Significance
Proteins, which constitute roughly half of the cell dry mass, are extremely diverse. By counting the protein copy number of each gene in the genome, we obtain the proteome—a comprehensive picture of a cell's biochemical machinery. The proteome reflects physiology, structure, metabolic capacities, and many other aspects of the cell's lifestyle. Here, we visualize quantitative proteome data using a graphical tool we call proteomaps, where proteins are shown as polygons whose sizes indicate the abundances. Proteins involved in similar cellular functions are arranged in adjacent locations, creating regions whose areas give insight into the relative investment in each functional class. Proteins or protein classes that dominate the proteomap indicate demanding cellular processes and promising targets for further research.
Keywords: Voronoi treemap, functional classification, mass spectrometry, cell resource allocation, cellular economy
Abstract
Proteomics techniques generate an avalanche of data and promise to satisfy biologists' long-held desire to measure absolute protein abundances on a genome-wide scale. However, can this knowledge be translated into a clearer picture of how cells invest their protein resources? This article aims to give a broad perspective on the composition of proteomes as gleaned from recent quantitative proteomics studies. We describe proteomaps, an approach for visualizing the composition of proteomes with a focus on protein abundances and functions. In proteomaps, each protein is shown as a polygon-shaped tile, with an area representing protein abundance. Functionally related proteins appear in adjacent regions. General trends in proteomes, such as the dominance of metabolism and protein production, become easily visible. We make interactive visualizations of published proteome datasets accessible at www.proteomaps.net. We suggest that evaluating the way protein resources are allocated by various organisms and cell types in different conditions will sharpen our understanding of how and why cells regulate the composition of their proteomes.
In recent years, novel methodologies have realized biologists' long held desire (1) to measure relative and absolute protein abundances on a proteome-wide scale in a variety of model organisms (2–14). Proteome datasets are often collected to address questions such as the degree of correlation between mRNA and protein levels or to what extent certain proteins change in response to an applied stimulus. However, these accumulated protein levels can also help us answer a simpler, more mundane question: what exactly is in a proteome?
Proteins and, by extension, genes perform numerous biological functions ranging from the catalysis of chemical reactions to the formation of physical cell structures and the processing of environmental signals. The fraction of the genome occupied by certain types of genes (e.g., metabolic or signaling) is often referenced to highlight the impact of that category. This logic is all the more compelling when discussing the proteome: given the extremely crowded environment of the cell (15, 16) and the amount of energy and carbon resources required to make proteins (17), we expect a general selection pressure against high protein expression, especially in microorganisms (18–21). It is therefore of great interest for molecular biologists to know which proteins and functional categories are most abundant: That is, in which proteins does a cell invest the bulk of its carbon, nitrogen and polymerization resources, reducing power, and ATP (22)?
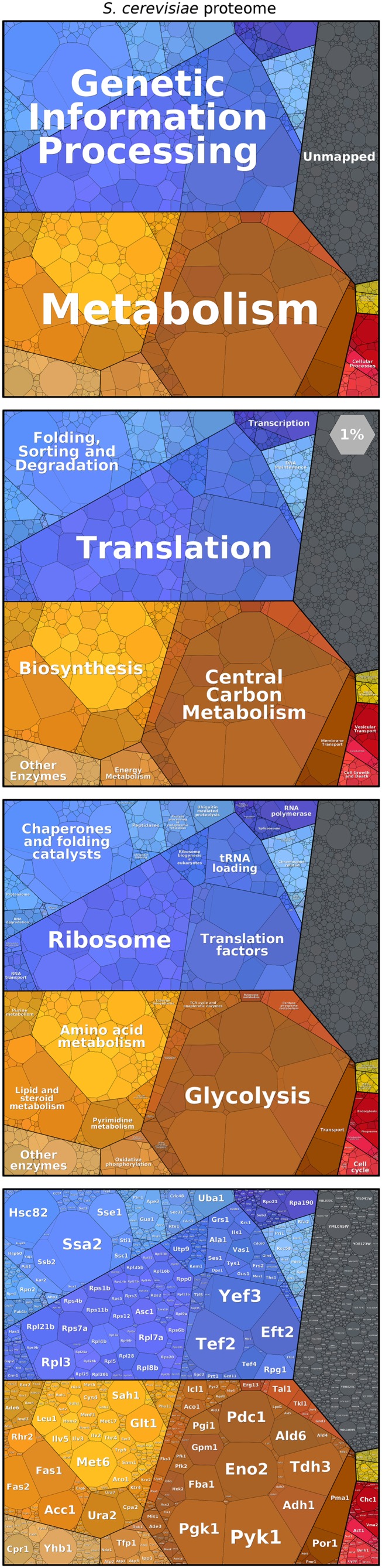
A proper visualization can be helpful to address this question and to explore and compare the structure of proteomes. Here, we introduce proteomaps, which depict the composition of a proteome hierarchically in various levels of granularity from general functions to single proteins (Fig. 1). To emphasize highly expressed proteins, each protein is associated with a polygon-shaped tile whose size is proportional to that protein’s abundance. Although treemaps have already been used to encode expression changes by colors (23–25), we encode protein abundance directly by size. We display mass fractions: i.e., protein copy numbers weighted by the chain lengths, thus showing the amino acid investment for protein production and maintenance. Functionally related proteins are placed in common subregions to show the functional makeup of a proteome at a glance. In interactive proteomaps (available at www.proteomaps.net), tiles are linked to further information about the proteins.
Fig. 1.
Proteomap of the budding yeast S. cerevisiae based on data from ref. 14. Every tile (small polygon) represents one type of protein. Tiles are arranged and colored according to the hierarchical KEGG pathway maps such that larger regions correspond to functional categories. The diagrams show three hierarchy levels (top three panels) and the level of individual proteins (Bottom). Tile sizes represent the mass fractions of proteins (protein abundances obtained by mass spectrometry, multiplied by protein chain lengths). Color code: blue, genetic information processing; brown, metabolism; red, cellular processes; green, signaling. Proteins—mostly low-abundance ones—that do not map to any category are shown in a gray area. The light-gray hexagon illustrates the area that covers 1% of the proteome.
Our approach complements the popular representation of abundances using data tables, which can be sorted to give quantitative information but lack some major advantages of visual perception. Common visualizations are based on stacked bar graphs or pie charts of measured abundances. These inherently one-dimensional representations suffer from strong limitations in comparison with our 2D maps. For example, proteins with abundances around one percent are easily visible in proteomaps whereas they become hard to make out in pie or bar plots (www.proteomaps.net/diagrams). Another advantage is the flexibility of arranging the proteins and their categories in a 2D plane compared with stacking them along a line.
Because proteins carry out most of the primary tasks of life—processing of genetic information, metabolism, signaling, transport, etc.—visualizing the contents of the proteome gives us a snapshot of how a cell invests its resources for protein production within a given environment and growth stage. In this study, we examine the composition of four model organisms’ proteomes with an eye toward understanding the similarities and differences among them.
High-throughput technologies enabling proteome-wide mapping of protein abundances range from fluorescent microscopy to mass spectrometry (MS), with the latter being the most common and highly promising. These methods produce the data that we visualize here. Each method has its strengths but also harbors caveats that should be noted. For example, due to their hydrophobicity, membrane-bound proteins may be underrepresented in MS due to problems with quantitative extraction using water-based solvents. In methods based on proteins tagged with fluorophores, the expression, localization, or functionality of proteins may be affected. Low abundance proteins might remain below the detection limit, and highly abundant proteins can be hard to measure due to detector saturation. Moreover, systematic biases can be caused by the size or physico-chemical properties of each protein: for instance, very large proteins or protein complexes may disappear from the sample during initial centrifugation. Cancer cell lines, which are often analyzed as examples of mammalian cells, might not reliably represent noncancerous primary tissue. These caveats should be taken into account when attempting to interpret the data. We proceed to show how proteomaps help highlight commonalities and point to key differences between species. Finally, we discuss how a high-level understanding of the proteome composition can help direct efforts to underresearched, highly abundant proteins and resource-consuming cellular processes.
Results
The Big Picture: Metabolism, Translation, and Folding Dominate the Proteome.
Cells contain thousands of different proteins of various functions. On the one hand, we are interested in understanding which individual proteins are abundant; but, on the other hand, we also want to understand protein levels in context. For example, are enzymes in the same metabolic pathway expressed at similar levels? Proteomaps allow us to inspect the proteome at several levels of granularity. Using functional gene classifications [e.g., the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps] (26), we can represent the contents of a proteome hierarchically by grouping proteins into pathways and then into higher-level categories, and so forth. This hierarchy is displayed in Fig. 1: each protein is represented by a polygon-shaped tile; proteins belonging to the same category share similar colors and are placed in adjacent locations to form larger regions. This arrangement makes it easy to spot protein categories that are the major components of a proteome. In the original KEGG pathway maps, many proteins were assigned to more than one category. For the proteomap, a unique annotation is chosen for each protein (as discussed in Methods).
The proteomap presented in Fig. 1 shows the proteome of the yeast Saccharomyces cerevisiae, measured using mass spectrometry (14). Each polygon represents the mass fraction of the protein within the proteome (i.e., the protein copy number, multiplied by the protein chain length). At the broadest level of functional resolution, the map is dominated by metabolic enzymes (orange-brown) and by proteins performing the steps of the central dogma leading from DNA to proteins (“genetic information processing,” in blue). Within the category of genetic-information processing, the ribosomal proteins followed by chaperones and translation factors make up the most prominent fractions (even though these categories contain fewer genes than the categories of genome replication and transcription). Metabolism is usually the largest constituent of the proteome, with glycolysis and amino acid metabolism being the biggest contributors to the category. For example, in S. cerevisiae grown on glucose, glycolytic enzymes are extremely highly expressed, occupying 15–20% of the proteome although they make up less than 1% of the genome.
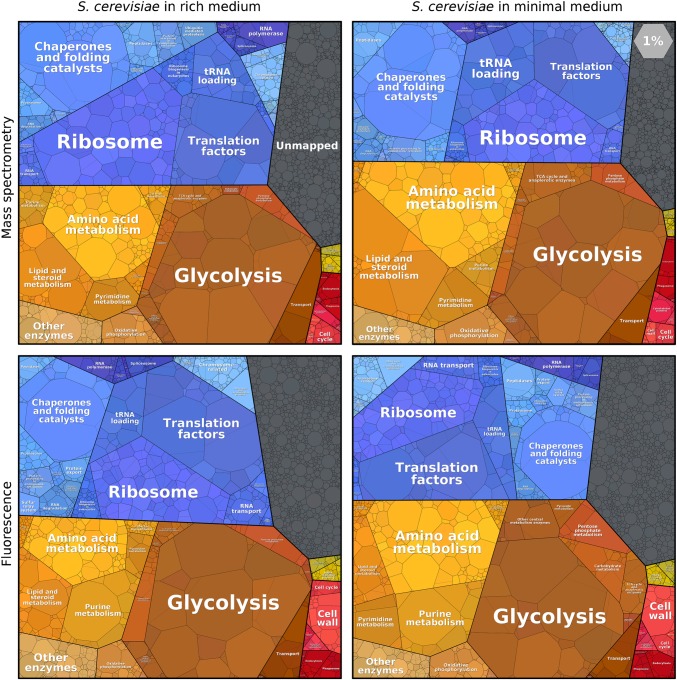
How do these observations change with different growth media or different measurement methods? Fig. 2 shows the proteome of S. cerevisiae, measured via fluorescent reporters (2) or mass spectrometry (5, 14) and, in each case, during growth on minimal or rich media. In all four cases, the functional groups that occupy the largest fractions of the proteome are the same: glycolysis, amino acid metabolism, ribosomes, translation factors, and chaperones. This remarkable similarity vividly shows that even significant changes in the physiology of the cell and the abundance of single proteins create only a limited shift in the allocation of protein resources in the grand scheme.
Fig. 2.
Proteomaps of the budding yeast S. cerevisiae, compared across nutrient conditions and measurement methods. Yeast cells were grown in a rich (Left) or minimal medium (Right). In a minimal medium, the proteome fraction of amino acid biosynthesis enzymes is higher. Protein abundances were measured by mass spectrometry (Upper) in rich (14) and in minimal (5) media, or by fluorescence of GFP-tagged proteins (Lower) in YEPD versus SD media (2).
Expression levels can vary greatly between individual proteins in the same functional group. Glycolysis contains many of the most abundant proteins, with enzymes like enolase constituting 2–4% of the proteome. Other glycolytic enzymes, however, are much less abundant. In contrast, the ribosomal proteins are expressed at comparable levels, as might be expected given the stoichiometric assembly of the ribosome. Absolute protein copy numbers can be plotted instead of mass fractions (www.proteomaps.net) to interrogate whether multiprotein complexes are expressed stoichiometrically.
Proteomaps Highlight Proteome Composition Conservation and Species-Specific Trends.
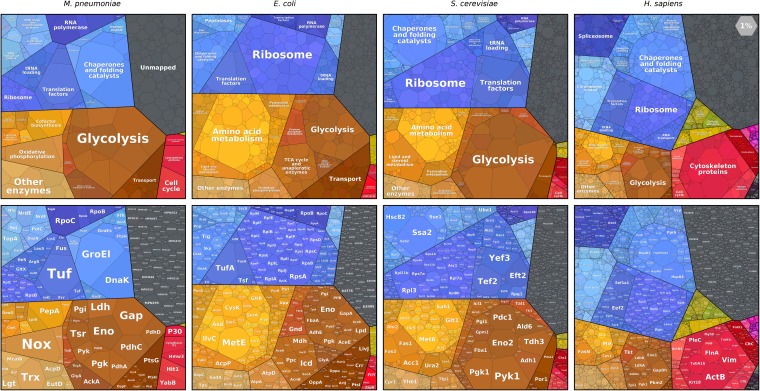
In this study, we include cells from four well-studied species: Mycoplasma pneumoniae, Escherichia coli, S. cerevisiae, and Homo sapiens cell lines. These cells' volumes span five orders of magnitude (from about 0.1 μm3 to over 1,000 μm3), and their growth rates differ considerably (characteristic doubling times ranging from <1 h to about a day). They represent various modes of life ranging from obligate parasitism to multicellularity and vary considerably in shape and number of compartments. Comparing the proteomes of these very different species may tell us which features of the proteome are conserved throughout evolution and, on the other hand, in which cellular functions the cell's investment changes dramatically.
Looking at the major functional categories in the composition of these four proteomes, one finds that, within genetic information processing (Fig. 3, blue), the total amount of protein dedicated to translation is 2–15 times larger than the amount invested in transcription or in DNA maintenance (including the replication machinery, histones, and DNA repair). In metabolism (Fig. 3, orange-brown), glycolytic enzymes consistently require a larger fraction of the proteome than the TCA cycle and oxidative phosphorylation, which, under aerobic conditions, are the major source of energy for many organisms. Although it contains only a handful of genes, glycolysis is a larger constituent of the proteome than almost any other pathway.
Fig. 3.
Proteomaps of several model organisms. (Upper) Proteomaps labeled by functional categories. (Lower) The same diagrams, with gene names. Protein abundances shown are for the tiny human pathogen M. pneumoniae (7), E. coli growing at a rate of 0.48 1/h (13), S. cerevisiae (14), and an H. sapiens HeLa cell line (11).
In contrast, some of the functional categories that dominate the focus of research laboratories are not nearly as well-represented in the proteome. For example, the genes involved in cell signaling (Fig. 3, “environmental signal transduction,” in green) occupy about 4% of a human HeLa cell line proteome and below 1% in S. cerevisiae and E. coli. Thus, signaling proteins are examples of systems that constitute a small fraction of the proteome but have an outsized effect on the organism’s behavior.
In all organisms considered here, metabolic proteins and the proteins implementing the central dogma are the two dominant constituents of the proteome, with the cytoskeleton and similar cellular processes representing a third major contributor in human cell lines. In all cases, signaling proteins make up a small fraction. The fraction associated with nonmapped proteins (i.e., proteins that are not linked to our functional hierarchy, possibly because of unknown function) ranges from about 10% in the well-annotated E. coli and S. cerevisiae proteomes to about 20% in the less thoroughly mapped M. pneumoniae and H. sapiens.
The areas occupied by different functional groups of proteins change drastically between organisms, often matching known physiological differences between them. Ribosomal proteins make up a large fraction of the proteomes of all four organisms, but the exact percentage varies greatly among them, ranging from less than 5% of the proteome in M. pneumoniae and human cell lines to about 10–20% in the faster-growing E. coli and S. cerevisiae. This trend across cell types could be associated with their different growth rates, as suggested by studies comparing ribosome abundance in different microbial growth conditions (27, 28) and by growth rate-dependent proteomes of E. coli (13) (see maps on www.proteomaps.net).
The total protein concentration in the cytosol is fairly stable (typically 200 gr/L) (16). However, there are also membrane proteins, such as transporters, and DNA-associated proteins, like histones, whose concentrations on membranes or along the DNA are dictated by geometric or physiological constraints and whose mass fraction should therefore vary with membrane area and genome size per cell volume. Indeed, transporters, although prone to various extraction biases, show a trend where they make up several percent of the proteome in M. pneumoniae and E. coli, while accounting for 1–2% in S. cerevisiae and much less in the H. sapiens cell lines. This difference might stem from the fact that the ratio of outer membrane surface area to cell volume is smaller for larger cells.
Some functional categories are particularly pronounced in certain organisms in accordance with their different cell structures or modes of life. As can be seen in Fig. 3, a HeLa cell devotes a much larger fraction of its proteome to cytoskeletal proteins (more than 15%) than E. coli (0.3%). It is striking how 17% of S. cerevisiae’s proteome is devoted to glycolysis, possibly a result of many years of selection for increased ethanol production.
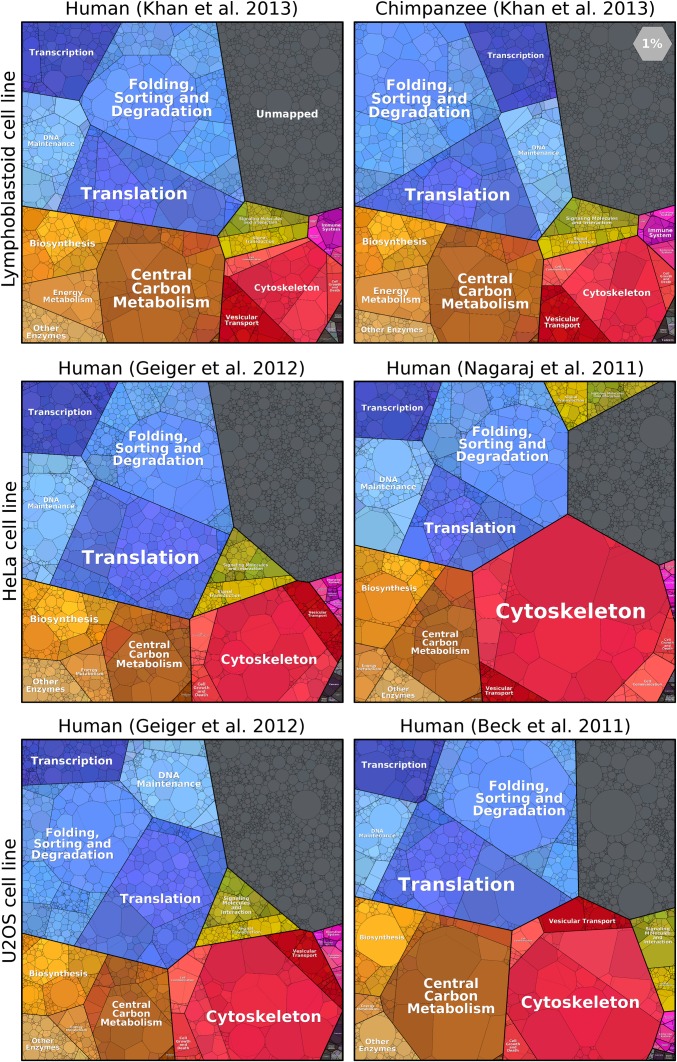
Using proteomaps, one can easily distinguish between cells from different domains of life. How large are differences among cell lines from the same organism? Does the composition of the proteome differ more when comparing two human cell lines or when comparing human and chimpanzee cells of the same type? Fig. 4 shows a comparison across three different cell lines. We find it striking how similarly proteome resources are allocated among cellular functions. The proteomaps of lymphoblastoid cells from human and chimpanzee are almost identical, even more so than the already very similar proteomes of various human cell lines. Differences between independent measurements of the same cell line are also shown for HeLa and U2OS cells. Many previous analyses focused on proteins that are expressed at relatively low levels, such as signaling proteins, where differences are pronounced. However, proteomaps reveal that functional categories and even dominant individual proteins are strongly conserved in terms of abundance. Differences and similarities at finer levels of functionality and at the single protein level can be analyzed in detail on the proteomaps website. As a follow-up to the comparison reported here, one can analyze cells from different tissues and between cell lines and primary cells.
Fig. 4.
Proteomes of primate cell lines. The Top row shows that lymphoblastoid cells from humans and chimpanzees display very similar proteomes (12). Proteomes of human HeLa cells are shown in the Middle row (10, 11), and human U2OS cells are compared at the Bottom (9, 11).
Discussion
Individual proteins can confer benefits to the cell in various ways, by catalyzing a chemical reaction, transporting an essential substrate, or transmitting signals that reflect the state of the environment. However, proteins also incur various costs: Proteins are made using precious carbon, nitrogen, sulfur, reducing power and energy resources, they require ribosomes for their continued synthesis, and they occupy volume in the crowded intracellular space (16). These general costs are roughly independent of the protein's identity and approximately proportional to its weight. Nevertheless, expressing a protein can have other more protein-specific effects that add to the costs, such as protein misfolding, perturbing the membrane integrity, creating an imbalance in the cell redox or energy state, etc. Such protein-specific costs are not captured by the visualization presented here.
Classical molecular biology studies often consider a protein important if knocking out its gene dramatically affects the behavior or viability of the cell. This approach often focuses efforts on regulatory proteins, such as transcription factors, which tend to have low expression levels. Theoretical analysis of metabolic enzymes (29) suggests an alternative interpretation of importance via the concept of relative marginal benefit that is predicted to be proportional to protein levels. Taking a quantitative proteomics viewpoint and observing how a cell invests its protein resources can help identify abundant proteins that are pivotal in certain environments but have unknown or poorly characterized function. Therefore, we propose that, all else being equal, highly abundant proteins are promising candidates for research efforts.
In the near future, proteome data will become available for many cell types and growth conditions. Proteomaps can also be applied to RNA transcript data, to phosphoproteome data, or—more generally—to the complete mass composition of a cell (including all types of macromolecules and small molecules). Furthermore, beyond molecular abundances, other genome-wide quantitative properties can easily be visualized. We suggest that proteomaps can help researchers achieve a clearer picture of similarities and differences in cell composition and the allocation of cellular resources across organisms, cell types, and growth conditions.
Methods
Proteome Tree Maps Visualization.
To generate proteomaps, we modified the algorithm for the construction of Voronoi treemaps described in ref. 23 to present polygons with variable sizes. The algorithm was implemented in the Paver software (DECODON), which is available at www.decodon.com/paver.html or upon request from the authors. Example maps on www.proteomaps.net can be browsed interactively; individual protein tiles are linked to protein information on the KEGG website (www.genome.jp/kegg/).
In the proteomaps shown here, we visualize three levels of functional categories and a level of individual proteins. To create a proteomap, a total area is first divided into polygons representing the top-level categories. These polygons are constructed from a Voronoi diagram, where the polygons' areas were chosen to represent copy numbers weighted by protein chain lengths (the investment in terms of amino acids, also termed the mass fraction). The top-level areas are then subdivided into subcategories, and the procedure is repeated down to the level of individual proteins. When several orthologous proteins exist in the same proteome, e.g., isozymes such as the two enolases Eno1 and Eno2 in yeast glycolysis, they share one subdivided polygon.
Proteins that do not have a functional category annotation are lumped in a subclass labeled “Not mapped.” Mass fractions smaller than 1/1,500,000 of the whole proteome (corresponding to 4 pixels within an area of 2,500 × 2,500 pixels in size) are excluded. The arrangement of categories and proteins over the area is kept as consistent as possible between proteomaps. To ensure a similar layout across datasets, a template proteomap can be used to initialize proteomaps for other datasets at the highest hierarchy level. However, due to differences in protein abundances, congruent arrangements cannot always be fully achieved. Colors are used for association within functional categories and have no quantitative meaning. Specifically, small variations in color are used to differentiate among detailed functional categories within the same broad functional category: e.g., shades of blue within “Genetic Information Processing” (Fig. 1).
Protein Abundance Data Sources and Gene Mapping.
Protein data were taken from the original publications and from the proteome database PaxDb (pax-db.org) (30). Criteria for choosing datasets to be included were as follows: a high proteome coverage; quantitative values that are proportional to abundance, ideally reported as absolute numbers; and refraining from biases such as mixed cell types or a known strong misrepresentation of cellular compartments or functions. All proteomes had been quantified by mass spectrometry, except for the data from ref. 2, which were quantified by fluorescence of GFP-tagged proteins. To assign proteins to functional categories, systematic gene names (ORF names) were annotated with KEGG Orthology identifiers (26). Protein chain lengths were obtained from Uniprot. Proteins of unknown length, due to mapping issues, were assigned a standard length of 350 amino acids.
Protein Functional Hierarchy and Category Assignment.
KEGG pathway maps were chosen as a basis for our functional gene hierarchy because of their clearly layered structure, which shows protein functions in different categories on a comparable degree of resolution. Proteins are assigned to functions via Kegg Orthology (KO) IDs, which makes them comparable between organisms.
In the KO, as in most other gene-classification schemes, the same protein can be assigned to multiple functional categories. However, a major limitation of all hierarchical visualization methods, including our use of Voronoi Treemaps, is that they require a tree-like hierarchy: i.e., multiple assignments are not allowed (23, 31). This inherent drawback forced us to assign multifunctional proteins to only one bottom-level category, preferably to the one corresponding to their principle task. We are aware that this assignment can depend on the researcher and does not fully reflect the nature of biological multifunctionality. In general, we defined a default priority order between the functional categories and assigned each KO ID to the bottom-level category with the highest priority. For instance, assignments to “Transcription” would override assignments to “Metabolism,” and therefore the protein RpoB was placed within “RNA polymerase” and not “Purine metabolism.” The default choice can be overridden by manual assignments. Moreover, we found that, for consistency with the literature, some functional categories had to be added, renamed, or restructured. The customized version of the KEGG hierarchy can be downloaded from www.proteomaps.net.
Because each KO ID appears, on average, in about two pathway categories, our priority order can create a bias toward certain categories. To quantify this bias, we randomized the priority order and computed median values and uncertainty ranges for category areas arising from different possible protein annotations. We found that random reassignments had only a small effect on the overall category areas and that none of our qualitative observations changed substantially.
The KEGG hierarchy proved useful for the present purpose, but proteomaps can also be produced with other classification trees such as TIGRFams, the original KEGG pathway maps, the Munich Information Center for Protein Sequences (MIPS) Functional Catalogue, The SEED (www.theseed.org), Riley scheme-derived classification systems, and many more (32, 33). Ontologies such as the widely used and flexible Gene Ontology (GO) (geneontology.org) (34) are typically directed acyclic graphs rather than a tree. In the GO, many nonterminal nodes are connected to several higher-level terms, and terminal terms are located at different distances from the root; for some genes, the GO contains more than 10 hierarchy levels. We found that proteomaps with a compact 3-level hierarchy are useful for visual comprehension. Thus, adapting the GO requires adaptation beyond the scope of this study. Nevertheless, we supply an example of a proteomap based on the GO for the curious reader in www.proteomaps.net/go.
When generating a proteomap, there are two required inputs: a hierarchy file and a data table. The hierarchy file is written in a simple textual format that can be easily edited with any text or spreadsheet editor. Layers can be added or removed, and genes can be moved between categories to reflect new discoveries. This format can also be used to introduce another level of organization to a proteomap: e.g., to display proteins that typically form complexes as clusters.
Supplementary Material
Acknowledgments
We thank Tamar Geiger, Uri Moran, Niv Antonovsky, Naama Barkai, Arren Bar-Even, Hermann-Georg Holzhütter, Leeat Keren, Rob Phillips, and Noa Rippel for helpful discussions. We further thank Henry Mehlan and Julia Schüler for help in solving problems with the preliminary version of the Voronoi tessellation algorithm and are grateful for the support of DECODON (Greifswald, Germany) providing the final version of Paver. This work was supported by the German Research Foundation (Ll 1676/2-1 and SFB Transregio 34/Z1). R.M. is the incumbent of the Anna and Maurice Boukstein Career Development Chair and is supported by the European Research Council (260392-SYMPAC), the Israel Science Foundation (Grant 750/09), the Helmsley Charitable Foundation, the Larson Charitable Foundation, the Estate of David Arthur Barton, the Anthony Stalbow Charitable Trust, and Stella Gelerman, Canada.
Footnotes
Conflict of interest statement: J.B. is working as research scientist at the Institute for Microbiology of the Ernst-Moritz-Arndt University of Greifswald and as chief scientist with Decodon GmbH and has financial interest in the company that commercializes software tools for proteomics, including Proteomaps.
*This Direct Submission article had a prearranged editor.
References
- 1.Pedersen S, Bloch PL, Reeh S, Neidhardt FC. Patterns of protein synthesis in E. coli: A catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978;14(1):179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 2.Newman JRS, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 3.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 4.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25(1):117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 5.de Godoy LMF, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 6.Ishihama Y, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kühner S, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326(5957):1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck M, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11:M111.014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan Z, et al. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342(6162):1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valgepea K, Adamberg K, Seiman A, Vilu R. Escherichia coli achieves faster growth by increasing catalytic and translation rates of proteins. Mol Biosyst. 2013;9(9):2344–2358. doi: 10.1039/c3mb70119k. [DOI] [PubMed] [Google Scholar]
- 14.Nagaraj N, et al. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012;11:M111.013722. doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276(14):10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 16.Dill KA, Ghosh K, Schmit JD. Physical limits of cells and proteomes. Proc Natl Acad Sci USA. 2011;108(44):17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips R, Milo R. A feeling for the numbers in biology. Proc Natl Acad Sci USA. 2009;106(51):21465–21471. doi: 10.1073/pnas.0907732106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436(7050):588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 19.Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178(3):1653–1660. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139(7):1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomala K, Korona R. Evaluating the fitness cost of protein expression in Saccharomyces cerevisiae. Genome Biol Evol. 2013;5(11):2051–2060. doi: 10.1093/gbe/evt154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen R, Gerstein M. Analysis of the yeast transcriptome with structural and functional categories: Characterizing highly expressed proteins. Nucleic Acids Res. 2000;28(6):1481–1488. doi: 10.1093/nar/28.6.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernhardt J, Funke S, Hecker M, Siebourg J. 2009. Visualizing gene expression data via Voronoi treemaps. Sixth International Symposium on Voronoi Diagrams, ed Anton F (IEEE Computer Society, Washington, DC), pp 233–241.
- 24.Otto A, et al. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat Commun. 2010;1:137. doi: 10.1038/ncomms1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto A, Bernhardt J, Hecker M, Becher D. Global relative and absolute quantitation in microbial proteomics. Curr Opin Microbiol. 2012;15(3):364–372. doi: 10.1016/j.mib.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marr AG. Growth rate of Escherichia coli. Microbiol Rev. 1991;55(2):316–333. doi: 10.1128/mr.55.2.316-333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122(3):855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klipp E, Heinrich R. Competition for enzymes in metabolic pathways: Implications for optimal distributions of enzyme concentrations and for the distribution of flux control. Biosystems. 1999;54(1-2):1–14. doi: 10.1016/s0303-2647(99)00059-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang M, et al. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11(8):492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nat Rev Genet. 2008;9(7):509–515. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- 32.Rentzsch R, Orengo CA. Protein function prediction: The power of multiplicity. Trends Biotechnol. 2009;27(4):210–219. doi: 10.1016/j.tibtech.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Rison SC, Hodgman TC, Thornton JM. Comparison of functional annotation schemes for genomes. Funct Integr Genomics. 2000;1(1):56–69. doi: 10.1007/s101420000005. [DOI] [PubMed] [Google Scholar]
- 34.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]