Significance
The organization of metazoan membranes into functional domains is a key feature of their physiology. A subset of these domains, known as “membrane rafts,” has been implicated in a large variety of cellular processes; however, the molecular mechanisms by which raft domains regulate cell function remain elusive. Here, we demonstrate that membrane raft association is a necessary and sufficient sorting signal for cell-surface localization of a specific single-pass membrane protein, linker for activation of T-cells, and that this raft association is governed by the properties of the protein transmembrane domain. These results begin to define the physical bases for protein raft affinity and establish lateral membrane domains as mediators of protein sorting in mammalian cells.
Keywords: membrane domain, endocytosis, trafficking, phase separation, microdomains
Abstract
The lipid raft hypothesis proposes lateral domains driven by preferential interactions between sterols, sphingolipids, and specific proteins as a central mechanism for the regulation of membrane structure and function; however, experimental limitations in defining raft composition and properties have prevented unequivocal demonstration of their functional relevance. Here, we establish a quantitative, functional relationship between raft association and subcellular protein sorting. By systematic mutation of the transmembrane and juxtamembrane domains of a model transmembrane protein, linker for activation of T-cells (LAT), we generated a panel of variants possessing a range of raft affinities. These mutations revealed palmitoylation, transmembrane domain length, and transmembrane sequence to be critical determinants of membrane raft association. Moreover, plasma membrane (PM) localization was strictly dependent on raft partitioning across the entire panel of unrelated mutants, suggesting that raft association is necessary and sufficient for PM sorting of LAT. Abrogation of raft partitioning led to mistargeting to late endosomes/lysosomes because of a failure to recycle from early endosomes. These findings identify structural determinants of raft association and validate lipid-driven domain formation as a mechanism for endosomal protein sorting.
Recent advances in superresolution microscopy (1), lipid analysis (2, 3), and plasma membrane (PM) isolation (4, 5) have confirmed the coexistence of lipid-driven, fluid domains in biological membranes. The relatively ordered domains, known as “membrane rafts,” have been proposed to be involved in protein sorting (6), viral/pathogen trafficking (3, 7), and PM signaling in a variety of contexts (8). However, despite the increasing evidence confirming the existence of dynamic, nanoscopic membrane rafts, the functional consequences of this phenomenon remain speculative because of the limitations of the previously used methods for defining raft association, i.e., the resistance of membrane components to solubilization by nonionic detergents (9).
Lipid-mediated domains have been implicated as a mechanism for protein sorting in the latter stages of the secretory pathway (trans-Golgi network to the PM) (2, 6, 10–12), with analogous pathways mediating endosomal sorting/recycling (13, 14). Raft lipids (i.e., sterols and sphingolipids) are significantly enriched at the PM (15–17), and recent observations confirm that these lipids also are enriched in sorting vesicles destined for the PM (2, 11). For proteins, several specific cytosolic signals exist for adapter/coat-mediated sorting between cellular organelles (18); in parallel, protein–lipid interactions through hydrophobic transmembrane domains (TMDs) also have been shown to regulate trafficking. For example, a strong correlation exists between the TMD length of bitopic proteins and their organelle specificity (19, 20), with longer TMDs targeting proteins to the PM and shorter TMDs found in the endoplasmic reticulum (ER), Golgi apparatus, and endocytic organelles. These findings suggest cargo sorting in the secretory and endocytic pathways, with proteins containing longer TMDs, together with sphingolipids and cholesterol, being specifically trafficked to the PM, although the mechanism for this observation remains unresolved.
One possibility for sorting of specific lipid classes along with proteins containing longer TMDs is lateral segregation and coalescence of ordered domains, followed by either domain-induced (21) or cytoskeleton-assisted (22) budding of raft-enriched transport vesicles. Proteins using this “raft pathway” would not require cytosolic sorting signals but rather would be recruited to transport vesicles by their raft affinity, i.e., their propensity to interact with specific lipids, ordered domains, or other raft-embedded proteins. Because ordered phases in lipid model systems consistently have been shown to be 0.6–1.5 nm thicker than disordered domains (23, 24), raft-associated transmembrane (TM) proteins would be predicted to have longer TMDs. TMD length-dependent protein sorting between coexisting lipid domains has been addressed experimentally only recently by measuring partitioning of an oligomeric toxin (perfringolysin O) with multiple (35–40) TM segments in synthetic, phase-separated liposomes (25). Whether these observations extend to single-pass TM proteins in biological membranes is unknown.
To evaluate the role of lipid-driven raft domains as a mechanism for subcellular protein sorting, we quantitatively compared the raft association of 30 TM protein variants with their subcellular localization. To quantify raft partitioning of the constructs comprising single-pass TM proteins with varying TMD lengths and sequences, we used giant PM vesicles (GPMVs). GPMVs are cell-detached PM blebs whose protein (26) and lipid (27) diversity mirrors that of the native PM. These PM vesicles separate into coexisting liquid phases (4) with different order (28), which recruit membrane components in accordance with their predicted raft affinity, i.e., saturated lipids, glycosphingolipids (29), glycosylphosphatidyl inositol-anchored proteins (4), and palmitoylated proteins (30) partition to the ordered phase, denoted here as the “raft phase.” Most importantly, these vesicles provide a platform for repeatable, direct, and quantitative analysis of raft partitioning (30), allowing investigation of the structural determinants of raft association and its effect on protein function. We find that perturbation of raft partitioning by three independent means (decreasing TMD length, mutation of palmitoylation sites, and TMD sequence manipulation) perturbed subcellular localization, leading to missorting of PM proteins to late endosomes and lysosomes because of a failure to recycle nonraft proteins from early endosomes (EEs). These results confirm the presence of a raft-mediated recycling route in nonpolarized cells, begin to define the molecular parameters for protein association with raft domains, and suggest an explanation for the accumulation of proteins with longer TMDs at the PM.
Results and Discussion
TMD Length Is a Determinant of Raft-Phase Partitioning.
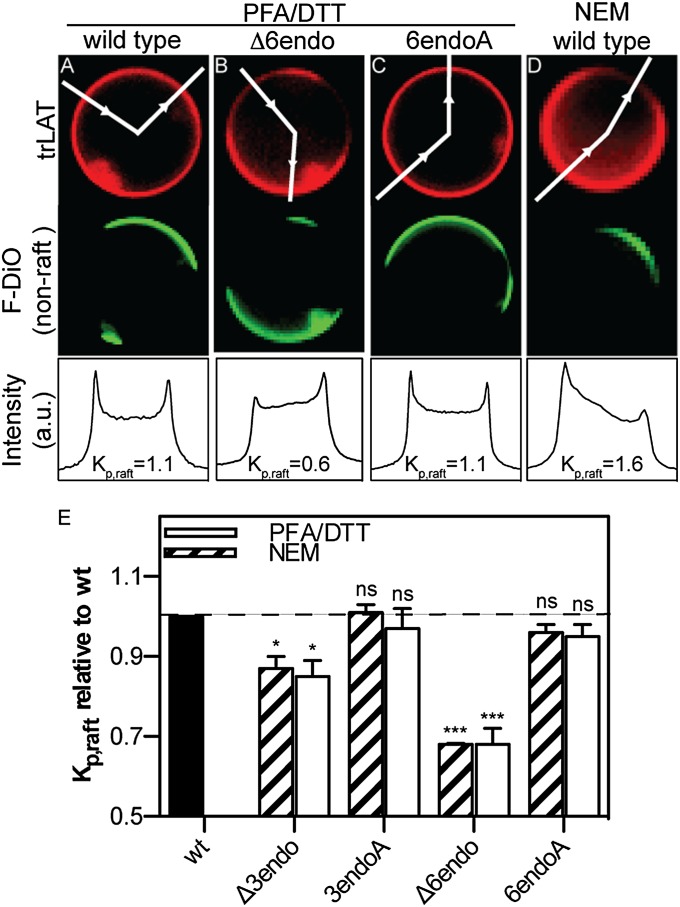
The aim of our study was to determine the effect of raft partitioning on subcellular protein trafficking and localization. Although few data exist on the structural determinants of protein partitioning to lipid rafts, recent observations in synthetic membranes have confirmed the long-standing hypothesis that TMD length is a determinant of raft association. Thus, we produced 13 mutants (Table SI) of a model single-pass TM protein [the TMD of linker for activation of T-cells (LAT) fused to a C-terminal monomeric RFP (mRFP), LAT-TMD-mRFP, hereafter “trLAT” (30)] with various TMD lengths in an attempt to generate variants with varying raft affinities. Raft association was assayed by quantifying the relative partitioning of these constructs between coexisting raft/nonraft phases in GPMVs (4, 5). Fig. 1 A–D shows exemplary images of GPMVs isolated from cells transiently transfected with trLAT variants and the quantification scheme used to calculate raft partitioning. Before vesicle isolation, cells were stained with an unsaturated lipid marker, F-DiO, with strong preference for disordered (nonraft) lipid domains (31). Phase separation in isolated vesicles was observed by the strong segregation of this fluorescent marker into the nonraft phase (Fig. 1 A–D, Middle Row). The partitioning of trLAT variants then was quantified by an established protocol (30, 31), i.e., measuring the ratio of fluorescent protein intensity in the two phases to yield Kp,raft, the raft-phase partition coefficient (Fig. 1 A–D, Bottom Row). Using this method, we readily observed striking differences between the mutants. For example, although both trLAT-WT (Fig. 1A) and a substitution mutant with six C-terminal (interfacing with the endoplasmic leaflet of the PM) residues of the TMD mutated to alanines (trLAT-6endoA) (Fig. 1C) partitioned approximately equally between the coexisting liquid phases (Kp,raft = 1.1 for both vesicles shown), the mutant with the same six amino acids deleted from the endoplasmic region of the TMD (trLAT-Δ6endo; Fig. 1B) was highly enriched in the disordered phase (Kp,raft = 0.6).
Fig. 1.
Raft partitioning of trLAT is disrupted by truncating the TMD. (A–D) Representative images of GPMVs isolated from cells expressing WT (A and D), -Δ6endo (B), and -6endoA (C) versions of trLAT. (Top Row) Protein (red) channel. (Middle Row) Unsaturated marker FAST-DiO (F-DiO; green) to visualize the nonraft phase. (Bottom Row) Normalized line scans of the protein intensity along the white lines in the images in the top row with the two peaks corresponding to raft and nonraft trLAT intensity, respectively. Background-subtracted ratios of these two intensities yield raft partition coefficients, Kp,raft. (E) Kp,raft normalized to trLAT-WT demonstrates that partitioning behavior is independent of preparation but is dependent on TMD length. Data are shown as mean ± SEM from three to six independent experiments; *P < 0.05; ***P < 0.001; nsP > 0.05, one-sample t tests for differences from unity.
The influence of TMD length is evident in the representative data in Fig. S1, which show partition coefficients and experimental deviations for a typical experiment representing 10–15 vesicles per mutant per condition. As previously observed (30), partitioning was a function of vesicle isolation agents, with paraformaldehyde (PFA)/DTT as the vesiculation agents (Fig. 1 A–C) generally yielding lower Kp,raft values than the non–cross-linking, nonreducing N-ethyl maleimide (NEM) (compare trLAT-WT distribution in Fig. 1 A and D). This effect has been attributed previously to the reduction of protein acylation by DTT (30) and to a greater-order difference between coexisting phases in the PFA/DTT preparation (31). Although raft-phase partitioning was consistently ∼50% higher in NEM-derived vesicles than in those derived from PFA/DTT (Fig. S1), the trends for mutant partitioning are consistent among the different preparations, with significant differences observed between trLAT-WT and shorter-TMD variants. Comparing different preparations directly by normalizing to the partitioning of trLAT-WT for that preparation clearly shows that mutants with shorter TMDs (-Δ3endo and -Δ6endo in Fig. 1E) have significantly lower raft affinity than substitution controls with normal-length TMDs (-3endoA and -6endoA).
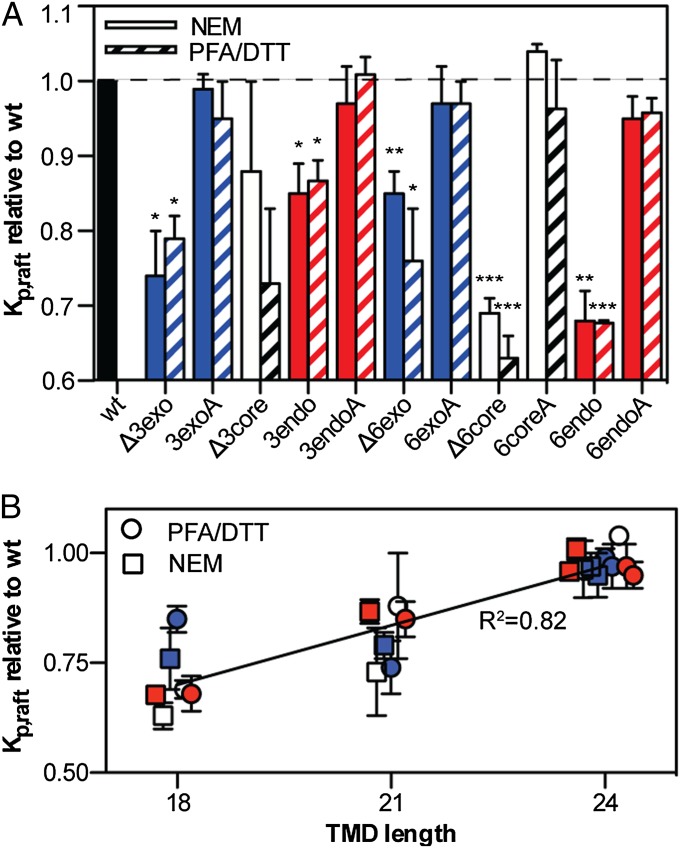
These observations were mirrored in deletions/substitutions from the other parts of the LAT TMD [the hydrophobic core (Fig. S1 A–C), the exoplasmic (Fig. S1 D–F), and the endoplasmic (Fig. S1 G–I) face]. Of the six deletion mutants tested, only one was not significantly less raft-preferring than WT (Fig. 2A). In contrast, all substitution mutants with normal-length (24-aa) TMDs were statistically equivalent to trLAT-WT. No partitioning perturbation was observed with alanine substitutions of the branched/bulky hydrophobic amino acids of the exoplasmic region of the TMD (i.e., -3exoA and -6exoA), in contrast to the effect of alanine substitution of analogous residues of the influenza spike glycoprotein hemagglutinin, which reduces detergent resistance (32). When Kp,raft and TMD length are compared across all preparations, a highly significant dependence of raft partitioning on TMD length is observed (Fig. 2B). For this model protein (trLAT), raft association is reduced by ∼5% for every amino acid shorter than the WT TMD. These findings are qualitatively consistent with a minimization of free energy caused by hydrophobic mismatch between the apolar bilayer core and the hydrophobic residues of the TM α-helix (33); i.e., longer TMDs would prefer to partition into the thicker raft phase to minimize the hydrophobic surfaces exposed to the aqueous environment. Finally, it is worth noting that none of the trLAT mutants (including several not described here) was measured to have greater raft-phase affinity than the WT construct. This observation suggests that the structure of the LAT TMD is evolved to ensure the partitioning of this important immune system signal transducer to its proper membrane subdomain.
Fig. 2.
Raft partitioning of trLAT is strongly dependent on TMD length. (A) Kp,raft normalized to trLAT-WT for deletion or alanine substitutions of either three or six amino acids from the exoplasmic (blue), endoplasmic (red), or core (black) region of the TMD. Data are shown as mean ± SEM from three to five independent experiments per construct; significance values are one-sample t tests for differences from unity (*P < 0.05 ; **P < 0.01; ***P < 0.001; P > 0.05 if no symbol). Mutants with shortened TMDs were significantly less raft-associated than WT in both NEM- (striped bars) and PFA/DTT (empty bars)-derived GPMVs. (B) Kp,raft data from all mutants/preparations normalized to WT show the strong dependence (linear regression R2 = 0.82 ; P = 1.4 × 10−8) of raft partitioning on TMD length regardless of GPMV preparation conditions. Color-coding is as in A. Points are offset for clarity.
TMD Mutations Do Not Affect Palmitoylation.
Because posttranslational modification of TM proteins by a saturated fatty acid (S-acylation, or “palmitoylation”) has been identified previously as a key determinant of raft partitioning (30, 34, 35), we quantified the palmitoylation level of several trLAT constructs at the PM using acyl–biotinyl exchange (ABE) on isolated GPMVs. To control for the inherent and significant variation in the efficiency of ABE, we normalized the palmitoylation of each trLAT construct to that of endogenous LAT in each preparation. As validation of this technique, the palmitoylation of trLAT-WT was nearly identical to that of endogenous LAT (Fig. S2). None of the tested TMD truncation mutants had palmitoylation levels statistically different from trLAT-WT (Fig. S2B). These observations suggest that, for these constructs, palmitoylation at the PM was not affected dramatically by TMD truncations, and therefore that raft partitioning was dependent on hydrophobic matching between the TMD and surrounding bilayer. It is important to emphasize that these observations do not definitively exclude the possibility of small changes in the palmitoylation levels of trLAT constructs or their impact on the differences in raft partitioning shown in Fig. 2. The limitations of current methodologies do not permit exact quantification of palmitoylation levels; thus whether TMD truncation influences raft partitioning directly (because of hydrophobic mismatch) or indirectly (e.g., via partial perturbation of palmitoylation) remains to be elucidated. Regardless of the mechanism of perturbation, our panel of constructs comprises a continuous range of raft affinities, allowing us to evaluate quantitatively the relationship between raft partitioning and subcellular trafficking.
TMD Length Determines PM Localization.
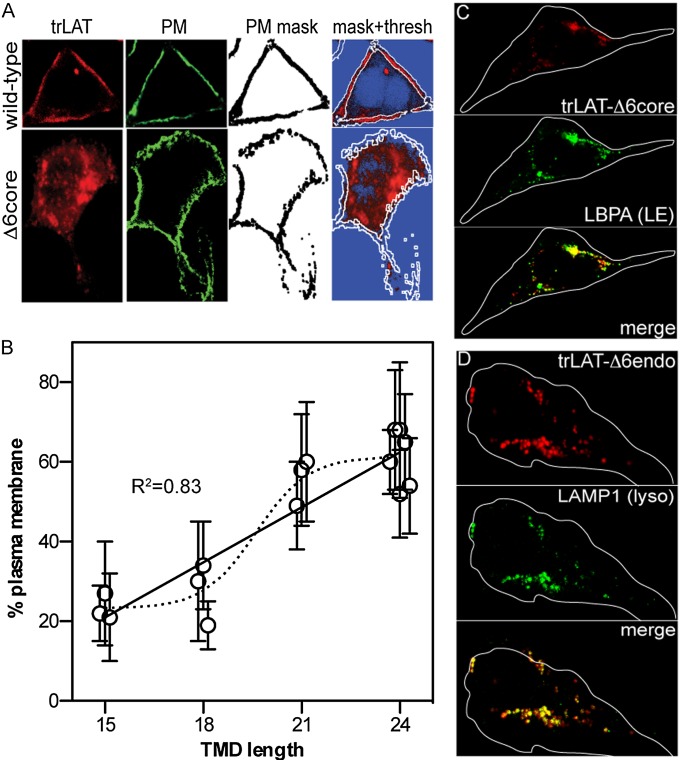
TMD length has been related previously to the subcellular localization of TM proteins, with shorter TMDs preferring the ER/Golgi/endosomal membranes (average TMD length ∼18 aa in vertebrates) and longer TMDs preferring the PM (average TMD length ∼24 aa) (20). We verified this bioinformatic analysis experimentally by quantifying the PM localization of the trLAT TMD mutants using an automated image-processing protocol. Briefly, PM proteins of cells expressing trLAT mutants were nonspecifically biotinylated by a membrane-impermeable, amine-reactive sulfo-NHS-biotin and then were fixed. The PMs were stained using fluorescein-streptavidin, yielding bright, uniform, and highly specific PM staining (second column from left in Fig. 3A and center images in Fig. S3). A mask created from this PM image was overlaid onto the trLAT image (white borders in Fig. 3A mask+thresh images), and the integral intensities inside and outside the PM mask were compared to yield a %PM value for each protein of interest.
Fig. 3.
TMD length determines subcellular localization. (A) Example of the automated image analysis protocol for quantifying PM localization of trLAT-WT and a non-PM mutant (-Δ6core). A mask (third column from left) is created from the thresholded image of the PM stain (green; second column from left), which is overlaid on the thresholded image of the trLAT in the far-right column (blue pixels are below threshold). %PM is calculated by comparing the integrated intensity inside and outside the PM mask. (B) PM localization is significantly dependent on TMD length (linear regression: R2 = 0.83; P = 9.5 × 10−5; dashed line represents a sigmoidal fit to the data). Data are shown as mean ± SD from 15 cells per mutant from at least three different experiments. Points are offset on x axis for clarity. (C and D) Non-PM TMD mutants are sorted to punctate, perinuclear structures, which are positive for (C) the late endosomal marker LBPA and (D) the lysosomal marker LAMP1 and thus are degradative late endosomes/lysosomes.
Quantitative PM localization derived using this automated protocol corresponded well to visual predictions, with PM-localized proteins (e.g., trLAT-WT; Fig. 3A) yielding %PM of 70–80%, whereas mutants with clearly internal accumulation (e.g., -Δ6core) yielded values of 20–30%. We observed a perturbation of PM localization with reduced TMD length: all variants with TMDs of 18 aa had little PM signal (Fig. 3A and Fig. S3), whereas the variants with TMDs of 24 aa were mostly PM localized (21-aa variants had both PM and intracellular signal). Thus, PM localization clearly depended on TMD length (Fig. 3B and Fig. S4). It is difficult from this dataset to distinguish the exact quantitative relationship between these two parameters, i.e., whether a linear relationship (shown as solid line in Fig. 3B) or the step-change between 18 and 21 aa (denoted by the dashed sigmoidal fit in Fig. 3B) would be a more appropriate fit to the data. Nevertheless, these findings are in qualitative agreement with the bioinformatic predictions (20) and clearly identify TMD length as a determinant of PM localization for trLAT. To evaluate the subcellular location of proteins that did not localize strongly at the PM, we performed costaining experiments on fixed cells with various organelle markers and found that such variants (e.g., -Δ6core and -Δ6endo) were sorted instead to perinuclear lysobisphosphatidic acid-positive (LBPA+; Fig. 3C) and lysosomal-associated membrane protein 1-positive (LAMP1+; Fig. 3D) late endocytic compartments (i.e., multivesicular bodies and lysosomes, respectively). Thus, TMD mutations that affect raft partitioning also lead to mistargeting from the PM to degradative organelles.
Raft Partitioning Is a Central Determinant of LAT PM Localization.
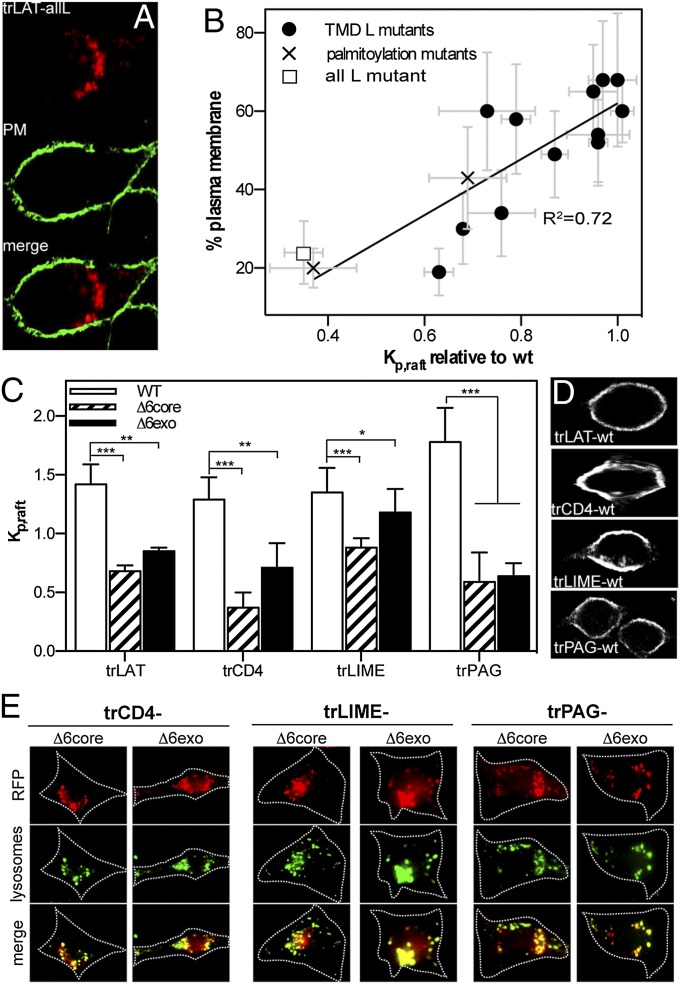
A trivial prediction of the observations in Figs. 2 and 3 is a positive relationship between raft partitioning of TMD mutants and their PM localization, and this prediction is confirmed by the filled circles in Fig. 4B. To establish the causal relationship between raft association and PM localization independent of TMD length [which has been shown previously to be related to organelle sorting (19, 20)], we examined raft partitioning and localization of two sets of trLAT mutants with normal-length (24 aa) TMDs. First, we mutated all TMD amino acids (except the palmitoylation sites) to leucines in an attempt to remove any physicochemical identity that might be required for raft-phase partitioning. Indeed, this trLAT-all leucine (hereafter, trLAT-allL) construct was nearly entirely excluded from the raft phase of GPMVs (Fig. 4B, white square and Fig. S5A), despite being efficiently palmitoylated (Fig. S5B). Correspondingly, this construct also was largely absent from the PM, instead accumulating in perinuclear endosomes (Fig. 4 A and B). Quantification of the relationship between raft partitioning and PM localization of trLAT-allL showed very good concordance with that established by the TMD-length mutants (Fig. 4B). This relationship also extended to two palmitoylation site mutants (C26A and C29A; crosses in Fig. 4B), which previously have been shown to reduce raft-phase association and PM localization (30, 36, 37). Thus, perturbation of raft partitioning by three distinct means (TMD truncation, deletion of palmitoylation sites, and removal of physicochemical identity) had a common result: failure of the constructs to reach their proper destination in the PM.
Fig. 4.
Raft partitioning is a determinant of PM localization. (A) A trLAT mutant (-allL) designed to remove physicochemical identity from the TMD was localized to punctate, perinuclear endo/lysosomes, similar to TMD truncation mutants. (B) PM localization was tightly dependent on Kp,raft for all trLAT constructs, including TMD truncations/substitutions (filled circles), palmitoylation site mutations (C26A and C29A) (crosses), and the allL mutant (open square) (linear regression: R2 = 0.72 ; P = 5.8 × 10−5). (C) Raft-dependent PM localization also was observed for several other proteins (trCD4, trLIME, and trPAG/Cbp). Wild-type TMDs (white bars) partitioned at parity with trLAT-WT, whereas TMD truncations (hatched and solid bars) significantly reduced raft association for all proteins. Data shown are mean ± SD for 10–15 vesicles per sample and are representative of at least two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (D and E) Subcellular localization followed the trend of LAT, with raft-partitioning WT constructs sorted to the PM (D), whereas truncated TMDs were nonraft and accumulated in lysosomes (E).
To investigate whether raft affinity is necessary for PM localization of full-length LAT, the TMD of LAT-GFP (fgLAT) was replaced by a non–raft-targeting TMD (allL). This fgLAT-allL construct was excluded almost entirely from the raft phase in GPMVs (Fig. S6 D and E), confirming that the TMD is the major determinant of raft partitioning for the full-length protein. More importantly, fgLAT-allL was missorted to endo/lysosomes (Fig. S6C), as is consistent with all nonraft trLAT variants. Thus, because no features of native LAT rescue PM localization in the absence of TMD-mediated raft association, we conclude that raft partitioning likely is necessary for cell-surface expression of LAT. Next, we investigated whether raft partitioning in particular, rather than protein–protein interactions mediated by specific residues of the trLAT construct, is sufficient for sorting. To this end, a “minimal” LAT construct (mrLAT; see SI Materials and Methods) was shown to maintain the raft affinity and PM localization (Fig. S6 A and B) of both trLAT and full-length LAT (Fig. S6 C–E). This construct contains only 29 residues of LAT, 24 of which have been mutated to alanine in various trLAT constructs (see substitution mutants in Table S1) with strong agreement between raft partitioning and PM localization (Fig. 4B). These strong correlations between PM localization and raft partitioning imply a causal (i.e., necessary and sufficient) relationship.
The same relationships among TMD length, raft-phase partitioning, and PM localization were observed for three other proteins. TMD constructs based on known raft proteins [CD4 (38), LIME (39), and PAG (40)] partitioned to the raft domain in GPMVs at parity with trLAT (Fig. 4C, white bars) and localized strongly to the PM (Fig. 4D). In contrast, truncation mutants generated by deletion of six residues from the TMD had greatly reduced raft-phase association (Fig. 4C, hatched and filled bars) and PM localization, instead accumulating in lysosomes (Fig. 4E). Thus, the raft-dependent localization observed for LAT is applicable to other proteins.
Raft-Mediated PM Recycling from Early Endosomes.
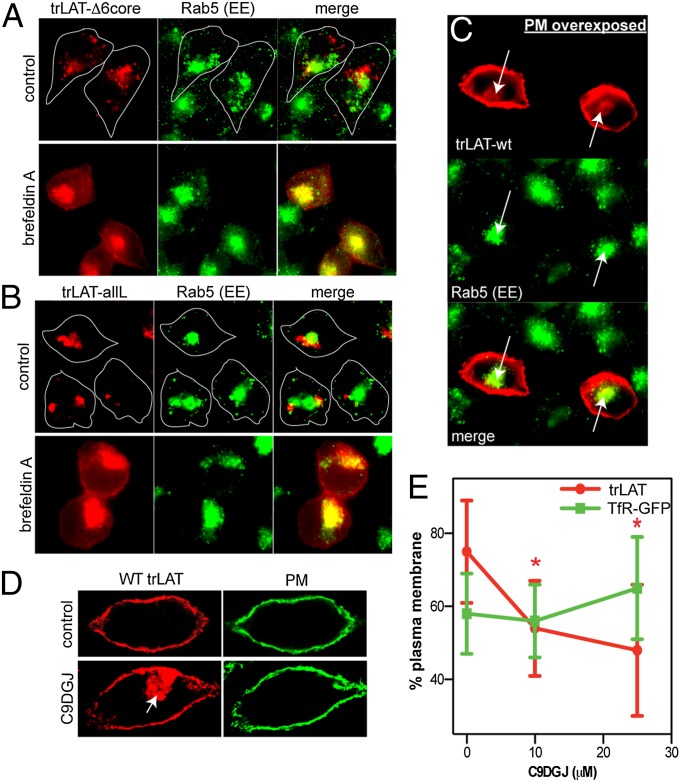
To investigate the mechanisms of aberrant localization resulting from failure to partition into raft domains, we treated cells with inhibitors of various trafficking machinery and assayed steady-state trLAT localization. Inhibition of dynamin-mediated endocytosis (SI Materials and Methods) had no observable effect on the distribution of any LAT variants. In contrast, interfering with endosomal maturation dramatically and specifically altered the localization of nonraft trLAT variants. Brefeldin A (BFA) inhibits the Arf1-dependent assembly of the COPI coat complex, with the most widely recognized result being the collapse of the Golgi apparatus into the ER (41). However, this compound also affects the organization of the endosomal system, effectively blocking progression from early to late endosomes (41, 42). Bafilomycin A (Baf) similarly blocks early-to-late endosome progression but does so by a different mechanism–inhibiting luminal acidification (43). Both BFA (Fig. 5 A and B and Fig. S7) and Baf (Fig. S8A) induced a striking redistribution of trLAT-Δ6endo and -allL from the lysosomes to the PM and an internal compartment identified as the early endosome (EE) by Rab5 staining. PM localization of trLAT-WT was not affected by either drug (Fig. S8B). Thus, the trafficking defect of nonraft LAT variants is endocytic, not secretory or biosynthetic. Moreover, the capacity of nonraft constructs to localize to the PM excludes the possibility that the steady-state lysosomal accumulation of these variants is caused by misfolding and/or aggregation, because arrival of TM proteins at the PM necessitates transit through the secretory pathway, and that transit in turn requires proper folding and membrane insertion.
Fig. 5.
Trafficking defect of nonraft variants involves failure in recycling from EEs. (A and B) Perturbation of endosomal traffic with BFA caused redistribution of both trLAT-Δ6core (A) and -allL (B) to the PM and Rab5+ EEs. (C) EE localization of trLAT-WT could be observed in unperturbed cells upon overexposure of PM signal. (D and E) Raft perturbation by inhibition of glycosphingolipid synthesis by C9DGJ significantly reduced PM localization of trLAT (D) but not TfR-GFP (E). The effect is quantified as in Fig. 3 A and B and is shown as mean ± SD from 15 cells per construct; *P < 0.05 compared with vehicle control; for TfR, P > 0.5 for all treatments.
All raft-partitioning constructs localize to the PM (Fig. 4), whereas abrogation of raft partitioning leads to lysosomal accumulation via the EE, suggesting that membrane rafts provide a mechanism for protein recycling from endosomes to the PM (44). This hypothesis is supported by previous observations of raft protein and lipid enrichment in early/recycling endosomes (13) and concomitant depletion of these components in more mature endosomes (14). A prediction of the hypothesis is that raft-associated variants also should transit the EE, and we observed this transit as weak intracellular staining (colocalized with Rab5) visible when the PM signal for trLAT-WT was overexposed (arrows in Fig. 5C). Finally, perturbation of raft domains would be expected to lead to endosomal accumulation of constructs dependent on raft domains for recycling. We interfered with raft composition by inhibiting the first committed enzyme in glycosphingolipid synthesis (glucosylceramide synthase) by N-nonyl-deoxygalactonojirimycin (C9DGJ), which dramatically reduces cellular glycosphingolipid and cholesterol levels, perturbing raft-dependent trafficking in cell lines (12, 45–47) and in vivo (48). Consistent with these observations and our hypothesis of raft-dependent EE-to-PM recycling, C9DGJ induced a significant and dose-dependent reduction of trLAT-WT PM localization (Fig. 5 D and E) with a concomitant accumulation in internal compartments with the characteristic morphology and localization of EE (arrow in Fig. 5D). This mislocalization was not observed for the non–raft-partitioning transferrin receptor (TfR)-GFP (Fig. 5E) (30), suggesting a specific perturbation of raft-mediated traffic. Together with the strong dependence of PM localization on raft-phase partitioning (Fig. 4), these results confirm the role of lipid-driven, ordered domains in protein recycling to the PM.
Conclusion
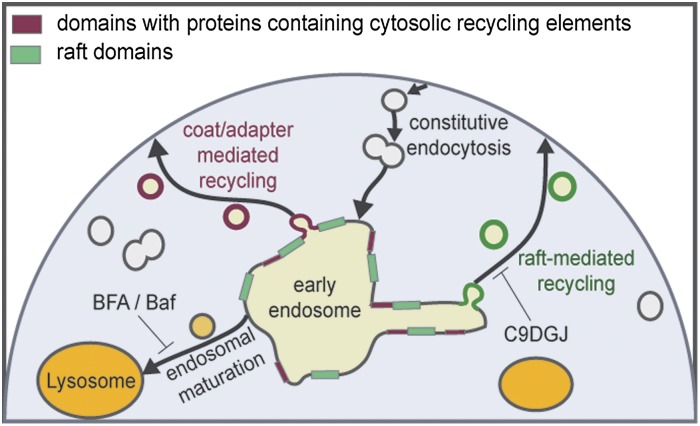
Our observations support a model of parallel, nonexclusive mechanisms for protein and lipid recycling to the PM (Fig. 6), analogous to the basolateral and apical sorting routes in polarized epithelial cells (18, 44). Both these routes are likely responsible for the recently described accumulation of proteins containing long TMDs at the PM of eukaryotes (20). One pathway is mediated by coat-and-adapter assemblies that recruit proteins through specific proteinaceous sorting motifs (Fig. 6, red bars and circles) (49). For this pathway, no involvement of specific lipid species has yet been described, although the TMDs of these proteins would still need to match the physical properties of the highly cholesterol-enriched PM. The other mechanism involves the lipid-driven condensation of membrane domains (green bars in Fig. 6) to form transport carriers (green circles in Fig. 6) enriched in raft lipids and raft-associated proteins and is supported by our observation of a causal relationship between raft association and PM recycling. In our observations, proteins with neither protein- nor lipid-mediated sorting signals are destined for the lysosome, possibly by bulk endosomal maturation. It is important to emphasize that although our scheme depicts the EE as the sole cellular sorting station, the separation of raft from nonraft components also may happen at other sites in the endosomal/recycling pathway, e.g., the PM and/or recycling endosomes.
Fig. 6.
Scheme of raft-dependent recycling. The relationship between raft association and subcellular localization supports the hypothesis of parallel recycling routes to the PM in nonpolarized cells, with one of these routes mediated by protein partitioning to lipid-driven raft domains (green rectangles) that promote the formation of raft-enriched recycling vesicles (green circles). Domains containing proteins with cytosolic sorting signals for coat/adapter protein-mediated recycling [not investigated in this work but established in literature (18, 44, 49)] are shown in red. Proteins with neither sorting signal are routed to lysosomes (yellow) by endosomal maturation. Although in the scheme the EE is pictured as the sole sorting station, the separation of raft from nonraft components also may happen in other endocytic compartments, including recycling endosomes and the PM.
Materials and Methods
GPMVs were isolated from rat basophilic leukemia (RBL) cells as described previously (4, 5). Constructs based on the trLAT backbone (30) were transfected into RBLs using Nucleofection (Lonza). Variants were constructed by custom DNA synthesis (Genscript). Detailed materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Humboldt Foundation and the Cancer Prevention and Research Institute of Texas (R1215) for funding support (for I.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404582111/-/DCSupplemental.
References
- 1.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457(7233):1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 2.Klemm RW, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185(4):601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerl MJ, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol. 2012;196(2):213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104(9):3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sezgin E, et al. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc. 2012;7(6):1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 6.Schuck S, Simons K. Polarized sorting in epithelial cells: Raft clustering and the biogenesis of the apical membrane. J Cell Sci. 2004;117(Pt 25):5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- 7.Mañes S, del Real G, Martínez-A C. Pathogens: Raft hijackers. Nat Rev Immunol. 2003;3(7):557–568. doi: 10.1038/nri1129. [DOI] [PubMed] [Google Scholar]
- 8.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 9.Lingwood D, Simons K. Detergent resistance as a tool in membrane research. Nat Protoc. 2007;2(9):2159–2165. doi: 10.1038/nprot.2007.294. [DOI] [PubMed] [Google Scholar]
- 10.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68(3):533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 11.Surma MA, Klose C, Klemm RW, Ejsing CS, Simons K. Generic sorting of raft lipids into secretory vesicles in yeast. Traffic. 2011;12(9):1139–1147. doi: 10.1111/j.1600-0854.2011.01221.x. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimori T, Keller P, Roth MG, Simons K. Different biosynthetic transport routes to the plasma membrane in BHK and CHO cells. J Cell Biol. 1996;133(2):247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagescu R, et al. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell. 2000;11(8):2775–2791. doi: 10.1091/mbc.11.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lusa S, et al. Depletion of rafts in late endocytic membranes is controlled by NPC1-dependent recycling of cholesterol to the plasma membrane. J Cell Sci. 2001;114(Pt 10):1893–1900. doi: 10.1242/jcs.114.10.1893. [DOI] [PubMed] [Google Scholar]
- 15.Orci L, et al. Heterogeneous distribution of filipin—cholesterol complexes across the cisternae of the Golgi apparatus. Proc Natl Acad Sci USA. 1981;78(1):293–297. doi: 10.1073/pnas.78.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange Y, Swaisgood MH, Ramos BV, Steck TL. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J Biol Chem. 1989;264(7):3786–3793. [PubMed] [Google Scholar]
- 17.Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27(17):6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 18.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9(11):833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14(19):4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142(1):158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipowsky R. Domain-induced budding of fluid membranes. Biophys J. 1993;64(4):1133–1138. doi: 10.1016/S0006-3495(93)81479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roux A, et al. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 2005;24(8):1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Sáez AJ, Chiantia S, Schwille P. Effect of line tension on the lateral organization of lipid membranes. J Biol Chem. 2007;282(46):33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- 24.Saslowsky DE, et al. Placental alkaline phosphatase is efficiently targeted to rafts in supported lipid bilayers. J Biol Chem. 2002;277(30):26966–26970. doi: 10.1074/jbc.M204669200. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, London E. Altering hydrophobic sequence lengths shows that hydrophobic mismatch controls affinity for ordered lipid domains (rafts) in the multitransmembrane strand protein perfringolysin O. J Biol Chem. 2013;288(2):1340–1352. doi: 10.1074/jbc.M112.415596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott RE. Plasma membrane vesiculation: A new technique for isolation of plasma membranes. Science. 1976;194(4266):743–745. doi: 10.1126/science.982044. [DOI] [PubMed] [Google Scholar]
- 27.Fridriksson EK, et al. Quantitative analysis of phospholipids in functionally important membrane domains from RBL-2H3 mast cells using tandem high-resolution mass spectrometry. Biochemistry. 1999;38(25):8056–8063. doi: 10.1021/bi9828324. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser HJ, et al. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci USA. 2009;106(39):16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengupta P, Hammond A, Holowka D, Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim Biophys Acta. 2008;1778(1):20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci USA. 2010;107(51):22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levental I, Grzybek M, Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc Natl Acad Sci USA. 2011;108(28):11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheiffele P, Roth MG, Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997;16(18):5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouritsen OG, Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys J. 1984;46(2):141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;2006(359):re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 35.Levental I, Grzybek M, Simons K. Greasing their way: Lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49(30):6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Trible RP, Samelson LE. LAT palmitoylation: Its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998;9(2):239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 37.Hundt M, et al. Palmitoylation-dependent plasma membrane transport but lipid raft-independent signaling by linker for activation of T cells. J Immunol. 2009;183(3):1685–1694. doi: 10.4049/jimmunol.0803921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fragoso R, et al. Lipid raft distribution of CD4 depends on its palmitoylation and association with Lck, and evidence for CD4-induced lipid raft aggregation as an additional mechanism to enhance CD3 signaling. J Immunol. 2003;170(2):913–921. doi: 10.4049/jimmunol.170.2.913. [DOI] [PubMed] [Google Scholar]
- 39.Hur EM, et al. LIME, a novel transmembrane adaptor protein, associates with p56lck and mediates T cell activation. J Exp Med. 2003;198(10):1463–1473. doi: 10.1084/jem.20030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brdicka T, et al. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J Exp Med. 2000;191(9):1591–1604. doi: 10.1084/jem.191.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lippincott-Schwartz J, et al. Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67(3):601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 42.Recchi C, Chavrier P. V-ATPase: A potential pH sensor. Nat Cell Biol. 2006;8(2):107–109. doi: 10.1038/ncb0206-107. [DOI] [PubMed] [Google Scholar]
- 43.Bayer N, et al. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: Implications for viral uncoating and infection. J Virol. 1998;72(12):9645–9655. doi: 10.1128/jvi.72.12.9645-9655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Boulan E, Kreitzer G, Müsch A. Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol. 2005;6(3):233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 45.Keller P, Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J Cell Biol. 1998;140(6):1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esfahani M, et al. Cholesterol regulates the cell surface expression of glycophospholipid-anchored CD14 antigen on human monocytes. Biochim Biophys Acta. 1993;1149(2):217–223. doi: 10.1016/0005-2736(93)90204-d. [DOI] [PubMed] [Google Scholar]
- 47.Lipardi C, Nitsch L, Zurzolo C. Detergent-insoluble GPI-anchored proteins are apically sorted in fischer rat thyroid cells, but interference with cholesterol or sphingolipids differentially affects detergent insolubility and apical sorting. Mol Biol Cell. 2000;11(2):531–542. doi: 10.1091/mbc.11.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, et al. Apicobasal domain identities of expanding tubular membranes depend on glycosphingolipid biosynthesis. Nat Cell Biol. 2011;13(10):1189–1201. doi: 10.1038/ncb2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: Structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.