Significance
Although progress has been made in the characterization of p53 in regulating metabolism, very little is known about the signaling pathways involved in this regulation in response to stress in vivo. Here we show that p53 controls hepatic fatty acid oxidation in mice in response to fasting. Disruption of ribosome protein (RP)-mouse double minute (Mdm)2 binding in Mdm2C305F mice results in fasting-induced hepatosteatosis. A full-dosage of p53 and an intact RP-Mdm2-p53 pathway are required for the induction of malonyl coA decarboxylase (MCD), a critical regulator of fatty acid oxidation. Thus, the RP-Mdm2-p53 pathway functions as a key regulator of hepatic lipid homeostasis in response to nutrient deprivation stress, a function that has implications in organismal survival and tumor suppression.
Abstract
The tumor suppressor p53 has recently been shown to regulate energy metabolism through multiple mechanisms. However, the in vivo signaling pathways related to p53-mediated metabolic regulation remain largely uncharacterized. By using mice bearing a single amino acid substitution at cysteine residue 305 of mouse double minute 2 (Mdm2C305F), which renders Mdm2 deficient in binding ribosomal proteins (RPs) RPL11 and RPL5, we show that the RP–Mdm2–p53 signaling pathway is critical for sensing nutrient deprivation and maintaining liver lipid homeostasis. Although the Mdm2C305F mutation does not significantly affect growth and development in mice, this mutation promotes fat accumulation under normal feeding conditions and hepatosteatosis under acute fasting conditions. We show that nutrient deprivation inhibits rRNA biosynthesis, increases RP–Mdm2 interaction, and induces p53-mediated transactivation of malonyl-CoA decarboxylase (MCD), which catalyzes the degradation of malonyl-CoA to acetyl-CoA, thus modulating lipid partitioning. Fasted Mdm2C305F mice demonstrate attenuated MCD induction and enhanced malonyl-CoA accumulation in addition to decreased oxidative respiration and increased fatty acid accumulation in the liver. Thus, the RP–Mdm2–p53 pathway appears to function as an endogenous sensor responsible for stimulating fatty acid oxidation in response to nutrient depletion.
The dynamic process of cell growth and division is under constant surveillance. As one of the primary “gatekeepers” of the cell, p53 plays a major role in sensing and responding to a variety of internal and external stressors to maintain cellular homeostasis. In addition to its conventional roles in promoting cell cycle arrest, senescence, and apoptosis, p53 has recently been shown to regulate metabolism through the transcriptional activation of genes involved in glucose transport, glycolysis, oxidative phosphorylation (OXPHOS), and glutamine hydrolysis as well as in the activation of genes upstream of the mammalian target of rapamycin and autophagic pathways (reviewed in ref. 1). Virtually all cancers show metabolic changes that result in enhanced glucose consumption and elevated glycolytic activity known as the Warburg effect (2). Because cells continuously undergo metabolic perturbations as a result of constantly changing physiological and environmental cues such as daily feeding/fasting cycles, p53 may act in this context as a metabolic stress regulator altering cellular metabolic programs under nonlethal or “low-stress” conditions.
Recent studies have shown that inhibition of ribosomal biogenesis can activate p53 through the ribosomal protein (RP)-mediated suppression of mouse double minute 2 (Mdm2) in the RP–Mdm2–p53 stress response pathway (3). Detailed analysis revealed that several RPs require the Mdm2 central zinc finger motif for efficient Mdm2 binding. Interestingly, cancer-associated MDM2 mutations have been reported to affect the central zinc finger motif (4, 5) and can specifically disrupt RP binding (6). To study the physiological functions of the RP–Mdm2–p53 pathway, we generated mice carrying a single human cancer-associated phenylalanine substitution at cysteine residue 305, which is structurally critical for the central zinc finger of Mdm2 (Mdm2C305F). The C305F mutation specifically disrupts Mdm2 binding to RPL11 and RPL5 (6) and increases the rate at which Mdm2C305F mice develop oncogenic c-Myc–induced lymphomas (7).
Here, we study the metabolic alterations of Mdm2C305F mice in the context of acute dietary restriction and report that nutrient depletion inhibits ribosome biogenesis and induces RP–Mdm2 interaction. As a result, p53 becomes activated and induces malonyl-CoA decarboxylase (MCD), an enzyme that stimulates malonyl-CoA turnover and enhances carnitine palmitoyltransferase (CPT) activity, thereby augmenting mitochondrial fatty acid uptake and oxidative respiration. To our knowledge, our results demonstrate for the first time in an animal model that the RP–Mdm2–p53 signaling pathway is critically involved in lipid homeostasis by coordinating the appropriate physiological response to nutrient deprivation.
Results
Mdm2C305F Mice Have Altered Body Composition and Basal Metabolism.
General characterization of the Mdm2C305F mice showed that they are viable to adulthood, fertile, and exhibit no clinical or pathological symptoms. The Mdm2C305F mice gained weight at a similar rate to their WT counterparts when maintained on a normal diet (Fig. S1 A and B). Furthermore, the Mdm2C305F mice showed a similar daily food intake to WT mice (Fig. S1C). Intriguingly, although the overall body weight was similar to the WT mice, body composition analysis revealed that the Mdm2C305F mice maintain a higher percentage of body fat (Fig. 1A) and a correspondingly lower percentage of lean mass (Fig. 1B) than age-matched WT mice, indicating that the Mdm2C305F mice may harbor a predisposition to obesity. To determine the basal metabolic rate, we analyzed energy expenditure in male mice fed a normal diet. During dark and light phases, O2 consumption (Fig. 1C), CO2 production (Fig. 1D), and heat generation (Fig. S1D) were all reduced in the Mdm2C305F mice relative to WT littermates. Because basal metabolic rate reflects mitochondrial activity, we directly tested mitochondrial OXPHOS [measured as oxygen consumption rate (OCR)] and glycolysis [measured as extracellular acidification rate (ECAR)] in liver homogenates. Basal oxygen consumption, which was measured in the presence of succinate and ADP, and the maximal oxygen consumption, which was measured in the presence of the mitochondrial uncoupler carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone (FCCP), were significantly reduced in the Mdm2C305F mice (Fig. 1E). In contrast, lactate levels were higher in Mdm2C305F mouse livers (Fig. 1F), indicating that the reduced OXPHOS was compensated by enhanced glycolytic energy production in Mdm2C305F mice. Simultaneously measuring respiration and glycolysis in mouse embryonic fibroblasts (MEFs) by using a Seahorse analyzer showed similar results whereby Mdm2C305F MEFs have lower OXPHOS activity and higher glycolysis activity (Fig. 1 G and H). The observed decreased energy expenditure, increased body fat gain, and impaired mitochondrial OXPHOS in the Mdm2C305F mice suggest that the RP–Mdm2–p53 pathway plays an important role in energy homeostasis under unstressed normal feeding conditions.
Fig. 1.
Mdm2C305F mice exhibit altered body composition and basal metabolism under unstressed conditions. Percentage of fat (A) and lean mass (B) was determined in 15-wk-old WT (n = 36) and Mdm2C305F (n = 19) mice fed a normal chow diet by echo-MRI (fat percentage, P = 0.0002; lean mass percentage, P = 0.003). (C and D) Indirect calorimetry of WT (black line) and Mdm2C305F (red line) mice fed a chow diet ad libitum. (C) VO2, (D) carbon dioxide release (i.e., VCO2). The white background indicates light hours, and the shaded background indicates dark hours. (E) OCR in the liver mitochondria of WT and Mdm2C305F mice. (F) Lactate level in the livers of WT and Mdm2C305F mice (n = 3 per genotype). (G) Linear graph showing OCR in WT and Mdm2C305F MEFs in the presence of oligomycin (1 μM), FCCP (0.5 μM), antimycin A (2 μM), and rotenone (1 μM). (H) Linear graph showing the ECAR in WT and Mdm2C305F MEFs in the presence of glucose (4.5 g/L) and oligomycin (1 μM). Error bars denote SEM.
Mdm2C305F Mice Lose More Body Weight upon Fasting.
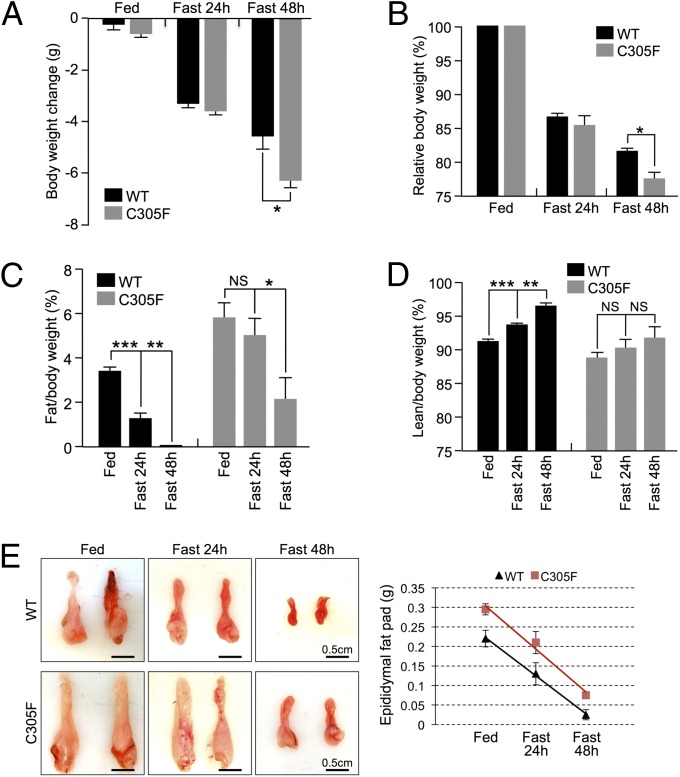
To further understand the role of the RP–Mdm2–p53 pathway in response to stress, we analyzed the Mdm2C305 mice when subjected to the dietary stress of acute fasting. Previous experiments have shown that, in laboratory animals, fasting induces a significant increase in spontaneous ambulatory activity and voluntary exercise, an effect referred to as “fasting-induced foraging activity” (8). Consistent with this observation, WT mice showed increased in-cage foraging activity upon fasting, whereas, in contrast, the Mdm2C305F mice showed no significant increase in foraging activity in response to fasting (Fig. S2). Interestingly, despite the decreased ambulatory activity of the Mdm2C305F mice, the Mdm2C305F mice lost significantly more body weight than their WT counterparts (Fig. 2 A and B). To further investigate the difference in body weight loss, we analyzed the body composition of the mice by quantitative magnetic resonance spectroscopy. Our results show that, although the Mdm2C305F mice lost more overall bodyweight, these mice were relatively resistant to fasting-induced fat loss (Fig. 2C), as they demonstrated a preference for the consumption of lean mass for their energy supply (Fig. 2D). To determine whether the inhibition of fat breakdown is caused by reduced lipolysis, we examined the epididymal fat pads of WT and Mdm2C305F mice. Consistent with higher levels of body fat, the Mdm2C305F mice showed 30% larger epididymal fat pads than the WT mice (Fig. 2E); however, fasting resulted in a similar rate of decrease in the epididymal fat pad size in the Mdm2C305F and WT mice, suggesting that lipolysis occurs at a normal rate in the mutant mice (Fig. 2E).
Fig. 2.
Fasted Mdm2C305F mice show accelerated loss of lean mass. (A–D) WT and Mdm2C305F mice were fed a chow diet ad libitum (Fed) or were fasted for 24 or 48 h (n = 5 per genotype). Body composition was determined by echo-MRI. (A) Effect of fasting on body weight change. (B) Fasted body weight normalized to body weight before fasting. (C) Percentage of fat in WT and Mdm2C305F mice under the indicated nutrient conditions. (D) Percentage of lean mass in WT and Mdm2C305F mice under the indicated nutrient conditions. (E) Changes in the size (Left) and weight (Right) of the epididymal fat pads from WT and Mdm2C305F mice under the indicated nutrient conditions. Error bars denote the SEM.
Fasted Mdm2C305F Mice Develop Hepatosteatosis.
To gain insight into the mechanisms leading to decreased body fat loss in fasted Mdm2C305F mice, we examined the livers because the liver is one of the primary organs involved in energy metabolism during fasting. Under ad libitum feeding conditions, the size and appearance of the Mdm2C305F mouse livers were similar to the livers of WT mice (Fig. 3 A and C). However, following fasting, whereas livers from the WT mice appeared normal in color and gross appearance (Fig. 3B), livers from Mdm2C305F mice were paler and displayed a mottled appearance (Fig. 3D) indicative of increased fat content. Oil red O-stained liver sections showed a small amount of lipid droplets in the livers of WT and Mdm2C305F mice under fed conditions (Fig. 3E, Top). After a 24-h period of fasting, a marked accumulation of lipid droplets in the WT and Mdm2C305F livers was observed (Fig. 3E, Middle), which is characteristic of the temporary fasting-induced accumulation of lipids in the liver (9, 10). After a 48-h period of fasting, the number and size of lipid droplets in the WT mouse livers returned to the basal level, whereas lipid droplet accumulation continued in the Mdm2C305F mouse livers (Fig. 3E, Bottom). Consistent with the lipid droplet accumulation in Mdm2C305F mouse livers, we observed a significant increase in liver triacylglyceride (TAG) levels in fasted Mdm2C305F mouse livers (Fig. 3F). These data indicate that Mdm2C305F mouse livers have an attenuated ability to metabolize fatty acids. The excess TAG accumulation in the Mdm2C305F mouse liver is consistent with hepatosteatosis, a consequence of metabolic syndrome (11). Collectively, these data suggest that the RP–Mdm2–p53 pathway is involved in maintaining hepatic lipid homeostasis.
Fig. 3.
Fasted Mdm2C305F mice develop hepatosteatosis. (A–D) WT and Mdm2C305F mice were fed a normal chow diet or were fasted for 48 h. The mice were killed, and the livers were collected. (Scale bars: 2.5 mm.) (E) Hepatic lipid droplets were visualized by oil red O staining on livers obtained from mice fed a normal chow diet or fasted for the indicated time. (F) Elevated TAG in Mdm2C305F mouse livers upon fasting (n = 3). Error bars denote SEM.
Fasting-Induced Fatty Acid Oxidation Is Impaired in Mdm2C305F Mice.
To further understand the metabolic alterations in the fasted mice, an integrated metabolomics study was conducted based on high-resolution NMR (1H NMR) spectroscopy. When fed ad libitum, spectra based on liver lysates obtained from WT and Mdm2C305F mice largely overlapped (Fig. 4A, filled symbols). Fasting resulted in a metabolic shift in the WT and Mdm2C305F mice; however, the WT and the Mdm2C305F metabolite spectra were notably different, producing distinct principal component analysis (PCA) plots (Fig. 4A, open symbols). These results suggest that fasting triggers unique global metabolic alterations in WT and Mdm2C305F mice.
Fig. 4.
Mdm2C305F mice demonstrate reduced FAO and mitochondrial dysfunction. (A) WT and Mdm2C305F mice were fed a chow diet ad libitum (Fed) or were fasted (Fast) for 48 h. Metabolomic analysis was performed on WT and Mdm2C305F mouse livers by H1-NMR. NMR spectra were acquired and were subjected to unsupervised PCA (WT, n = 4; Mdm2C305F, n = 3). (B) WT and Mdm2C305F mice were treated as indicated in A. The major metabolic perturbations associated with the cluster PCA plots were identified from orthogonal partial least-squares loading coefficient plots. The identified metabolites are shown as individual peaks. β-HB and glucose are indicated with black arrows. (C) Serum β-HB was determined in WT and Mdm2C305F mice fed a normal chow diet ad libitum or fasted for 24 or 48 h (WT, n = 10; Mdm2C305F, n = 8). (D) Fatty acid β-oxidation was examined in liver homogenates from WT and Mdm2C305F mice fed a normal chow diet ad libitum or fasted for 24 h (n = 6). The total β-oxidation activity was defined as the rate of [1-14C]palmitate conversion to CO2 and acid-soluble metabolites. (E) RCR of liver mitochondria isolated from WT and Mdm2C305F mice fed a normal chow diet ad libitum or fasted overnight (WT fed, n = 9; Mdm2C305F fed, n = 9; WT fast, n = 5; Mdm2C305F fast, n = 5). RCR was determined based on the ratio of the OCR in the presence of FCCP and the OCR in the presence of oligomycin. (F) WT and Mdm2C305F MEFs were cultured in normal DMEM or glucose-free DMEM supplemented with 10% FBS for 24 h, and then the cells were seeded into Seahorse XF24 plates (6 × 104 per well), and the ECAR was recorded upon sequential injection of glucose (4.5 g/L) and oligomycin (1 μM). The maximal glycolytic activity was determined by the addition of oligomycin. Error bars denote the SEM. *, **, and *** are used to indicate statistical significance corresponding to P values < 0.05, 0.01, and 0.001, respectively. NS indicates no significant difference.
Upon further analysis of the metabolic profiles, fasted WT and Mdm2C305F mouse livers differed significantly in the levels of key individual metabolites. Specifically, β-hydroxybutyrate (β-HB), a ketone body derived from fatty acid oxidation (FAO; β-oxidation), accumulated in WT but not Mdm2C305F mouse livers, whereas glucose, which is derived mostly from the digestion of lean tissue and muscle during prolonged starvation, was more abundant in the Mdm2C305F mouse livers (Fig. 4B). Additionally, fasted Mdm2C305F mice demonstrated high levels of muscle autophagy (Fig. S3A) and increased expression of glucose-6-phosphatase (Fig. S3B). These results are consistent with the observation that Mdm2C305F mice tend to lose more muscle mass during nutrient deprivation (Fig. 2).
β-HB accumulation is a direct consequence of FAO during prolonged fasting and can serve as a hepatic and serum biomarker of starvation (12). Ketogenesis, a process that produces β-HB during FAO (13), was fully induced in fasted WT mice as indicated by high levels of serum β-HB (Fig. 4C). In contrast, ketogenesis was reduced in fasted Mdm2C305F mice (Fig. 4C), indicative of attenuated FAO. Notably, even under fed conditions Mdm2C305F mice generated less β-HB than WT mice, suggesting that the RP–Mdm2–p53 pathway may play a role in fatty acid catabolism during daily feeding/fasting cycles.
To directly measure fatty acid metabolism, we analyzed long-chain FAO (LC-FAO) in mouse liver homogenates using [1-14C]palmitate (a 16-carbon saturated fatty acid) as a substrate. As shown in Fig. 4D, fasting resulted in a significant increase in the rate of maximal LC-FAO in WT but not in Mdm2C305F mouse livers. Accordingly, fasted WT mice showed enhanced mitochondrial OXPHOS relative to Mdm2C305F mice (Fig. 4E), and fasted Mdm2C305F MEFs demonstrated higher glycolytic activity (Fig. 4F). Altogether, our data support a model whereby the RP–Mdm2–p53 pathway regulates global metabolic homeostasis during nutrient deprivation in part through the modulation of hepatic FAO.
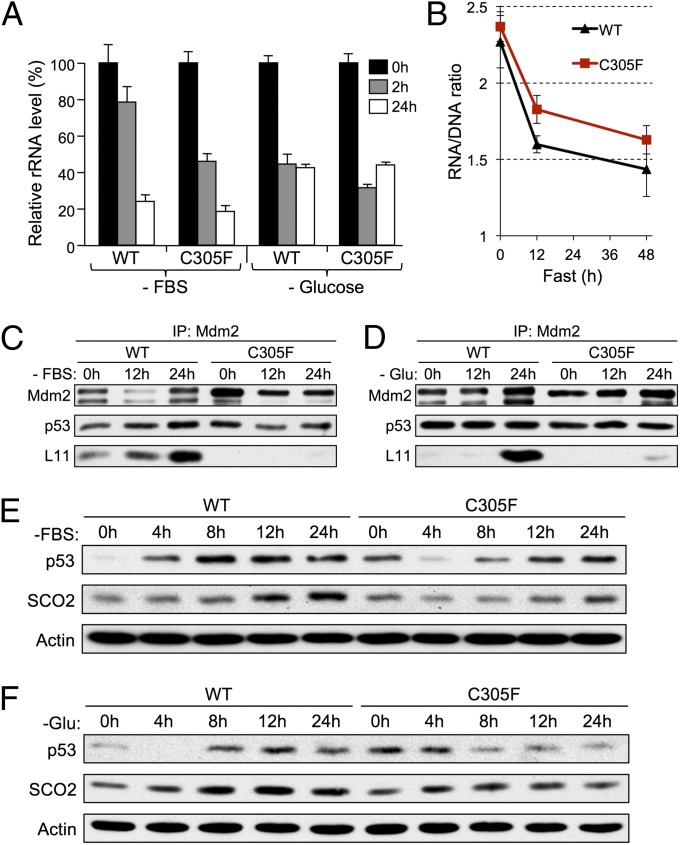
Nutrient Depletion Inhibits rRNA Synthesis, Induces RP–Mdm2 Binding, and Activates p53.
Previous studies have demonstrated that the cells grown in serum-depleted medium show increased RP–Mdm2 binding and p53 activation (14). To investigate whether nutrient depletion inhibits ribosome biosynthesis in WT and Mdm2C305F MEFs, we examined nascent rRNA levels. MEFs were pulse-labeled with [3H]methionine, after which total cellular RNA was isolated and probed for the amount of radioactivity, which was used as a surrogate marker for newly synthesized rRNA precursors. Upon serum depletion or glucose starvation, rRNA synthesis was reduced to ∼20% and 40%, respectively (Fig. 5A). The dynamics and extent of inhibition of rRNA synthesis were similar in WT and Mdm2C305F MEFs, indicating that the C305F mutation does not affect rRNA synthesis. Next, we examined the RNA/DNA ratio in mouse livers, an assay that has been used to measure rRNA synthesis in fasted rats (15). Consistent with the MEF cell data, the RNA/DNA ratio in WT and Mdm2C305F mouse livers decreased similarly during the first 12 h of fasting (Fig. 5B), indicating that nutrient deprivation rapidly and profoundly affects rRNA synthesis. Next, we examined RPL11–Mdm2 binding and p53 activation in nutrient-deprived MEFs. Serum starvation and glucose deprivation augmented RPL11–Mdm2 binding in WT but not Mdm2C305F MEFs (Fig. 5 C and D). As a result, p53 was stabilized and activated to a greater degree in WT MEFs compared with Mdm2C305F MEFs (Fig. 5 E and F). Consistent with the increase in p53 activity, SCO2, a known p53 metabolic target (16), was induced in WT but not in Mdm2C305F MEFs. We have consistently observed that the p53 protein level is higher in untreated Mdm2C305F MEFs than in WT MEFs, which was reported in one of our previous studies (7); the nature of the elevated basal level of p53 in the mutant cells remains unclear. Nonetheless, our results show that nutrient deprivation in WT and Mdm2C305F MEFs results in the inhibition of rRNA biosynthesis, which results in RPL11–Mdm2 binding and activation of p53 in WT but not Mdm2C305F cells.
Fig. 5.
Nutrient depletion inhibits rRNA synthesis, induces L11–Mdm2 binding, and activates p53. (A) WT and Mdm2C305F MEFs were pulse-labeled with [methyl-3H]methionine, and then the total RNA was isolated. The radioactivity of the RNA was measured by using a scintillation counter. The bar graph represents the average of three experiments performed in duplicate. (B) Ratio of total liver RNA to DNA during fasting. Total RNA and DNA were extracted simultaneously with TRIzol reagent from 200 mg of liver pieces from WT and Mdm2C305F mice under the indicated treatment conditions. (C and D) WT and Mdm2C305F MEFs were treated with (C) serum starvation (-FBS) or (D) glucose starvation (-Glu) for the indicated times. Cell lysates were then immunoprecipitated with anti-Mdm2 antibody (2A10) and immunoblotted for Mdm2, p53, and L11. (E and F) WT and Mdm2C305F MEFs were treated as in C and D for the indicated times. Cell lysates were analyzed by immunoblotting for total p53 and SCO2. Error bars denote the SEM.
Nutrient Depletion Induces RP–Mdm2–p53 Pathway-Dependent Activation of MCD.
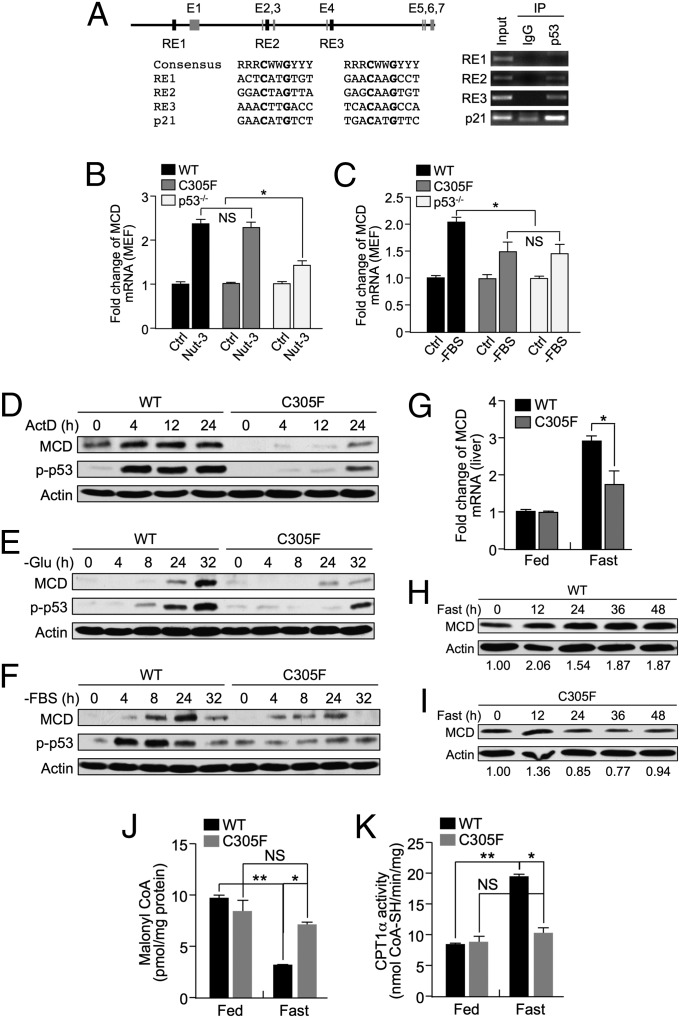
We reasoned that the phenotype displayed by the Mdm2C305F mice is likely a result of altered p53 function. By using an inducible p53ERTAM MEF cell system, we performed microarray analysis for p53 target genes. One robust candidate identified by microarray analysis was MCD (Fig. S4A). MCD catalyzes the conversion of malonyl-CoA, a key fatty acid precursor and a FAO blocker, into acetyl-CoA and carbon dioxide. A rate-limiting step to LC-FAO in the liver is the transfer of fatty acids from the cytosol into the mitochondria by CPT1α, an outer mitochondrial membrane enzyme that regulates the entry and subsequent oxidation of fatty acids by the mitochondria (17). Importantly, CPT1α is allosterically inhibited by malonyl-CoA (13, 18). Thus, MCD functions as a central regulator of LC-FAO by regulating the intracellular levels of malonyl-CoA to control the balance between lipid and glucose metabolism. To confirm the microarray results and to test the ability of p53 to directly activate MCD, a ChIP assay was performed to probe several putative p53 response elements present in the MCD promoter region (Fig. 6A). Of the three putative p53 response elements tested, RE2 and RE3 were robustly bound by p53, suggesting a direct regulatory mechanism. Furthermore, MCD transcript expression was induced in a p53-dependent manner in MEFs treated with nutlin-3 (Fig. 6B) or nutrient deprivation (Fig. 6C). Notably, in Mdm2C305F MEFs, MCD was induced by nutlin-3 to a similar extent relative to WT MEFs. However, upon nutrient deprivation, Mdm2C305F MEFs failed to induce MCD expression, indicating that the C305F mutation specifically affects nutrient stress signaling. We observed a slight induction of MCD in p53-null MEFs upon nutrient deprivation (Fig. 6C), as well as a slight increase in p53-null MEFs treated with nutlin-3 (Fig. 6B), suggesting that MCD can be partially induced in a p53-independent manner. Furthermore, MCD protein expression was greatly induced in WT MEFs but only slightly induced in Mdm2C305F MEFs under conditions that inhibit ribosome biosynthesis including actinomycin D (Fig. 6D) and nutrient depletion (Fig. 6 E and F). Consistent with a p53-dependent effect, MCD induction correlated with levels of total p53 (Fig. 5 E and F) and S-18 phospho-p53 (Fig. 6 D–F). We observed a similar p53-dependent induction of MCD in the p53 isogenic human cancer cell line HCT116 (Fig. S4B).
Fig. 6.
Nutrient depletion induces RP–Mdm2–p53 pathway-dependent activation of MCD and augments CPT1 activity. (A) p53 binds to the Mcd gene promoter. (Left) Location and sequence of putative p53 response elements (RE1, RE2, and RE3). (Right) p53 binds to RE2 and RE3 as determined by a ChIP assay. The p53 response element found within the p21 promoter was included as a positive control. (B and C) Quantitative real time-PCR (qRT-PCR) analysis of MCD from WT, Mdm2C305F, and p53−/− MEFs treated with (B) nutlin-3 (10 μM) for 12 h or (C) serum starvation for 6 h. (D–F) Ribosomal biosynthesis stress activates p53 and induces MCD in MEF cells. WT and Mdm2C305F MEFs were treated with (D) 5 nM actinomycin D, (E) glucose starvation (-Glu), or (F) serum starvation (-FBS) for the indicated time, and the total cell lysates were analyzed by immunoblotting. (G) qRT-PCR analysis of MCD mRNA expression in livers of WT and Mdm2C305F mice fed a chow diet ad libitum (Fed) or fasted (Fast) for 12 h (n = 3 for each treatment). (H and I) WT and Mdm2C305F mice were fasted for the indicated amount of time. Immunoblotting shows the MCD expression level in liver homogenates. Quantitative relative expression of MCD was normalized to β-actin. (J) Malonyl-CoA levels in livers isolated from WT and Mdm2C305F mice (n = 3) fed a normal chow diet or fasted for 12 h were determined by HPLC and MS. (K) WT and Mdm2C305F mice were fed a normal chow diet or were fasted for 12 h (n = 3). CPT1α activity from liver homogenates was then measured spectrophotometrically. Error bars denote the SEM. *, **, and *** are used to indicate statistical significance corresponding to P values < 0.05, 0.01, and 0.001, respectively. NS indicates no significant difference.
To determine the effect of nutrient deprivation on MCD expression in mice, young male mice were fasted, after which livers were collected and examined for MCD mRNA and protein expression. MCD mRNA (Fig. 6G) and protein (Fig. 6 H–I) expression was induced in the livers of fasted WT mice but not in Mdm2C305F mice. Furthermore, MCD protein levels peaked around 12 h of fasting, which is a characteristic expression pattern for genes that are involved in FAO (19). MCD catalyzes the degradation of malonyl-CoA, thereby activating CPT1α and increasing liver LC-FAO (20). Consistent with the pattern of MCD induction in fasted mice, we observed a significant decrease of malonyl-CoA (Fig. 6J) and an increase in CPT1α activity (Fig. 6K) in WT but not Mdm2C305F mouse livers. Altogether, our data indicate that the RP–Mdm2–p53 pathway regulates nutrient depletion stress-stimulated FAO at least partially through the p53-dependent induction of MCD. The resulting induction of MCD functions to decrease intracellular malonyl-CoA levels, thereby increasing CPT1α activity.
Reduction of p53 Dosage Mimics Hepatosteatotic Phenotype Observed in Mdm2C305F Mice.
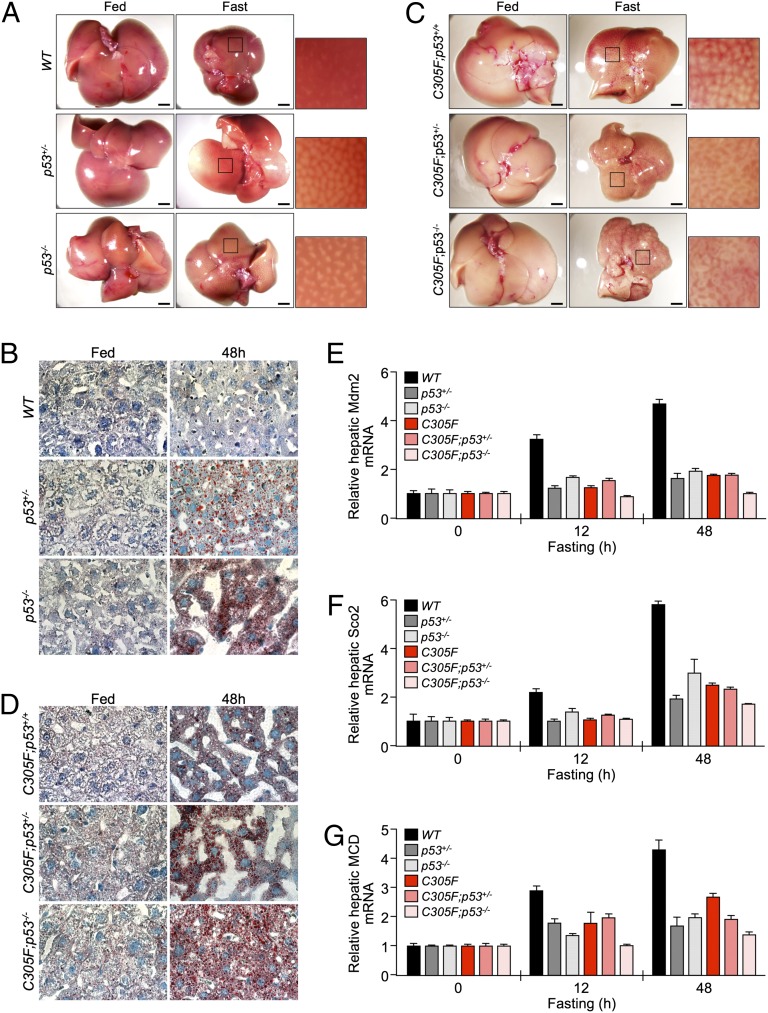
To confirm that the hepatosteatotic phenotype observed in fasted Mdm2C305F mice is a p53-dependent phenomenon, p53+/− and p53−/− mice were fasted, after which the livers were harvested and analyzed for lipid droplet accumulation. As shown in Fig. 7A, livers from fasted p53+/− and p53−/− mice showed a mottled appearance reminiscent of the appearance of livers in fasted Mdm2C305F mice. Oil red O staining of liver sections obtained from fasted p53+/− and p53−/− mice also resembled fasted Mdm2C305F mouse livers with respect to the accumulation of lipid droplets (Fig. 7B). To determine whether there is an additive effect between the Mdm2C305F mutation and p53 deletion, we bred Mdm2C305F mice into p53+/− and p53−/− backgrounds to examine the effect of p53 deletion on fasting-induced hepatosteatosis. Fasted Mdm2C305F;p53+/− and Mdm2C305F;p53−/− mice exhibited a mottled appearance (Fig. 7C) and lipid droplet accumulation (Fig. 7D) that was similar to the fasted Mdm2C305F mice, suggesting that the observed hepatosteatotic phenotype from fasted Mdm2C305F mice is p53-dependent. This observation was further corroborated based on p53 target gene expression in mouse liver extracts. We observed a slight but noticeable increase in liver lipid staining in Mdm2C305F;p53+/− and Mdm2C305F;p53−/− livers compared with p53+/− and p53−/− livers, suggesting that there is a possible p53-independent function for the Mdm2C305F mutation in the regulation of liver lipid metabolism. Transcript levels from the p53 target genes Mdm2, Sco2, and Mcd were significantly induced in fasted WT mouse livers but were only slightly induced in livers from p53+/−, p53−/−, Mdm2C305F, Mdm2C305F;p53+/−, and Mdm2C305F;p53−/− mice (Fig. 7 E–G). Importantly, the degree by which the gene expression of Mdm2, Sco2, and Mcd was reduced was similar among all five mutant mice when fasted, suggesting that reducing the p53 dosage by half is sufficient to attenuate much of the effect observed from fasting-induced p53 activation.
Fig. 7.
Reduction of p53 dosage mimics hepatosteatotic phenotype observed in fasted Mdm2C305F mice. (A and C) Representative images of livers harvested from mice with the indicated genotype fed a normal chow diet or fasted for 48 h. (Scale bars: 2.5 mm.) (B and D) Hepatic lipid droplets were visualized by oil red O staining on liver sections obtained from mice with the indicated genotypes fed a normal chow diet or fasted for the indicated amount of time. (E–G) qRT-PCR analysis of hepatic Mdm2 (E), SCO2 (F), and MCD (G) mRNA expression in mice with indicated genotypes fed a chow diet ad libitum (Fed) or fasted (Fast) for the indicated amount of time (n = 3 for each treatment). Error bars denote the SEM.
Discussion
Housekeeping Function for the RP–Mdm2–p53 Pathway in Energy Homeostasis.
The results presented here demonstrate that, in mice, nutrient restriction elicits a p53 response that is mediated by the RP–Mdm2 interaction. Moreover, the RP–Mdm2 interaction is essential for regulating hepatic metabolism for efficient energy utilization. Starvation-mediated p53 induction requires an intact RP–Mdm2–p53 pathway, as demonstrated by a lack of p53 activity following nutrient deprivation in the RP-binding deficient Mdm2C305F mutant. As a transcription factor, p53 serves as a point of convergence responsible for integrating a variety of upstream stress signals, including DNA damage, oncogene overexpression, hypoxic insults, and the inhibition of ribosome biosynthesis. Much attention has been devoted to dissecting how these upstream signals trigger p53 activation and how p53 modulates the expression of downstream genes to induce growth arrest, senescence, and apoptosis. Recent studies have pointed to the regulation of energy metabolism as a new theme for p53 function and have provided explanations to many previously puzzling observations. One such observation is that p53 is found almost throughout the animal kingdom, including in many ancient organisms predating the appearance of cancer (21). A plausible idea for the widespread presence of p53 in animals could be that p53 initially evolved as a regulator of energy homeostasis and later evolved to assume a role as a general stress sensor. In the present study, Mdm2C305F mice are indistinguishable from WT mice in terms of growth and development; however, even under normal unstressed conditions, Mdm2C305F mice clearly show altered basal metabolism (Fig. 1). We have found that p53−/− MEFs accumulate more lipid droplets than WT MEFs when cultured in regular medium (Fig. S5A). Furthermore, when fed with normal chow diet, the livers from p53−/− mice contain more lipid droplets than the livers from WT mice (Fig. S5B). Whether p53 acts as a stress sensor under day-to-day “unstressed” conditions depends on how “stress” is defined. We speculate that periodic feeding and fasting cycles in mice even fed ad libitum could generate small fluctuations in the rate of ribosome biosynthesis, resulting in the initiation of a signal that is relayed through the RP–Mdm2 interaction to p53. Oscillations in p53 activity can contribute to fine-tuning the metabolic response according to the energy requirements of the organism; fasting represents an extreme case of the normal energy fluctuation that occurs between meals. In the sense of responding to the daily feeding/fasting cycle, p53 can be viewed as a housekeeping gene responsible for maintaining the energy balance to favor fitness and survival.
RP–Mdm2–p53 Pathway Controls FAO Under Nutrient Shortage Stress.
p53 regulates metabolism through a number of pathways; however, the complexity of p53-dependent metabolic regulation remains incompletely understood (22). Several studies have shown that p53 can reduce overall glycolytic function through mechanisms including the suppression of glucose transporters GLUT1/4 (23), the inhibition of phosphoglycerate mutase activity (24), and the induction of TIGAR (25). A recent study has shown that, in response to nutrient stress, p53 up-regulates FAO by inducing the expression of Lpin (26). To complement the function of inhibiting glycolysis, p53 increases OXPHOS through the transactivation of SCO2, an essential component of mitochondrial complex IV (16), and glutaminase 2, which promotes ATP production and suppresses oxidants (27, 28). Mitochondrial FAO plays a critical role in generating ATP when the glucose supply is low. Given the overall protective function of p53 in an organismal context, it is not surprising that p53 promotes FAO during glucose starvation (29). By using the Mdm2C305F mouse model, we have shown that the RP–Mdm2–p53 pathway plays a central role in sensing nutrient depletion stress and in modulating energy output. Based on our results, we propose a model in which changes in nutrient availability impinge on ribosome biosynthesis, leading to an increase in RP–Mdm2 interaction and subsequent activation of p53. By inducing the expression of MCD, a key enzyme for malonyl-CoA turnover and CPT1α activation, p53 promotes long-chain fatty acid uptake by the mitochondria. Together with the induction of SCO2, p53 promotes FAO and maintains metabolic homeostasis during nutrient shortage stress (Fig. 8).
Fig. 8.
RP–Mdm2–p53 pathway forms a crucial link between nutrient deprivation and cellular energy homeostasis. Nutrient deprivation induces lipolysis in adipocytes generating free fatty acids (FFAs), which enter the liver to serve as a direct energy source or as a precursor for ketone body formation. In normal mouse liver, nutrient depletion causes ribosomal stress by inhibiting rRNA biosynthesis, which in turn inhibits Mdm2 (indicated by the smaller letters) and activates p53 to induce MCD expression. MCD facilitates the transport of FFAs into the mitochondrial matrix through CPT1α to be oxidized to acetyl-CoA by β-oxidation. Acetyl-CoA is then oxidized via the TCA cycle to produce energy and “reducing equivalents” (NADH and FADH2), which are used for ATP synthesis by OXPHOS. p53 also induces the expression of SCO2, a crucial component of the electron transport chain, to promote OXPHOS. Together, the action of the RP–Mdm2–p53 pathway maintains liver lipid homeostasis during the fasting state. However, in Mdm2C305F mouse livers, ribosomal stress in the form of nutrient deprivation cannot activate p53 (grayed out), which results in the accumulation of FFAs in the liver and hepatosteatosis.
The present study focuses on defining how nutrient deprivation influences cellular responses through the RP–Mdm2–p53 pathway. However, overnutrition represents another type of nutrient stress, and whether the RP–Mdm2–p53 pathway plays a role during nutrient overload is an interesting question for future studies. Given the emergence of obesity as a worldwide epidemic, it seems equally imperative to determine the cellular stress responses associated with nutrient overabundance. Future research focused on clarifying the role of the RP–Mdm2–p53 pathway as a metabolic stress response pathway and additional roles that this pathway might play in the p53-dependent modulation of nutrient partitioning and energy production provides an exciting opportunity to expand upon the myriad roles of p53 in regulating cellular homeostasis.
Experimental Procedures
Animals and Treatments.
WT and Mdm2C305F mice were bred and maintained in standard cages on a 12:12 h light/dark cycle with ad libitum access to regular chow and water. All mice were handled in strict accordance with our protocol (10-045) as approved by the institutional animal care and use committee at the University of North Carolina Animal Care Facility. Body weight was measured weekly. All mice used for experiments were between 9 and 11 wk of age unless otherwise specified. For the fasting studies, mice were housed individually with ad libitum access to water for 12 h (7:00 PM to 7:00 AM), 24 h, or 48 h.
Metabolic Analysis.
Body water, body fat, and lean mass were analyzed by using an EchoMRI-100 quantitative magnetic resonance whole-body composition analyzer (Echo Medical Systems) according to the instructions from the manufacturer.
For metabolic studies, mice were placed individually in the metabolic chambers of an Oxymax system (Columbus Instruments) for 120 h, and readings were taken after a 24-h acclimation period. Physical activity, food consumption, OCR (VO2), carbon dioxide production rate (VCO2), and respiratory exchange ratio were obtained. VO2, VCO2, and respiratory exchange ratio were measured every 26 min during a 48-h period before or after fasting and were normalized to the body weight. Activity was measured on x, y, and z axes by counting interruptions in the infrared beams.
Hepatic TAG and β-HB (Stanbio) levels were measured by using colorimetric commercial kits according to the manufacturer’s instructions. Malonyl-CoA content was assessed from whole liver extracts. After separation, purification, and elution, lipid metabolite extracts were separated by HPLC, and individual and total lipid species were analyzed by liquid chromatography and tandem MS.
Metabolomic Analysis.
Freshly isolated mouse livers were used for metabolomic analysis. SI Experimental Procedures provides a more detailed protocol.
Mitochondrial Activity and Glycolytic Flux.
OCR and ECAR were evaluated by using a Seahorse XF24 Analyzer (Seahorse Bioscience). For MEFs, 6 × 104 cells were seeded 24 h before the assay. The basal level of OCR and ECAR were measured in XF assay medium containing 4.5 g/L glucose followed by the addition of oligomycin (1 μM), which allowed for the assessment of the maximal glycolytic capacity (i.e., ECAR) or by the sequential addition of oligomycin (1 μM), FCCP (0.5 μM; maximal OXPHOS), and antimycin A (2 μM) plus rotenone (1 µM) to determine the OCR. For animal experiments, mitochondria were isolated from mouse livers, and 7.5 µg of mitochondria was transferred into each well of an XF24 plate immediately after extraction. Then, the OCR and ECAR were measured at basal levels in the presence of succinate (5 mM) as well as after the sequential addition of ADP (100 μM), oligomycin (1 μM), FCCP (0.5 μM), and antimycin A (2 μM). Mitochondrial respiration control ratio (RCR) was evaluated based on the OCR-FCCP/OCR-oligomycin ratio.
Oil Red O Staining.
Mouse liver tissues were fixed in 10% (vol/vol) neutral formalin for 24 h and then were submerged in 20% sucrose for 2–3 d. The liver pieces were then frozen in optimal cutting temperature compound, and 5-µm-thick tissue sections were cut. Oil red O staining was conducted according to the standard protocol used in the Histology Research Core Facility at the University of North Carolina (UNC).
Fatty Acid β-Oxidation in Liver.
Freshly isolated liver was homogenized with a motor-driven Teflon pestle and glass mortar in ice-cold SET buffer (250 mM sucrose, 1 mM EDTA, 10 mM Tris⋅HCl, and 2 mM ATP, pH 7.4).Two milligrams of each liver homogenate was incubated with BSA-conjugated palmitate (200 µM sodium palmitate plus 8 µM [1-14C]palmitate at a final concentration of 0.1 µCi per reaction mixture) in a mixture containing 125 mM sucrose, 24 mM K2HPO4, 200 mM KCl, 2.5 mM MgCl2, 2.5 mM l-carnitine, 0.25 mM malic acid, 20 mM Tris⋅HCl, 2.5 mM DTT, 0.25 mM NAD+, 4 mM ATP, and 0.125 mM CoA (pH 7.4) at 37 °C for 30 min in a customized two-well system: one well contained the reaction mixture with the liver homogenates, and the adjoining well contained 1 N NaOH. The reaction was terminated by adding 70% perchloric acid to the assay well, and then the plate was incubated for 1 h to drive the CO2 into the NaOH-containing well. Radioactivity of acid-soluble metabolites in the supernatant of the reaction mixture and CO2 was determined by liquid scintillation. FAO was quantified by using the following formula: [(dpm − BL)/SA]/[gram of liver (wet weight) × time (in hours) of reaction mixture incubation], where dpm stands for disintegrations per minute, BL represents the disintegrations per minute of the blank wells, and SA is the fatty acid-specific radioactivity.
Determination of CPT1 Activity.
The activity of CPT1 was measured spectrophotometrically by using a modified version of a previously described method (30). In brief, a 50-mg liver piece was homogenized in a buffer (0.25 M sucrose, 1 mM EDTA, and 10 mM Tris⋅HCl, pH 7.5) supplemented with protease inhibitor and PMSF, followed by centrifugation at 300 × g at 4 °C for 10 min, after which the supernatant was further purified by a second centrifugation at 12,000 × g at 4 °C for 15 min to pellet the mitochondria. Fifty microliters of the precleared supernatant was mixed with 50 μL of Tris-5,5′-dithio-bis-(2-nitrobenzoic acid) buffer [116 mM Tris, 2.5 mM EDTA, 2 mM Tris-5,5′-dithio-bis-(2-nitrobenzoic acid), and 0.2% Triton X-100, pH 8.0] with or without malonyl-CoA (10 µM) in individual wells of a 96-well plate. The mixture was preincubated for 5 min at room temperature, and then 50 μL of 1 mM palmitoyl-CoA was added to the wells. The reaction was initiated by adding 5 μL of l-carnitine solution (1.2 mM dissolved in 1 M Tris, pH 8.0), and the release of coenzyme A (CoA-SH) from palmitoyl-CoA was followed spectrophotometrically at 412 nm for 180 s by using a SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices). A CoA-SH (Sigma-Aldrich) standard curve (1–200 nmol per well) was generated. The activity of CPT1, defined as the amount of nmol of CoA-SH released per minute per milligram protein, was calculated by subtracting CPT2 activity (with malonyl-CoA) from the total CPT activity (without malonyl-CoA).
Microarray Analysis.
Microarray analysis was performed as previously described (31). Briefly, total RNA was isolated from MEFs that were untreated or were treated with 4-hydroxytamoxifen for 24 h by using an RNeasy kit (Qiagen). Total RNA was amplified and labeled, and microarray hybridization was performed at the UNC genomics and bioinformatics core facility. Genes that were significantly up- or down-regulated were identified by using significance analysis of microarrays.
ChIP Assay.
A ChIP assay was performed by using the QuikChIP kit (IMGENEX) according to the manufacturer’s suggested protocol. Briefly, MEFs were seeded in 10-cm dishes at a density of 1.5 × 104 cells per square centimeter. Twelve to twenty-four hours after plating, cells were cross-linked with formaldehyde (1%) at 37 °C for 10 min and incubated with glycine (0.125 M) for 5 min. Cells were washed with ice-cold PBS solution and lysed in SDS lysis buffer. Cell lysates were sonicated four times for 10 s each time with a Branson Sonifier. Insoluble material was removed by centrifugation at 12,000 × g at 4 °C for 10 min. Chromatin extracts were precleared with salmon sperm DNA/protein A/G agarose and then were incubated with a goat polyclonal antibody specific for p53 (FL393; Santa Cruz) or IgG control (Life Technologies) overnight at 4 °C. Reactions were then incubated for 1 h with salmon sperm DNA/protein A/G agarose beads. The beads were then washed several times, and the chromatin complex was eluted with the provided elution buffer. The eluates were further incubated at 65 °C overnight in the presence of 200 mM of NaCl to reverse the crosslinks followed by incubation with an RNase A solution and then a proteinase K solution. The DNA was purified by phenol/chloroform extraction. PCR was then performed in the presence of 5% DMSO. The primers used for the PCR amplification step were as follows: RE1 forward, 5′-CTGGACTGGCTGGCTCTTGTCC-3′; RE1 reverse, 5′-CTCTGCCTCGCTTCCGGCTC-3′; RE2 forward, 5′-GTGCACTCCAGGAAGCCCCTAG-3′; RE2 reverse, 5′-CCTCCTGCCTCGGCCTTCC-3′; RE3 forward, 5′-CCTTACAGGCCAAGCAAACACTTGAG-3′; RE3 reverse, 5′-GTGATAGCACGGACTCCCAGAATAC-3′; p53 (p21Cip1) forward, 5′-GAGACCAGCAGCAAAATCG-3′; p53 (p21Cip1) reverse, 5′-CAGCCCCACCTCTTCAATTC-3′.
Statistical Analysis.
Data were analyzed by using Graph Pad 5.0 software. Repeated-measures ANOVA was performed to evaluate the differences in O2 consumption, CO2 production and heat generation between WT and Mdm2C305F mice (SAS 9.3; SAS Institute). P values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Yue Xiong, Hilary Clegg, Koji Itahana, Yoko Itahana, Kim Gooding, and Brittany Daughtry for their helpful advice and technical assistance. This research was supported by National Institutes of Health Grants CA100302, CA127770, and CA167637; an American Cancer Society Grant (to Y.Z.), UNC Center for Environmental Health and Susceptibility Grant P30ES010126 (to J.M.); fellowships from the UNC Genetics and Molecular Biology Training Grant (to P.L. and L.T.); a Career Award in Biomedical Science from the Burroughs Wellcome Fund (to Y.Z.), a Howard Temin Award from the National Cancer Institute (to Y.Z.); a Scholar Award from the Leukemia and Lymphoma Society (to Y.Z.); and National Institutes of Health Grant DK056350 to the UNC Chapel Hill Nutrition Obesity Research Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315605111/-/DCSupplemental.
References
- 1.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9(10):691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlott T, et al. Point mutations and nucleotide insertions in the MDM2 zinc finger structure of human tumours. J Pathol. 1997;182(1):54–61. doi: 10.1002/(SICI)1096-9896(199705)182:1<54::AID-PATH815>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 5.Tamborini E, et al. Analysis of the molecular species generated by MDM2 gene amplification in liposarcomas. Int J Cancer. 2001;92(6):790–796. doi: 10.1002/ijc.1271. [DOI] [PubMed] [Google Scholar]
- 6.Lindström MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol. 2007;27(3):1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macias E, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18(3):231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overton JM, Williams TD. Behavioral and physiologic responses to caloric restriction in mice. Physiol Behav. 2004;81(5):749–754. doi: 10.1016/j.physbeh.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 9.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: The PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96(13):7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjornvad CR, Elnif J, Sangild PT. Short-term fasting induces intra-hepatic lipid accumulation and decreases intestinal mass without reduced brush-border enzyme activity in mink (Mustela vison) small intestine. J Comp Physiol B. 2004;174(8):625–632. doi: 10.1007/s00360-004-0452-2. [DOI] [PubMed] [Google Scholar]
- 11.Machado M, Cortez-Pinto H. Non-alcoholic steatohepatitis and metabolic syndrome. Curr Opin Clin Nutr Metab Care. 2006;9(5):637–642. doi: 10.1097/01.mco.0000241677.40170.17. [DOI] [PubMed] [Google Scholar]
- 12.Wieland O. Ketogenesis and its regulation. Adv Metab Disord. 1968;3:1–47. doi: 10.1016/b978-1-4831-9943-6.50007-4. [DOI] [PubMed] [Google Scholar]
- 13.McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 14.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23(12):2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch CA, Hiatt HH. Turnover of liver ribosomes in fed and in fasted rats. J Biol Chem. 1966;241(24):5936–5940. [PubMed] [Google Scholar]
- 16.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 17.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281(49):37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 18.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 19.Lane MD, Wolfgang M, Cha SH, Dai Y. Regulation of food intake and energy expenditure by hypothalamic malonyl-CoA. Int J Obes (Lond) 2008;32(suppl 4):S49–S54. doi: 10.1038/ijo.2008.123. [DOI] [PubMed] [Google Scholar]
- 20.Chien D, Dean D, Saha AK, Flatt JP, Ruderman NB. Malonyl-CoA content and fatty acid oxidation in rat muscle and liver in vivo. Am J Physiol Endocrinol Metab. 2000;279(2):E259–E265. doi: 10.1152/ajpendo.2000.279.2.E259. [DOI] [PubMed] [Google Scholar]
- 21.Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: Gazing through an evolutionary lens. Nat Rev Cancer. 2009;9(10):758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 22.Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med (Berl) 2011;89(3):237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64(7):2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 24.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65(1):177–185. [PubMed] [Google Scholar]
- 25.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 26.Assaily W, et al. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44(3):491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 27.Hu W, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107(16):7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA. 2010;107(16):7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzzai M, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67(14):6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 30.Derdak Z, et al. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J Hepatol. 2013;58(4):785–791. doi: 10.1016/j.jhep.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deisenroth C, Thorner AR, Enomoto T, Perou CM, Zhang Y. Mitochondrial Hep27 is a c-Myb target gene that inhibits Mdm2 and stabilizes p53. Mol Cell Biol. 2010;30(16):3981–3993. doi: 10.1128/MCB.01284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.