Significance
The discovery of lytic polysaccharide monooxygenases (LPMOs) has profoundly changed our understanding of the enzymatic conversion of recalcitrant polysaccharides, such as cellulose. Although in-depth studies of fungal cellulolytic LPMOs have been reported, the structures and functions of their bacterial counterparts with no detectable sequence similarity remain largely elusive. We present the structures of a conserved pair of bacterial cellulose-active LPMOs supplemented with extensive functional characterization. The structural data allow a thorough comparative assessment of fungal and bacterial LPMOs, providing insight into the structural basis of substrate specificity and the oxidative mechanism (C1/C4 oxidation). Importantly, we show that this LPMO pair acts synergistically when degrading cellulose, a finding that may help explain the occurrence of multiple LPMOs in a single microbe.
Keywords: GH61, CBM33
Abstract
For decades, the enzymatic conversion of cellulose was thought to rely on the synergistic action of hydrolytic enzymes, but recent work has shown that lytic polysaccharide monooxygenases (LPMOs) are important contributors to this process. We describe the structural and functional characterization of two functionally coupled cellulose-active LPMOs belonging to auxiliary activity family 10 (AA10) that commonly occur in cellulolytic bacteria. One of these LPMOs cleaves glycosidic bonds by oxidation of the C1 carbon, whereas the other can oxidize both C1 and C4. We thus demonstrate that C4 oxidation is not confined to fungal AA9-type LPMOs. X-ray crystallographic structures were obtained for the enzyme pair from Streptomyces coelicolor, solved at 1.3 Å (ScLPMO10B) and 1.5 Å (CelS2 or ScLPMO10C) resolution. Structural comparisons revealed differences in active site architecture that could relate to the ability to oxidize C4 (and that also seem to apply to AA9-type LPMOs). Despite variation in active site architecture, the two enzymes exhibited similar affinities for Cu2+ (12–31 nM), redox potentials (242 and 251 mV), and electron paramagnetic resonance spectra, with only the latter clearly different from those of chitin-active AA10-type LPMOs. We conclude that substrate specificity depends not on copper site architecture, but rather on variation in substrate binding and orientation. During cellulose degradation, the members of this LPMO pair act in synergy, indicating different functional roles and providing a rationale for the abundance of these enzymes in biomass-degrading organisms.
The enzymatic conversion of plant biomass is an issue of major scientific and commercial interest. Although this process originally was thought to involve only hydrolytic enzymes, such as cellulases, we now know that oxidative enzymes called lytic polysaccharide monooxygenases (LPMOs) play an important role (1). Using powerful oxidative chemistry (2, 3), LPMOs cleave glycosidic bonds in polysaccharides that are inaccessible to cleavage by hydrolytic enzymes, such as endoglucanases and cellobiohydrolases. By increasing substrate accessibility, LPMOs boost the overall efficiency of enzymatic degradation of insoluble polysaccharides (4–6). Indeed, the latest generation of commercial cellulase mixtures for processing of lignocellulosic biomass benefits from the presence of LPMOs (7).
In the CAZy database (8), LPMOs are classified into the auxiliary activity (AA) families 9 (AA9; previously known as GH61), 10 (AA10; previously known as CBM33), and 11 (AA11) (9). Families AA9 and AA11 comprise fungal enzymes, whereas family AA10 comprises enzymes from all domains of life. Hereinafter, these three families are referred to as LPMO9, LPMO10, and LPMO11, respectively. Members of these families share low sequence identity, but have similar Ig-like folds with a flat substrate-binding surface. The solvent-exposed active site contains two histidines that coordinate a copper ion in a histidine brace (2, 10, 11). The role of the copper ion is to reduce dioxygen, which requires electrons from an external electron donor. The reduced dioxygen likely abstracts a hydrogen from the substrate, which eventually leads to cleavage of the β-1,4 glycosidic linkage (12–14). As a result of this reaction, a carbon in the scissile glycosidic bond is oxidized. Some LPMOs exclusively oxidize C1, others exclusively oxidize C4, and a third group can oxidize either C1 or C4 (13, 15). The latter LPMO group can generate “double-oxidized” products that are formed when a polysaccharide chain is cleaved twice, once with C1 oxidation and once with C4 oxidation (13). The reaction products are either δ-1,5 lactones, which may be hydrated to form aldonic acids (C1 oxidation) (5, 16), or 4-ketoaldoses, which may be hydrated to form gemdiols (C4 oxidation) (17). Notably, the existence of other types of non-C1 oxidation has been suggested (10, 12). Based on previous studies (15, 17) and experimental evidence presented here, and for the sake of simplicity, only C4 oxidation is considered hereinafter.
The abundance of LPMOs in the genomes of biomass-degrading organisms (18), expression data (19), and functional studies (5, 6) demonstrate the great importance of LPMOs in biomass processing. Thus, studying the structure and function of these enzymes is of interest. Fungal LPMO9s have been relatively well studied and are known to act on cellulose and oxidize C1, C4, or both (11, 13). In contrast, little is known about bacterial cellulose-degrading LPMO10s. Oxidative cleavage of cellulose has been described for only two enzymes, both oxidizing C1 (16, 20), and no structural information is available. All LPMO10s with known structures act on chitin and oxidize C1. This lack of information limits our understanding of bacterial cellulose degradation as well as LPMO functionality and diversity.
The dominant bacterial genus responsible for aerobic biomass decomposition in soil is the Gram-positive Streptomycetes (21). A recent secretome/transcriptome study showed that Streptomyces sp. SirexAA-E (ActE) secretes a plethora of enzymes targeting carbohydrates, including abundantly expressed LPMO10s (19). Two of its six LPMOs (SACTE_3159 and SACTE_6428) were up-regulated and secreted during growth on pure cellulosic substrates or plant biomass, and three other LPMOs were up-regulated when chitin served as the substrate (19). Homologs of SACTE_3159 and SACTE_6428 in the well-studied cellulolytic bacterium Thermobifida fusca YX, known as E7 and E8, are also up-regulated during growth on cellulose (22). Homologs of this pair of putative LPMOs also exist among the seven LPMOs encoded in the Streptomyces coelicolor A3 (2) genome (ScLPMO10B and CelS2, respectively; the formal name of CelS2 is ScLPMO10C). Previous studies have indicated that CelS2 is coexpressed with a cellulase (23), and we previously showed that CelS2 is a C1-oxidizing LPMO acting in synergy with cellulases (16). The enzymatic properties of other bacterial LPMOs putatively acting on cellulose, and the functional significance of the coexpression of CelS2-ScLPMO10B-like pairs of LPMOs, have not yet been described.
Here we show that ScLPMO10B (and E7) are C1- and C4-oxidizing LPMOs that complement the activity of C1-oxidizing CelS2 (and E8), yielding synergy when combined in a reaction. We also present the X-ray crystallographic structures of both S. coelicolor enzymes, that is, the first structures of bacterial cellulose-degrading LPMOs. The analysis and comparison of these structures is supported by studies of copper binding by isothermal titration calorimetry (ITC), determination of the redox potentials (E°), and analysis of copper coordination by electron paramagnetic resonance (EPR) spectroscopy.
Results
Enzyme Activity.
Five LPMOs were produced and characterized: CelS2 from S. coelicolor (comprising an LPMO domain and a family 2 carbohydrate-binding module termed CBM2), the N-terminal LPMO domain of CelS2 (CelS2-N), the N-terminal domain of E8 (E8-N), and the single-domain proteins ScLPMO10B and E7. Studies with phosphoric acid swollen cellulose (PASC) and Avicel found that CelS2, CelS2-N, and E8-N generated C1-oxidized products only, whereas ScLPMO10B and E7 showed a different product profile (Fig. 1 and SI Appendix, Fig. S1). Chromatographic peak assignments from a recent in-depth study of a C4-oxidizing LPMO9 (17) allowed identification of the additional products generated by these latter two enzymes as C4-oxidized (4-ketoaldoses) and double (C4/C1)-oxidized cello-oligosaccharides (SI Appendix, Fig. S2). MS data support this conclusion (Fig. 1).
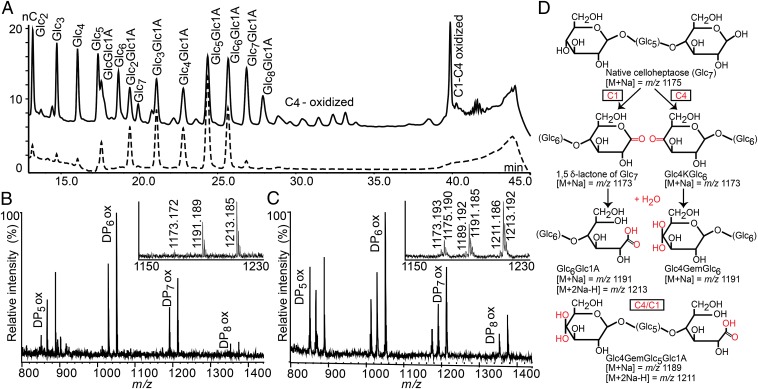
Fig. 1.
Comparison of the two cellulose-active LPMOs from S. coelicolor in PASC degradation. (A) High-performance anion-exchange chromatography product profile for CelS2-N (dotted chromatogram) and ScLPMO10B (solid chromatogram). Peaks were annotated using native and C1-oxidized standard cello-oligosaccharides (16), and products were compared with those obtained previously with C4-oxidizing NcLPMO9C (17) (SI Appendix, Fig. S2). (B and C) MALDI-TOF MS analysis of products generated by CelS2 (B) and ScLPMO10B (C), with sodium saturation. (Insets) Details of the heptamer ion clusters. (D) Possible products in these clusters. Both B and C show the lactone or ketoaldose (1173), the aldonic acid or gemdiol (4-ketoaldose + water:1191), and the sodium adduct of the aldonic acid sodium salt (1213). C also shows native Glc7 (1175), the double-oxidized heptamer (1189), and the sodium adduct of the sodium salt of the double-oxidized heptamer (1211). Here 100% relative intensity represents 4.2 × 104 arbitrary units (a.u.) for the full spectra and 2.8 × 104 a.u. for the spectra in the Insets. Similar results for the E7/E8 enzyme pair are shown in SI Appendix, Fig. S1.
To verify the presence of double-oxidized products, we degraded soluble products formed by ScLPMO10B, E7 or CelS2-N (negative control) with a cellobiohydrolase, followed by C1 oxidation by cellobiose dehydrogenase from Myriococcum thermophilum (MtCDH). As expected, peaks assigned as double-oxidized disappeared after cellobiohydrolase treatment, whereas double-oxidized products, resulting from C1 oxidation of C4-oxidized fragments, were detected again after subsequent treatment with MtCDH (SI Appendix, Fig. S3). Taken together, these data clearly show that ScLPMO10B and E7 have mixed activity, yielding C1- and C4-oxidized products. Double-oxidized products and native oligomers alike are the result of two oxidative cleavages in the same polysaccharide chain (one C1 and one C4) with different outcomes, whereas for all LPMOs, native oligomers also result from oxidative cleavage near an original chain end (24).
ScLPMO10B and E7, but not CelS2 and E8-N, also showed activity on squid pen β-chitin (SI Appendix, Fig. S4), whereas no products were obtained from crab shell α-chitin. The product profiles differed from those previously described for chitin-active LPMOs (5, 9), in that considerable amounts of partially deacetylated oligomers were produced. This indicates that the enzymes also (or preferably) act on deacetylated regions of the substrate, which notably resemble cellulose rather than fully acetylated parts.
Product formation over time was assessed by incubating CelS2, CelS2-N, and ScLPMO10B with PASC, individually or in combination. Clear synergistic effects were observed after CelS2 and ScLPMO10B were combined in the same reaction (Fig. 2 and SI Appendix, Fig. S5). A similar effect was seen when E7 and E8 were combined (SI Appendix, Fig. S6).
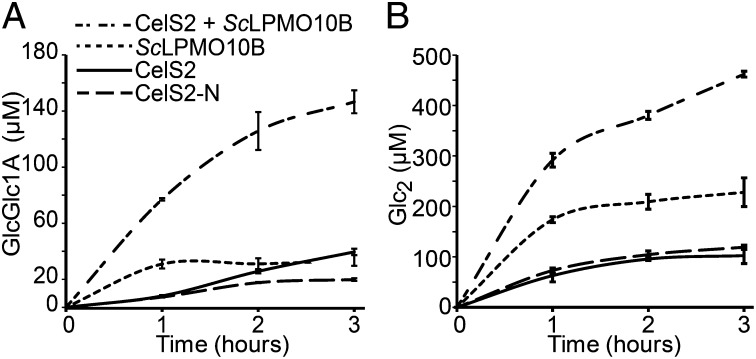
Fig. 2.
Time course of released oxidized (A) and native (B) cellobiose after incubation of 1 µM LPMO (CelS2, CelS2-N, or ScLPMO10B) or a mixture of 0.5 µM CelS2 and 0.5 µM ScLPMO10B with 2 g/L PASC and 2 mM ascorbic acid in 20 mM ammonium acetate buffer (pH 6.0). Before analysis, oligomeric products generated by the LPMOs (Fig. 1A) were converted to shorter fragments by treatment with a cellobiohydrolase to facilitate product quantification. In the sample with the highest sugar concentration (i.e., CelS2 + ScLPMO10B; 3 h), the amounts of cellobionic acid (GlcGlc1A) and cellobiose (Glc2) represent 2.4% ± 0.2% and 7.5% ± 0.1% conversion, respectively, of the substrate. Glc2Glc1A was an additional minor end product, and the Glc2Glc1A: GlcGlc1A ratio was approximately constant. Control experiments (dose–response curves for the individual LPMOs) are shown in SI Appendix, Fig. S5. SDs were calculated based on three independent reactions. Similar results for the T. fusca LPMO pair are shown in SI Appendix, Fig. S6.
Three-Dimensional Structures of CelS2 and ScLPMO10B.
The structure of ScLPMO10B was determined to 2.1 Å using zinc single-wavelength anomalous diffraction. The partially refined structure was then used as a search model for molecular replacement to obtain the structures of ScLPMO10B with zinc (1.4 Å) and copper (1.3 Å), as well as the structure of CelS2-N with copper (1.5 Å). Statistics for diffraction data and structure refinement are summarized in SI Appendix, Table S1. Both ScLPMO10B and CelS2 have the Ig-like β-sandwich fold observed in other LPMO structures, which also includes the sequence-disparate family AA9 LPMOs (Fig. 3 and SI Appendix, Fig. S7). The family AA10 LPMOs consist of a distorted β-sandwich comprising two β-sheets, one containing three antiparallel strands (S1, S4, and S7) and the other containing four antiparallel strands (S5, S6, S8, and S9). Additional strands adorn the four-stranded sheet in either an antiparallel (S2) or parallel (S3) fashion. The β-sandwich contains several conserved aromatic residues, potentially contributing to the electron transfer pathway necessary for reduction of the copper ion.
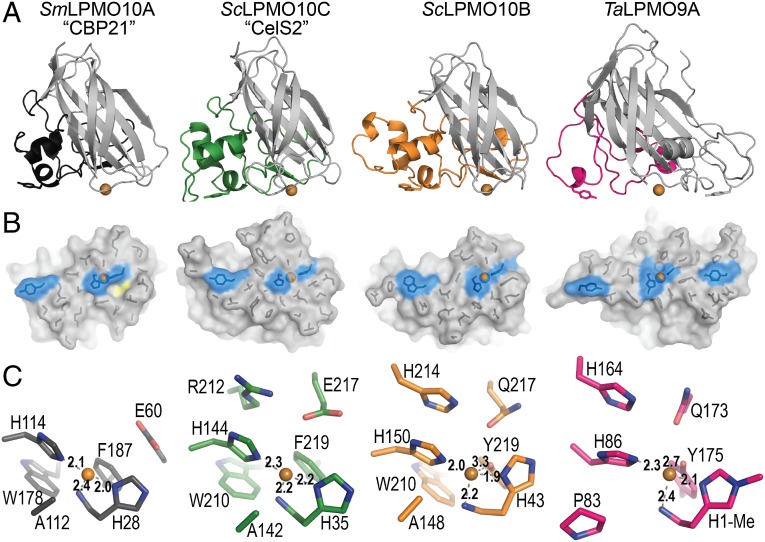
Fig. 3.
Structural comparison of four LPMOs. (A) Cartoon representation of CBP21 (black; chitin-active, C1-oxidizing, PDB ID code 2BEM), CelS2 (green; cellulose-active, C1-oxidizing), ScLPMO10B (orange; a C1/C4-oxidizer of cellulosic substrates, also capable of C1 oxidation of β-chitin), and TaLPMO9A (pink; a C1/C4-oxidizer of cellulosic substrates, PDB ID code 2YET). The L2 loop is colored, and the rest of the enzyme is in gray. Residues with aromatic rings lying parallel to the putative substrate-binding surface are shown as sticks. Metal ions are shown as orange spheres. (B) Surface projection of the proposed substrate-binding surface, related to Fig. 3A by a 90° rotation along the horizontal axis. The side chains of residues forming the relatively flat surface are shown as black sticks. Metal ions are shown as orange spheres; coordinating histidine residues and residues with aromatic rings parallel to the surface are in blue. The yellow area in CBP21 indicates a characteristic cavity on the surface of chitin-active LPMO10s (SI Appendix, Fig. S8). (C) LPMO active sites, showing residues within 6 Å of the respective metal ions. Figures were created in PyMOL (32).
Structural diversity in both AA10 and AA9 families is confined mainly to the region between strands S1 and S3, known as loop 2 (L2 loop) in LPMO9s. This region forms a large protuberance, which in LPMO10s contains several helices and contributes to at least 50% of the putative substrate-binding surface. As shown in Fig. 3, the L2 loops of the two cellulose-active S. coelicolor LPMOs are larger than the L2 loop of the archetypal chitin-active SmLPMO10A (CBP21). A similarly small L2 loop is present in chitin-active EfLPMO10A (4AO2) (25), whereas the L2 loop in BaLPMO10A (2YOY, which probably is chitin-active) (3, 26) is larger. Both S. coelicolor LPMOs have two disulfide bridges in the L2 region (SI Appendix, Fig. S7). One of these disulfide bridges links helix H1 to helix H1.1, which is conserved in CBP21, and the other tethers the L2 loop to the four-stranded β-sheet (via strand S9). The BaLPMO10A and EfLPMO10A structures do not contain any disulfide bridges. Relative to CBP21, CelS2 and (by inference from sequence alignment) E8 (SI Appendix, Fig. S7) have an insertion between strand S6 and S7 that contributes to the putative binding surface and corresponds to an area designated the LS loop in LPMO9s (27).
Aromatic residues are often involved in enzyme–carbohydrate interactions and are indeed found on the surface of LPMO9s in conformations (i.e., rings parallel to the binding surface) that suggest a role in substrate binding (13, 27). The two S. coelicolor LPMO10s have only one aromatic residue with a ring parallel to the binding surface, in structurally equivalent positions, Tyr79 in CelS2 and Trp88 in ScLPMO10B (Fig. 3). This single aromatic residue is conserved and positioned similarly in essentially all LPMO10s with known structures, including Tyr54 in CBP21 (2BEM) (28), Trp58 in EfLPMO10A (4A02) (25), and Trp50 in BaLPMO10A (2YOW) (3). The binding surfaces of chitin-active LPMO10s have a cavity close to the catalytic center that has been postulated to accommodate dioxygen (3); this cavity is absent in the cellulose-active LPMO10s (Fig. 3 and SI Appendix, Fig. S8).
Copper Site.
The copper site of ScLPMO10B is highly similar to that of the C1/C4 cellulose-oxidizing LPMO9A from Thermoascus aurantiacus (TaLPMO9A) (10) and exhibits an octahedral coordination geometry with Jahn–Teller distortion (Fig. 3). Three equatorial ligands form a “histidine brace” (10) composed of His43, His150, and the N-terminal amino group (His43). An acetate ion occupies the solvent-facing axial position, while its other oxygen extends partially into the fourth equatorial position (SI Appendix, Fig. S9). The axial position on the protein-facing side is occupied by the hydroxyl group of Tyr219 at a distance of 3.3 Å. A third histidine, His214, stabilizes His150 through π–π interaction, as seen in TaLPMO9A (His164 stabilizes His86) (10). Ala148, which is highly conserved in LPMO10s, has been suggested to restrict access to the solvent-facing axial position (3) and is present in ScLPMO10B as well.
In contrast, the copper site of CelS2 is similar to that of chitin-active LPMO10s (3, 25, 28) (Fig. 3), clearly showing that the active site features found in ScLPMO10B, and first described for TaLPMO9A (10), are not essential for activity on cellulose. Minor differences between CBP21 and CelS2 occur directly outside the copper coordination shell; Glu60 (CBP21, strand 2) and Glu217 (CelS2, strand 9) are at structurally equivalent positions but located at different positions in the protein sequences. In CelS2, this glutamate has a hydrogen bond with Arg212, which fills in the cavity typically seen in the chitin-active LPMOs (Fig. 3 and SI Appendix, Fig. S8). Interestingly, the eight molecules in the asymmetric unit show variation in copper coordination and ligand bond lengths that may reflect variation in the oxidation state of the copper (SI Appendix, Fig. S10); see Hemsworth et al. (11) for a discussion. All chains (A–H) show density (refined as a water molecule) in the equatorial position located 1.6–2.0 Å from the copper and in close contact with Glu217 (2.7 Å). Chains E and G have additional density (refined as water) positioned 2.4–2.9 Å from the copper, askew from the axial position. Electron density has been observed in the same position for EfLPMO10A containing oxidized copper (Cu2+) (PDB ID code 4ALC) (11). We note that the average distance between the density in the equatorial position (refined as water) and copper is 1.8 Å (SI Appendix, Fig. S10), and that this distance corresponds to that of the Cu(II)-oxyl species described in a recently suggested catalytic mechanism for LPMOs (1.8 Å) (14) and in other studies (1.8–1.9 Å) (29–31).
Metal Binding, Redox Potential, and EPR Spectroscopy.
ITC-measured dissociation constants were 31 nM for CelS2-Cu2+ and 12 nM for ScLPMO10B-Cu2+ (SI Appendix, Fig. S11 and Table 1). Redox potentials of 242 ± 7 mV for CelS2 and 251 ± 15 mV for ScLPMO10B were determined as described previously (2) and shown in SI Appendix, Fig. S12. Combining the redox potentials and dissociation constants for Cu2+ in three thermodynamic relationships (SI Appendix, Fig. S12) allowed estimation of the dissociation constants for reduced copper (Cu+), resulting in values of 1.1 nM for CelS2 and 0.3 nM for ScLPMO10B.
Table 1.
Thermodynamic parameters obtained from ITC experiments performed in triplicates for binding of Cu2+ to apo-CelS2 or apo-ScLPMO10B at pH 5.5 and t = 10 °C
| LPMO | Kd, nM | ∆G°, kcal/mol | ∆H°, kcal/mol | −T∆S°, kcal/mol |
| CelS2 | 31 ± 6 | −9.7 ± 0.1 | −6.4 ± 0.6 | −3.3 ± 0.6 |
| ScLPMO10B | 12 ± 6 | −10.3 ± 0.3 | −9.6 ± 1.3 | −0.7 ± 1.3 |
Copper coordination was studied by EPR spectroscopy (SI Appendix, Fig. S13). The EPR spectra were simulated; the estimated spin Hamiltonian parameters are summarized in Table 2. The g and ACu tensors reflect the active site copper coordination environment, and of these the gz and tensors could be modeled with greatest accuracy (3). The gz and tensors of CelS2 (gz = 2.267; = 153 ∙ 10−4 cm−1) and ScLPMO10B (gz = 2.270; = 158 ∙ 10−4 cm−1) were similar and resemble those reported previously for cellulose-active TaLPMO9A (gz = 2.27; = 162 ∙ 10−4 cm−1) (10). Interestingly, chitin-active LPMO10s showed a lower tensor (SI Appendix, Fig. S14).
Table 2.
Spin Hamiltonian parameters*
| Parameter | Cu(II) buffer | ScLPMO10B | CelS2 |
| gx | 2.059 | 2.020 | 2.015 |
| gy | 2.059 | 2.090 | 2.102 |
| gz | 2.270 | 2.270 | 2.267 |
| AxCu† | 12.3 | 5.0 | 11.7 |
| AyCu† | 12.3 | 10.0 | 17.0 |
| AzCu† | 165 | 158 | 153 |
Assuming collinear g and ACu tensors in all simulations.
(10−4 cm−1).
Discussion
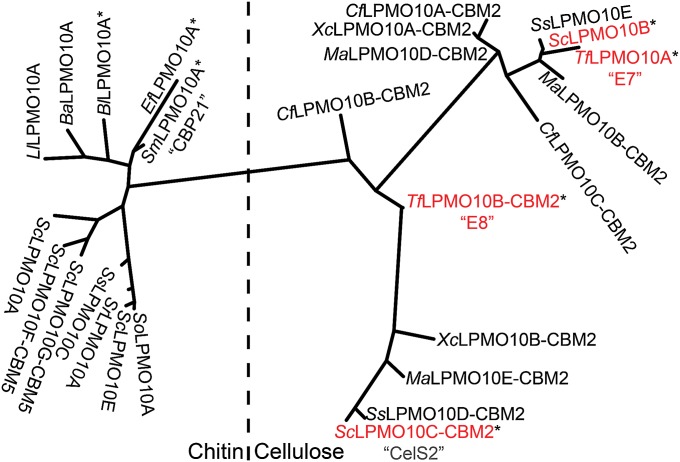
All of the LPMO10s that were characterized before the present study are C1 oxidizers (5, 16, 20, 25), whereas the family of cellulose-active fungal LPMO9s contains strict C1 oxidizers, strict C4 oxidizers, and enzymes that can oxidize both C1 and C4 (15). Here we show that the cellulolytic enzyme systems of S. coelicolor and T. fusca also are equipped with more than one LPMO type, a strict C1 oxidizer (CelS2 and E8) and a C1/C4 oxidizer (ScLPMO10B and E7). Importantly, the two enzymes display synergy when acting on cellulose (Fig. 2 and SI Appendix, Fig. S6), providing a possible explanation for the occurrence of a multitude of LPMOs in biomass-degrading microorganisms. Interestingly, sequence analysis shows that this pair of enzymes is common in other cellulolytic actinomycetes, including Cellulomonas, Micromonospora, Streptomyces, Thermobifida, and Xylanimonas (Fig. 4).
Fig. 4.
Phylogeny of six S. coelicolor LPMO10s and a selection of other LPMO10s, selected on the basis of literature data documenting up-regulation during growth on biomass (19), binding to chitin (26, 33, 34), synergism with glycoside hydrolases (35), and/or substrate cleavage and oxidation (5, 16, 20, 25). Proteins for which substrate degradation and oxidation has been demonstrated are labeled with an asterisk.
The two crystal structures presented here and the data on redox properties and metal binding allow, for the first time to our knowledge, structural and functional comparison of LPMOs belonging to the same family but having different substrate specificities. Whereas phylogenetic analysis of LPMO10s with known or inferred activities on chitin and cellulose shows clear separation between the two substrate specificities (Fig. 4), the structural and functional data reveal few pronounced differences. The chitin- and cellulose-active LPMO10s have similar redox potentials and copper affinities [data for chitin from Aachmann et al. (2) and Hemsworth et al. (3)], and the cores of their copper-binding sites, the histidine braces, are essentially identical. The copper-binding sites of CelS2 and chitin-active LPMO10s, such as CBP21, are remarkably similar (Fig. 3), whereas the copper-binding site of ScLPMO10B is clearly different, resembling that of LPMO9s. On the other hand, the EPR spectrum of ScLPMO10B is very similar to that of CelS2.
Two clear findings stand out. First, chitin-active and cellulose-active LPMO10s seem to be separated by their AzCu (SI Appendix, Fig. S14), and the values suggest a more distorted axial geometry in the former than in the latter (3, 20). It should be noted, however, that the recently discovered chitin-active LPMO11 from Aspergillus oryzae is grouped with the cellulose-active LPMOs according to the Peisach–Blumberg plot (SI Appendix, Fig. S14) (9). Second, the substrate-binding surface of chitin-active LPMO10s consistently contains a cavity adjacent to the active site (Fig. 3 and SI Appendix, Fig. S8) that is lacking from the cellulose-active enzymes, potentially accommodating an N-acetyl group.
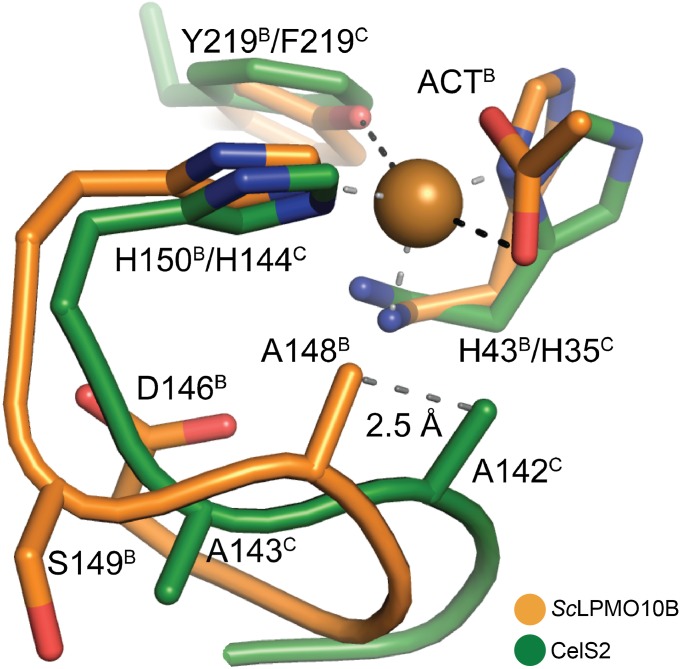
The conserved active site alanine in LPMO10s (Fig. 3) has been postulated to prevent ligands from binding in the solvent-facing axial position, making dioxygen activation more likely in the equatorial position (3, 11). Conversely, binding of dioxygen in the solvent-facing axial position has been proposed for a C1/C4-oxidizing LPMO9 (14), which lacks this alanine (10). Indeed, C1-oxidizing CelS2 seems incapable of ligand coordination in the solvent-facing axial position (SI Appendix, Fig. S10). In contrast, ScLPMO10B does coordinate a ligand (acetate) in this position, despite the presence of the alanine (SI Appendix, Fig. S9). Importantly, comparison of the structures presented here shows that the loop hosting the conserved alanine adopts different conformations in the two enzymes (Fig. 5). In ScLPMO10B, Asp146 forces the main chain of the loop into a conformation that positions Ala148 2.5 Å away from the position of the corresponding alanine (Ala142) in CelS2, allowing sufficient space for a ligand in the solvent-facing axial position. Asp146 is conserved in E7 and in other LPMO10s that phylogenetically cluster with E7 and ScLPMO10B (Fig. 4 and SI Appendix, Fig. S7). Importantly, closer inspection of available LPMO9 structures revealed a similar scenario (SI Appendix, Fig. S15); LPMO9s with C4- and C1/C4-oxidizing activity have an open solvent-facing axial coordination site, whereas LPMO9s known to be strict C1 oxidizers have a tyrosine, preventing optimal axial access to the copper ion (SI Appendix, Fig. S15). Thus, the ability to bind a ligand in the axial position could be a determinant of C4-oxidizing activity.
Fig. 5.
Position of the conserved active site alanine in CelS2 (green) and ScLPMO10B (orange). Black and gray dashed lines indicate the axially and equatorially coordinated ligands, respectively, in ScLPMO10B. ACT indicates the acetate ion; the skewed equatorial contact between the acetate and the copper is not drawn (SI Appendix, Fig. S9). Note the 2.5-Å relative shift in position of Ala142/148 between CelS2 and ScLPMO10B; in the former, the alanine (Ala142) is much closer to the acetate oxygen.
When the first combined functional and structural data on cellulose-active LPMO9s and chitin-active LPMO10s became available, it was suggested that differences in the copper-containing catalytic centers of these enzymes (i.e., between these families) could yield variable oxidative power, which in turn could affect the ability to cleave, for example, cellulose (11). Our data show no obvious correlation between the geometry of the copper center and substrate specificity. In fact, including recent data on a LPMO11, it would seem that LPMOs use a continuum of active site configurations, with the histidine brace being the key conserved element. All in all, it seems that substrate specificity depends on variations that are more remote from the active site and that affect substrate binding and positioning, as well as possibly electron transfer in the enzyme–substrate complex. The cavity observed in the binding surface of chitin-active LPMO10s supports this notion. On this note, one might expect some substrate promiscuity, as we indeed observed for ScLPMO10B. Clearly, additional data on enzyme–substrate interactions, supplementing data from modeling and NMR studies (2, 13, 27), are needed to gain more insight into the issue of substrate specificity.
Materials and Methods
Detailed information for all experimental procedures is provided in SI Appendix, Materials and Methods.
Enzyme Activity.
LPMOs were expressed heterologously in Escherichia coli and purified by chitin-affinity, ion-exchange, and size-exclusion chromatography. Substrate degradation was analyzed by MALDI-TOF MS and high-performance anion-exchange chromatography. Standard reactions were set up with 2 g/L substrate (PASC or β-chitin), 1 µM LPMO, and 2 mM ascorbate in 20 mM ammonium acetate buffer (pH 6.0) in a shaking incubator at 50 °C.
Metal Binding by ITC.
Dissociation constants and thermodynamic data for binding of Cu2+ to ScLPMO10B and CelS2 were obtained by measuring the heat produced by titrating 4-µL aliquots of metal ion solution (120–150 µM) to 5 µM apo-LPMO in 20 mM Mes (pH 5.5) in a 1.42-mL reaction cell of a VP-ITC system (MicroCal) at 10 °C.
Determination of Redox Potential and Dissociation Constant.
The cell potential for the LPMO-Cu2+/LPMO-Cu+ redox couple was determined by monitoring a reaction between reduced N,N,N′,N′-tetramethyl-1,4-phenylenediamine (TMPred) and LPMO-Cu2+. The calculated E° value was then combined with ITC data to estimate the dissociation constant for Cu+ as described previously (2).
EPR Spectroscopy.
Resting-state EPR spectra were recorded for Cu2+-charged ScLPMO10B and CelS2 in Pipes buffer (pH 6.0) using a Bruker EleXsyS 560 SuperX instrument equipped with an ER 4122 SHQE SuperX high-sensitivity cavity and a liquid nitrogen cooled cold finger. The instrument settings were 0.5 mW microwave power, 5 G modulation amplitude, and a temperature of 77 K.
Protein Crystallization and Data Collection.
Details and statistics for protein crystallization, data collection, structure determination, and refinement are provided in SI Appendix, Materials and Methods and Table S1.
Supplementary Material
Acknowledgments
We thank Roland Ludwig for the purified MtCDH, Gabriele Cordara for assistance with data collection, the European Synchrotron Radiation Facility and Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung staff for help and beamtime, and Gregg Beckham and Seonah Kim (National Renewable Energy Laboratory, Golden, CO) for helpful discussions. This work was funded by Norwegian Research Council Grants 190965, 214138, and 214613 and by the Norwegian Academy of Science and Letters Vista Program Grant 6505. A.S.A.’s work on structural methods is supported by the US Department of Energy’s Integrated Diffraction Analysis Technologies program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4OY6, 4OY7, and 4OY8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402771111/-/DCSupplemental.
References
- 1.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VGH. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aachmann FL, Sørlie M, Skjåk-Bræk G, Eijsink VGH, Vaaje-Kolstad G. NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc Natl Acad Sci USA. 2012;109(46):18779–18784. doi: 10.1073/pnas.1208822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemsworth GR, et al. The copper active site of CBM33 polysaccharide oxygenases. J Am Chem Soc. 2013;135(16):6069–6077. doi: 10.1021/ja402106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaaje-Kolstad G, Horn SJ, van Aalten DM, Synstad B, Eijsink VGH. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J Biol Chem. 2005;280(31):28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- 5.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 6.Harris PV, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry. 2010;49(15):3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 7.Cannella D, Jørgensen H. Do new cellulolytic enzyme preparations affect the industrial strategies for high solids lignocellulosic ethanol production? Biotechnol Bioeng. 2014;111(1):59–68. doi: 10.1002/bit.25098. [DOI] [PubMed] [Google Scholar]
- 8.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10(2):122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Beeson WT, Phillips CM, Cate JH, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134(2):890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Beeson WT, 4th, Phillips CM, Marletta MA, Cate JH. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure. 2012;20(6):1051–1061. doi: 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Ståhlberg J, Sandgren M, Paton RS, Beckham GT. Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc Natl Acad Sci USA. 2014;111(1):149–154. doi: 10.1073/pnas.1316609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vu VV, Beeson WT, Phillips CM, Cate JH, Marletta MA. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc. 2014;136(2):562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 16.Forsberg Z, et al. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011;20(9):1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaksen T, et al. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289(5):2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berka RM, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29(10):922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- 19.Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci Rep. 2013;3:1030. doi: 10.1038/srep01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg Z, et al. Comparative study of two chitin-active and two cellulose-active AA10-type lytic polysaccharide monooxygenases. Biochemistry. 2014;53(10):1647–1656. doi: 10.1021/bi5000433. [DOI] [PubMed] [Google Scholar]
- 21.Hodgson DA. Primary metabolism and its control in streptomycetes: A most unusual group of bacteria. Adv Microb Physiol. 2000;42:47–238. doi: 10.1016/s0065-2911(00)42003-5. [DOI] [PubMed] [Google Scholar]
- 22.Adav SS, Ng CS, Arulmani M, Sze SK. Quantitative iTRAQ secretome analysis of cellulolytic Thermobifida fusca. J Proteome Res. 2010;9(6):3016–3024. doi: 10.1021/pr901174z. [DOI] [PubMed] [Google Scholar]
- 23.Garda AL, Fernández-Abalos JM, Sánchez P, Ruiz-Arribas A, Santamaría RI. Two genes encoding an endoglucanase and a cellulose-binding protein are clustered and co-regulated by a TTA codon in Streptomyces halstedii JM8. Biochem J. 1997;324(Pt 2):403–411. doi: 10.1042/bj3240403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westereng B, et al. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE. 2011;6(11):e27807. doi: 10.1371/journal.pone.0027807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaaje-Kolstad G, et al. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J Mol Biol. 2012;416(2):239–254. doi: 10.1016/j.jmb.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Chu HH, Hoang V, Hofemeister J, Schrempf H. A Bacillus amyloliquefaciens ChbB protein binds beta- and alpha-chitin and has homologues in related strains. Microbiology. 2001;147(Pt 7):1793–1803. doi: 10.1099/00221287-147-7-1793. [DOI] [PubMed] [Google Scholar]
- 27.Wu M, et al. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the Basidiomycota fungus Phanerochaete chrysosporium. J Biol Chem. 2013;288(18):12828–12839. doi: 10.1074/jbc.M113.459396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaaje-Kolstad G, Houston DR, Riemen AH, Eijsink VGH, van Aalten DM. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J Biol Chem. 2005;280(12):11313–11319. doi: 10.1074/jbc.M407175200. [DOI] [PubMed] [Google Scholar]
- 29.Yoshizawa K, Kihara N, Kamachi T, Shiota Y. Catalytic mechanism of dopamine beta-monooxygenase mediated by Cu(III)-oxo. Inorg Chem. 2006;45(7):3034–3041. doi: 10.1021/ic0521168. [DOI] [PubMed] [Google Scholar]
- 30.Huber SM, et al. Generating Cu(II)-oxyl/Cu(III)-oxo species from Cu(I)-alpha-ketocarboxylate complexes and O2: In silico studies on ligand effects and C-H-activation reactivity. Chemistry. 2009;15(19):4886–4895. doi: 10.1002/chem.200802338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crespo A, Martí MA, Roitberg AE, Amzel LM, Estrin DA. The catalytic mechanism of peptidylglycine alpha-hydroxylating monooxygenase investigated by computer simulation. J Am Chem Soc. 2006;128(39):12817–12828. doi: 10.1021/ja062876x. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL, Lam JW. PyMOL: A communications tool for computational models. Abstr Pap Am Chem S. 2005;230:U1371–U1372. [Google Scholar]
- 33.Kolbe S, Fischer S, Becirevic A, Hinz P, Schrempf H. The Streptomyces reticuli alpha-chitin–binding protein CHB2 and its gene. Microbiology. 1998;144(Pt 5):1291–1297. doi: 10.1099/00221287-144-5-1291. [DOI] [PubMed] [Google Scholar]
- 34.Schnellmann J, Zeltins A, Blaak H, Schrempf H. The novel lectin-like protein CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridis gene and binds specifically to crystalline alpha-chitin of fungi and other organisms. Mol Microbiol. 1994;13(5):807–819. doi: 10.1111/j.1365-2958.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 35.Vaaje-Kolstad G, Bunaes AC, Mathiesen G, Eijsink VGH. The chitinolytic system of Lactococcus lactis ssp. lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of alpha- and beta-chitin. FEBS J. 2009;276(8):2402–2415. doi: 10.1111/j.1742-4658.2009.06972.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.