Significance
Cleidocranial dysplasia (CCD) is a hereditary human skeletal disease. Mutations in Runt-related transcription factor 2, which functions as a heterodimer with core binding factor β (Cbfβ), are found in most individuals with CCD. It has been suspected that Cbfβ may be responsible for other CCD cases. The pathogenesis of CCD and the role of Cbfβ in postnatal skeletogenesis remain unclear. There has been no animal model to study this disease. We demonstrate that ablation of Cbfβ in various skeletal cells results in severe craniofacial and skeletal dysplasia with the phenotype recapitulating clinical features of CCD. The findings from this study of Cbfβ in the skeleton provide insight into the role of Cbfβ in postnatal skeletogenesis and pathogenesis of CCD, which may assist in developing new therapies for CCD and osteoporosis.
Keywords: osteoblast differentiation, chondrocyte differentiation, ossification, growth plate formation, endochondral bone formation
Abstract
The pathogenesis of cleidocranial dysplasia (CCD) as well as the specific role of core binding factor β (Cbfβ) and the Runt-related transcription factor (RUNX)/Cbfβ complex in postnatal skeletogenesis remain unclear. We demonstrate that Cbfβ ablation in osteoblast precursors, differentiating chondrocytes, osteoblasts, and odontoblasts via Osterix-Cre, results in severe craniofacial dysplasia, skeletal dysplasia, abnormal teeth, and a phenotype recapitulating the clinical features of CCD. Cbfβf/fOsterix-Cre mice have fewer proliferative and hypertrophic chondrocytes, fewer osteoblasts, and almost absent trabecular bone, indicating that Cbfβ may maintain trabecular bone formation through its function in hypertrophic chondrocytes and osteoblasts. Cbfβf/fCollagen, type 1, alpha 1 (Col1α1)–Cre mice show decreased bone mineralization and skeletal deformities, but no radical deformities in teeth, mandibles, or cartilage, indicating that osteoblast lineage-specific ablation of Cbfβ results in milder bone defects and less resemblance to CCD. Activating transcription factor 4 (Atf4) and Osterix protein levels in both mutant mice are dramatically reduced. ChIP assays show that Cbfβ directly associates with the promoter regions of Atf4 and Osterix. Our data further demonstrate that Cbfβ highly up-regulates the expression of Atf4 at the transcriptional regulation level. Overall, our genetic dissection approach revealed that Cbfβ plays an indispensable role in postnatal skeletal development and homeostasis in various skeletal cell types, at least partially by up-regulating the expression of Atf4 and Osterix. It also revealed that CCD may result from functional defects of the Runx2/Cbfβ heterodimeric complex in various skeletal cells. These insights into the role of Cbfβ in postnatal skeletogenesis and CCD pathogenesis may assist in the development of new therapies for CCD and osteoporosis.
Core binding factor (Cbf) plays crucial roles during skeletal development and hematopoiesis. Cbf consists of two subunits: Cbf alpha (Cbfα) and Cbf beta (Cbfβ). Runt-related transcription factor 2 (Runx2) has been shown to be critical for the differentiation of osteoblasts and skeletal development. Cleidocranial dysplasia (CCD) mainly stems from Runx2 deficiency and is characterized by hypoplastic/aplastic clavicles, patent fontanelles, supernumerary teeth, and short stature (1–3). Notably, there were two case reports of CCD patients with no RUNX2 mutation involving 16q22 large deletions, suggesting CBFβ haploinsufficiency might be involved in cranial facial defects (4, 5). Although great progress has been made toward understanding the function of Runx2 and Cbfβ during the past 20 y, their function in postnatal skeletogenesis and in the pathogenesis of related diseases (e.g., CCD) still remains unclear.
Runx2-I/Cbfβ and Runx2-II/Cbfβ double transgenic mice had enhanced inhibition of osteoblast maturation, resulting in severe osteopenia and fragility in adult mice (6). There is a lack of genetic evidence establishing a specific effect of the missing Runx2/Cbfβ complex in postnatal skeletogenesis and the pathogenesis of CCD due to the fact that Runx2−/− mice die at birth and Cbfβ−/− mice die at midgestation (7, 8) and CbfβGFP/GFP knock-in mice and transgenic rescued Runx2−/− mice [Cbfβ−/− Tg(Tek-GFP/Cbfb mice and Cbfβ−/− Tg(Gata1/Cbfb) mice] die soon after birth (9–11). To investigate the role of Cbfβ in postnatal skeletal development and the pathogenesis of CCD, we used Osterix-Cre (Osx-Cre) mice, which specifically delete Cbfβ flanked by loxP (Cbfβf/f) in osteoblast precursors, differentiating chondrocytes, osteoblasts, and odontoblasts (12, 13), and Col1α1-Cre mice, which specifically delete Cbfβ in the osteoblast lineage (14). In this study, through the powerful approach of conditional knockout (cKO) for genetic dissection, we are able to provide unique insight into the pathogenesis of CCD and reveal multiple functions of Cbfβ required for skeletal development.
Results
Cbfβf/fOsx-Cre Mice Have Decreased Ossification and Skeletal Deformities, Resulting in a CCD-Like Phenotype.
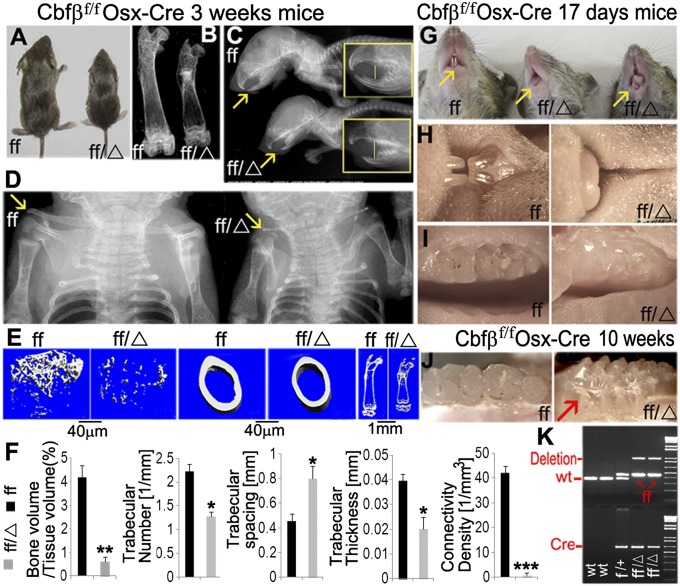
Cbfβf/fOsx-Cre mice survived into adulthood, but the homozygote mice displayed severe postnatal skeletal defects. Three-wk-old Cbfβf/fOsx-Cre mice were of a dispoportionately short stature compared with their wild-type (WT) cohorts (Fig. 1A). X-ray analysis of femurs from 3-wk-old Cbfβf/fOsx-Cre mice showed decreased growth and reduced ossification (Fig. 1B). Cbfβf/fOsx-Cre mice also exhibited underdeveloped mandibles and mandibular retrognathism, resulting in a large gap inside the oral cavity and causing an anterior open bite, and severely affected teeth (Fig. 1C). X-ray analysis revealed hypoplasia/aplasia of clavicles and underdeveloped long bones, leading to severe deformities in the mutant mice (Fig. 1D). These skeletal defects persisted in 3- and 10-wk-old Cbfβf/f Osx-Cre mice (Fig. S1). Classical CCD is caused by RUNX2 haploinsufficiency. Our results showed that there was a milder CCD phenotype in Cbfβ cKO heterozygous mice and a more severe CCD phenotype in Cbfβ cKO homozygous mice (Fig. S1E). Microcomputed tomography (µCT) scan of femurs revealed a drastic decrease in bone density and an almost complete lack of trabecular bone in the mutant mice (Fig. 1 E and F). Moreover, incisors were completely absent (Fig. 1G, middle mouse) or severely underdeveloped in 17-d-old Cbfβf/f Osx-Cre mice (Fig. 1G, right mouse). Higher magnification further illustrated the underdeveloped incisors (Fig. 1H) and underdeveloped molars in Cbfβf/fOsx-Cre mice (Fig. 1I). The teeth defects were still apparent in 10-wk old mutant mice, which also exhibited a supernumerary teeth-like phenotype, a clinical feature of CCD (Fig. 1J). The genotypes of the mice were confirmed by PCR (Fig. 1K) from tail snip DNA. These results showed that Cbfβ ablation in osteoblast precursors results in many clinical features of CCD, including short stature, hypoplastic/aplastic clavicles, and dental anomalies.
Fig. 1.
Cbfβf/f Osx-Cre mice have decreased bone mineralization and skeletal deformities, resulting in a CCD-like phenotype. (A) Photographic analysis of 3-wk-old Cbfβf/f Osx-Cre (ff/Δ) mice and WT (ff) mice. (B–D) X-ray analysis of (B) femurs, (C) mandibles, and (D) clavicles. Yellow arrows in C and D indicate that Cbfβf/f Osx-Cre mice have a severe anterior open bite and mandibular retrognathism as well as hypoplastic/aplastic clavicles, respectively. (E and F) The µCT scans (E) and quantification (F) show that bones from Cbfβf/f Osx-Cre mice are smaller and less mineralized than those of WT. (G and H) Photographic analysis (G) and high magnification (H) of incisor tooth development. Yellow arrows indicate normal (left mouse), stunted (middle mouse), or abnormal (right mouse) tooth development. (I) High-magnification analysis of molar tooth development. (J) Photographic analysis shows defective, supernumerary-like teeth (red arrow) in Cbfβf/f Osx-Cre mice compared with WT. (K) All mice were genotyped by PCR from tail snip DNA.
Cbfβf/fOsx-Cre Mice Have Shortened Limbs, Decreased Bone Ossification, and Disproportionately Defective Skull, Calveria, and Mandible.
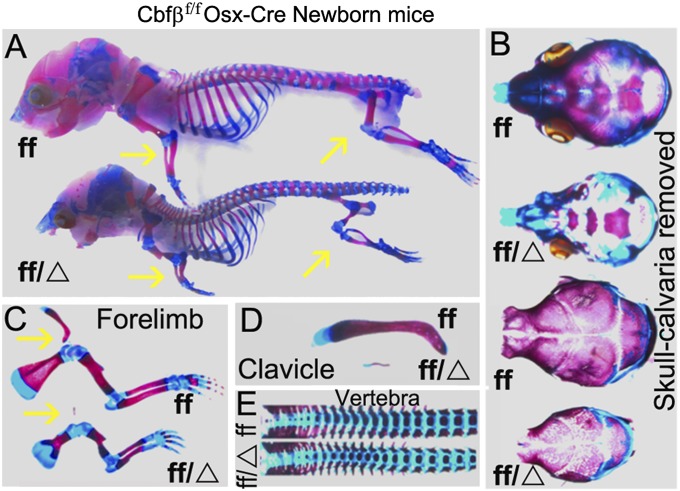
Alizarin red and Alcian blue staining showed that, except for the vertebrae and sternum, every bone in 6-d-old Cbfβf/fOsx-Cre mice was severely underdeveloped (Fig. 2A). However, some bones showed greater defects than other bones. The skull, calveria, and mandible were not only undercalcified in the mutant mice, but also displayed patent fontanelles (Fig. 2B). Moreover, the forelimbs (Fig. 2C) and clavicles (Fig. 2D) were severely affected in the mutant mice, whereas, again, the vertebrae were largely unaffected (Fig. 2E). The mandibula, hyoid bone, thyroid cartilage, and cricoid cartilage in the newborn Cbfβf/fOsx-Cre mice were also underdeveloped due to decreased ossification (Fig. S2). Further, the underdevelopment of the hyoid bone was still apparent in 6-d-old Cbfβf/fOsx-Cre mice. Overall, the skeleton of the mutant mice was severely underdeveloped compared with normal littermates. The data suggest that the process of bone ossification was delayed in mutant mice. Notably, Cbfβf/fOsx-Cre mice had patent fontanelles, another common characteristic of CCD.
Fig. 2.
Cbfβf/f Osx-Cre mice have shortened limbs, decreased bone ossification, and disproportionately defective skull, calveria, and mandible. (A) Whole-mount skeletal was stained by Alizarin red and Alcian blue staining of newborn Cbfβf/f Osx-Cre (ff/Δ) and WT (ff) mice. Yellow arrows show reduced bone ossification in the limbs of the mutant mice. (B–E) The skull (B), forelimbs (C), clavicles (D), and vertebrae (E) of the mutant mice display decreased ossification. Yellow arrows in C show that the mutant mice have less calcified trabecular bone adjacent to the hypertrophic zone.
Cbfβf/fOsx-Cre Newborn Tibiae Have Impaired Endochondral and Intramembranous Bone Ossification, and Goldner’s Trichrome Staining Revealed a Decrease in Osteoblasts Numbers Through Histomorphometric Analysis.
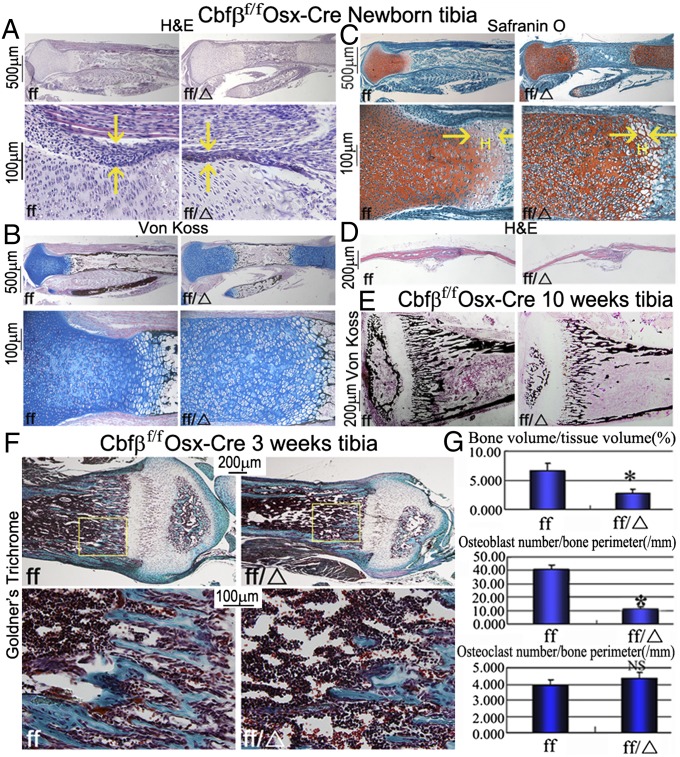
To further analyze the growth retardation observed in the Cbfβf/fOsx-Cre mice, we performed hematoxylin/eosin (H&E), Von Kossa, Alcian blue, and Safranin O staining on tibia from newborn mice. Newborn Cbfβf/fOsx-Cre mice had decreased ossification and endochondral bone formation (Fig. 3 A and B). Newborn Cbfβf/fOsx-Cre mice had decreased cartilage and underdeveloped growth zones, which may partially stem from the reduction in chondrocytes of the hypertrophic region of the growth plate in mutant mice (Fig. 3 A–C). Furthermore, we found that the columns of proliferating and hypertrophic chondrocytes were less organized in Cbfβf/fOsx-Cre mice (Fig. 3C). We also found a decrease in intramembranous bone formation in Cbfβf/fOsx-Cre mice as assessed by H&E staining (Fig. 3D). Finally, Von Kossa and Fast red staining revealed a substantial decrease in calcium and a significantly shorter medullary cavity in 10-wk-old Cbfβf/fOsx-Cre mice compared with control mice (Fig. 3E). Collectively, these results demonstrate that Cbfβf/fOsx-Cre mice survive to adulthood and recapitulate the clinical features of CCD. Goldner’s Trichrome staining revealed a decrease in bone density in Cbfβf/fOsx-Cre (Fig. 3F) and Cbfβf/fCol1α1-Cre mice (Fig. S3B). Notably, there was a significant decrease in bone volume and fewer osteoblasts in the Cbfβf/fOsx-Cre mice (Fig. 3G), but the number of osteoclasts was not significantly affected (Fig. 3G). The tartrate-resistant acid phosphatase staining confirmed that the number of osteoclasts was not significantly affected in Cbfβf/fOsx-Cre mice (Fig. S3A). Goldner’s Trichrome staining also showed that there was a significant decrease in bone volume and fewer osteoblasts (Fig. S3B), but the number of osteoclasts was not significantly affected in Cbfβf/fCol1α1-Cre tibia sections (Fig. S3 B and C).
Fig. 3.
Cbfβf/fOsx-Cre newborn tibiae have impaired endochondral bone ossification, and Goldner’s Trichrome staining revealed a decrease in osteoblast numbers. (A–C) H&E staining (A), Von Kossa and Alcian blue staining (B), and Safranin O staining (C) of tibiae from newborn Cbfβf/f Osx-Cre (ff/Δ) and WT (ff) mice. Yellow arrows in A and C indicate that Cbfβf/f Osx-Cre mice have a reduced bone collar and fewer hypertrophic chondrocytes in the growth plate, respectively. (D) H&E staining shows defective intramembranous bone formation in newborn mutant mice compared with WT. (E) Von Kossa staining of tibia from 10-wk-old Cbfβf/f Osx-Cre and WT bones. (F) Goldner’s Trichrome stain of hard-tissue sections of tibia from 3-wk-old Cbfβf/f Osx-Cre (ff/Δ) and WT (ff) mice. For histological detail of trabeculae, the bottom row shows a higher magnification of areas in yellow boxes. (G) Quantification of the data shown in F.
Cbfβf/fCol1α1-Cre Mice Have Decreased Bone Mineralization and Skeletal Deformities but No Radical Deformities in Teeth, Mandibles, or Cartilage.
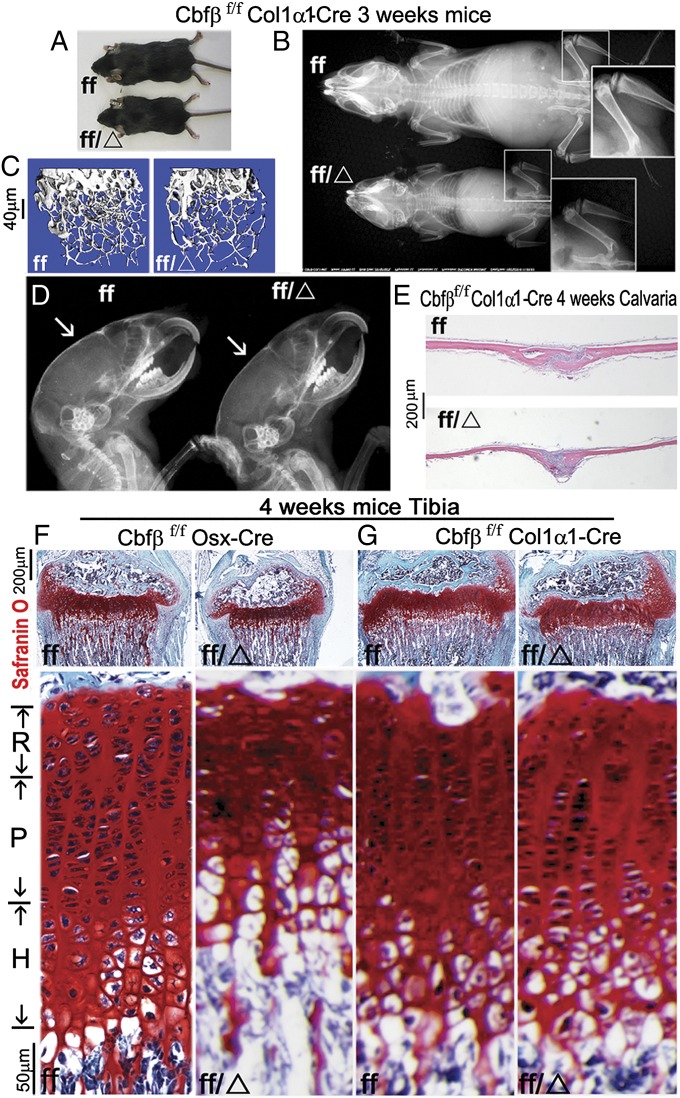
Because Cbfβf/fOsx-Cre mice delete the Cbfβ gene in both osteoblasts and hypertrophic chondrocytes, we generated Cbfβf/fCol1α1-Cre mice to observe the impact of Cbfβ specifically in the osteoblast lineage during postnatal bone development (Fig. 4). Similar to the Cbfβf/fOsx-Cre mice (Fig. 1), bone formation was severely inhibited in Cbfβf/fCol1α1-Cre mice, leading to shorter stature and decreased bone density compared with WT mice (Fig. 4 A–C). Unlike Cbfβf/fOsx-cre mice (Fig. 1), the clavicle (Fig. 4B), mandibles and teeth (Fig. 4D), and intramembranous bone formation (Fig. 4E) were not dramatically affected in Cbfβf/fCol1α1-Cre mice. Safranin O staining revealed that there is a shortened growth plate, a decreased and disorganized proliferative zone, and a lack of hypertrophic chondrocytes in Cbfβf/f Osx-Cre mutant mice (Fig. 4F). However, the growth plate development in Cbfβf/f Col1α1-Cre mutant mice is normal compared with the WT mice control (Fig. 4G).
Fig. 4.
Cbfβf/fCol1α1-Cre mice have decreased bone mineralization and skeletal deformities, but no radical deformities in teeth, mandibles, or cartilage. (A–D) Photographic analysis (A), X-ray analysis focusing on femoral bone density (B), µCT scans of femurs (C), and X-ray analysis of calvaria (D) of 3-wk-old Cbfβf/f Col1α1-Cre (ff/Δ) and WT (ff) mice. White arrows in D show that the calvaria are less calcified in the mutant mice. (E) H&E staining shows that intramembranous bone formation was not dramatically affected in Cbfβf/fCol1α1-Cre mice compared with WT. (F and G) Safranin O staining of tibia and comparison of growth plate development among 4-wk-old Cbfβf/f Osx-Cre mutant, Cbfβf/fCol1α1-Cre mutant, and WT mice.
Cbfβ Deficiency Affects Chondrocyte Proliferation and Maturation in Cbfβf/fOsx-Cre Mice.
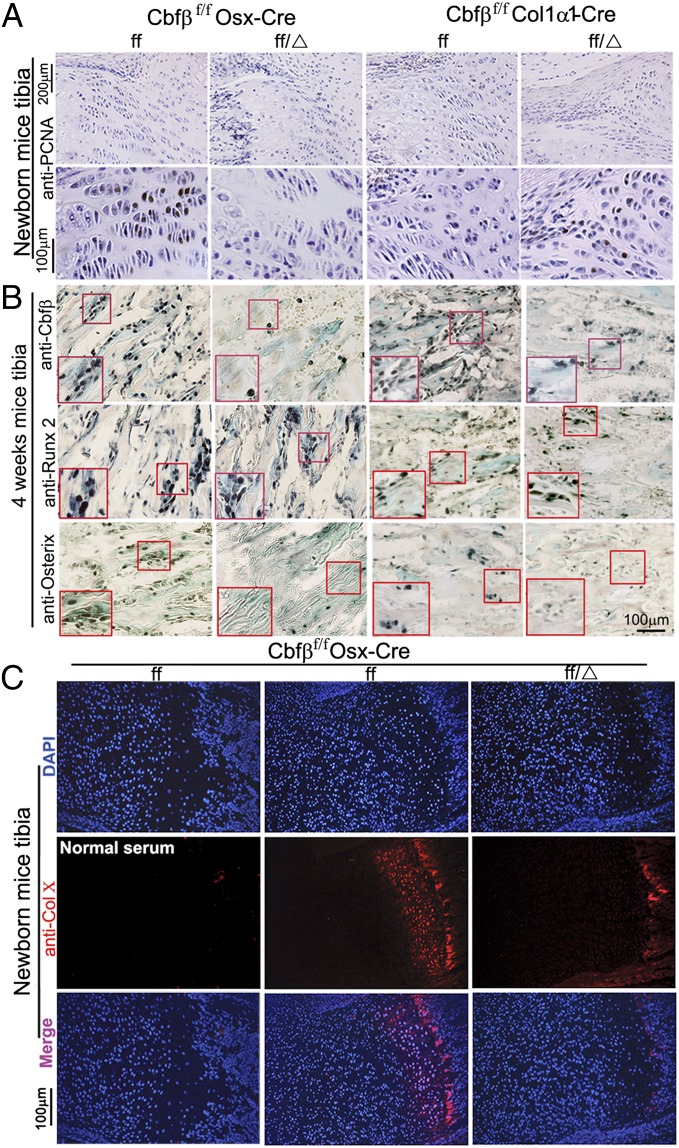
Further examination of the impact of the Cbfβ deletion in chondrocytes through proliferating cell nuclear antigen (PCNA) staining revealed that there is a reduction in proliferative chondrocytes in both the resting and proliferation zones of the Cbfβf/fOsx-Cre but not in the Cbfβf/fCol1α1-Cre mice (Fig. 5A). This is due to the fact that Cbfβf/f Osx-Cre mice excise the Cbfβ gene in odontoblasts, osteoblasts, and chondrocytes, whereas Cbfβf/fCol1α1-Cre mice only delete Cbfβ in the osteoblast lineage. Importantly, this finding indicates that Cbfβ is required for chondrocyte differentiation and the subsequent growth plate formation during postnatal skeletal development. As several studies indicate that Cbfβ interacts with Runx1, Runx2, and Runx3 during skeletal development, we examined whether the Cbfβ deficiency affected the expression of Runx1, Runx2, and Runx3. Immunohistochemistry analysis showed that there is a reduction in the expression of Osx and Cbfβ in Cbfβf/fOsx-Cre and Cbfβf/f Col1α1-Cre mice but that Cbfβ deletion did not affect the level of Runx2 expression (Fig. 5B). Immunostaining analysis of newborn Cbfβf/fOsx-Cre and WT mice femur showed that Runx1 expression was mainly detected in the proliferative zone, Runx2 expression was mainly detected in the hypertrophic zone, and Runx3 expression was mainly detected in hypertrophic and prehypertrophic chondrocytes (Fig. S4 A–C). Interestingly, expression of Runx1, Runx2, and Runx 3 showed no variation between WT and mutant mice (Fig. S4 A–C). Immunostaining analysis of newborn mice showed a drastic reduction in the expression of Collagen X (ColX), which is a marker of hypertrophic chondrocytes, in the growth plates of Cbfβf/fOsx-Cre mice (Fig. 5C). These results demonstrate that chondrocyte proliferation and maturation are dependent on Cbfβ’s function.
Fig. 5.
Cbfβ deficiency decreases chondrocyte proliferation, reduces expression of Cbfβ and Osterix, and impairs chondrocyte hypertrophy in Cbfβf/f Osx-Cre mice. (A and B) PCNA staining for cellular proliferation (A) and immunohistochemistry (IHC) staining with anti-Cbfβ, anti-Runx2, and anti-osterix antibodies (B) of tibial paraffin sections from 4-wk-old Cbfβf/f Osx-Cre (ff/Δ), Cbfßf/f Col1α1-Cre (ff/Δ), and WT (ff) mice. (Insets) The magnified images of the red boxed areas, which show a decrease in chondrocyte proliferation in Cbfβf/f Osx-Cre mice and reduced expression of Cbfβ and Osterix in both mutant mice compared with WT. (C) Immunofluorescence staining of the tibia from newborn Cbfβf/f Osx-Cre (ff/Δ) and WT (ff) mice.
Cbfβ Deficiency in Primary Calvarial Cells Cultured from Cbfβf/f Osx-Cre and Cbfβf/fCol1α1-Cre Mice Inhibits Osteoblastogenesis.
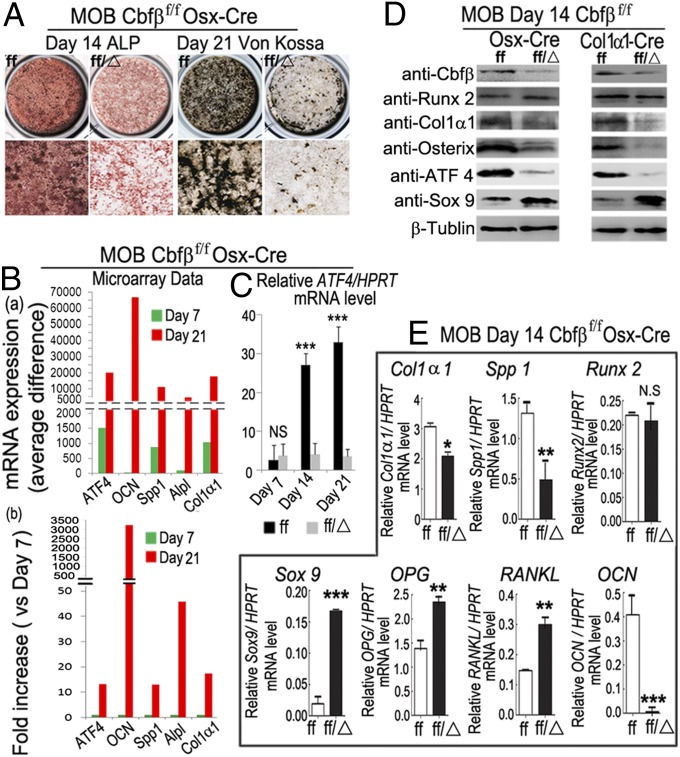
We investigated the impact of Cbfβ deletion on osteoblastogenesis. Calvarial cells from Cbfβf/fOsx-Cre (Fig. 6A) and Cbfβf/fCol1α1-Cre mice (Fig. S5A) after 14 d of culture showed reduced alkaline phosphatase (ALP), indicating a decreased number of osteoblasts in the mutant cells. The reduction in mineralization observed in Cbfβf/fOsx-Cre was characterized by Von Kossa staining after 21 d of culture (Fig. 6A). GeneChip analysis indicated that Atf4 mRNA expression in mouse C57/BL6 WT calvarial cells on day 7 and day 21 of osteoblastogenesis is similar to that of secreted phosphoprotein 1 (Spp1), ALP liver/bone/kidney (Alpl), and Col1α1, whereas osteocalcin (OCN) has mRNA expression levels that were much higher (Fig. 6B). Further microarray data analysis revealed that Atf4 mRNA expression in mouse calvarial cells increased more than 10-fold on culture day 21 (Fig. 6B). The dramatic changes in Atf4 mRNA expression were confirmed by quantitative RT-PCR using Cbfβf/fOsx-Cre, Cbfβf/fCol1α1-Cre, and WT mouse calvarial cells during osteoblastogenesis (Fig. 6C, Fig. S5 B and C, and Fig. S6). Western blot was used to analyze the expression of several key factors that influence osteoblast functions in Cbfβf/fOsx-Cre and Cbfβf/fCol1α1-Cre mice. Cbfβ deficiency reduced the expression of Cbfβ, Osx, Col1α1, and Atf4, but not Runx2 (Fig. 6D). In contrast, the expression of SRY (sex determining region Y)-box 9 (Sox9) was increased in Cbfβ-deficient calvaria-derived osteoblasts (Fig. 6D). Taken together, these results demonstrate that Cbfβ deletion impacts chondrocyte and osteoblast differentiation by affecting the expression of critical downstream genes at the mRNA level. Notably, on day 14, Cbfβf/fOsx-Cre calvarial cells still expressed some Col1α1 and Spp1 but not OCN (Fig. 6E). These data indicate that osteoblasts generated from Cbfβf/fOsx-Cre mice were maintained in an immature stage and prevented from differentiation into mature osteoblasts, which are critical for bone formation.
Fig. 6.
Cbfβ deficiency in primary calvarial cells cultured from Cbfβf/fOsx-Cre and Cbfβf/fCol1α1-Cre mice inhibits osteoblastogenesis. (A) Calvarial cells from Cbfβf/f Osx-Cre (ff/Δ) and WT (ff) newborn mice were applied to osteoblastogenesis assays. (B) GeneChip analysis of the expression of Atf4, Ocn, Spp1, Alpl, and Col1α1 in mouse calvarial cells. (C) qPCR analysis of the mRNA expression level of Atf4 in mouse calvarial cells after culturing in the osteoblast differentiation media. (D) Protein expression levels were analyzed by Western blot analysis. (E) qPCR analysis of mRNA expression levels of Col1α1, Spp1, Runx2, Sox9, OPG, RANKL, and OCN in calvaria-derived osteoblasts from Cbfβf/f Osx-Cre (ff/Δ) and WT (ff) mice. Results are expressed as mean ± SD, n ≧ 6 in each group. *P < 0.05, **P < 0.01, ***P < 0.005.
Cbfβ Up-Regulates Osterix and Atf4 Expression Directly by Associating with Their Promoters.
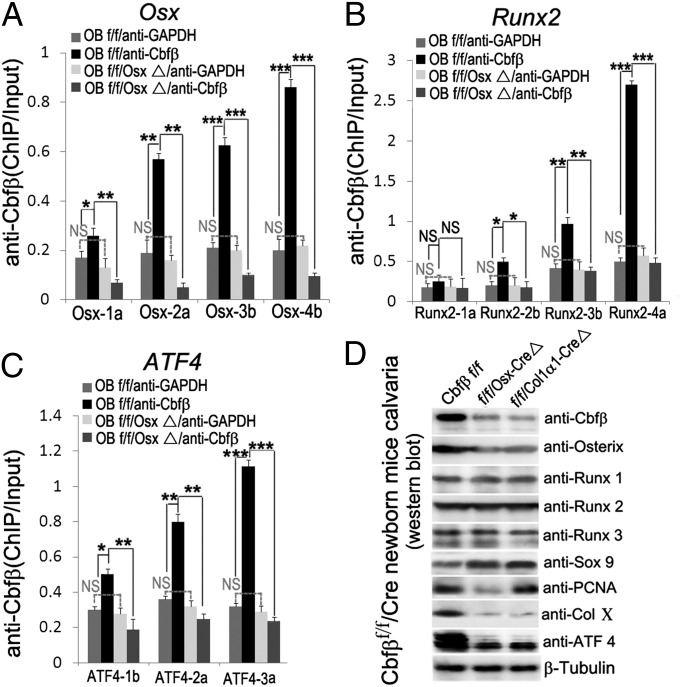
To determine if the Runx/Cbfβ complex binds to Osx, Runx2, and Atf4 promoters, chromatin immunoprecipitation (ChIP) assay was performed. We analyzed the respective promoter regions and designed primers accordingly. We found several Runx binding sites in the Osx promoter region (–3000/+80), Runx2 promoter region (–3000/+80), and Atf4 promoter region (–4000/+80) (Fig. S7). ChIP analysis was performed using the anti-Cbfβ antibody, and the DNA was pulled down, amplified, and analyzed using primers as shown in Fig. S7. The ChIP input value using each primer represents the binding efficiency of an adjacent region around the location of the primer pair. We found that Osx primer 4b resulted in the highest value, indicating that Runx binding sites 13–15 in the Osx promoter region should be the most efficient (Fig. 7A). In addition, Runx2 primer 4a showed that Runx binding sites 9 and 10 in the Runx2 promoter region may be most efficient, whereas Atf4 primer 3a showed that Runx binding site 5 in the Atf4 promoter region may be the most efficient (Fig. 7 B and C). The promoter luciferase assay showed that luciferase activity driven by Atf4 promoters is very low in the absence of Cbfβ (Fig. S8). Luciferase activity was highest when driven by the longest Atf4 promoter fragment (–4051/+80) and less high when driven by the Atf4 promoter fragments (–3109/+80 and –1676/+80) in WT cells, but not in the mutant cells. Interestingly, about 70% of the full Luciferase activity driven by the longest Atf4 promoter fragment (–4051/+80) still remains with the Atf4 promoter fragment (–500/+80) in WT cells, but not in the mutant cells. This indicates that the Atf4 promoter region (–500/+80) containing Runx binding sites 5–7 is critical for Cbfβ regulation of Atf4 gene expression (Figs. S7 and S8). Consistently, the primer 3a amplifying regions near Runx binding sites 5–7 in the Atf4 promoter gave out the highest value in ChIP assays (Fig. 7C), indicating that the Cbfβ/RUNX complex may bind Runx binding sites 5–7 to up-regulate Atf4 gene expression. There was also a decrease in luciferase expression driven by the Runx2 promoter (–1580/+80 and –900/+80) in the absence of Cbfβ. However, Runx2 expression does not vary between WT and mutant cells (Fig. 7D and Fig. S6A). This indicates that there may be some other mechanism driving the steady expression of Runx2 in Cbfβ knockout mice and cells, probably working on a distant end of the Runx2 promoter (before –1580). In conclusion, we believe that Cbfβ directly associates with the Osx and Atf4 promoter regions and regulates their expression. To confirm these Western and real-time RT-PCR results in calvarial culture experiments, we used protein directly isolated from calvaria (Fig. 7D and Fig. S6). The Western blot result is consistent with that from the calvarial culture experiment result (Fig. 6D and Fig. 7D).
Fig. 7.
Cbfβ regulates Osx, Runx2, and Atf4 expression by directly associating with their promoters. (A–C) ChIP analysis of Cbfβ binding to the (A) Osx promoter, (B) Runx2 promoter, and (C) Atf4 promoter in calvaria-derived osteoblasts using primers as indicated on the x axes. Results are presented as ChIP/Input. Results are presented as mean ± SD, n ≧ 6 in each group. *P < 0.05, **P < 0.01, ***P < 0.005. (D) Western blot analysis of the expression of Cbfβ, Osx, Runx1, Runx2, Runx3, Sox9, PCNA, ColX, and Atf4 in the calvaria of newborn Cbfβf/fOsx-Cre, Cbfβf/fCol1α1-Cre, and WT mice. β-tubulin is used as the loading control.
Discussion
CCD May Result from a Functional Defect of the Runx2/Cbfβ Heterodimeric Complex in Various Cell Types.
Our data show that Cbfβf/fOsx-Cre mice provide a unique CCD model recapitulating most of the characteristics of human CCD (i.e., wide/open fontanels, midface retrusion, abnormal dentition, severe clavicular hypoplasia, and hand/paw abnormalities). The CCD phenotype of Cbfβf/fOsx-Cre mice indicates that multiple functions of Cbfβ are required for skeletal development and homeostasis in postnatal skeletogenesis. The fact that the Runx2 protein level was not changed in Cbfβf/fOsx-Cre cKO mice and that Cbfβf/fOsx-Cre mice exhibit a CCD-like phenotype (Fig. 1) supports the notion that CCD may result from a functional defect of the Runx2/Cbfβ heterodimeric complex in various cell types. It also indicates that, in terms of the pathogenesis of CCD, Cbfβ deficiency may be equivalent to RUNX2 haploinsufficiency as it relates to the function of the Runx2/Cbfβ complex in skeletogenesis.
Indispensable Role of Hypertrophic Chondrocytes in Endochondral Bone Formation.
The role of hypertrophic chondrocytes in endochondral bone formation is a long-standing question. Our results showed that there is a shortened growth plate, a decreased and disorganized proliferative zone, and a reduction in hypertrophic chondrocytes in Cbfβf/fOsx-Cre mutant mice. However, growth plate development in Cbfβf/fCol1α1-Cre mutant mice is normal compared with the WT control. These phenotypes of the mutant mice provided a unique opportunity to address the role of hypertrophic chondrocytes in endochondral bone formation. Our results indicate that Cbfβ maintains trabecular bone formation through its function in chondrocytes. Based on the severe endochondral bone defects in Cbfβf/fOsx-Cre mutant mice (Figs. 1 and 3) and the mild endochondral bone defects in Cbfβf/fCol1α1-Cre mutant mice (Fig. 4), we conclude that the role of hypertrophic chondrocytes in endochondral bone formation is indispensable. We hypothesize that the lack of trabecular bone is not the result of a lack of osteoblast precursors and preosteoblasts but may be a result of a lack of the factor(s) (such as Ihh) secreted by prehypertrophic or hypertrophic chondrocytes, although the mechanism of this dependence remains to be identified.
Cbfβ Plays an Indispensable Role in Postnatal Skeletal Development and Homeostasis by Up-Regulating the Expression of Atf4 and Osterix.
Cbfβ deficiency reduced the expression of several key factors that mediate osteoblast formation and/or function (e.g., Osx and Atf4). This suggests that Cbfβ may have a role in promoting the commitment of osteoblast precursors into the osteoblast lineage. Yang et al. reported that ATF4 is a critical regulator of osteoblast differentiation and function (15). However, how ATF4 is regulated at the transcriptional regulation level remains unclear. Our study shows that Cbfβ associates with the promoter regions of Osx and Atf4 and highly up-regulates the expression of Atf4 at the transcriptional regulation level as shown by ChIP assay, microarray analysis, quantitative PCR (qPCR) analysis, and promoter reporter assay. We also found that Cbfβ is crucial for the later stages of chondrocyte differentiation as its deletion affects chondrocyte maturation and the formation of the growth plate.
The Clinical Features of CCD Are Recapitulated in Cbfβf/fOsx-Cre Mice.
Because Runx2 functions as a heterodimer with CBFβ, it has been suspected that Cbfβ may be responsible for some cases of CCD. Although no CBFβ mutation has yet been identified in classical CCD patients, our Cbfβf/fOsx-Cre mouse models support the notion that search genetic alterations in the CBFβ gene may be responsible for CCD in those patients with no RUNX2 mutation. Our results are in agreement with that of previous studies reporting that Runx2 deficiency causes an arrest in tooth development (16) and that Osx is necessary for odontoblast differentiation (13). These findings provide great insight into the pathogenesis of CCD and the role of Cbfβ in both postnatal skeletal and tooth development. The insights resulting from this study may assist in the development of novel treatments for CCD and other bone diseases.
Materials and Methods
Animal Experimentation and Generation of Cbfβ cKO Mice.
Cbfβf/f mice (Jackson Laboratory, strain name B6.129P2-Cbfβtm1Itan/J) were crossed with skeletal tissue cell (including osteoblast precursors, osteoblasts, chondrocytes, and odontoblasts)-specific Osx-cre mice (12) [Tg(Sp7-tTA,tetO-EGFP/cre)1Amc, Mouse Genome Informatics] or osteoblast-specific Col1α1-Cre mice. Their progeny were crossed with Cbfβf/f mice to obtain Cbfβf/fCol1α1-Cre mice or Cbfβf/fOsx-Cre mice. In our study, we only use one copy of Osx-Cre (Cbfβf/f Osx-Cre/+) in the cKO mutation. We used Cbfβf/f mice and Osx-Cre/+ mice as controls. Mouse C57/BL6 WT calvarial cells were also used. All research procedures using mice were approved by the University of Alabama at Birmingham (UAB) Animal Care and Use Committee and conformed to the National Institutes of Health guidelines.
Skeletal Analysis, Tissue Preparation, and Histology Stains.
Histomorphometric samples were processed as nondecalcified hard-tissue sections. Bone parameters were quantified via 6 µm sections obtained from 3-wk-old mice. For paraffin sections, samples were decalcified and dehydrated in ethanol, cleared in xylene, embedded in paraffin, and sectioned at 6 μm with Leica microtome and mounted on Superfrost Plus slides (Fisher). Histological analysis was performed including staining with Alcian blue, safranin O, and H&E using paraffin sections.
ChIP and Promoter Analyses.
Cells were derived from calvaria of newborn WT mice and mutant mice. Osx, Runx2, and Atf4 promoter sequences were analyzed for putative Runx binding sites with PROMO3.0 (http://alggen.lsi.upc.es/) using version 8.3 of the TRANSFAC database. ChIP was performed using monoclonal anti-Cbfβ antibody (sc-20693X) and DNA extraction, and qPCR was performed. The Osx, Runx2, and Atf4 promoters were amplified using PCR from BAC clones provided by the BACPAC Resource Center at Children's Hospital Oakland Research Institute. These amplified fragments were cleaved and ligated into the pGL3 vector from Promega. The calvarial cells were cotransfected with each construct with the amount of 100 ng per well as well as pSV–β-galactosidase construct with the amount of 50 ng per well (Promega) and incubated for 6–8 h. The culture medium was replaced with osteogenic medium and cultured for 2 d. Luciferase activity was measured using a Steady-Glo luciferase assay system (Promega cat. no. E2510). pSV–β-galactosidase activity was measured using a β-galactosidase Enzyme Assay system (Promega cat. no. E2000).
Statistical Analysis.
All data are presented as the mean ± SD (n ≧ 6). Statistical significance was assessed using Student t test. P values < 0.05 were considered significant. Data are expressed as mean ± SD, n ≧ 6, *P < 0.05, **P < 0.01, ***P < 0.001. The results are representative of at least four individual experiments. The analyses of the data were performed with the SPSS 16.0 software (SPSS Incorporation).
Please see SI Materials and Methods for additional details.
Supplementary Material
Acknowledgments
We appreciate the assistance provided by the Center for Metabolic Bone Disease at the University of Alabama at Birmingham (Grant P30 AR046031). We are also grateful for the assistance from the Small Animal Phenotyping Core, Metabolism Core, and Neuroscience Molecular Detection Core Laboratory at the University of Alabama at Birmingham (Grant P30 NS0474666). This work was supported by National Institutes of Health Grants AR-44741 and AR-055307 (both to Y.-P.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310617111/-/DCSupplemental.
References
- 1.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 2.Mundlos S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 3.Lee B, et al. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16(3):307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 4.Goto T, et al. Large fontanelles are a shared feature of haploinsufficiency of RUNX2 and its co-activator CBFB. Congenit Anom (Kyoto) 2004;44(4):225–229. doi: 10.1111/j.1741-4520.2004.00043.x. [DOI] [PubMed] [Google Scholar]
- 5.Khan A, Hyde RK, Dutra A, Mohide P, Liu P. Core binding factor beta (CBFB) haploinsufficiency due to an interstitial deletion at 16q21q22 resulting in delayed cranial ossification, cleft palate, congenital heart anomalies, and feeding difficulties but favorable outcome. Am J Med Genet A. 2006;140(21):2349–2354. doi: 10.1002/ajmg.a.31479. [DOI] [PubMed] [Google Scholar]
- 6.Kanatani N, et al. Cbf beta regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296(1):48–61. doi: 10.1016/j.ydbio.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki K, et al. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93(22):12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida CA, et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32(4):633–638. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 10.Kundu M, et al. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32(4):639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 11.Miller J, et al. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32(4):645–649. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- 12.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, et al. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88(10):904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224(2):245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117(3):387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 16.D’Souza RN, et al. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126(13):2911–2920. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.