Significance
Noninvasive prenatal testing (NIPT) using fetal DNA in maternal plasma has been rapidly adopted worldwide. Current NIPT for fetal chromosomal disorders are based on the counting of DNA molecules in maternal plasma. Here, we show that plasma DNA-based molecular diagnostics can also be built around DNA fragment size, instead of count. First, we demonstrate that the fetal DNA fraction in maternal plasma can be rapidly measured by size analysis, even simply using microchip-based capillary electrophoresis. Second, we show that plasma DNA size analysis can be used for the detection of multiple types of fetal chromosomal aneuploidies with high accuracy. This strategy has many potential diagnostic applications, e.g., in oncology and transplantation monitoring.
Keywords: size profiling, fetal aneuploidy, next-generation sequencing, Down syndrome, Turner syndrome
Abstract
Noninvasive prenatal testing using fetal DNA in maternal plasma is an actively researched area. The current generation of tests using massively parallel sequencing is based on counting plasma DNA sequences originating from different genomic regions. In this study, we explored a different approach that is based on the use of DNA fragment size as a diagnostic parameter. This approach is dependent on the fact that circulating fetal DNA molecules are generally shorter than the corresponding maternal DNA molecules. First, we performed plasma DNA size analysis using paired-end massively parallel sequencing and microchip-based capillary electrophoresis. We demonstrated that the fetal DNA fraction in maternal plasma could be deduced from the overall size distribution of maternal plasma DNA. The fetal DNA fraction is a critical parameter affecting the accuracy of noninvasive prenatal testing using maternal plasma DNA. Second, we showed that fetal chromosomal aneuploidy could be detected by observing an aberrant proportion of short fragments from an aneuploid chromosome in the paired-end sequencing data. Using this approach, we detected fetal trisomy 21 and trisomy 18 with 100% sensitivity (T21: 36/36; T18: 27/27) and 100% specificity (non-T21: 88/88; non-T18: 97/97). For trisomy 13, the sensitivity and specificity were 95.2% (20/21) and 99% (102/103), respectively. For monosomy X, the sensitivity and specificity were both 100% (10/10 and 8/8). Thus, this study establishes the principle of size-based molecular diagnostics using plasma DNA. This approach has potential applications beyond noninvasive prenatal testing to areas such as oncology and transplantation monitoring.
In the plasma of pregnant women, cell-free fetal DNA is present in a large background of maternally derived DNA (1). Cell-free DNA molecules are mainly short fragments of less than 200 bp (2, 3). Early work based on real-time quantitative PCR has shown that fetal DNA is generally shorter than maternally derived DNA (2). Subsequently, researchers have taken advantage of such a size difference to enrich for fetal DNA in maternal plasma samples for noninvasive prenatal testing (4–6).
More recently, the development of paired-end massively parallel sequencing has allowed the size distributions of fetally and maternally derived DNA to be studied at a single-base resolution (7, 8). Both the size distributions of fetally and maternally derived DNA exhibit a series of peaks, including a major peak at 166 bp, a smaller peak at 143 bp, and a 10-bp periodicity below 143 bp (8). The most distinctive difference between fetal and maternal DNA in maternal plasma is that fetal DNA shows a reduced proportion of molecules of 166 bp and an increased proportion of molecules of less than 150 bp (8). In this study, we outlined the theoretical basis and explored the implementation of using molecular size analysis of plasma DNA as a diagnostic approach. We demonstrated the feasibility of this approach using two important applications in the field of noninvasive prenatal testing, namely, for measuring fetal DNA fraction in maternal plasma and detecting fetal aneuploidy.
The fetal DNA fraction in maternal plasma is an important parameter that affects the accuracy of cell-free DNA-based prenatal tests (9, 10). In particular, false-negative results could occur in samples with low fetal DNA fractions (9). In addition, a number of laboratories have incorporated the fetal DNA fraction into their diagnostic algorithms (6, 11). However, the added complexity of such a measurement means that such a practice has not been universally adopted. We reasoned that as fetal DNA is shorter than maternal DNA (2), plasma samples with higher fetal DNA fractions would have higher proportions of short DNA molecules. Thus, we proposed that it might be possible to estimate the fetal DNA fraction by measuring the overall size distribution of maternal plasma DNA.
The noninvasive prenatal detection of fetal chromosomal aneuploidies is the most rapidly adopted clinical use of noninvasive prenatal testing (12, 13). In pregnancies with aneuploid fetuses, the extra or missing copy of fetal chromosome would alter the proportional representation of the affected chromosome in the maternal plasma (14). Hence, massively parallel sequencing has been used for counting the number of sequences in maternal plasma that have originated from the affected chromosome (14–19). We reasoned that as fetal DNA is shorter than maternal DNA in maternal plasma (2), the presence of an extra fetal chromosome in fetal trisomy would shorten the size distribution of DNA in maternal plasma derived from that chromosome. For example, in a pregnant woman carrying a trisomy 21 fetus, the additional chromosome 21 would release an extra dosage of shorter fetal-derived DNA into the maternal plasma, resulting in an increase in the proportion of short DNA from chromosome 21. The reverse would take place in case of fetal monosomy, such as monosomy X for a fetus suffering from Turner syndrome. Therefore, we proposed that it might be possible to assess the fetal chromosome dosage by detecting the increased or reduced proportion of short fragments from the aneuploid chromosome in maternal plasma.
Results
Paired-End Sequencing of Maternal Plasma DNA.
Two sample sets were used in this study. The first sample set included 144 maternal plasma samples that had been analyzed in two previous studies (17, 20). These included 60 cases each with a euploid fetus, 36 cases each with a trisomy 21 fetus, 27 cases each with a trisomy 18 fetus, and 21 cases each with a trisomy 13 fetus. The second sample set included 26 maternal plasma samples from 16 cases each with a euploid female fetus and 10 cases each with a monosomy X fetus. The median gestational ages at the time of maternal blood sampling for the first and second sample sets were 13.0 wk (interquartile range 12.6–14.0), and 13.5 wk (interquartile range 13.2–13.9), respectively. All blood samples were collected before performing any invasive procedures.
All maternal plasma DNA samples were analyzed by paired-end massively parallel sequencing. For the first sample set, we obtained a median of 4.7 million (ranging from 1.8 million to 13.5 million) alignable and nonduplicated reads per sample for subsequent analysis. For the second sample set, a median of 35 million (ranging from 24.8 million to 44.4 million) reads per sample was used for subsequent analysis.
Size Distribution of DNA in Maternal Plasma Samples with Different Fetal DNA Fractions.
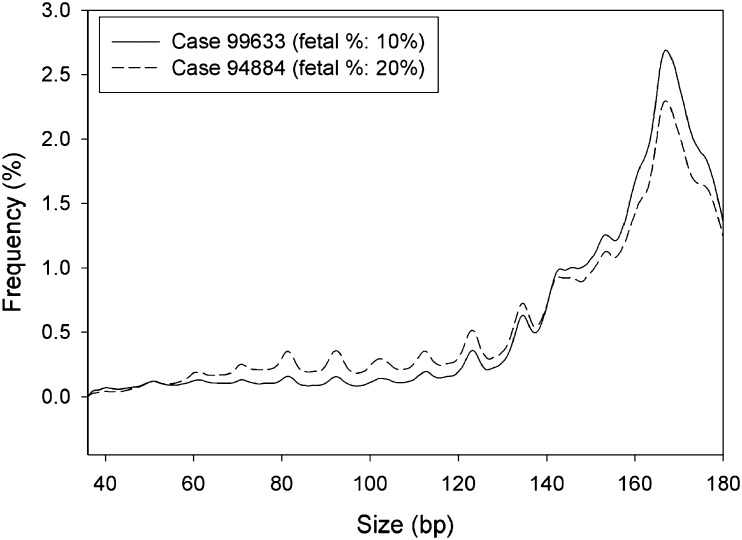
Because fetal DNA is generally shorter than maternal DNA (2), we hypothesized that maternal plasma samples with a higher fetal DNA fraction would have a higher proportion of short plasma DNA fragments. To confirm this hypothesis, we first determined the size distributions of plasma DNA molecules for two pregnant women each carrying a male fetus (from the first sample set). The size of each sequenced plasma DNA molecule was deduced from the start and end coordinates of the paired-end reads. The plasma sample with a higher fetal DNA fraction had a higher proportion of short fragments of less than 150 bp and a lower proportion of fragments of 166 bp compared with the sample with a lower fetal DNA fraction (Fig. 1).
Fig. 1.
Size distributions of DNA molecules in two maternal plasma samples with different fetal DNA fractions. Solid line represents the sample (case 99633) with a lower fetal DNA fraction, whereas dashed line represents the sample (case 94884) with a higher fetal DNA fraction.
Fetal DNA Fraction Estimation Through Size Analysis of Maternal Plasma DNA by Sequencing.
We further postulated that if the relative proportions of short and long DNA fragments in the maternal plasma were correlated with the fetal DNA fraction, we would be able to determine the fetal DNA fraction in maternal plasma by maternal plasma DNA size analysis. Thus, we analyzed plasma samples from 73 pregnant women, each carrying a male fetus (from the first sample set). We performed paired-end sequencing and determined the overall plasma DNA size distribution for each maternal plasma DNA sample (Dataset S1). A size ratio indicating the relative proportions of short and long DNA fragments was calculated for each sample using the following equation.
where P(100−150) denotes the proportion of sequenced fragments with sizes ranging from 100 bp to 150 bp; and P(163−169) denotes the proportion of sequenced fragments with sizes ranging from 163 bp to 169 bp.
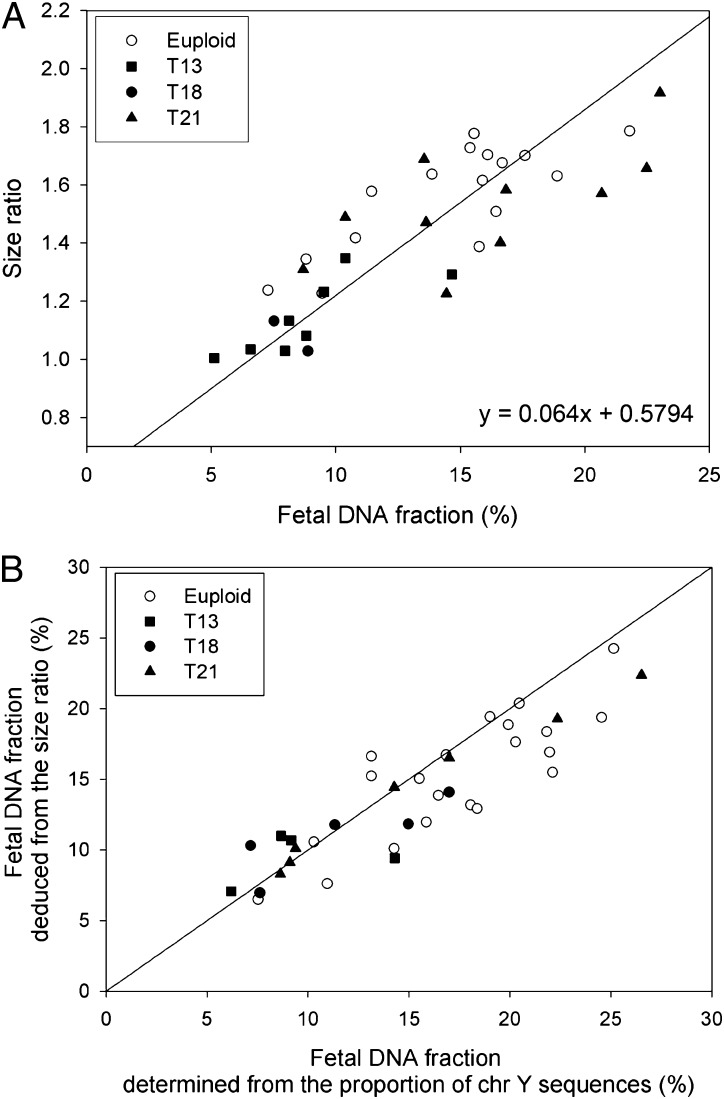
The 73 samples were randomly divided into two groups, namely a training group and a validation group, containing 36 and 37 samples, respectively. We first examined the relationship between the size ratio and the fetal DNA fraction with the training group. The fetal DNA fraction was determined from the proportion of chromosome Y sequences in the maternal plasma samples. We observed a positive correlation between the size ratio and the fetal DNA fraction (r = 0.827, P < 0.0001, linear regression) (Fig. 2A).
Fig. 2.
(A) Correlation between size ratios and fetal DNA fractions for the 36 samples in the training group. The fetal DNA fraction is determined from the proportion of chromosome Y sequences in the maternal plasma samples. The solid line is the linear regression line through the data, and the corresponding regression equation is given on the graph. (B) Plot of the fetal DNA fraction deduced from the size ratio using the above regression equation against that determined from the proportion of chromosome Y sequences for the 37 samples in the validation group.
Then, we deduced the fetal DNA fractions of the 37 maternal plasma samples of the validation group from the size ratios using the regression equation obtained from the training group. The size-deduced fetal DNA fractions were highly concordant with those determined from the proportion of chromosome Y sequences (Fig. 2B). The median absolute difference between the two values was 2.3% (interquartile range: 0.5–3.5%).
Fetal DNA Fraction Estimation Using Size Information Obtained from Electrophoresis of Maternal Plasma DNA Sequencing Libraries.
We further explored if the size information obtained from microchip-based capillary electrophoresis (using a Bioanalyzer) of the maternal plasma DNA sequencing library could be used to deduce the fetal DNA fraction in maternal plasma.
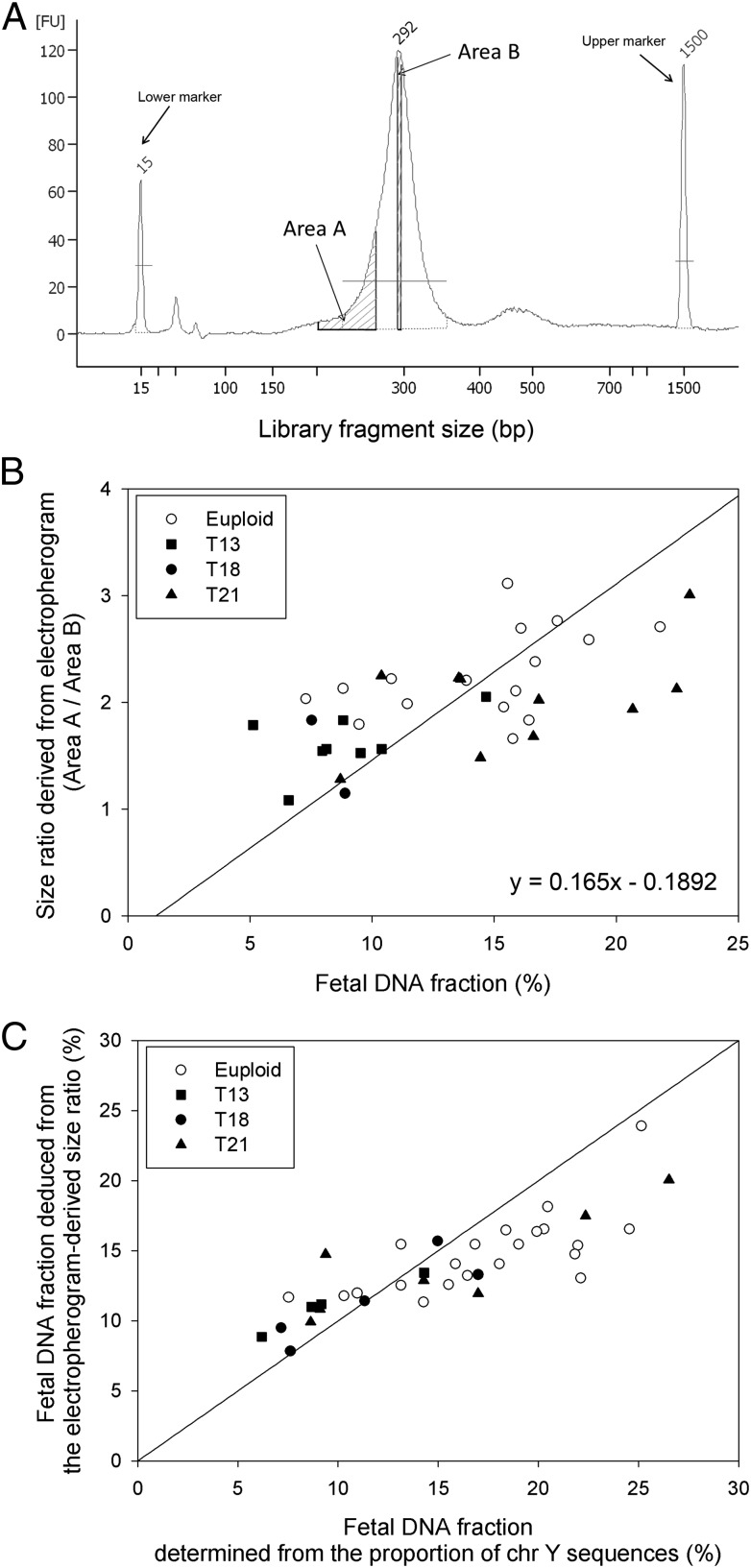
As a quality control step before sequencing, we checked the size distribution of each adaptor-ligated DNA library with the Bioanalyzer. A representative electropherogram of a maternal plasma DNA sequencing library obtained from the Bioanalyzer is shown in Fig. 3A. Areas A and B on the electropherogram correspond to library fragment sizes of 200–265 bp and 285–290 bp, respectively. During DNA sequencing library preparation, adaptors with a total size of 122 bp were ligated to both ends of each plasma DNA molecule. Therefore, areas A and B actually represent DNA fragments with sizes of 78–143 bp and 163–168 bp, respectively. Electropherograms of the 73 plasma DNA sequencing libraries from pregnant women with a male fetus (from the first sample set) were analyzed. A size ratio was calculated for each maternal plasma DNA sample by dividing area A by area B.
Fig. 3.
(A) A representative Bioanalyzer electropherogram of a maternal plasma DNA sequencing library. Areas A and B correspond to library fragment sizes of 200–265 bp and 285–290 bp, respectively. The library fragment size is the size of the plasma DNA molecule plus the size of the sequencing adaptor (122 bp). (B) Correlation between size ratios (area A/area B) derived from electropherograms and fetal DNA fractions for the 36 samples in the training group. The fetal DNA fraction is determined from the proportion of chromosome Y sequences in the maternal plasma samples. The solid line is the linear regression line through the data, and the corresponding regression equation is given on the graph. (C) Plot of the fetal DNA fraction deduced from the electropherogram-derived size ratio using the above regression equation against the fetal DNA fraction determined from the proportion of chromosome Y sequences for the 37 samples in the validation group.
We first examined the relationship between the size ratio obtained from the electropherogram and the fetal DNA fraction with the training group. We found a positive correlation between the size ratio obtained from electropherogram and the fetal DNA fraction (r = 0.610, P < 0.0001, linear regression) (Fig. 3B).
Then, we deduced the fetal DNA fractions of the 37 maternal plasma samples of the validation group from the size ratios using the regression equation obtained from the training group. The size-deduced fetal DNA fractions were highly concordant with those determined from the proportion of chromosome Y sequences (Fig. 3C). The median difference between the two values was 2.3% (interquartile range: 1.4–4.0%).
Fetal Trisomies 21, 18, and 13 Detection by Size Analysis of Maternal Plasma DNA by Sequencing.
In a previous study, we have demonstrated that the entire fetal genome is represented in the maternal plasma, and fetal DNA is present in a constant relative proportion to maternal DNA across the genome (8). When there is an extra or a missing copy of a fetal chromosome, the relative proportions of fetal and maternal DNA for the affected chromosome would be altered. We postulated that this would also alter the size distribution of plasma DNA for the affected chromosome. Based on this reasoning, chromosome-specific size profiling of maternal plasma DNA can potentially be used for fetal aneuploidy detection.
We first tested this size profiling method for the detection of the three most common autosomal aneuploidies, namely trisomy 21, trisomy 18, and trisomy 13, with the first sample set. We constructed the size distribution of maternal plasma DNA individually for each chromosome using only those DNA fragments originating from that chromosome (Dataset S1). We then calculated the difference in the proportion of short DNA fragments between the target chromosome and the reference chromosomes (all autosomes except chromosomes 21, 18, and 13), denoted by ∆FchrN, using the following equation.
where P(≤ 150)chrN denotes the proportion of sequenced fragments originating from the target chromosome with sizes ≤ 150 bp and P(≤ 150)chrRef denotes the proportion of sequenced fragments originating from the reference chromosomes with sizes ≤ 150 bp.
Next, we randomly assigned twenty cases with a euploid fetus in the first sample set as reference controls. We determined the mean values and SDs of ΔFchr21, ΔFchr18, and ΔFchr13 of these reference controls and calculated a size-based z-score for each of the three target chromosomes for each test sample using the following equation.
where ΔFchrN_sample is the ΔFchrN for the test sample, meanΔFchrN_ref is the mean ΔFchrN of the reference samples, and SDΔFchrN_ref is the SD of the ΔFchrN of the reference samples.
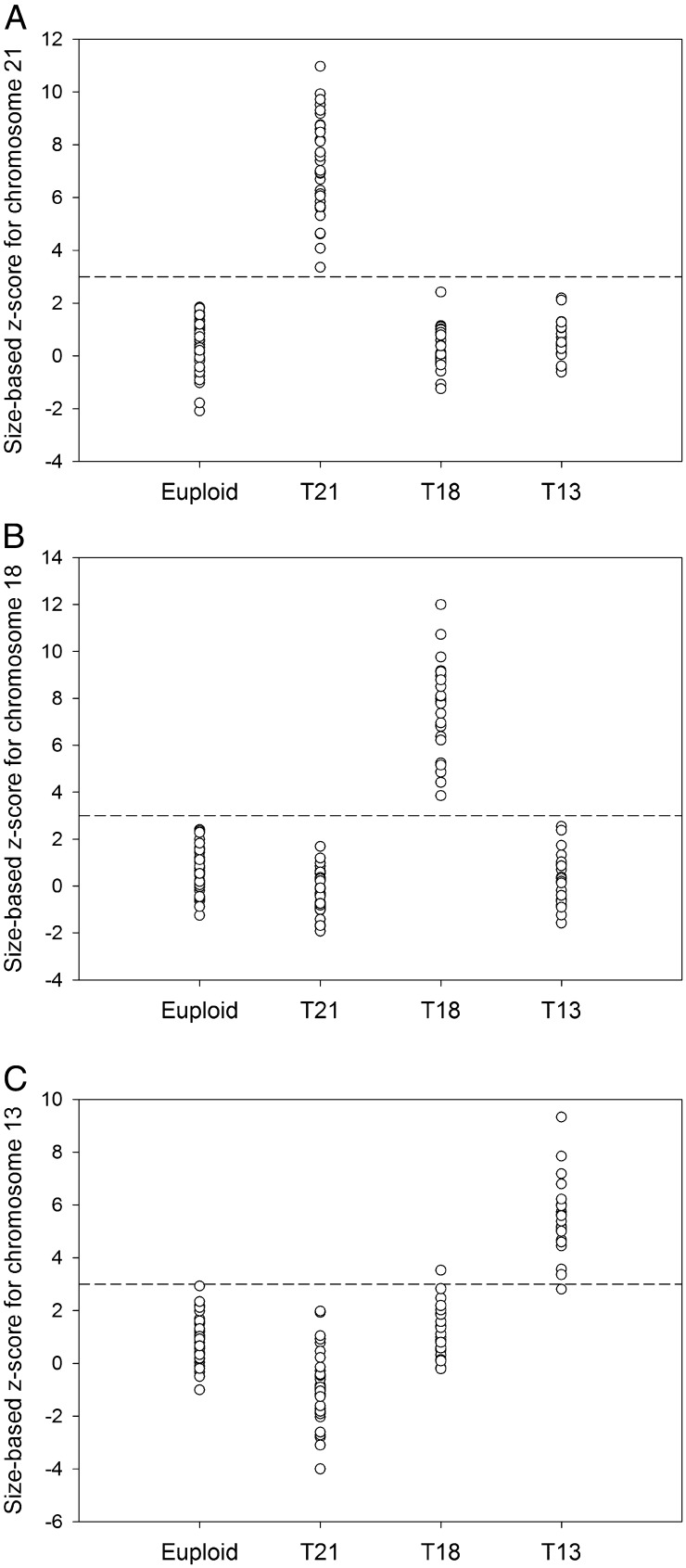
Using a size-based z-score cutoff value of >3, all 36 trisomy 21 cases and 88 nontrisomy 21 cases were correctly identified (Fig. 4A and Table S1). All 27 trisomy 18 cases and 97 nontrisomy 18 cases were correctly identified (Fig. 4B and Table S1). Twenty out of 21 trisomy 13 cases (sensitivity: 95.2%) and 102 out of 103 nontrisomy 13 cases (specificity: 99%) were correctly identified (Fig. 4C and Table S1).
Fig. 4.
Size-based z-scores for (A) chromosome 21, (B) chromosome 18, and (C) chromosome 13 for the 124 test samples in the first sample set (which include 40 euploid, 36 trisomy 21, 27 trisomy 18, and 21 trisomy 13 cases). Dashed line in each panel indicates the z-score cutoff value of 3.
The same set of samples was also analyzed for fetal trisomies 21, 18, and 13 using the tag counting method. All cases were correctly identified (Table S1).
Fetal Monosomy X Detection.
Thus far, results from a number of studies have suggested that the performance of fetal sex chromosomal aneuploidy detection by maternal plasma DNA analysis is less robust than for detecting an autosomal aneuploidy such as Down syndrome (15, 21). Here, we explored if the size profiling method could be used for detecting monosomy X (Turner syndrome) with the second sample set (Dataset S2).
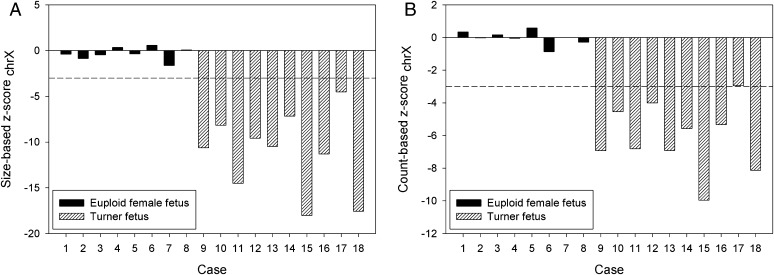
We randomly assigned eight pregnant cases carrying euploid female fetuses from the second sample set as reference controls. The remaining eight pregnancies carrying a euploid female fetus and 10 pregnancies each carrying a monosomy X fetus were the test cases. First, we confirmed that there was no significant amount of chromosome Y sequences in the 18 test cases, indicating that these fetuses did not carry a chromosome Y. Then, we assessed the fetal chromosome X dosage using both the tag counting and the size profiling methods. We used a z-score of <−3 as the cutoff for indicating monosomy X. Using the size profiling method, both the sensitivity and specificity for detecting monosomy X were 100% (Fig. 5A and Table S1). Using the tag counting method, the sensitivity and specificity were 90% and 100%, respectively (Fig. 5B and Table S1).
Fig. 5.
(A) Size-based and (B) count-based z-scores for chromosome X of the 18 cases in the second sample set, including 8 cases with euploid female fetuses and 10 cases with monosomy X fetuses. Another 8 cases with euploid female fetuses from the second sample set were used as reference controls. Dashed lines indicate the z-score cutoff value of −3.
Discussion
In this study, we have outlined the principle and demonstrated the realization of a size-based approach for molecular diagnostics using plasma DNA. First, we showed that one could estimate the fetal DNA fraction in maternal plasma based on the overall size distribution of maternal plasma DNA. The measurement of plasma DNA size distribution can be performed by both paired-end massively parallel sequencing and capillary electrophoresis of sequencing libraries. Furthermore, we demonstrated the use of maternal plasma DNA size analysis for the noninvasive prenatal detection of multiple types of fetal aneuploidies, including trisomies 21, 18, and 13, as well as monosomy X.
Fetal DNA fraction in maternal plasma is a crucial factor affecting the accuracy of maternal plasma DNA-based prenatal tests (9, 10). Currently, there are three main categories of fetal-specific markers for measuring the fetal DNA fraction, namely Y chromosomal markers (22), polymorphic markers (8, 23), and DNA methylation markers (24–26). The first category of markers would only be applicable to pregnancies involving male fetuses. The second category would require multiple polymorphic markers to achieve a broad population coverage. Although DNA methylation markers are potentially applicable to all pregnancies, it involves either bisulfite conversion (25) or methylation-sensitive restriction enzyme digestion (24, 26). A relatively simple measurement method for fetal DNA fraction applicable to all pregnancies, regardless of fetal sex and genetic makeup, would streamline the day-to-day operation of noninvasive prenatal tests. The size-based method described in this study represents an approach that could potentially fulfill this role. In particular, the size analysis using capillary electrophoresis of sequencing libraries is a very attractive approach for quick and reasonably accurate estimation of the fetal DNA fraction with virtually no additional cost, because this is one of the standard quality control steps for massively parallel sequencing (27).
Previously, it has been shown that besides the major peak at 166 bp, plasma DNA also exhibits a smaller peak at 143 bp (8). This 143-bp peak is more prominent in fetal DNA compared with the maternal counterpart. Therefore, during our initial investigation with the training set, we had tested a number of combinations of size ranges for the fetal fraction calculation, including (i) 100–150 bp and 163–169 bp, (ii) <150 bp and 163–169 bp, (iii) 140–146 bp and 163–169 bp, (iv) 140–154 bp and 163–169 bp, and (v) <150 bp and ≥150 bp. We had also explored a number of size cutoffs for fetal aneuploidy detection, including 140 bp, 145 bp, 150 bp, 155 bp, and 160 bp. Among them, 100–150 bp and 163–169 bp gave the best performance for fetal DNA fraction calculation, and a cutoff of 150 bp gave the best performance for fetal aneuploidy detection. Hence, these size ranges and size cutoff were used in this study.
Many current NIPT protocols adopt a lower cutoff of 3–4% fetal DNA fraction for fetal aneuploidy detection (18). In future studies, comparison of the performance of this size-based approach and the conventional approaches would need to include more plasma samples in the low concentration range.
The ability of the size-based approach to detect fetal chromosomal aneuploidies is an interesting development. Because the same sequencing data can be used for both the size profiling and the tag counting analyses, one may combine the results from two types of analyses for arriving at a final test result. One possibility is to combine results from these two types of analyses using an “OR” algorithm. Thus, a plasma sample is classified as positive if either the size-based or the count-based approach gives a positive result. An alternative is to combine results from these two types of analyses using an “AND” algorithm, where a plasma sample is classified as positive only if both the size-based and the count-based approaches give positive results. From our current dataset, we saw an indication that one could potentially improve the sensitivity of fetal aneuploidy detection with the OR algorithm (e.g., where the sensitivity for detecting trisomy 13 has increased from 95.2 to 100%), or reduce false positives with the AND algorithm (e.g., a small reduction in false positive rate for trisomy 13 detection) (Table S1). The actual diagnostic impact of such algorithms would require future validation using large sample cohorts. At present, we would predict that the AND algorithm and its associated enhancement in specificity would be of value if one wishes to expand the clinical spectrum of noninvasive prenatal testing, e.g., for chromosomal microdeletions or even obtaining a genomewide molecular karyotype (28–31). It is also possible that an even higher level of diagnostic specificity could be achieved by combining size, count, and DNA methylation information (32).
Finally, size-based molecular diagnostics could be extended to the analysis of cell-free DNA in other clinical contexts, such as cancer (33–35) and transplantation (36). Indeed, previous studies have shown a size difference between tumor-derived and noncancer cell-derived DNA (37, 38), as well as between donor-derived and recipient-derived DNA (39). In particular, it has been shown in a transplantation model that nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma (39). It is currently unclear how much one could extrapolate such results to an individual who has not previously undergone transplantation, e.g., a healthy pregnant woman. The striking association between the size distribution in plasma and the fetal DNA fraction suggests that in healthy pregnant women, the proportion of DNA from the nonfetal and nonhematopoietic sources is likely to be relatively minor or stable. Nonetheless, it would be important for future studies to explore the correlation between the plasma DNA size profile and the fetal DNA fraction in pregnant women suffering from various pregnancy-associated disorders, e.g., preeclampsia. Indeed, plasma DNA size profiling might even have diagnostic or monitoring implications for pregnant women with such disorders. In conclusion, size profiling of plasma DNA provides valuable biological information and has exciting diagnostic applications.
Materials and Methods
Details of the sample collection and processing are in SI Text.
Sequencing Library Preparation.
Indexed DNA libraries were constructed with the Paired-End Sequencing Sample Preparation Kit (Illumina) and the Multiplexing Sample Preparation Oligonucleotide Kit (Illumina) (20). The adaptor-ligated DNA was enriched by an 18-cycle PCR. Before sequencing, we checked the size distributions of the adaptor-ligated libraries using the DNA 1000 Kit (Agilent) with a 2100 Bioanalyzer (Agilent).
DNA Sequencing.
We sequenced all libraries in the first sample set using a 2-plex sequencing protocol on a Genome Analyzer IIx (Illumina). Among the 33 samples in the second sample set, three of them were sequenced with one lane on a HiSeq 2000 sequencer (Illumina), and the remaining 30 samples were sequenced using a 4-plex protocol. We performed 36 cycles and 50 cycles of paired-end sequencing for the first and second sample sets, respectively. An additional 7 cycles of sequencing were performed to decode the index sequence on each sequenced DNA molecule. For consistency to other samples in terms of read depth, we only used one-fourth of the reads from the one-lane data of the three samples for downstream analyses.
Sequence Alignment.
Sequences from each lane were assigned to the corresponding samples based on the six-base index sequences, with one mismatch being allowed in the index read sequences. All sequenced reads were aligned to the non-repeat-masked human reference genome (NCBI Build 36.1/hg18) (http://genome.ucsc.edu) using the Short Oligonucleotide Alignment Program 2 (SOAP2) (http://soap.genomics.org.cn/). For the first sample set, we did not allow any nucleotide mismatch in the sequencing reads, whereas for the second sample set, we allowed up to two nucleotide mismatches for each member of the paired-end reads due to a longer read length used. Only paired-end reads with both ends aligned to the same chromosome with the correct orientation, spanning an insert size of ≤600 bp, were used for downstream analyses. All but one duplicated reads with identical start and end coordinates were filtered.
Calculation of Fetal DNA Fractions from the Proportion of Chromosome Y Sequences.
In pregnancies carrying a male fetus, the fetal DNA fraction (f) in a maternal plasma sample can be determined from the proportion of reads aligned to chromosome Y (%chrY) as previously described (20). Details are in SI Text.
Detection of Chromosome Y.
We set a cutoff value of %chrY, below and above which represented the absence and presence of chromosome Y in the sample, respectively. We determined the mean and the SD of the %chrY of the eight euploid female fetus cases from the second sample set, and calculated the cutoff value which was defined as the mean plus three SDs of the %chrY of the eight euploid female fetus cases.
Tag Counting Analysis.
We determined the mean values and SDs of the genomic representation of the tested chromosome (GRchrN) of the reference controls, and calculated a count-based z-score for each chromosome of each test sample using the following equation (14):
where GRchrN_sample is the GRchrN of the sample, mean GRchrN_ref is the mean GRchrN of the reference samples, and SD GRchrN_ref is the standard deviation of the GRchrN of the reference samples.
Supplementary Material
Footnotes
Conflict of interest statement: R.W.K.C. and Y.M.D.L. have support from Sequenom, Inc. for the submitted work. R.W.K.C. and Y.M.D.L. are consultants to, and hold equities in, Sequenom, Inc. K.C.A.C. holds equities in Sequenom, Inc. The technology reported in this paper is covered by US Patent 8,620,593 and US Patent Application 2013/0237431.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406103111/-/DCSupplemental.
References
- 1.Lo YMD, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Chan KCA, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50(1):88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 3.Chan KCA, Yeung SW, Lui WB, Rainer TH, Lo YMD. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin Chem. 2005;51(4):781–784. doi: 10.1373/clinchem.2004.046219. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, et al. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem. 2004;50(6):1002–1011. doi: 10.1373/clinchem.2003.029835. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, et al. Detection of paternally inherited fetal point mutations for beta-thalassemia using size-fractionated cell-free DNA in maternal plasma. JAMA. 2005;293(7):843–849. doi: 10.1001/jama.293.7.843. [DOI] [PubMed] [Google Scholar]
- 6.Lun FMF, et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(50):19920–19925. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010;56(8):1279–1286. doi: 10.1373/clinchem.2010.144188. [DOI] [PubMed] [Google Scholar]
- 8.Lo YMD, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 9.Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013;33(7):667–674. doi: 10.1002/pd.4126. [DOI] [PubMed] [Google Scholar]
- 10.Lo YMD, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci USA. 2007;104(32):13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: Evaluation for trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206(4):319. doi: 10.1016/j.ajog.2012.01.030. e1-9. [DOI] [PubMed] [Google Scholar]
- 12.Greene MF, Phimister EG. Screening for trisomies in circulating DNA. N Engl J Med. 2014;370(9):874–875. doi: 10.1056/NEJMe1401129. [DOI] [PubMed] [Google Scholar]
- 13.Soothill PW, Lo YMD. Royal College of Obstetricians and Gynaecologists 2014. Non-invasive Prenatal Testing for Chromosomal Abnormality Using Maternal Plasma DNA, Scientific Impact Paper 15 (Royal College of Obstetricians and Gynaecologists, London). Available at www.rcog.org.uk/files/rcog-corp/SIP_15_04032014.pdf. Accessed March 4, 2014.
- 14.Chiu RWK, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(51):20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bianchi DW, et al. MatErnal BLood IS Source to Accurately diagnose fetal aneuploidy (MELISSA) Study Group Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 16.Buysse K, et al. Reliable noninvasive prenatal testing by massively parallel sequencing of circulating cell-free DNA from maternal plasma processed up to 24h after venipuncture. Clin Biochem. 2013;46(18):1783–1786. doi: 10.1016/j.clinbiochem.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Chen EZ, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS ONE. 2011;6(7):e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13(11):913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi DW, et al. CARE Study Group DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 20.Chiu RWK, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazloom AR, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat Diagn. 2013;33(6):591–597. doi: 10.1002/pd.4127. [DOI] [PubMed] [Google Scholar]
- 22.Lun FMF, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008;54(10):1664–1672. doi: 10.1373/clinchem.2008.111385. [DOI] [PubMed] [Google Scholar]
- 23.Chu T, Bunce K, Hogge WA, Peters DG. A novel approach toward the challenge of accurately quantifying fetal DNA in maternal plasma. Prenat Diagn. 2010;30(12-13):1226–1229. doi: 10.1002/pd.2656. [DOI] [PubMed] [Google Scholar]
- 24.Chan KCA, et al. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem. 2006;52(12):2211–2218. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- 25.Chim SS, et al. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci USA. 2005;102(41):14753–14758. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nygren AO, et al. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem. 2010;56(10):1627–1635. doi: 10.1373/clinchem.2010.146290. [DOI] [PubMed] [Google Scholar]
- 27.Liao GJ, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem. 2011;57(1):92–101. doi: 10.1373/clinchem.2010.154336. [DOI] [PubMed] [Google Scholar]
- 28.Jensen TJ, Dzakula Z, Deciu C, van den Boom D, Ehrich M. Detection of microdeletion 22q11.2 in a fetus by next-generation sequencing of maternal plasma. Clin Chem. 2012;58(7):1148–1151. doi: 10.1373/clinchem.2011.180794. [DOI] [PubMed] [Google Scholar]
- 29.Peters D, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011;365(19):1847–1848. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92(2):167–176. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu SCY, et al. Noninvasive prenatal molecular karyotyping from maternal plasma. PLoS ONE. 2013;8(4):e60968. doi: 10.1371/journal.pone.0060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lun FMF, et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin Chem. 2013;59(11):1583–1594. doi: 10.1373/clinchem.2013.212274. [DOI] [PubMed] [Google Scholar]
- 33.Chan KCA, et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59(1):211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 34.Chan KCA, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci USA. 2013;110(47):18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroun M, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 36.Lo YMD, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351(9112):1329–1330. doi: 10.1016/s0140-6736(05)79055-3. [DOI] [PubMed] [Google Scholar]
- 37.Chan KCA, Leung SF, Yeung SW, Chan AT, Lo YMD. Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clin Cancer Res. 2008;14(13):4141–4145. doi: 10.1158/1078-0432.CCR-08-0182. [DOI] [PubMed] [Google Scholar]
- 38.Diehl F, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng YWL, et al. Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: A transplantation model. Clin Chem. 2012;58(3):549–558. doi: 10.1373/clinchem.2011.169318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.