Significance
Immunodeficient mice are important tools to define stem cells that drive malignancies (cancers). Primary myelofibrosis (PMF) is a chronic myeloproliferative neoplasm that can progress to malignant leukemia. In a study to define PMF stem cells in transplanted mice, we observed a high incidence of mouse leukemia. We show that endogenous retrovirus (ERV), whose replication is unrestricted in immunodeficient mice, are pathogenic in the PMF-xenograft microenvironment, likely because of increased numbers of proliferating mouse cells stimulated by PMF-derived cells. Proliferating cells are targets of retroviral transformation and spontaneous mutations, and thus susceptible to leukemia induction. These results substantiate the importance of paracrine mechanisms in PMF disease and expose the presence of replicating ERVs in mice commonly used to model human diseases.
Abstract
The compound immunodeficiencies in nonobese diabetic (NOD) inbred mice homozygous for the Prkdcscid and Il2rgnull alleles (NSG mice) permit engraftment of a wide-range of primary human cells, enabling sophisticated modeling of human disease. In studies designed to define neoplastic stem cells of primary myelofibrosis (PMF), a myeloproliferative neoplasm characterized by profound disruption of the hematopoietic microenvironment, we observed a high frequency of acute myeloid leukemia (AML) in NSG mice. AML was of mouse origin, confined to PMF-xenografted mice, and contained multiple clonal integrations of ecotropic murine leukemia virus (E-MuLV). Significantly, MuLV replication was not only observed in diseased mice, but also in nontreated NSG controls. Furthermore, in addition to the single ecotropic endogenous retrovirus (eERV) located on chromosome 11 (Emv30) in the NOD genome, multiple de novo germ-line eERV integrations were observed in mice from each of four independent NSG mouse colonies. Analysis confirmed that E-MuLV originated from the Emv30 provirus and that recombination events were not necessary for virus replication or AML induction. Pathogenicity is thus likely attributable to PMF-mediated paracrine stimulation of mouse myeloid cells, which serve as targets for retroviral infection and transformation, as evidenced by integration into the Evi1 locus, a hotspot for retroviral-induced myeloid leukemia. This study thus corroborates a role of paracrine stimulation in PMF disease progression, underlines the importance of target cell type and numbers in MuLV-induced disease, and mandates awareness of replicating MuLV in NOD immunodeficient mice, which can significantly influence experimental results and their interpretation.
The classic myeloproliferative neoplasms (MPN) are a heterogenous group of chronic disorders characterized by cellular proliferation of one or more myeloid lineages, which is thought to arise from a mutated hematopoietic stem cell (HSC) (1, 2). Progression of the diseases from a neoplastic (precancerous) to an aggressive malignant (leukemia) stage is quite variable in rate and incidence, but forebodes a poor prognosis. The aim of current research is to define the neoplastic stem cells that are predicted to maintain the disease, and as such are important therapeutic targets, but also to understand the step-wise but nonlinear process leading to overt malignancy (3, 4). Such endeavors are most advanced for chronic myeloid leukemia, the most common MPN that is characterized by the BCR/ABL translocation. In vivo and in vitro studies have demonstrated that the chronic myeloid leukemia neoplastic stem cell maintaining the chronic disease shares many characteristics of normal HSC; furthermore, their downstream progenitors are the likely targets of transformation events that generate the malignant clone in the acute phase (3, 5).
For the other three classic MPNs [i.e., polycythemia vera, essential thrombocytosis, and primary myelofibrosis (PMF)], neither the functional phenotype of the neoplastic stem cell nor the origin of the malignant stem cell is known. Notably, these MPNs share a driving mutation—JAK2-V617F, occurring in ∼95% of cases of polycythemia vera and 50–60% of both essential thrombocytosis and PMF—but their clinical symptoms are quite distinct, suggesting additional events that define disease variability (6, 7). Of special interest is PMF, which has the worst prognosis and is characterized by bone marrow (BM) fibrosis and an increased risk of transformation to acute myeloid leukemia (AML) (around 20–30%) (8).
Immunodeficient mice are crucial tools for identifying cancer stem cells and dissecting the molecular mechanisms—genetic, epigenetic, or environmental—that drive the evolution of neoplastic to malignant stem cell states (9). Mice of the nonobese diabetic (NOD)-PrkdcscidIl2rgnull [NOD-Scid-γ (NSG)] strain are severely immunodeficient and are often used for xenograft transplantations (10). The genetic background of NSG mice is provided by the NOD/ShiLt (NOD) inbred mouse strain. As with all mice, the NOD genome contains multiple copies of endogenous retrovirus (ERV) related to the γ-genus of exogenous retrovirus, typified by the family of murine leukemia virus (MuLV) (11, 12). The majority of these ERVs are defective because of the irreversible accumulation of mutations; however, this inevitable extinction is counteracted by adaptive mechanisms that work to maintain active ERVs, which are an important source of somatic and genomic diversity (13, 14). A good example is the recent and recurrent entry of ecotropic (E)-MuLVs into the Mus musculus germ line, as shown by their insertional polymorphism (15) and evidence of novel insertions in viremic mouse strains (16).
In the course of xenograft experiments to define the PMF neoplastic stem cell, a high incidence of mouse AML was observed in NSG mice. We reasoned that this may be a result of activated ERVs and thus sought to unravel the mechanism. The fact that AML was limited to PMF-xenografted mice suggested a causal interplay. Our work demonstrates that replicating MuLVs in NSG mice are instrumental in leukemia induction, but that PMF-engrafted cells likely provide a receptive cell milieu for retroviral transformation. Active replication of MuLV in NSG mice observed in several independent mouse colonies, leading to both somatic and germ-line integrations and thus genetic mutations, has major implications for scientists using these mice.
Results
Induction of AML in PMF-Transplanted NSG Mice.
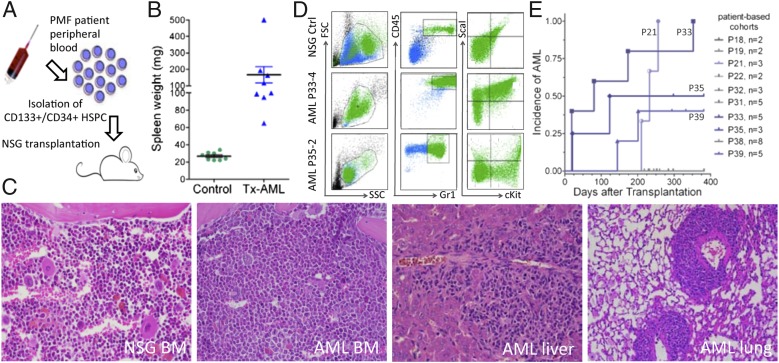
In experiments designed to evaluate the capacity of mobilized hematopoietic stem and progenitor cells (HSPC) from patients to engraft and reproduce PMF disease in NSG xenografted mice (Fig. 1A), we observed that several mice developed AML, characterized by splenomegaly, BM hyperplasia with >20% blasts, infiltration of nonhematopoietic tissue, and myeloid sarcomas (Fig. 1 B and C). Furthermore, the AML was transplantable to secondary recipients (Fig. S1). FACS analysis confirmed mouse origin (mCD45+) and myeloid progenitor phenotype (Gr1+ or Kit1+Sca1+) of the leukemic cells (Fig. 1D); the phenotype was also confirmed with the CD11b myeloid marker. Of a total of 38 mice transplanted with HSPC derived from 10 independent patients, 13 mice receiving cells from 4 patients developed AML with variable latencies. Human-derived HPSC were consistently detectable in all engrafted mice (range 2–10% in BM), but the risk of disease development was singly correlated to the patient sample itself, as illustrated by AML incidence rates from the independent mouse cohorts (each cohort representing a single patient) (Fig. 1E). These results were observed over a 2-y span and could not be traced to offspring of specific breeding pairs. No conditioning of mice (e.g., radiation, immunosuppression) before transplantation was performed in any experiment. AML induction could not be correlated with any specific clinical symptom or JAK2-V617F burden of the patient samples (Table S1). Finally, AML was never observed in NSG mice inoculated with saline solution or with HSPC isolated from either normal BM or umbilical cord blood and monitored from 4 to 12 mo (n = 102 mice) (Table S2).
Fig. 1.
Characterization of AML induced in PMF-transplanted NSG mice. (A) Schematic model of the approach used to identify neoplastic stem cells in PMF. (B) Spleen weights of either nontreated NSG controls (n = 8) or mice succumbing to AML-induction after transplantation (Tx) of PMF HSPC (n = 8). The mean and SEM is shown. (C) Histological analysis of a typical AML mouse. The heterogenous and hypocellular BM section of a control NSG mouse (far Left) contrasts with the homogenous and hypercellular BM of an AML mouse. Infiltrating AML cells (dark staining) were observed in both liver and lung of diseased mice (H&E staining; objective 40×) (D) FACS analysis of BM cells from an untreated NSG mouse and two AML mice. (E) Cumulative incidence of AML induction in mouse cohorts, each receiving HSPC from distinct patient samples. Geometric symbols indicate mice with overt AML symptoms, whereas short vertical lines indicate healthy mice that were culled for analysis to determine the percentage of human engrafted cells. The patient sample corresponding to a specific geometric symbol is indicated in the graph.
Both Normal and Leukemic NSG Cells Release Retrovirus.
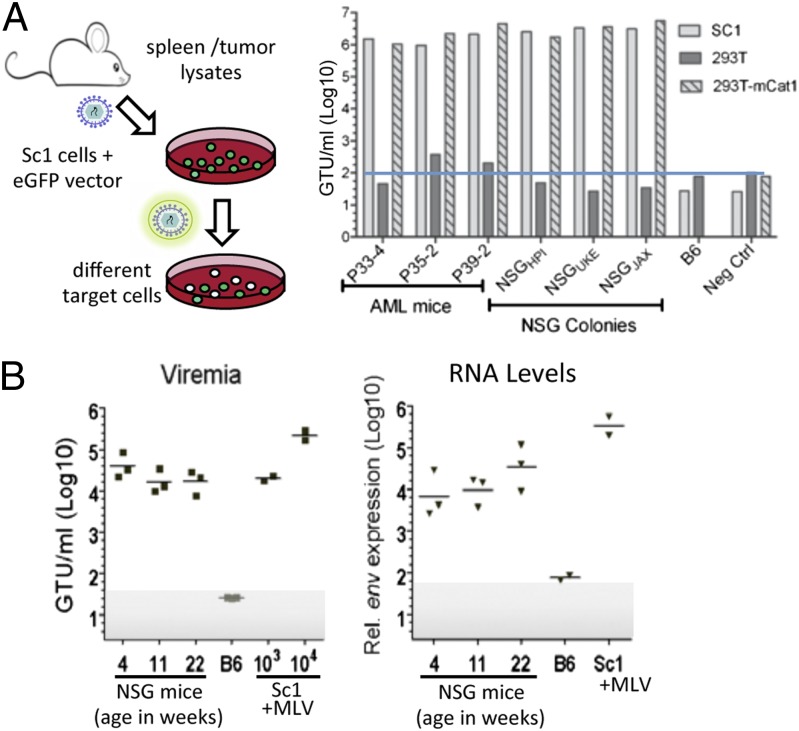
Spontaneous AML induction in mice has been reported in a few mouse strains and has been attributed to insertional mutagenesis by infectious E-MuLV recombinants of ERV (17). The NOD mouse strain carries a single eERV proviral copy (designated Emv30), and thus we investigated the possibility that leukemia induction could be a result of activation of the Emv30 provirus. Using a mobilization assay (Fig. 2A), infectious MuLV could readily be detected in spleens from three independent AML mice. Host-range analysis determined that the MuLV infects mouse but not human cells, a characteristic of ecotropic retrovirus; definitive proof of an ecotropic host range was obtained by demonstrating that expression of the E-MuLV cellular receptor (mCat1) in human 293T cells conferred virus infectivity (Fig. 2A).
Fig. 2.
Detection of MuLV in NSG mice. (A) Mobilization assay used to detect MuLV in mice. The titers of MuLV/SF91-GFP pseudotypes are expressed as GFP-transfer units (GTU) per milliliter of supernatant determined on mouse SC1 or human 293T cells, as indicated. The lower limit of the assay was determined by mock infections (medium alone) for each target cell, and is denoted with a blue line for 293T cell infections. (B) Quantitative assays to determine viremia and MuLV expression in NSG mice. (Left) Plasma dilutions were cultivated with indicator cells and titered after 1 wk. Virus stocks with known titers (103 and 104 GTU/mL) were assayed in parallel to confirm linearity and sensitivity of assay. (Right) Quantitative RT-PCR was used to detect MuLV spliced env message in liver cells. SC1 cells producing E-MuLV served as positive control. Shaded area denotes lower limits of assays.
Somewhat surprisingly, E-MuLV could also be detected in serum or supernatant from homogenized spleens of untreated NSG control mice, but not untreated C57BL/6 (B6) mice (negative control) (Fig. 2B). To determine if the observed viremia in the NSG mice was a characteristic of the specific mouse colony (NSGHPI) used for these experiments, mice from two other NSG colonies (NSGUKE and NSGJAX) were also analyzed. In all cases, replication-competent E-MuLV could be readily detected (Fig. 2A). Interestingly, viremia was observed as early as 4 wk after birth and viral titers remained rather constant up to 22 wk, as measured by quantitative mobilization assays and assessment of viral mRNA (Fig. 2B).
De Novo Germ-Line MuLV Integrations in NSG Mice and Multiple Clonal Integrations in Leukemic Cells.
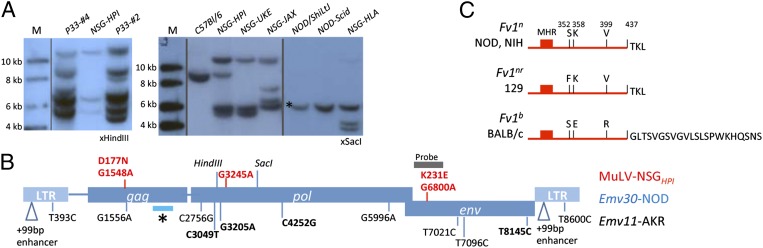
The presence of infectious E-MuLV adds credence to the hypothesis that AML is induced by retroviral infection. For further verification, Southern blot analysis was performed on DNA isolated from spleens of control, untreated NSG mice, and diseased mice (Fig. 3A). Notably, spleens from AML mice showed multiple (>10) proviral integration sites, which were judged to be clonal based on digestion of DNA with restriction enzymes to identify shared insertion sites. Multiple clonal integrations are typical of leukemia/lymphoma induced by retroviral infections. Somewhat surprisingly, however, untreated NSG mice analyzed showed up to four or five proviral integration sites, in contrast to the single provirus (Emv30) detected in DNA isolated from NOD or NOD.CB17-Prkdcscid (NOD-Scid) mice. Distinct proviral integration patterns could be detected in mice from independent NSG colonies. The NSGHPI colony was derived from the NSGUKE colony and bred separately for 2 y, a fact reflected in the presence of both common and distinct proviral integration sites (Fig. 3A). De novo ERV integrations are consistent with the high levels of constitutive viremia in the NSG strain (16).
Fig. 3.
Analysis of somatic and germ-line MuLV integrations and host restriction mechanisms. (A) Southern blot analysis of genomic DNA isolated from indicated mice. (B) Schematic representation of the Emv30 provirus and sequence variations between the infectious E-MuLV isolated from NSGHPI mice (in red) or the putative Emv11 provirus from AKR mice (in black). Nonsynonomous base changes are indicated in bold. Asterisk denotes the Fv1 interaction domain within the gag capsid protein. (C) Depiction of the carboxyl terminus of the Fv1 factor encoded by the indicated alleles. Sequence analysis of the Fv1 locus in NOD mice demonstrated 100% identity with the Fv1n allele of NIH 3T3 mice. The major homology region (MHR) and critical amino acids (single letter code) are indicated (20).
Emv30-Linked Provirus Is Replication-Competent and Is Not Restricted by the Fv1 Locus in NOD Mice.
MuLV replication is effectively inhibited by an intact adaptive immune system in adult mice, but also by several mechanisms that occur at the intracellular level. For example, spontaneous inactivating mutations of the ERV provirus accumulate with time, which negatively impacts replication. A second potent inhibitor is the Fv1 restriction factor, which blocks retroviral replication by interfering with viral RNA transport to the nucleus (18). Fv1 allelic variants differentially inhibit MuLV subtypes that differ in key sequences in the Gag capsid protein.
To determine if Emv30 had incurred defects leading to its inactivation, and thus requiring recombination events to resurrect an infectious MuLV, sequence data of the Emv30 locus was compiled and compared with sequences obtained from infectious MuLV isolated from NSGHPI, and with the Emv11 and Emv2 loci of AKR and B6 inbred mice, respectively (Fig. 3B and Table S3). The Emv30 provirus contains complete ORFs in all three genes (gag, pol, env) and shows >99% homology to the infectious E-MuLV isolated from NSGHPI mice. A total of three G-to-A transitions were found in the infectious E-MuLV compared with Emv30, two of which led to conserved amino acid substitutions and one of which was silent. These mutations are not found in the replication-competent ERV isolated from Emv11, and thus their mutation would not be predicted to be necessary for replication. G-to-A transition is a frequent mutation found during retrovirus replication and has been linked to antiviral APOBEC3 activity (19). Thus, we conclude that the Emv30 provirus is intact and no recombination events were necessary for the observed replication.
High sequence identity between Emv30 (NOD) and either Emv11 (AKR) or Emv2 (B6) (98% and 99%, respectively) reflects a recent common ancestor (Fig. 3B and Table S3). The most salient difference between the different Emv proviruses was the presence of a large duplication of the transcriptional enhancer region within the LTR of the highly virulent AKR-MuLV derived from the Emv11 provirus, which was not observed in the proviral sequence of Emv30, Emv2, or the E-MuLV isolated from NSGHPI. Notably, all analyzed proviral genomes showed 100% identity within the gag coding region determining Fv1 susceptibility. Emv11 and Emv2 produce N-tropic virions and thus are sensitive to the Fv1-restriction factor isoforms encoded by the Fv1b alleles found in B6 and BALB/c mouse strains but can productively infect mice with Fv1n alleles, prototypically found in the NIH inbred mouse strain. To determine the allelic status of the Fv1 locus in NOD mice, the locus was amplified by PCR and sequenced, which confirmed an Fv1nn genotype (Fig. 3C). Thus, we can conclude that the Emv30 locus harbors a replication-competent E-MuLV that is not inhibited by Fv1 resistance in the NOD background.
PMF Xenografts, However, Not MuLV-NSGHPI Viral Isolates, Induce Myeloproliferation and Splenomegaly in NSG Mice.
The experiments described above confirm active replication of MuLV (originating from the Emv30 locus) in all NSG mice analyzed to date, but do not explain the rather high incidence of AML induction in distinct cohorts of PMF-engrafted mice. We thus examined MuLV isolated from AML tumor samples for recombination or mutational events that may influence virulence. Primers designed to flank the enhancer region were used to screen virus-infected bulk cultures or individual clones (n = 96) for LTR variants; no such duplication was observed. Sequence analysis of 24 clones also showed no sequence variations. Thus, the pathogenesis observed in the PMF-transplanted mice could not be attributed to increased virulence because of alterations in the LTR enhancer.
The generation of de novo MuLV variants through recombination with inactive polytropic ERVs (present with about 40 copies per genome) is also a common mechanism that contributes to retrovirus pathogenicity. Indeed, such recombination events were evident by the very low but reproducible ability of NSG virus isolates to infect human 293T cells, a characteristic of polytropic but not ecotropic retrovirus (Fig. 1A). Characterization of a provirus isolated from human 293T cells infected with supernatant from suspended AML splenic cells confirmed a recombinant MuLV containing polytropic pol-env sequences (Fig. S2). However, PCR amplification of genomic DNA from untreated NSG spleens demonstrated similar polytropic recombinants. Thus, we conclude that recombination events leading to a polytropic (P)-MuLV are not unique to AML mice, and thus most likely do not have a major impact on disease induction.
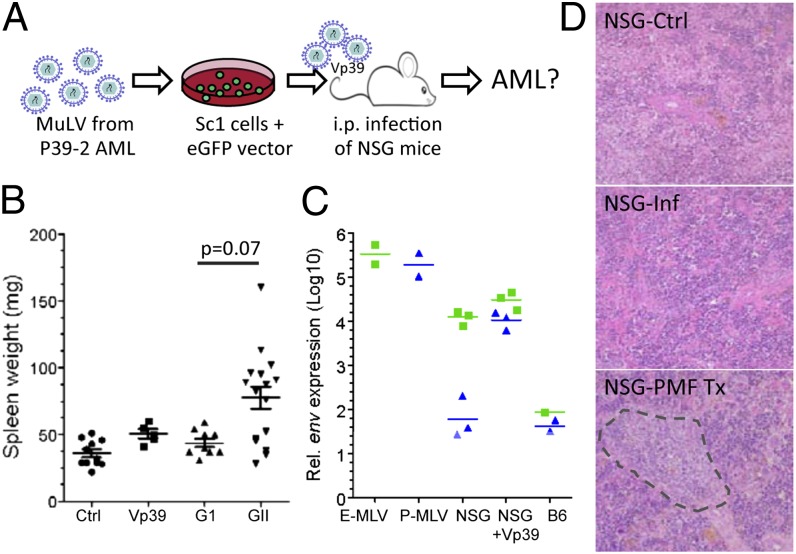
As a final proof that a unique recombination event had not occurred to induce a more virulent retrovirus, we inoculated newborn NSG mice with high-titer viral supernatants (>5 × 106 GFP-transfer-units/mL) isolated from SC1 cell cultures infected with virus from leukemic spleens (Fig. 4A). Over a 340-d observation period, no disease induction was noted. Spleen weights and the hypocellular architecture resembled that of uninfected NSG controls (Fig. 4B). Quantitative RT-PCR was performed to measure MuLV levels in infected mice. Interestingly, E-MuLV transcripts in livers of infected mice were slightly higher than the control uninfected NSG mice; however, the level of P-MuLV transcripts was increased by two orders-of-magnitude (Fig. 4C). The high level of P-MuLV in the infected mice is likely attributable to its amplification by E-MuLV pseudotyping in SC1 cells (21). Taken together, these results exclude the possibility that AML induction is caused by spontaneous generation of a more virulent E-MuLV or P-MuLV in the NSGHPI mouse strain, thus adding support to the hypothesis that the PMF xenograft contributes to AML induction.
Fig. 4.
Characterization of MuLV isolates in vivo. (A) Approach used to determine if infectious MuLVs isolated from AML cells are responsible for AML induction. (B) Spleen weights of NSG mice from untreated controls (n = 12), infected with MuLV (Vp39) isolated from AML cells (n = 5), or transplanted (Tx) with either group I (G1; n = 11) or group II (G2; n = 14) patient samples. Mean and the SEM are shown. The P value for the observed difference between spleen weights was calculated by using a two-tailed unpaired t-test. (C) Quantitative assay to determine E-MuLV and P-MuLV transcript levels in infected NSG mice. Mice were analyzed up to 340 d after infection. Quantitative RT-PCR was performed on RNA isolated from livers or, as positive controls, from SC1 cells producing E-MuLV or 293T cells producing P-MuLV. (D) Representative histology sections demonstrating splenic hypocellularity in NSG controls or infected NSG mice. In contrast, the spleen of PMF-Tx mouse is hypercellular and large foci of myeloid cells are frequently detected (H&E staining, objective 20×).
In addition to viral determinants, it is well established that host determinants that regulate availability of known target cells for transformation are also key factors in modulating MuLV pathogenicity. To determine if the PMF xenograft contributed to the leukemia induction by stimulating the myeloid compartment, mice from all NSG engrafted cohorts (i.e., each cohort receiving samples from independent PMF patients) were closely analyzed. Notably, significantly increased spleen weights, which could not be attributed directly to engrafted human cells (<1–15% of total cell number), were observed in >70% of all mice from six cohorts (group II) but not in mice from four other cohorts (group I) (Fig. 4B). All AML mice were in cohorts belonging to group II (Table S1). These results are consistent with the hypothesis that an underlying characteristic of the patient sample itself is a critical parameter in inducing host myeloproliferation. Histological analysis of spleen from these mice showed clusters of proliferating cells of the myeloid lineage (Fig. 4D). Interestingly, this was often in close association with megakaryocytes, which were of both mouse and human origin. We predict that the expanded myeloid cell population provides prime targets for MuLV-NSG infection, which consequently leads to AML induction through stochastic integration near cooperating oncogenes.
De Novo Retroviral Integration Events Contribute to Disease Induction.
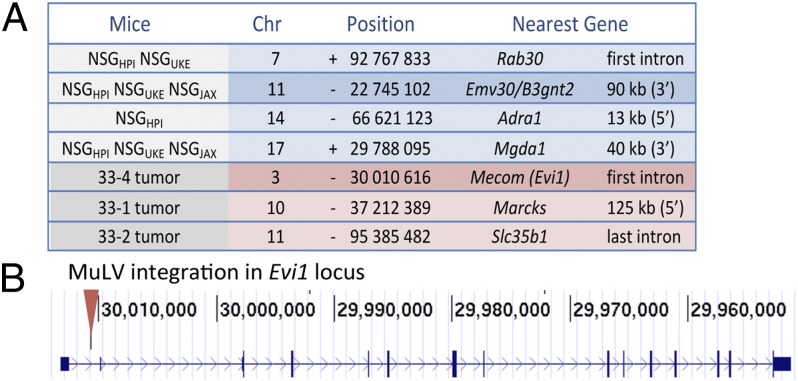
As a final proof of principle that MuLV-NSG contributed to AML induction, we cloned the proviral integration sites in three AML samples, and subsequently assessed their presence in normal tissue from different NSG strains. As shown in Fig. 5A, four integration sites were isolated that were found in normal NSGHPI tissue, including an integration site on chr11, corresponding to the original Emv30 locus. Significantly, all NSG strains analyzed (NSGHPI, NSGUKE, and NSGJAX) also shared an integration site on chr17, whereas an integration site on chr7 was found in exclusively in the NSGHPI and NSGUKE strains. Notably, three other integration sites were isolated and confirmed to be unique to AML tissue, including an integration site in the first intron of the ecotropic viral integration site 1 (Evi1) locus. Evi1 was first identified in MuLV-induced AML in mice and its human counterpart is targeted by a recurrent translocation event in human AML and by retroviral vector integrations in gene therapy trials (22).
Fig. 5.
Characterization of MuLV/ERV integration sites in NSG mice and AML samples. (A) Location of integration sites identified in untreated or AML NSG mice. Nucleotide and strand is that directly upstream of the 5′ LTR. The closest gene and relative position to the provirus are indicated. (B) Position of the MuLV integration site within the Evi1 locus mapped to the B6 genome (GRCm38/mm10 assembly), which cluster with other known retroviral integrations annotated in the Retrovirus and Transposon tagged Cancer Gene Database (http://variation.osu.edu/rtcgd). None of the other integrations mapped to known common integration sites.
Discussion
MuLV and their endogenous counterparts, ERVs, are well-established tools for unraveling mechanisms of hematological diseases. In this study, the rather unexpected induction of leukemia by MuLV replication in a xenograft model of PMF underlines the pivotal role of a permissive cellular milieu in transformation. Notably, PMF is unique among other MPNs, in that the neoplastic stem cells or their clonal derivatives directly alter the microenvironment, which in a ratcheting process leads to disease progression (23). The study presented here carries this idea one step further by demonstrating that the presence of PMF cells supports susceptibility of normal cells to transformation, which has important implications for understanding the origin and evolution of the malignant stem cell in PMF. Furthermore, this study has exposed the presence of a replicating and mutagenic ERV in immunodeficient NOD mouse models.
Mechanisms that control the detrimental effects of MuLV replication and pathogenesis during mammalian development have evolved through their long-standing coexistence. A coordinated B- and T-cell response is a well-established, pivotal defense mechanism against retroviral replication and pathogenesis (24–26). Innate immunity is also essential to control MuLV, which includes mechanisms that sense the virus and activate adaptive immunity (27), but also mechanisms that inhibit entry or transport to the nucleus, or interfere with reverse transcription accuracy and efficacy (13, 28). NSG mice lack cells of the adaptive immune system (B-, T- and NK-cells) and have several defects in classic innate functions, including an absent hemolytic complement system, reduced dendritic cell function, and defective macrophage activity (10). Our study also demonstrated that E-MuLV replication is not blocked by the Fv1 restriction factor, which is the most common and powerful restriction against MuLV in the murine system at the intracellular level (18); nor has the Emv30 provirus been subject to incapacitating mutations, reflecting its relatively new insertion into the NOD germ line and the low impact of APOCBEC3 restriction on this class of MuLVs (29). Thus, NOD mice differ strikingly from the common inbred mouse strains DBA/2, BALB/c, and B6, in which the eERV provirus is replication incompetent, restricted by the Fv1 factor, or both, respectively.
So, why is the MuLV produced by Emv30 apathogenic in NSG mice? The proximal agents of MuLV-induced disease are generally recombinant P-MuLV: viruses with an E-MuLV backbone and polytropic ERV-derived env. In addition, highly virulent P-MuLV recombinants are generated by incorporation of sequences from xenotropic ERV provirus (Xmv), in particular with the Bxv1/Xmv43 locus (30, 31), which is absent in the NOD genome (32). Thus, although infectious P-MuLV are generated in NSG mice, they are weakly pathogenic. Nevertheless, previous studies have linked the spontaneous development of T- or B-cell lymphomas in NOD-Scid mice to the Emv30 provirus (12), demonstrating the pathogenic potential of MuLVs derived from this locus. However, the transforming target cells (T and B cells) are absent in NSG mice because of inactivation of the Il2rg gene (33). Thus, the low virulence of Emv30-derived MuLV in NSG mice is likely a result of both the distinct ERV profile of the NOD genomic background, which normally contributes to the generation of virulent MuLVs, and the absence of proliferating lymphoid cells, which are susceptible targets of transformation. The concept that viral virulence is dependent on factors that modulate the number or susceptibility of host cells for transformation is clearly demonstrated by analysis of the host susceptibility factor Fv2 in Friend-MuLV disease models (34). In the PMF-xenograft NSG mouse model described here, stimulation of cells within the myeloid or stem cell compartment provides new targets for Emv30-linked transformation.
How does the PMF xenograft provide targets for ERV transformation? The most likely mechanism is stimulation of myeloproliferation by cytokines secreted by PMF cells. Earlier studies have reported increased circulating levels of several cytokines in PMF patient blood, including colony-stimulating factors, interleukins, and thrombopoietin. Indeed, several of the cytokines were shown to be prognostic for poor survival (35, 36). Notably, many of the highly expressed cytokines have been implicated in directly regulating the HSC compartment (37, 38). Thus, it is likely that cytokines—either singly or in synergy with other factors—induce HSPC expansion and myeloproliferation. Alternatively, stimulation of the myeloid compartment may be the result of an indirect effect, involving changes in the stroma cells by cytokines released by the PMF graft. Megakaryocytes and activated monocytes have been implicated as the source of cytokines that stimulate or disrupt the stroma cells that maintain the hematopoietic compartment (23, 39). It is notable that the majority of group II patients in our study, whose samples gave rise to mice with notable splenomegaly, have elevated leukocyte counts, which has been correlated with increased cytokine levels and poor prognosis (Table S1) (35). Clearly, a systematic evaluation of different cytokines released by PMF HSPCs and mature cells is needed to understand the key mechanisms that foster PMF disease and malignant transformation.
Is the observed leukemia induction relevant to the PMF disease? The malignant transformation observed in our xenograft model was induced by retrovirus transformation, as demonstrated by multiple and clonal integrations of MuLV in the tumor samples and an integration into the Evi1 locus, which has been shown to enhance transcription and promote leukemogenesis by interaction with several oncogenic pathways (22). Although post-PMF AML induction does not have a viral etiology, an underlying mechanism may be shared. The most critical observation is that leukemia induction was initiated in normal cells by paracrine signaling arising from the neoplastic cells. This finding leads to the prediction that the neoplastic and malignant stem cells can have an unrelated origin and evolve independently. This theory starkly contrasts to the more accepted model, in which neoplastic cells incur secondary mutations leading to their malignant transformation, as evidenced in leukemia models where a chronic or convert phase precedes malignant transformation (3, 40). Can such a model be envisaged in PMF transformation? The strongest support for such a model comes from mutational analysis of MPN cells. Despite the prevalence of JAK2-V617F mutations in PMF, and the widely accepted notion that they drive the neoplastic stem cell, close to half of AMLs arising from JAK2-V617V–positive PMF patients do not carry the JAK2 mutation (41, 42). Furthermore, mutations in the TET2 gene, which are implicated in AML transition, are found in hematopoietic cell clones distinct to those carrying the JAK2 mutation (43, 44). The idea that leukemic transformation may arise in a clone unrelated to the JAK2 neoplastic clone has been previously proposed (44). Our study not only supports this concept but also implicates paracrine stimulation as a contributing mechanism.
Finally, this study also should be interpreted as a warning signal to the many laboratories working with NSG mouse strains. First, de novo MuLV germ-line integrations may have serious repercussions on the genetic stability and physical reproductive fitness of these inbred strains. Second, replicating E-MuLV is likely to increase the likelihood of recombination events with polytropic or xenotropic ERV sequences, leading to recombinant MuLVs that can infect human xenografted cells. Indeed, it has recently been shown that the XMRV, a putative pathogenic human retrovirus, was generated through recombination events between two mouse ERVs and subsequently infected human tumor cells being passaged in the mouse (45). Backcrossing NSG mice to mice of related strains to eliminate Emv30 (46) would be an effective approach to eliminate unwanted infectious MuLVs in this widely used immunodeficient mouse model.
Materials and Methods
Mice and Xenotransplantation.
All NOD-related mice were obtained from The Jackson Laboratory between July 2009 and January 2013 and maintained in individually ventilated cages in specific pathogen-free facilities at the University Medical Center Hamburg-Eppendorf (UKE) or Heinrich Pette Institute (HPI). Animal experiments were approved by the Hamburg Ministry of Health and Consumer Protection. Female mice between 4 and 8 wk were used for xenotransplantations, in which HPSC isolated from PMF patient peripheral blood were introduced by intravenous injections (2.5 × 105 to 1 × 106 cells per mouse).
Patient Samples.
All patients agreed to biological material donation by informed written consent under an approved protocol from the Hamburg Ärztekammer. Details to patient clinical features are presented in Table S1.
Detailed description of the procedures used to characterize infectious MuLV, to isolate retroviral integration sites, and to assess morphological changes in engrafted or infected mice are available in SI Materials and Methods. See Table S4 for a list of primers and oligos used.
Supplementary Material
Acknowledgments
We thank M. Kuehl, U. Müller, G. Pilnitz-Stolze, S. Roscher, and members of the HPI Animal Facilities and the HPI and UKE FACS Core Facilities for excellent assistance. This work was supported in part by the Roggenbuck Foundation, German Research Foundation (FE 568/11-2), and Hamburger Krebsgesellschaft and Else-Kroner Fresenius Foundation. The HPI is financed by the German Federal Ministry of Health and the Freie und Hansestadt Hamburg.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ668269, KJ668270, and KJ668271).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401215111/-/DCSupplemental.
References
- 1.Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355(23):2452–2466. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson CH, Barroga CF, Vainchenker WP. Miscreant myeloproliferative disorder stem cells. Leukemia. 2008;22(11):2011–2019. doi: 10.1038/leu.2008.290. [DOI] [PubMed] [Google Scholar]
- 3.Sloma I, Jiang X, Eaves AC, Eaves CJ. Insights into the stem cells of chronic myeloid leukemia. Leukemia. 2010;24(11):1823–1833. doi: 10.1038/leu.2010.159. [DOI] [PubMed] [Google Scholar]
- 4.Valent P, et al. Heterogeneity of neoplastic stem cells: Theoretical, functional, and clinical implications. Cancer Res. 2013;73(3):1037–1045. doi: 10.1158/0008-5472.CAN-12-3678. [DOI] [PubMed] [Google Scholar]
- 5.Jamieson CH, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 6.Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118(7):1723–1735. doi: 10.1182/blood-2011-02-292102. [DOI] [PubMed] [Google Scholar]
- 7.Levine RL, Pardanani A, Tefferi A, Gilliland DG. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer. 2007;7(9):673–683. doi: 10.1038/nrc2210. [DOI] [PubMed] [Google Scholar]
- 8.Barbui T, et al. European LeukemiaNet Philadelphia-negative classical myeloproliferative neoplasms: Critical concepts and management recommendations from European LeukemiaNet. J Clin Oncol. 2011;29(6):761–770. doi: 10.1200/JCO.2010.31.8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 10.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7(2):118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 11.Coffin JM, Stoye JP, Frankel WN. Genetics of endogenous murine leukemia viruses. Ann N Y Acad Sci. 1989;567:39–49. doi: 10.1111/j.1749-6632.1989.tb16457.x. [DOI] [PubMed] [Google Scholar]
- 12.Prochazka M, Gaskins HR, Shultz LD, Leiter EH. The nonobese diabetic scid mouse: Model for spontaneous thymomagenesis associated with immunodeficiency. Proc Natl Acad Sci USA. 1992;89(8):3290–3294. doi: 10.1073/pnas.89.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65(21):3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10(6):395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43(1):26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins NA, Copeland NG. High frequency germline acquisition of ecotropic MuLV proviruses in SWR/J-RF/J hybrid mice. Cell. 1985;43(3 Pt 2):811–819. doi: 10.1016/0092-8674(85)90254-5. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32(1):166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 18.Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: Discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol. 2013;94(Pt 12):2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 19.Kim T, Mudry RA, Jr, Rexrode CA, 2nd, Pathak VK. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J Virol. 1996;70(11):7594–7602. doi: 10.1128/jvi.70.11.7594-7602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens A, et al. Retroviral capsid determinants of Fv1 NB and NR tropism. J Virol. 2004;78(18):9592–9598. doi: 10.1128/JVI.78.18.9592-9598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenke K, et al. Profound amplification of pathogenic murine polytropic retrovirus release from coinfected cells. J Virol. 2012;86(13):7241–7248. doi: 10.1128/JVI.00225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Métais JY, Dunbar CE. The MDS1-EVI1 gene complex as a retrovirus integration site: Impact on behavior of hematopoietic cells and implications for gene therapy. Mol Ther. 2008;16(3):439–449. doi: 10.1038/sj.mt.6300372. [DOI] [PubMed] [Google Scholar]
- 23.Lataillade J-J, et al. French INSERM and the European EUMNET Networks on Myelofibrosis Does primary myelofibrosis involve a defective stem cell niche? From concept to evidence. Blood. 2008;112(8):3026–3035. doi: 10.1182/blood-2008-06-158386. [DOI] [PubMed] [Google Scholar]
- 24.Old LJ, Boyse EA, Lilly F. Formation of cytoxic antibody against leukemias induced by Friend virus. Cancer Res. 1963;23:1063–1068. [PubMed] [Google Scholar]
- 25.Nair S, et al. Distinct roles of CD4+ T cell subpopulations in retroviral immunity: Lessons from the Friend virus mouse model. Retrovirology. 2011;8:76. doi: 10.1186/1742-4690-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young GR, et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491(7426):774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu P, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37(5):867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Wolf D, Goff SP. Host restriction factors blocking retroviral replication. Annu Rev Genet. 2008;42:143–163. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stavrou S, et al. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc Natl Acad Sci USA. 2013;110(22):9078–9083. doi: 10.1073/pnas.1217399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoye JP, Moroni C, Coffin JM. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65(3):1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoggan MD, O’Neill RR, Kozak CA. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: Characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J Virol. 1986;60(3):980–986. doi: 10.1128/jvi.60.3.980-986.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baliji S, Liu Q, Kozak CA. Common inbred strains of the laboratory mouse that are susceptible to infection by mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J Virol. 2010;84(24):12841–12849. doi: 10.1128/JVI.01863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2(3):223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 34.Ney PA, D’Andrea AD. Friend erythroleukemia revisited. Blood. 2000;96(12):3675–3680. [PubMed] [Google Scholar]
- 35.Tefferi A, et al. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: A comprehensive cytokine profiling study. J Clin Oncol. 2011;29(10):1356–1363. doi: 10.1200/JCO.2010.32.9490. [DOI] [PubMed] [Google Scholar]
- 36.Elliot MA, Yoon S-Y, Kao P, Li C-Y, Tefferi A. Simultaneous measurement of serum thrombopoietin and expression of megakaryocyte c-Mpl with clinical and laboratory correlates for myelofibrosis with myeloid metaplasia. Eur J Haematol. 2002;68(3):175–179. doi: 10.1034/j.1600-0609.2002.01534.x. [DOI] [PubMed] [Google Scholar]
- 37.de Graaf CA, Metcalf D. Thrombopoietin and hematopoietic stem cells. Cell Cycle. 2011;10(10):1582–1589. doi: 10.4161/cc.10.10.15619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuettpelz LG, Link DC. Regulation of hematopoietic stem cell activity by inflammation. Front Immunol. 2013;4:204. doi: 10.3389/fimmu.2013.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tefferi A. Pathogenesis of myelofibrosis with myeloid metaplasia. J Clin Oncol. 2005;23(33):8520–8530. doi: 10.1200/JCO.2004.00.9316. [DOI] [PubMed] [Google Scholar]
- 40.Greaves M. Cancer stem cells: Back to Darwin? Semin Cancer Biol. 2010;20(2):65–70. doi: 10.1016/j.semcancer.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Theocharides A, et al. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood. 2007;110(1):375–379. doi: 10.1182/blood-2006-12-062125. [DOI] [PubMed] [Google Scholar]
- 42.Campbell PJ, et al. Mutation of JAK2 in the myeloproliferative disorders: Timing, clonality studies, cytogenetic associations, and role in leukemic transformation. Blood. 2006;108(10):3548–3555. doi: 10.1182/blood-2005-12-013748. [DOI] [PubMed] [Google Scholar]
- 43.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 44.Beer PA, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115(14):2891–2900. doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 45.Paprotka T, et al. Recombinant origin of the retrovirus XMRV. Science. 2011;333(6038):97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serreze DV, et al. Emv30null NOD-scid mice. An improved host for adoptive transfer of autoimmune diabetes and growth of human lymphohematopoietic cells. Diabetes. 1995;44(12):1392–1398. doi: 10.2337/diab.44.12.1392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.