spatial information, we used a transgenic mouse with a FosGFP reporter (28, 29), and time-lapse two-photon in vivo imaging (30, 31). We modified the Morris water maze experimental design to distinguish between RSC patterns activated by two versions of this task that depend on different strategies. On each day, the animals were tested either on the marked platform (MP) task or on the unmarked platform (UP) task (Fig. 1A). For the UP task, we used the traditional Morris maze with its complex set of extra maze cues, and with the escape platform hidden from view. In contrast, in the MP task, the escape platform was marked with a metal rod, and a black curtain surrounding the pool obscured extra maze cues. This design allowed us to evaluate whether the FosGFP activation patterns we measured in RSC were significantly driven by stress, locomotion, motivation, etc. (imaged after both MP and UP sessions). If so, we would expect to see very little difference in the FosGFP activation patterns triggered by the two tasks. Both tasks require general navigation, are affected by stress, and require motivation, but only the UP task depends on spatial/contextual memory; performance in the MP task is solely determined by the position of the metal rod marking the escape platform. We allowed 24-h intervals between each task to allow the FosGFP protein to return to basal levels. Before the start of behavioral training, each mouse was implanted with a cranial window positioned directly over the dysgranular RSC (Methods). The chronic in vivo imaging technique we used allowed us to align and register image stacks from multiple sessions. Therefore, this enabled us to track the FosGFP levels for each imaged cell during the course of the experiment. We monitored in vivo FosGFP levels in cells of layer II of RSC in nine mice with implanted cranial windows. We acquired 1–4 fluorescent image stacks (total of 23 stacks) from the RSC of each animal, 90 min after each of four behavioral sessions: fourth unmarked platform (4UP), fourth marked platform (4MP), fifth unmarked platform (5UP) and fifth marked platform (5MP), (Fig. 1 A and B). Analysis of the search patterns suggests that during this stage of the experiment, animals usually start adopting spatial strategies in the water maze. We then identified the positions of FosGFP-positive cells (regions of interest, ROIs) in each image stack and measured their fluorescence intensity (Methods). During each session, between 50% and 60% of all of the neurons in layer II of dysgranular RSC (RSC-d) were activated (Fig. S1). We estimate that each cell within the field of view was detected at least once during the four in vivo sessions mentioned above. A total of 25,127 ROIs from 23 image stacks were registered. We first compared the activation response (defined by the level of FosGFP fluorescence) between UP and MP sessions. Fig. 1C shows FosGFP changes in a typical RSC cell, whereas Fig. 1D shows the average for all RSC cells analyzed. Both UP sessions (4UP and 5UP) evoked higher levels of FosGFP activation than the corresponding MP sessions (4MP and 5MP). The average response after session 4MP was 83.42% of the response observed after session 4UP, whereas the session 5MP response was 82.77% of the session 5UP response (Fig. 1D). This pattern of FosGFP responses, with both UP sessions evoking more robust activation than MP sessions, was observed in approximately 30% of all RSC cells analyzed (Fig. 1E, pattern 6). This effect was not merely related to the length of the behavioral session, because Fos activation showed no positive correlation with the time to reach the platform (Fig. S2). In the somatosensory (barrel) cortex (SSC), the patterns of FosGFP activation did not correlate with either the UP or MP tasks (Fig. S3). The FosGFP activation we imaged in the barrel cortex is probably due to somatosensory activation, such as whisking sides of the pool and the metal rod after completion of the trial. Imaging studies in the motor cortex (MC), another region used as a control, detected no substantial FosGFP activity in the superficial layers.
Fig. 1.
Two-photon (2P) in vivo imaging of FosGFP reporter during a modified water maze task. (A) Behavioral setup. Days 1, 3, 5, 7, and 9 show unmarked platform task, with platform location indicated by extra maze cues on the walls of the room. Days 2, 4, 6, 8, and 10 show marked platform task, with room cues obscured by curtain and the platform location marked by vertical rod attached to the center of the platform. Arrows indicate the release points. On each of the last 4 d (marked by a dashed rectangle), 90 min after the behavioral session, FosGFP was imaged using a 2P microscope. (B) Sample images of the same area of the RSC acquired after the last four behavioral sessions. Dashed circle marks the cell shown in C. (C) Example of typical changes in FosGFP level of a single RSC cell during the course of the experiment. For each corresponding UP/MP pair, ΔF was calculated to estimate FosGFP level related to the UP-dependent component of the task (ΔF4 = 4UP − 4MP, ΔF5 = 5UP − 5MP). (D) Summary of FosGFP activation in the RSC after alternating UP and MP sessions. UP sessions evoked higher expression of the reporter than the corresponding MP sessions. Nanimals = 9, NROIs = 25,127. (E) Summary of activation changes displayed by RSC cells during the imaging experiment. Each cell could follow one of eight patterns of response. Approximately 30% of the cells show higher FosGFP level in both UP sessions and lower in the MP sessions (pattern 6, marked in red) and therefore are potentially specific for the UP version of the task. Nanimals = 9, NROIs = 25,127. (F) High Fos level in session 4 is a good predictor of activity in session 5. Spatially specific cells that follow pattern 6 in E were divided into four quartiles according to the ΔF4 and ΔF5 values and a colocalization matrix was created. Cells that show highest ΔF in session 4 (Q1) are 50% more likely than chance to remain in the Q1 also for session 5. Matrix element [1,1] contains 9.27% of the entire population, statistically different from random distribution (*Z score = 2.37, P < 0.01). Nanimals = 9.
Next, we examined in detail the previously identified population of RSC cells that displayed higher FosGFP activity in both UP sessions imaged (shown as pattern 6 in Fig. 1E). We determined whether the relative levels of FosGFP in response to one UP session (4UP) would predict FosGFP responses in the subsequent UP session (5UP). To focus our analysis on the memory component of RSC activation specific for the UP task (and not on activation due to other behavioral determinants such as motivation, general navigation requirements, etc.), we first calculated the difference in fluorescence between each UP session and the corresponding MP session for each ROI (ΔF4 = 4UP − 4MP, ΔF5 = 5UP − 5MP, Fig. 1C). The resulting values of ΔF4 and ΔF5 should reflect more closely activation in RSC due to the spatial components specific for each UP session. Our analyses show that for each session the distribution of ΔF values is Gaussian like and clearly unimodal (Fig. S4). This distribution does not reveal a significant range of fluorescence changes. We therefore divided the ROIs into quartiles, according to their descending ΔF4 and ΔF5 values, and created a cross-correlation matrix (Fig. 1F). If the activation of FosGFP during subsequent sessions were random, and if there were no correlation between FosGFP levels in sessions 4 and 5, we should obtain a uniform distribution, where all 16 elements of the array would contain 6.25% of the cells. Instead, we observed a nonrandom distribution of the cells in the matrix. Importantly, 9.27% of all of the cells measured belonged to the top quartile in both ΔF4 and ΔF5 (represented by the matrix element [1,1] in Fig. 1F). This represents a 48% increase in the number of cells in that quartile compared with chance levels. Additionally, we performed a Monte-Carlo–type analysis to assess the statistical significance of such distribution. For each image stack we shuffled the relative positions of cells between ΔF4 and ΔF5 300 times to obtain random distributions of probabilities for each element of the matrix. Then, we compared experimentally obtained percentages with the SD for those randomized distributions, and calculated the Z score. For the element [1,1] the average Z was 2.37 (P < 0.05). These results showed that a population of neurons in layer II of RSC, consisting of cells with robust FosGFP activation, is indeed preferentially reactivated upon consecutive spatial sessions of the same type, a result consistent with the hypothesis that the RSC encodes spatial memory traces.
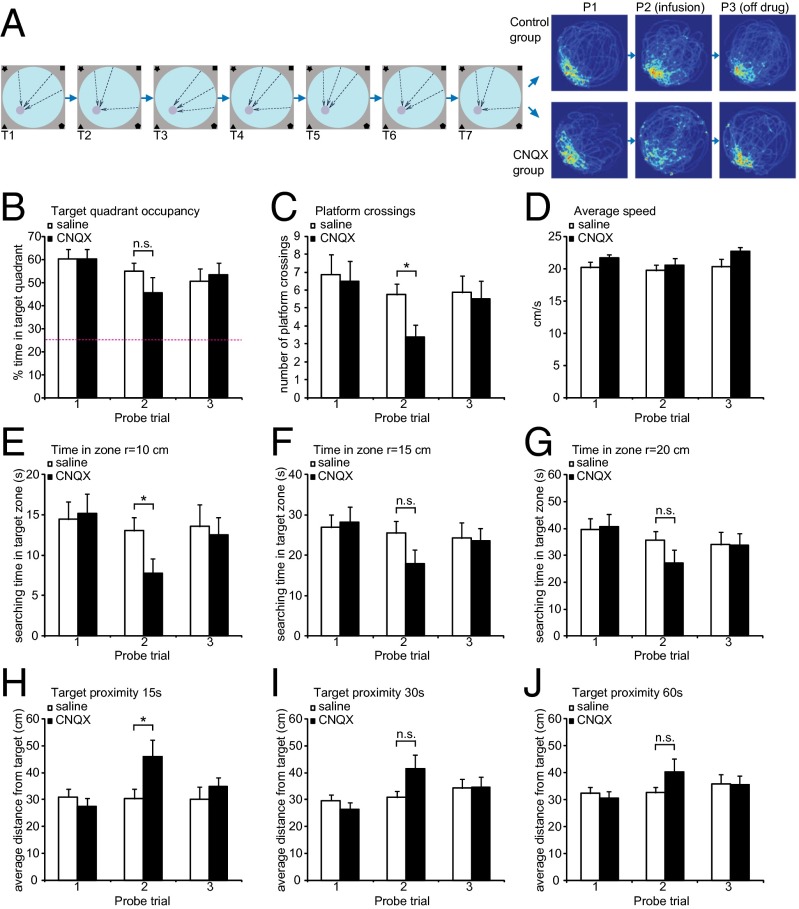
Previous lesion studies (11, 32) suggested that the RSC is needed for spatial navigation, but they do not address whether it is required for spatial memory retrieval in trained mice. To address this question, we trained mice with intact RSC and then reversibly inactivated it before a probe trial in the Morris maze to determine if this structure is needed during a spatial memory test in mice. Mice were implanted with bilateral cannuli that projected into the RSC and then allowed to recover for 2 wk (Methods). The mice were then trained on the standard hippocampus-dependent Morris water maze task, with three trials per day for 7 d followed by a probe trial. Based on the results, animals were assigned to two groups of equal performance (Fig. 2 A and B–J, first pair of columns). Ten minutes before the next probe trial (24 h after the first test), one group received an infusion of 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), a reversible AMPA receptor antagonist that blocks neurotransmission, and the control group received saline. This manipulation allowed us to silence neuronal activity specifically in RSC (Fig. S5). Analysis of this probe trial revealed a specific deficit in the group with CNQX inactivation in the RSC (CNQX group). It was related to the degree of precision of the direct approach toward the target. The target quadrant occupancy during the entire probe trial did not show a significant difference between groups (Fig. 2B). The observed lack of difference is attributable to a ceiling effect. Analyses of other more demanding probe trial measures revealed a deficit in the CNQX group: First, the number of target crossings was significantly lower in the CNQX group (Fig. 2C). Second, the time spent in a circular zone with a 10-cm radius (but not with a 15-cm or 20-cm radius) around the platform in the target quadrant was lower for the CNQX group (Fig. 2 D–G), a possible reflection of deficits in spatial memory. The average distance to the platform during the first 15 s of searching in the probe trial was significantly higher for the CNQX animals (Fig. 2H). Although the trend is still there, this difference is not statistically significant at either 30 s or 60 s (Fig. 2 I and J). It appears that inactivation of the RSC during the probe trial may interfere specifically with the ability to quickly find a direct route to the goal by using allothetic cues but not with the overall recognition of the target. Once the animals find the correct quadrant, they are able to remain in close proximity and continue the search there. It is possible that with more time (>15 s) the mice would be able to retrieve memory representations in other structures to support spatial specific searches for the platform. The effect of CNQX is reversible; when animals were retested in another probe trial 24 h after the CNQX infusion, no deficit was observed. Also, the deficit is specific to the hidden platform task. When the same group of animals was retrained using the visible platform task (Methods), RSC inactivation did not cause a deficit in the mice’s ability to navigate to the marked platform (Fig. S6), suggesting that the RSC is not essential for general navigation to a target, for swimming, motivation, visual perception, motor skills, and other behavioral components that are common between the two tasks. Instead, these results are consistent with the hypothesis that the RSC contains a memory trace for the contextual information required to find the hidden platform.
Fig. 2.
The effect of RSC inactivation on retrieval of spatial memory in Morris water maze. (A) Experimental design and density maps of performance during probe trials. Mice (n = 16) were implanted with cannuli and trained for 7 d with three trials per day. Dashed arrows represent release points during training sessions. Twenty-four hours after last training session animals were tested for memory retention. On the following day, mice were infused with CNQX (n = 8) or saline (n = 8) and probed again. Twenty-four hours later, another probe trial was performed. Heat maps depict time spent by the animals in each pixel during probe trials. (B–F) Performance measures during probe trials 1–3. (B) Target quadrant occupancy (percentage of time spent in the quadrant with the target). CNQX-infused animals are not significantly worse in general measure of memory retention. (C) Average number of platform crossings (swims over former platform location). CNQX group is significantly impaired when assessed by a specific measure of search specificity. (*P < 0.05). (D) Average speed (centimeters per second). (E–G) Percentage of time spent in the target zone, defined as circular region of r = 10 cm (H), 15 cm (I), or 30 cm (J) centered on the former platform location. (*P < 0.05). (H–J) Average distance from the target during the first 15 s (E), 30 s (F), and the entire probe trial (G). Statistical significance: *P < 0.05, paired t test.
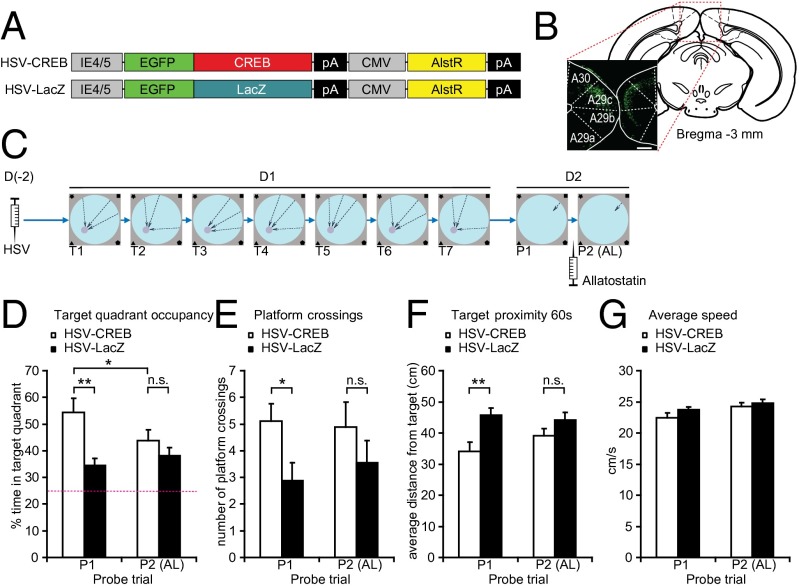
Previous studies demonstrated that overexpression of the transcription factor CREB can lead to enhancements in memory (33–35). If the RSC encodes spatial information, increasing CREB levels there should enhance spatial memory. To increase CREB levels specifically in a subpopulation of RSC neurons, we used an HSV-CREB viral vector (34, 36) (Fig. 3 A and B). An HSV vector expressing LacZ (HSV-LacZ) was used as control (Fig. 3C). To more easily detect enhancements of short-term spatial learning and memory, mice were trained in a single day with seven sessions of three trials each (37). Memory performance was tested in a probe trial given 24 h later. Analysis of target quadrant occupancy showed that control mice learned the task to some extent, spending approximately 35% of the time in the target quadrant. Remarkably, animals injected with HSV-CREB in the RSC outperformed their controls; similar results were found on other measures of spatial performance, including platform crossings and target proximity (Fig. 3 D–F, probe trial 1). In both measures, the performance of the mice injected with HSV-CREB in the RSC was better than controls.
Fig. 3.
HSV-CREB in the RSC affects learning in water maze. (A) Schematic diagram of vectors used for HSV production. (B) Expression of GFP in retrosplenial cortex following HSV infection. (Scale bar, 200 µm.) (C) Experimental procedure. Animals were injected with HSV expressing either CREB (n = 10) or LacZ (n = 10) and allowed to recover for 2 d. On the training day, mice were subjected to seven training sessions (T1–T7, each consisting of three trials), separated by 30 min. After 24 h, mice were probed for memory retention (probe trial 1, P1), infused with allatostatin, and then probed again (P2). (D–F) Performance measures during P1 and P2. (D) Target quadrant occupancy (percentage of time spent in the quadrant centered on former platform location). Mice infected with HSV-CREB spent more time in the target quadrant than HSV-LacZ animals (**P < 0.01). This effect is abolished by allatostatin infusion (P2). Inactivation of AlstR-expressing neurons impairs the performance of HSV-CREB animals (P1 vs. P2, *P < 0.05) whereas HSV-LacZ mice remain unaffected. (E) Target crossings (number of passes over the former location of the platform) during P1 and P2. CREB-HSV animals are more precise than CREB-LacZ mice (*P < 0.05). This effect is eliminated by allatostatin infusion. (F) Target proximity (average distance from former platform location) during P1 and P2. CREB-HSV animals search more closely to the target than CREB-LacZ mice (*P < 0.05). This effect is eliminated by allatostatin infusion. (G) Average swimming velocity during probe trials (centimeters per second).
The HSV vectors we used included the Drosophila allatostatin receptor (34), which can hypopolarize and inactivate mammalian neurons (38). To determine whether the RSC cells infected with the HSV-CREB vector were directly responsible for the spatial memory enhancement observed, we inactivated these cells using the allatostatin receptor as previously described (34). As control, we used a HSV-LacZ vector with the allatostatin receptor. As expected, analyses of target quadrant occupancy, platform crossings, and target proximity showed that infusion of the allatostatin peptide into the RSC did not affect performance of mice with the HSV-LacZ vector. However, this same treatment occluded the spatial learning enhancements observed previously in the mice with the HSV-CREB vector (Fig. 3 D–F, probe trial 2), reverting the performance to the level observed for controls. This result is consistent with the hypothesis that CREB enhanced spatial memory in the RSC, and that allatostatin inactivated the RSC cells underlying an important component of the network supporting spatial memory.
Discussion
The results presented here provide multiple lines of convergent evidence supporting the hypothesis that the retrosplenial cortex encodes and stores spatial information. First, our time-lapse in vivo two-photon imaging experiments identified a population of RSC neurons that is reliably reactivated during consecutive sessions of a demanding spatial task. Second, CNQX inactivation experiments showed that the RSC is critical for retrieval of recently formed spatial memory. Third, HSV-CREB experiments indicated that enhancing molecular memory processes in the RSC can enhance performance in spatial tasks. Finally, allatostatin inactivation of HSV-CREB expressing cells confirmed that these RSC cells are responsible for the spatial memory enhancement measured.
There is widespread evidence for using immediate early genes expression (such as Fos), as a marker for behaviorally relevant neuronal activation (39). Therefore, it is reasonable to propose that the FosGFP cellular patterns reported here reflect cell activation patterns in retrosplenial cortex. Our behavioral experiments showed that the repeated activation patterns uncovered in the RSC are responsive to spatial learning in the Morris water maze. Control imaging experiments in barrel cortex and motor cortex demonstrated that these repeated activation patterns are specific to the RSC and do not reflect general cortical activation patterns. In vivo electrophysiology studies in rat also show that the RSC develops context-specific firing patterns in response to spatial learning, although they are not related merely to directional information but rather to the entire context (24). Interestingly, the changes in firing rate are bidirectional, possibly facilitating discrimination of two competing contexts. This mechanism could be responsible for the differences in Fos expression observed between tasks in this study. Our estimates suggest that approximately 1% of layer II neurons of RSC cells participate in formation of a stable memory engram. The findings presented here suggest therefore that the RSC population code is sparsely distributed within layer II, similarly to the sparse codes uncovered in cortical regions that encode sensory information (40).
It is important to note that the in vivo chronic method used here to image Fos expression patterns allows for multiple observations of the same cell population. The flexibility of this approach made it possible for the inclusion of within-subject behavioral controls that were critical for our conclusions. For example, despite the many common elements between MP and UP sessions, the cell population repeatedly activated during UP sessions is less activated during the MP sessions, a finding that strengthened the conclusion that this repeated activation is connected with demanding spatial components of the Morris water maze. Recently, an alternative elegant transgenic technique was used to define a stable population of Fos-expressing cells in multiple brain regions following various behavioral manipulations (41, 42). This powerful approach, however, is limited to only two independent measurements, thus considerably restricting the number of possible behavioral manipulations. It is important to note that in the approach used here, the Fos protein is fused with GFP and is measured in all sessions by direct GFP fluorescence, with no need for signal amplification. Therefore, we were able to image and distinguish a wide range of FosGFP expression levels. Our analyses showed that out of the spectrum of cells expressing different levels of FosGFP, only those cells with robust expression were reliably reactivated during spatial learning, a result consistent with the hypothesis that these cells contribute to a memory trace for spatial information. This also suggests that high Fos expression in the RSC might either be associated with spatial processes or be required for these processes.
Beyond in vivo imaging studies reporting repeated activation in the RSC in response to spatial learning, our findings also showed that temporarily blocking the activity of the RSC with a pharmacological agent (CNQX) known to block glutamatergic transmission, disrupted performance in the spatial version of the Morris water maze. Although previous studies had shown that the RSC is needed for spatial learning (7, 43), these other studies mostly involved permanent lesions, which could have affected the acquisition phase of the task or disrupted the memory consolidation. Posttraining pharmacological inactivation studies carried out in rats did not reveal a clear effect on the performance in a standard spatial task (18). Our findings showed that inactivation of the mouse RSC specifically before retrieval can affect performance in the Morris water maze, a finding consistent with the idea that this structure already stores spatial memory traces at that time point.
Remarkably, expression in the RSC of a gene (CREB) known to specifically enhance memory in other structures and in other species (44), improved spatial performance. Inactivating the RSC neurons expressing the HSV-CREB gene with the allatostatin system occluded this memory enhancement, a result that directly implicates these neurons in spatial memory. Work in Aplysia, Drosophila, mice, and rats have shown that the transcription factor CREB has a key role in the stability of synaptic changes, and in protein-synthesis–dependent formation of long-term memory (45). Presumably, proteins like Fos, whose transcription is known to be CREB dependent, are involved in molecular and cellular processes that stabilize memory traces (27). Thus, our finding that increasing CREB levels in the RSC enhances spatial memory supports the hypothesis that the RSC encodes memory traces for spatial information.
However, what is the exact content of potential memory traces in the RSC? Electrophysiological recordings in freely moving animals indicate that in the absence of hippocampal inputs, the stellate cells in layer II of MEC lose their grid-firing properties and become exclusively modulated by the external head direction signal (46). Because the hippocampal and RSC projections converge in deep layers of MEC, and are further relayed to layers II and III (5), it is possible that the RSC provides the head direction input into MEC and contributes to grid and place cell firing as the animal moves through its environment. Thus, memory traces in the RSC might consist of a collection of heading sequences sufficient to trigger downstream spatial processes that guide locomotion and retrieve previously learned navigation paths. All together the results presented here indicate that as a part of the role of the RSC in spatial navigation, this structure may encode and store directional information that contributes to the formation of spatial maps in downstream structures such as the hippocampus.
Methods
Mice.
For all experiments, we used F1 hybrid mice from crosses between C57/Bl6T (Taconic) and 129Sv/J (Jackson Laboratories), 3–6 mo old. The FosGFP reporter line (28) was maintained in the C57/Bl6T background. For in vivo imaging experiments, it was crossed with the 129Sv/J line to obtain F1 hybrids. Mice were kept on a 12-h light/dark cycle. Experiments were conducted during the light phase. Food and water were available ad libitum. For all behavioral experiments, mice were handled for 10 d before the onset of training. Handling sessions were designed to mimic the experimental procedure and included transport from vivarium to the experimental room, appropriate light and sound changes, and individual handling for 2 min.
Surgical Procedures.
The mice were pretreated with atropine sulfate (0.1 mg/kg, i.p.) and anesthetized with a mixture of fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidin (0.5 mg/kg). Standard stereotactic surgical procedures were used. To prevent inflammation and provide analgesia, animals were treated with carprofen (5 mg/kg) for 48 h after surgery. Mice were allowed to recover for 2 wk. All procedures were approved by the Chancellor’s Animal Research Committee at the University of California, Los Angeles, in accordance with National Institutes of Health guidelines.
Cranial Window Implantation.
A modified previously described procedure (30) was used. Custom-made coverslips (round, 3-mm diameter) were used. Coverslips were cleaned in ethanol and sterilized. A circular region of skull 3 mm in diameter was carefully marked using stereotactic coordinates (RSC: bregma −3 mm, SSC: bregma −1, lateral +4, MC: bregma +1). The skull was thinned with a dental drill and removed. After stabilizing the surgical site with Gelfoam and saline, a coverslip was placed gently on the dura surface and fastened with Loctite 420 adhesive and dental acrylics to expose a circular window of approximately 2-mm diameter. Next, a titanium bar with threaded hole was attached to stabilize animals during imaging sessions.
Microscope.
An Olympus Fluoview 1000 two-photon scanning microscope was used. Several modifications were made to accommodate the whole animal under the objective lenses. A custom-made stage with heating pad and a head mount was installed. For GFP excitation, a MaiTai HP two-photon laser was used, with the pulse width of 100 fs. The power was adjusted to 200 mW at the back aperture of the objective lenses and kept constant between imaging sessions. The laser was tuned to 904 nm. A 20× 0.9 NA water immersion objective (Olympus) was used for imaging.
In Vivo Imaging.
Mice were trained for 10 d alternatively on two versions of the Morris maze. On days 1, 3, 5, 7, and 9 a standard spatial task was used. On days 2, 4, 6, 8, and 10 the platform was marked with a metal rod and the room cues were obscured with a black curtain. In vivo images were acquired 90 min after each of the last four behavioral sessions (two UP and two MP sessions on days 7–10). The animals were lightly anesthetized (50% of the dose used for surgery) and attached to the microscope head mount. During the first imaging session, after identifying the RSC-d based on bregma reference, and finding GFP-positive cells in layers II/III, stacks of 50–70 planes were acquired. The vertical spacing of planes was 2 μm, with single image dimensions of 1,024 × 1,024 pixels (0.67 μm per pixel). A single session took ∼30 min to complete and consisted of two to four stacks within the RSC-d. For subsequent imaging sessions, the previously imaged areas were recognized using unique patterns of vasculature. The resulting datasets consisted of four time points per stack.
Image Analysis.
A semiautomated blind procedure to identify cells and measure fluorescence intensity was developed. Image stacks from each session were aligned using Image Pro Plus software (Media Cybernetics). High-frequency noise was removed using a bandpass filter difference of Gaussians (31). Threshold was applied to the resulting images using the Otsu algorithm. After 3D object separation with a watershed algorithm, XYZ coordinates of FosGFP positive nuclei were obtained in 3D Constructor (Media Cybernetics). These coordinates were then used to read the average optical density (AOD) values from the original, unfiltered images using ImageJ, and a custom written plugin (Time Series Analyzer). For each cell, data from a circular region 15 pixels in diameter was obtained across the Z stack, and values from three central optical slices were averaged. The background was collected from Z planes that did not contain cells and subtracted. The AOD values obtained were then further analyzed using OriginPro 9 and Matlab.
Cannula Implantation.
For CNQX and HSV administration, custom-modified double guide cannulae (24 gauge; Plastics One) were used. The cannulae were implanted into retrosplenial cortex at the following stereotactic coordinates: bregma −2.9 mm, lateral ±0.25 mm, depth −0.5 mm. The cannulae were attached with Loctite adhesive and dental acrylics.
Behavior.
The Morris water maze apparatus was previously described (37). For training with the visible platform, the platform position was changed between sessions, the pool was surrounded with a black curtain, and a metal rod 10 cm in height was inserted into the platform to provide a single visual cue. Training trials lasted 60 s or until the mouse reached the platform. Upon finishing a trial, mice were allowed to remain on the platform for 15 s. For probe trials, an “atlantis” platform 20 cm in diameter was used and elevated after 60 s to allow escape. Behavioral training was conducted by a researcher blind to the experimental conditions. Data were collected and analyzed using the Water Maze system (Actimetrix).
Pharmacological Inactivation.
Mice (n = 16) were trained for 7 d on the Morris maze task with a single session per day consisting of three trials. One day after completion of the training, the animals were subjected to a single probe trial. Based on the results, animals were assigned to two groups (n = 8) matched for performance and swim speed. On the following day, each animal was briefly anesthetized with isoflurane to allow insertion of an injection cannula that protruded 0.5 mm below the edge of the guide cannula. A total of 0.65 μL of CNQX was infused into each hemisphere at the concentration of 1 mg/mL and rate of 0.065 μL/min. PBS was used as control. At 10 min after the infusion, the animals were subjected to another probe trial. Twenty-four hours later, the animals were tested for the third time. Three weeks later the same group of mice was retrained with the visible platform for 4 d. The latencies from the last training session were used to assign animals to two matched groups (n = 8). On the following day, CNQX or PBS was infused into the RSC and after 10 min, a single trial was conducted with the visible platform present.
HSV Infection.
HSV vectors with the allatostatin receptor (AlstR) and expressing either CREB (HSV-CREB vector) or LacZ (HSV-LacZ, control vector) were used. Genes of interest (CREB, LacZ, and AlstR) were cloned into a bicistronic HSV amplicon as previously described (34). To visualize transgene expression in cells infected, EGFP was fused to the 5′ end of the CREB or LacZ cDNA. CREB or LacZ was expressed from the IE 4/5 promoter and AlstR from the CMV promoter. Virus infusion started 7 d after surgery. Virus solution (total volume of 1.3 μL, titer 10−12) was delivered bilaterally to the RSC at a flow rate of 0.065 μL/min via two injection cannulas (Plastics One) attached by polyethylene tubing to Hamilton microsyringes mounted in an infusion pump (Harvard Apparatus). The infusion cannula was left in place for an additional 10 min.
CREB/Allatostatin Experiments.
Three days after HSV infusion, mice (n = 20) were subjected to a 1-d Morris water maze training protocol (37). The training consisted of seven sessions, three trials in each session. After completion of each session, mice were placed in their home cages and allowed to rest for 1 h. Twenty-four hours after completion of the last session, the animals were subjected to the first probe trial. After the probe trial, animals were anesthetized with isoflurane, and allatostatin (New England Biolabs) was infused into the RSC. The drug solution (10 μM, 1.3 μL) was delivered bilaterally at a flow rate of 0.065 μL/min through injection cannulas. The cannula was left in place for 2 min postinjection. Thirty minutes after injection, another probe trial was conducted.
Supplementary Material
Acknowledgments
We thank members of the A.J.S. laboratory, as well as P. Golshani, M. Mehta, A. Gdalyahu, and T. Stensola for helpful comments and discussions, and Katie Cai for invaluable technical help. This work was supported by Grants P50 MH077972 and R37 AG013622 from the National Institutes of Health (to A.J.S.) and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (A.J.S.). R.C. was partially supported by the European Union 7th Framework Programme Project BIO-IMAGINE: BIO-IMAGing in research INnovation and Education, GA 264173.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313222111/-/DCSupplemental.
References
- 1.Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297(5868):681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 2.Hafting T, Fyhn M, Molden S, Moser MB, Moser EI. Microstructure of a spatial map in the entorhinal cortex. Nature. 2005;436(7052):801–806. doi: 10.1038/nature03721. [DOI] [PubMed] [Google Scholar]
- 3.Cho J, Sharp PE. Head direction, place, and movement correlates for cells in the rat retrosplenial cortex. Behav Neurosci. 2001;115(1):3–25. doi: 10.1037/0735-7044.115.1.3. [DOI] [PubMed] [Google Scholar]
- 4.Jones BF, Witter MP. Cingulate cortex projections to the parahippocampal region and hippocampal formation in the rat. Hippocampus. 2007;17(10):957–976. doi: 10.1002/hipo.20330. [DOI] [PubMed] [Google Scholar]
- 5.Czajkowski R, et al. Superficially projecting principal neurons in layer V of medial entorhinal cortex in the rat receive excitatory retrosplenial input. J Neurosci. 2013;33(40):15779–15792. doi: 10.1523/JNEUROSCI.2646-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugar J, Witter MP, van Strien NM, Cappaert NL. The retrosplenial cortex: Intrinsic connectivity and connections with the (para)hippocampal region in the rat. An interactive connectome. Front Neuroinform. 2011;5:7. doi: 10.3389/fninf.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vann SD, Aggleton JP, Maguire EA. What does the retrosplenial cortex do? Nat Rev Neurosci. 2009;10(11):792–802. doi: 10.1038/nrn2733. [DOI] [PubMed] [Google Scholar]
- 8.Spiers HJ, Maguire EA. The neuroscience of remote spatial memory: A tale of two cities. Neuroscience. 2007;149(1):7–27. doi: 10.1016/j.neuroscience.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 9.Epstein RA, Parker WE, Feiler AM. Where am I now? Distinct roles for parahippocampal and retrosplenial cortices in place recognition. J Neurosci. 2007;27(23):6141–6149. doi: 10.1523/JNEUROSCI.0799-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maguire EA. The retrosplenial contribution to human navigation: A review of lesion and neuroimaging findings. Scand J Psychol. 2001;42(3):225–238. doi: 10.1111/1467-9450.00233. [DOI] [PubMed] [Google Scholar]
- 11.Vann SD, Aggleton JP. Extensive cytotoxic lesions of the rat retrosplenial cortex reveal consistent deficits on tasks that tax allocentric spatial memory. Behav Neurosci. 2002;116(1):85–94. [PubMed] [Google Scholar]
- 12.Vann SD, Kristina Wilton LA, Muir JL, Aggleton JP. Testing the importance of the caudal retrosplenial cortex for spatial memory in rats. Behav Brain Res. 2003;140(1–2):107–118. doi: 10.1016/s0166-4328(02)00274-7. [DOI] [PubMed] [Google Scholar]
- 13.Vann SD, Aggleton JP. Testing the importance of the retrosplenial guidance system: Effects of different sized retrosplenial cortex lesions on heading direction and spatial working memory. Behav Brain Res. 2004;155(1):97–108. doi: 10.1016/j.bbr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Vann SD, Aggleton JP. Selective dysgranular retrosplenial cortex lesions in rats disrupt allocentric performance of the radial-arm maze task. Behav Neurosci. 2005;119(6):1682–1686. doi: 10.1037/0735-7044.119.6.1682. [DOI] [PubMed] [Google Scholar]
- 15.Pothuizen HH, Aggleton JP, Vann SD. Do rats with retrosplenial cortex lesions lack direction? Eur J Neurosci. 2008;28(12):2486–2498. doi: 10.1111/j.1460-9568.2008.06550.x. [DOI] [PubMed] [Google Scholar]
- 16.Lukoyanov NV, Lukoyanova EA, Andrade JP, Paula-Barbosa MM. Impaired water maze navigation of Wistar rats with retrosplenial cortex lesions: Effect of nonspatial pretraining. Behav Brain Res. 2005;158(1):175–182. doi: 10.1016/j.bbr.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Cooper BG, Manka TF, Mizumori SJ. Finding your way in the dark: The retrosplenial cortex contributes to spatial memory and navigation without visual cues. Behav Neurosci. 2001;115(5):1012–1028. doi: 10.1037//0735-7044.115.5.1012. [DOI] [PubMed] [Google Scholar]
- 18.Cooper BG, Mizumori SJ. Retrosplenial cortex inactivation selectively impairs navigation in darkness. Neuroreport. 1999;10(3):625–630. doi: 10.1097/00001756-199902250-00033. [DOI] [PubMed] [Google Scholar]
- 19.Wesierska M, Adamska I, Malinowska M. Retrosplenial cortex lesion affected segregation of spatial information in place avoidance task in the rat. Neurobiol Learn Mem. 2009;91(1):41–49. doi: 10.1016/j.nlm.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Cooper BG, Mizumori SJ. Temporary inactivation of the retrosplenial cortex causes a transient reorganization of spatial coding in the hippocampus. J Neurosci. 2001;21(11):3986–4001. doi: 10.1523/JNEUROSCI.21-11-03986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiers HJ, Maguire EA. Thoughts, behaviour, and brain dynamics during navigation in the real world. Neuroimage. 2006;31(4):1826–1840. doi: 10.1016/j.neuroimage.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 22.Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science. 2004;304(5672):881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- 23.Tse D, et al. Schemas and memory consolidation. Science. 2007;316(5821):76–82. doi: 10.1126/science.1135935. [DOI] [PubMed] [Google Scholar]
- 24.Smith DM, Barredo J, Mizumori SJ. Complimentary roles of the hippocampus and retrosplenial cortex in behavioral context discrimination. Hippocampus. 2012;22(5):1121–1133. doi: 10.1002/hipo.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson S, Poorman CE, Marder TJ, Bucci DJ. Identification of functional circuitry between retrosplenial and postrhinal cortices during fear conditioning. J Neurosci. 2012;32(35):12076–12086. doi: 10.1523/JNEUROSCI.2814-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tse D, et al. Schema-dependent gene activation and memory encoding in neocortex. Science. 2011;333(6044):891–895. doi: 10.1126/science.1205274. [DOI] [PubMed] [Google Scholar]
- 27.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89(1):121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24(29):6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barth AL. Visualizing circuits and systems using transgenic reporters of neural activity. Curr Opin Neurobiol. 2007;17(5):567–571. doi: 10.1016/j.conb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trachtenberg JT, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420(6917):788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 31.Wang KH, et al. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126(2):389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 32.Harker KT, Whishaw IQ. Impaired spatial performance in rats with retrosplenial lesions: Importance of the spatial problem and the rat strain in identifying lesion effects in a swimming pool. J Neurosci. 2002;22(3):1155–1164. doi: 10.1523/JNEUROSCI.22-03-01155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. Molecular and cellular approaches to memory allocation in neural circuits. Science. 2009;326(5951):391–395. doi: 10.1126/science.1174519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci. 2009;12(11):1438–1443. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han JH, et al. Selective erasure of a fear memory. Science. 2009;323(5920):1492–1496. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 36.Barco A, Marie H. Genetic approaches to investigate the role of CREB in neuronal plasticity and memory. Mol Neurobiol. 2011;44(3):330–349. doi: 10.1007/s12035-011-8209-x. [DOI] [PubMed] [Google Scholar]
- 37.Cui Y, et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135(3):549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechner HA, Lein ES, Callaway EM. A genetic method for selective and quickly reversible silencing of Mammalian neurons. J Neurosci. 2002;22(13):5287–5290. doi: 10.1523/JNEUROSCI.22-13-05287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barry DN, Commins S. Imaging spatial learning in the brain using immediate early genes: Insights, opportunities and limitations. Rev Neurosci. 2011;22(2):131–142. doi: 10.1515/RNS.2011.019. [DOI] [PubMed] [Google Scholar]
- 40.Gdalyahu A, et al. Associative fear learning enhances sparse network coding in primary sensory cortex. Neuron. 2012;75(1):121–132. doi: 10.1016/j.neuron.2012.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317(5842):1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 42.Tayler KK, Tanaka KZ, Reijmers LG, Wiltgen BJ. Reactivation of neural ensembles during the retrieval of recent and remote memory. Curr Biol. 2013;23(2):99–106. doi: 10.1016/j.cub.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Keene CS, Bucci DJ. Contributions of the retrosplenial and posterior parietal cortices to cue-specific and contextual fear conditioning. Behav Neurosci. 2008;122(1):89–97. doi: 10.1037/0735-7044.122.1.89. [DOI] [PubMed] [Google Scholar]
- 44.Han JH, et al. Increasing CREB in the auditory thalamus enhances memory and generalization of auditory conditioned fear. Learn Mem. 2008;15(6):443–453. doi: 10.1101/lm.993608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benito E, Barco A. CREB’s control of intrinsic and synaptic plasticity: Implications for CREB-dependent memory models. Trends Neurosci. 2010;33(5):230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Bonnevie T, et al. Grid cells require excitatory drive from the hippocampus. Nat Neurosci. 2013;16(3):309–317. doi: 10.1038/nn.3311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.