Significance
Sterols and triterpenes are complex molecules that are synthesized from the isoprenoid pathway. The functions of sterols in plants have been studied extensively, but the role of triterpenes is less well understood. Here we investigate triterpene synthesis and regulation in diploid oat. We show that the genes for triterpene synthesis are regulated by an ancient root development process that is conserved across diverse plants. We further show that mutants in which the metabolism of the most common plant triterpene, β-amyrin, is blocked undergo a change early in the development of the root epidermis that leads to a “superhairy” root phenotype. Our findings shed light on triterpene synthesis and provide evidence for a role for the simple triterpene β-amyrin in plant development.
Keywords: cell specification, root development, hormones
Abstract
Sterols have important functions in membranes and signaling. Plant sterols are synthesized via the isoprenoid pathway by cyclization of 2,3-oxidosqualene to cycloartenol. Plants also convert 2,3-oxidosqualene to other sterol-like cyclization products, including the simple triterpene β-amyrin. The function of β-amyrin per se is unknown, but this molecule can serve as an intermediate in the synthesis of more complex triterpene glycosides associated with plant defense. β-Amyrin is present at low levels in the roots of diploid oat (Avena strigosa). Oat roots also synthesize the β-amyrin–derived triterpene glycoside avenacin A-1, which provides protection against soil-borne diseases. The genes for the early steps in avenacin A-1 synthesis [saponin-deficient 1 and 2 (Sad1 and Sad2)] have been recruited from the sterol pathway by gene duplication and neofunctionalization. Here we show that Sad1 and Sad2 are regulated by an ancient root developmental process that is conserved across diverse species. Sad1 promoter activity is dependent on an L1 box motif, implicating sterol/lipid-binding class IV homeodomain leucine zipper transcription factors as potential regulators. The metabolism of β-amyrin is blocked in sad2 mutants, which therefore accumulate abnormally high levels of this triterpene. The accumulation of elevated levels of β-amyrin in these mutants triggers a “superhairy” root phenotype. Importantly, this effect is manifested very early in the establishment of the root epidermis, causing a greater proportion of epidermal cells to be specified as root hair cells rather than nonhair cells. Together these findings suggest that simple triterpenes may have widespread and as yet largely unrecognized functions in plant growth and development.
Sterols and triterpenes are isoprenoids that are synthesized via the mevalonate pathway (1). Plant sterols are important structural components of membranes and also have roles in signaling (as steroidal hormones). In contrast, triterpenes are not regarded as essential for normal plant growth and development and often accumulate as conjugates with carbohydrates and other macromolecules, most notably as triterpene glycosides. Triterpene glycosides have important ecological and agronomic functions, contributing to pest and pathogen resistance and to food quality in crop plants. They also have a wide range of commercial applications in the food, cosmetics, pharmaceutical, and industrial biotechnology sectors (2–5).
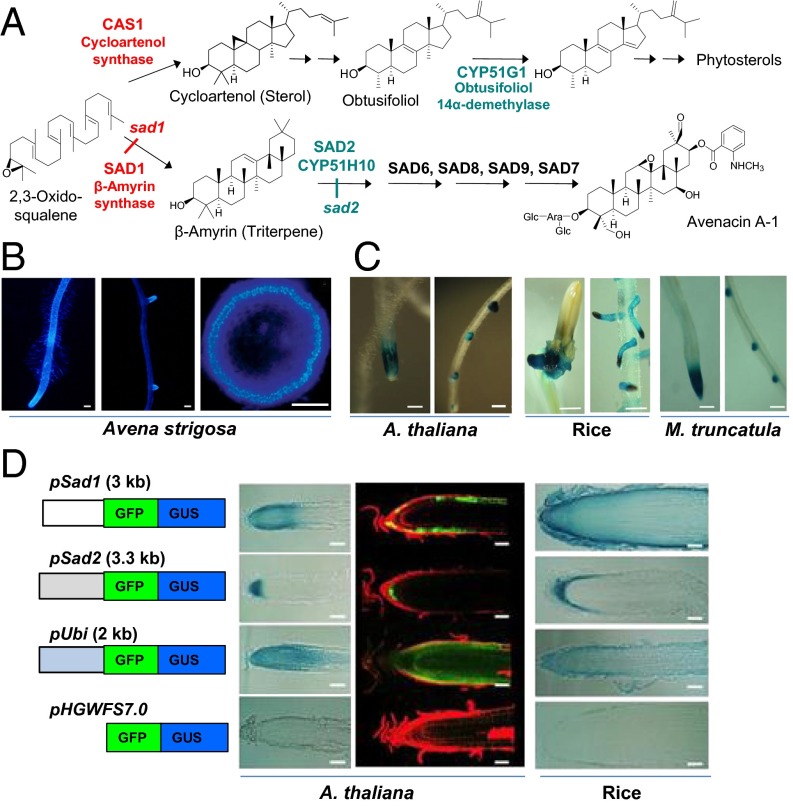
In plant sterol biosynthesis, 2,3-oxidosqualene is cyclized to cycloartenol, the precursor for essential primary sterols, by the oxidosqualene cyclase enzyme cycloartenol synthase (1). 2,3-Oxidosqualene can also be converted to alternative triterpene cyclization products, such as the major plant triterpene β-amyrin, by other oxidosqualene cyclases (1–5). Simple triterpenes are common, if not ubiquitous, in plants, but their function is unknown (5). In addition to existing in simple unmodified form, they often also serve as intermediates for the synthesis of more elaborate specialized metabolites, such as the antimicrobial triterpene glycoside avenacin A-1, which is produced by oats (Avena spp.) (Fig. 1A) (6). Avenacin A-1 is produced in the roots of oats and confers resistance to soil-borne diseases (7). The first committed step in avenacin biosynthesis is the cyclization of 2,3-oxidosqualene to the simple triterpene β-amyrin by the oxidosqualene cyclase, β-amyrin synthase (bAS1/SAD1) (8). β-Amyrin is then oxidized by the CYP450 enzyme CYP51H10 (SAD2) (9, 10), and further modified by a series of downstream enzymes to give the pathway end-product, avenacin A-1 (Fig. 1A) (11–14). Previously we have shown that the saponin-deficient 1 and 2 (Sad1 and Sad2) genes have been recruited from sterol biosynthesis by duplication and neofunctionalization of the genes for cycloartenol synthase (CAS1) and obtusifoliol 14 α-demethylase (CYP51G1), respectively (8, 9). Genes for primary sterol biosynthesis normally are expressed throughout different plant organs. In contrast, Sad1, Sad2, and other avenacin biosynthetic genes are expressed specifically in the epidermal cells of oat root tips, the site of accumulation of avenacin A-1 (8, 9, 11–13, 15). Interestingly, in the oat genome Sad1 and Sad2 are organized in a metabolic gene cluster together with other genes for avenacin biosynthesis (7, 9, 11–13, 15). There is evidence suggesting that physical clustering may facilitate regulation at the level of chromatin (16). However, very little is known about the regulation of avenacin biosynthesis at any level or indeed about the regulation of plant triterpene biosynthesis in general (5).
Fig. 1.
Sterols and avenacins are both synthesized from 2,3-oxidosqualene. (A) The first and second steps in the avenacin pathway are catalyzed by SAD1/bAS1 and SAD2/CYP51H10. These steps have been recruited from the sterol pathway enzymes CAS1 and CYP51G1, respectively. Other avenacin biosynthetic loci are indicated (7, 11, 12). (B) Fluorescence of avenacin A-1 in the root tip (Left) and lateral root primordia (Center) of A. strigosa (Scale bars: 200 μm.) (Right) Cross-section of a root showing that this fluorescence is localized in the epidermal cell layer. (Scale bar: 50 μm.) Reproduced with permission from ref. 16; Copyright American Society of Plant Biologists (www.plantcell.org). (C) GUS staining showing pSad1-reporter activity in A. thaliana, rice, and M. truncatula. (Scale bars: 100, 500, and 200 μm, respectively.) (D) Analysis of promoter activity in sections of A. thaliana and rice roots. (Scale bars: 50 μm and 100 μm, respectively). (Left) Constructs contained the Sad1, Sad2, or ubiquitin (pUbi) promoters fused to a GFP-GUS reporter; the promoterless construct pHGWFS7.0 was included as a control. The blue-stained sections (A. thaliana and rice) indicate GUS activity; in A. thaliana the green fluorescence indicates GFP expression, and the red fluorescence indicates propidium iodide staining of plant cell walls.
Here we show that the avenacin pathway genes Sad1 and Sad2 are under the control of an ancient regulatory mechanism associated with root development that is conserved across monocots and eudicots. We use promoter– reporter lines to investigate Sad1 promoter function in Arabidopsis thaliana and demonstrate that promoter activity is dependent on an L1 box motif, thus implicating sterol/lipid-binding class IV homeodomain leucine zipper (HD-ZIP IV) transcription factors (17) as potential regulators. We further demonstrate that the accumulation of elevated levels of β-amyrin results in a “superhairy” root phenotype in oats. Importantly, this effect impacts early in the establishment of the root epidermis, causing a greater proportion of epidermal cells to be specified as trichoblasts (specialized shorter cells that give rise to root hairs) rather than as atrichoblasts (longer cells that will not give rise to root hairs). Most determinants of root hair cells that have been characterized so far in plants act downstream of this process by regulating root hair formation (17–21). Our findings shed light on the regulation of triterpene synthesis and provide evidence for a role for the simple triterpene β-amyrin in plant development.
Results
Sad1 and Sad2 Are Under the Control of a Conserved Regulatory Mechanism Associated with Root Development in Higher Plants.
Avenacin A-1 has strong autofluorescence and can be visualized readily in the root tips and lateral roots of oat seedlings under UV illumination, specifically in the epidermal cells of the root meristems (Fig. 1B) (22). Previously, we have shown by mRNA in situ hybridization that the genes for avenacin biosynthesis are expressed specifically in this cell layer (8, 9, 11–13, 15). The physical clustering of the avenacin-pathway genes in oat is suggestive of regulation at the level of chromatin, a prediction supported by DNA FISH analysis of chromatin decondensation in this region (16). However, when the Sad1 promoter was fused to the β-glucuronidase (GUS) reporter gene and introduced into A. thaliana, the resulting GUS-expression pattern mirrored that of avenacin A-1 fluorescence in oats; i.e., GUS staining was detected in the root tips and lateral root initials (Fig. 1C). Similar results were obtained with rice and Medicago truncatula (Fig. 1C). This promoter therefore contains the information necessary for appropriate expression in the root tips and lateral root initials of diverse plant species, including both monocots and eudicots. Similar results were obtained with the Sad2 promoter in A. thaliana (Fig. S1A).
Further analysis using GUS and GFP reporters revealed that the Sad1 and Sad2 promoters drive reporter gene expression predominantly in the epidermal cells of the root tips in both A. thaliana and rice (Fig. 1D). Expression was also observed in the lateral root primordia (Fig. S1 A and B). Differences observed with the GUS and GFP reporters are likely to be caused by the diffusion of the GUS product into nonexpressing neighboring tissue. The activity of the Sad2 promoter (pSad2) in the root tip was more restricted than that of the Sad1 promoter (pSad1) and was localized primarily in the cells of the quiescent center and the epidermal stem cells (Fig. 1D). Interestingly in M. truncatula roots pSad1 activity was observed not only in primary roots and lateral roots but also in the meristematic regions of root nodules, suggesting similarities in the regulatory mechanisms within the root meristem and nodules (Fig. S1C).
These experiments led us to conclude that the Sad1 and Sad2 promoters are under the control of a highly conserved root development process that is present in both monocots and eudicots.
Analysis of the Sad1 Promoter.
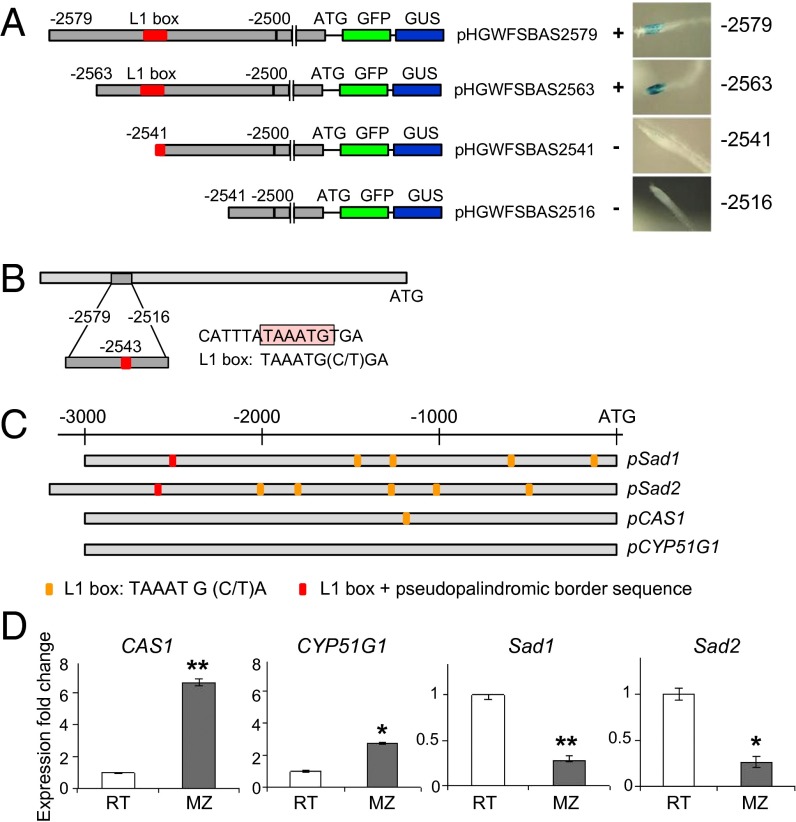
We next carried out deletion experiments using the 3-kb Sad1 promoter and A. thaliana as our expression system to identify promoter elements implicated in the regulation of gene expression. Preliminary experiments revealed that deletion of ∼500 bp from the 5′ end of the Sad1 promoter region resulted in the loss of GUS expression (Fig. S2A) and defined a region between −2598 and −2488 as critical for promoter function (Fig. S2B). Insertion of the 5′-most kilobase of the Sad1 promoter (the region from −2992 to −1874) directly in front of the GFP/GUS reporter genes (construct pHGWFSBAS2992–1847) resulted in GUS expression similar to that observed with the full 3-kb pSad1 region (Fig. S2B). Further fine deletion narrowed the critical region down to only 22 nucleotides (Fig. 2A). This region contains a pseudopalindromic sequence CATTTATAAATGTGA with an embedded L1 box motif [5′-TAAATG(C/T)A-3′] (Fig. 2B) (23). Pseudopalindromic sequences that overlap L1 boxes are binding sites for HD-ZIP IV proteins (24). Members of this family of transcription factors contain a steroidogenic acute regulatory protein-related lipid transfer (START) domain suggestive of sterol/lipid binding and are implicated in epidermis-related developmental processes (17). Seedlings transformed with a −2541 nucleotide construct disrupted in the L1 box motif showed very faint GUS staining in occasional individual cells, and the typical pSad1-expression pattern disappeared (Fig. 2A). Bioinformatics analysis revealed that a pseudopalindromic L1 box sequence also is present in the Sad2 promoter. The Sad1 and Sad2 promoters also have multiple core L1 box motifs. The promoters of the oat sterol biosynthesis genes CAS1 and CYP51G1 from which Sad1 and Sad2 have been recruited (8, 9, 15) do not contain pseudopalindromic L1 box sequences, although the CAS1 promoter has a single core L1 box motif (Fig. 2C).
Fig. 2.
Deletion experiments for pSad1 identify an L1 box motif that is required for promoter activity. (A) (Left) Promoter fragments (gray) are fused to the coding regions of GFP and GUS. The numbers indicate nucleotide positions relative to the translation start codon ATG. Fine deletion experiments indicate that the region between −2563 and −2541 is critical for GUS expression. This region contains an L1 box (indicated in red). (Right) GUS activity in 10-d-old A. thaliana seedlings is shown. (B) The L1 box and associated pseudopalindromic sequence identified in the Sad1 promoter sequence. (C) The presence or absence of full L1 boxes with pseudopalindromic sequences (red) and core L1 box sequences (yellow) in the promoter regions of Sad1, Sad2, CAS1, and CYP51G. (D) The genes for sterol and avenacin biosynthesis are inversely regulated at the level of transcription. qRT-PCR analysis of transcript levels in the terminal 1 mm of the root tips (RT) and in the maturation zone (0.5 cm) (MZ) of 3-d-old A. strigosa seedlings using gene-specific primers for CAS1, CYP51G1, Sad1, and Sad2. Transcript levels were normalized to those for EF1-α. Values are means ± SD (n = three plants). The data were analyzed using the comparative cycle threshold (CT) method (42). Relative transcript levels are shown. **P < 0.01; *P < 0.05 (unpaired two-tailed t test).
Expression of Sad1, Sad2, and the other avenacin biosynthesis genes is restricted to the epidermal cells of root tips and lateral root primordia (8–13, 15). We compared the relative transcript levels of Sad1, Sad2, and the corresponding sterol biosynthesis genes CAS1 and CYP51G1 in seedlings of diploid oat (Avena strigosa). CAS1 and CYP51G1 transcript levels were higher in the maturation zone than in the root tips, whereas the reverse was true for Sad1 and Sad2 transcript levels (Fig. 2D). These results are consistent with our earlier mRNA in situ experiments, which suggest that the sterol and avenacin pathways are spatially separated and inversely regulated at the level of transcription (16).
Experiments in which pSad1-GUS reporter lines of A. thaliana were crossed with mutants defective in auxin-related transport (25, 26) or signaling (27) gave normal GUS-expression patterns (Fig. S3). Homozygous mutant lines of the auxin response mutant MONOPTEROS, mpG12 (28), do not form primary root meristems during embryogenesis, and GUS expression was not seen in this mutant background. Occasionally mpG12 mutant seedlings produce adventitious roots (28), and GUS staining was detected in these rare adventitious roots (Fig. S3). Thus, Sad1 promoter activity is auxin independent but depends on the formation of the primary root meristem.
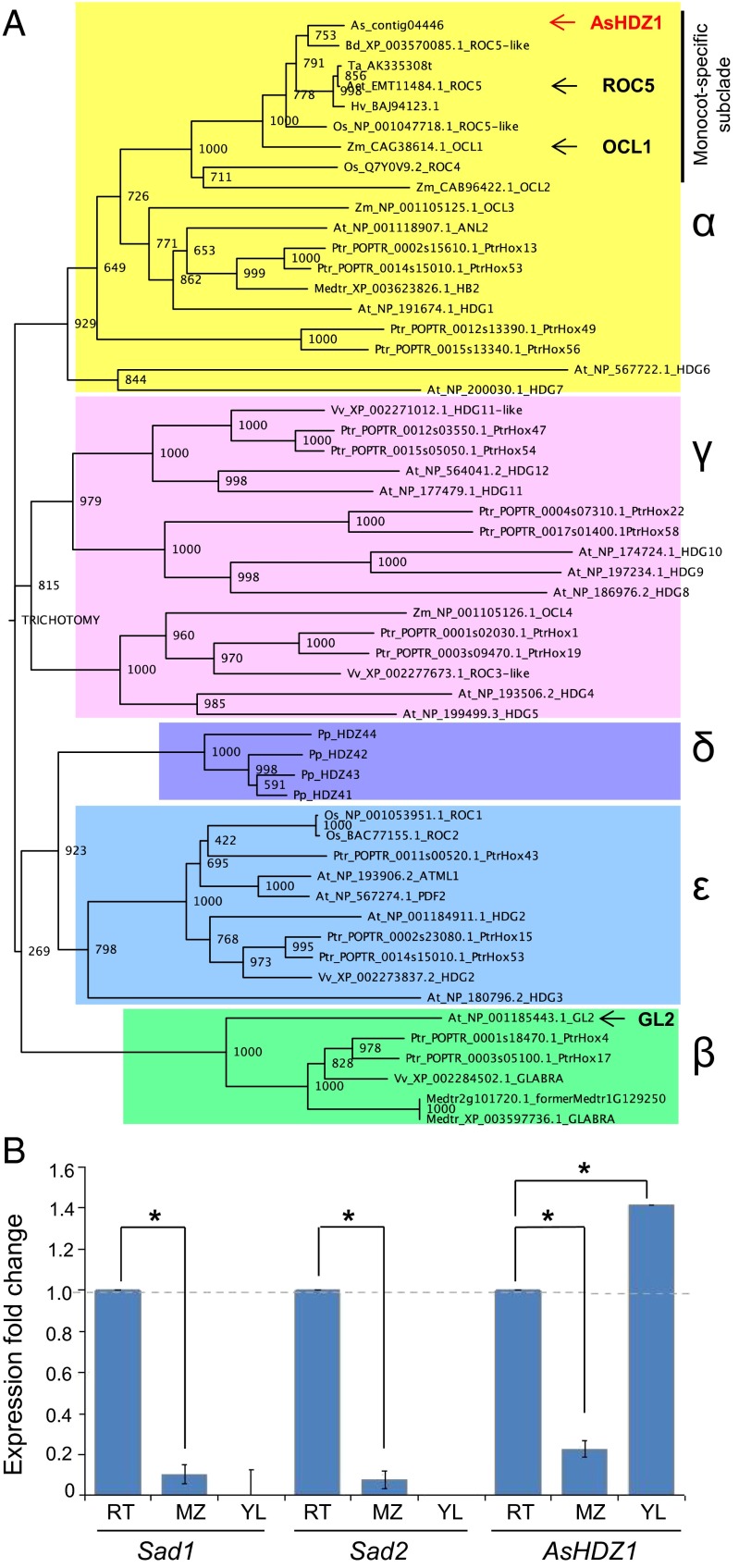
To investigate likely regulators of the avenacin-pathway genes, we generated a root tip transcriptome database for A. strigosa. Representation of root tip-expressed genes within this database was validated by searching for matches to each of the five cloned avenacin genes (SI Materials and Methods and Table S1). BLAST searches using all 16 A. thaliana HD-ZIP IV family members as probe sequences (24) identified multiple reads corresponding to a single class IV HD-ZIP sequence, AsHDZ1. AsHDZ1 shares close amino acid sequence similarity (e-values = 0) with HD-ZIP IV proteins from other cereals, including Triticum aestivum (AK335308), Hordeum vulgare (AK362919), ROC5 from rice (Oryza sativa; AB101648), a ROC5-like protein from Brachypodium distachyon (XM_003570037), and OCL1 from Zea mays (NM_001112023) (Fig. 3A). These sequences together form a monocot-specific subclade within the α-clade of the plant HD-ZIP IV superfamily (29). OCL1 and ROC5 are expressed in the epidermal cell layers of organs in monocots and are involved in regulation of processes such as the determination of epidermal cell fate, anthocyanin accumulation, and root development (30, 31). The wider α-clade includes the A. thaliana HD-ZIP IV proteins HDG1, HDG6, HDG7, and ANL2 (24, 32). GLABRA2 (GL2), a well-characterized negative regulator of root hair development in A. thaliana (17, 33), belongs to a different clade—the β-clade (Fig. 3A).
Fig. 3.
AsHDZ1 is an α-clade HD-ZIP IV transcription factor that is transcribed in root tips. (A) Phylogenetic tree of HD-ZIP IV amino acid sequences from the monocots oat (As), Brachypodium (Bd), wheat (Ta), barley (Hv), rice (Os), maize (Zm), and Aegilops (Aet), the dicots A. thaliana (At), poplar (Ptr), M. truncatula (Medtr), and grape (Vv), and the moss Physcomitrella patens (Pp). The tree was generated using maximum likelihood with standard settings and 1,000 bootstrap replicates. The different HD-ZIP IV clades (29) are color-coded. The oat HD-ZIP IV transcription factor AsHDZ1 is indicated in red. Other functionally characterized members of the monocot subgroup of the α-clade [OCL1 and ROC5 (30, 31)] and GL2 (from A. thaliana) are indicated. (B) AsHDZ1 shows a pattern of expression similar to that of the avenacin biosynthesis genes Sad1 and Sad2 in oat roots and is also expressed in the young leaves. qRT-PCR analysis of transcript levels of AsHDZ1, Sad1, and Sad2 in the root tips (RT), maturation zone (MZ), and young leaves (YL) of 3-d-old A. strigosa seedlings. Transcript levels were normalized to those for EF1-α. Values are means ± SD (three plants). Data were analyzed using the comparative CT method (42). Relative transcript levels are shown.*P < 0.05 (unpaired two-tailed t test).
AsHDZ1 has an expression pattern similar to that of Sad1 and Sad2 in roots (Fig. 3B). Unlike Sad1 and Sad2, however, this gene is also expressed in young leaves (as are OCL1 from Zea mays and ROC5 from rice) (25, 26). The expression patterns of the A. thaliana HD-ZIP IV genes belonging to the α-clade overlap, suggesting functional redundancy (24). HDG7 is a basal member of this clade and is expressed in roots. Experiments evaluating pSad1-GUS reporter activity in a homozygous hdg7 mutant background indicated that Hdg7 is dispensable for GUS expression (Fig. S4). Future experiments to identify regulators of pSad1 and pSad2 expression in A. thaliana are likely to require the generation of multiple combinatorial mutants for the α-clade HD-ZIP IV genes. Although it is clear that the Sad1 and Sad2 promoters are under the control of an ancient development process that is conserved across diverse plant species, it does not necessarily follow that the transcription factors involved are orthologous. The establishment of a TILLING platform (34) for diploid oat will open future opportunities to carry out functional analysis of AsHDZ1 in A. strigosa using reverse genetics approaches.
Accumulation of β-Amyrin Leads to a Change in Cell Fate in the Root Epidermis.
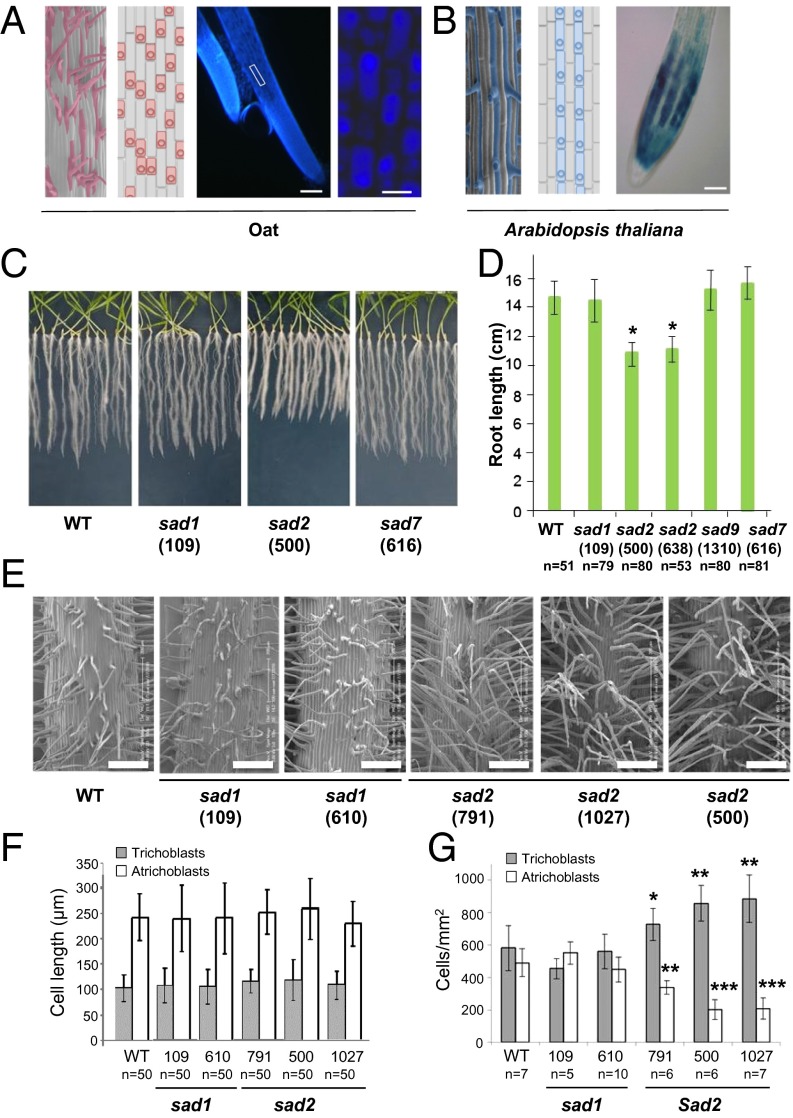
In most plant species the cells that will give rise to root hairs are predetermined (20). These predetermined trichoblasts are smaller than those that are destined to become atrichoblasts and differentiate into root hair cells in the differentiation zone of the root. Trichoblasts develop in one of two distinct patterns. They may alternate with the atrichoblasts along longitudinal cell files, as is the case in oat; alternatively, trichoblasts and atrichoblasts may develop in separate longitudinal files as in A. thaliana (Fig. 4A) (18, 19). Fluorescence microscopy indicates that the fluorescence attributable to avenacin A-1 is present predominantly in the atrichoblasts of oat root epidermal cells (Fig. 4A). Interestingly, investigation of pSad1 reporter lines revealed that GUS activity was detectable primarily in the atrichoblasts of A. thaliana (Fig. 4B). Thus, Sad1 is expressed in atrichoblasts, and this cell type-specific expression is conserved in A. thaliana.
Fig. 4.
Accumulation of β-amyrin triggers changes in root development. (A) (Left) Cryo-SEM image of wild-type A. strigosa root epidermis showing the root hair pattern. Hair cells are highlighted in pink in the cryo-SEM image at the far left. The patterning of trichoblast (pink) and atrichoblast (gray) cells before root hair emergence (modified from ref. 20) is illustrated in the adjacent diagram. (Right) The blue fluorescence in the maturation zone of oat roots is stronger in atrichoblasts than in trichoblasts. The white box indicates the magnified region shown in the panel at the far right. (Scale bars: 200 μm and 20 μm, respectively.) (B) cryo-SEM image of A. thaliana Col0 root epidermis (Left) and schematic view (Center) showing the epidermal cell patterning before root hair emergence; trichoblasts are indicated in blue and atrichoblasts in gray (modified from ref. 20). (Right) pSad1-driven GUS expression in A. thaliana roots is stronger in atrichoblasts than in trichoblasts, creating a striped pattern in the elongation zone. (Scale bar: 50 μm.) (C and D) Growth of wild-type, sad1, and sad2 lines of A. strigosa after 15 d on water agar. Results in D are represented as mean ± SE; *P < 0.0001 (unpaired two-tailed t test); n indicates number of seedlings analyzed. (E) Representative cryo-SEM images of the maturation zones of wild-type A. strigosa and the sad1 mutant #109 and #610, and sad2 mutant #791, #1027, and #500 A. strigosa lines. (Scale bars: 200 μm.) (F and G) Lengths of trichoblasts (one-way ANOVA: F5,294 = 2.01; P = 0.08) and atrichoblasts (one-way ANOVA: F5,294 = 1.66; P = 0.14) (F) and numbers of trichoblasts and atrichoblasts per square millimeter (G) in the maturation zones for the lines shown in E. ***P < 0.001; **P < 0.01; *P < 0.05 (unpaired two-tailed t test). Mutant #791 is a partial sad2 mutant (9).
The SAD1 product β-amyrin is widespread in plants (5). It is present at low levels in the roots of wild-type A. strigosa seedlings (∼1.5–3 μg/g dry weight) and at even lower but detectable levels in sad1 mutants (∼0.7–0.8 μg/g dry weight) (9, 35). sad2 mutants are unable to convert β-amyrin to avenacins (Fig. 1A) and thus accumulate considerably elevated levels of this triterpene (∼40–50 μg/g dry weight) (9, 35). sad2 mutants have a short root phenotype, whereas the wild type and other avenacin biosynthesis mutants (11, 12) do not (Fig. 4 C and D). The short root phenotype therefore is associated with the accumulation of abnormal levels of β-amyrin and not with the loss of the entire pathway (sad1 mutants) or the accumulation of later pathway intermediates (sad7 and other downstream mutants). A partial sad2 mutant (no. 791) that accumulates intermediate levels of avenacin A-1 and β-amyrin (Fig. S5) (7, 9, 35) has an intermediate root-length phenotype (Fig. S6).
Cryo-scanning electron microscopy (cryo-SEM) revealed that the roots of sad2 mutants have a superhairy phenotype (Fig. 4E). The lengths of the trichoblast and atrichoblast cells in the various mutant lines were unaltered as compared with the wild type (Fig. 4F). However, the sad2 mutants had significantly more root hair cells per unit area than the wild-type or sad1 lines. Importantly, these cells have the shorter dimensions typical of trichoblast cells, i.e., they were predetermined to become hair cells (Figs. 4G and 5). Thus, the accumulation of elevated levels of β-amyrin triggers a change in cell specification in roots. The intermediate sad2 mutant #791 had an intermediate phenotype (Fig. 4 E and G). Thus, the accumulation of elevated levels of β-amyrin in oat roots results in a greater proportion of epidermal cells becoming trichoblasts. This effect is distinct from the actions of most genetically characterized root hair determinants, including GL2, which act after cell specification to regulate root hair growth (17–21).
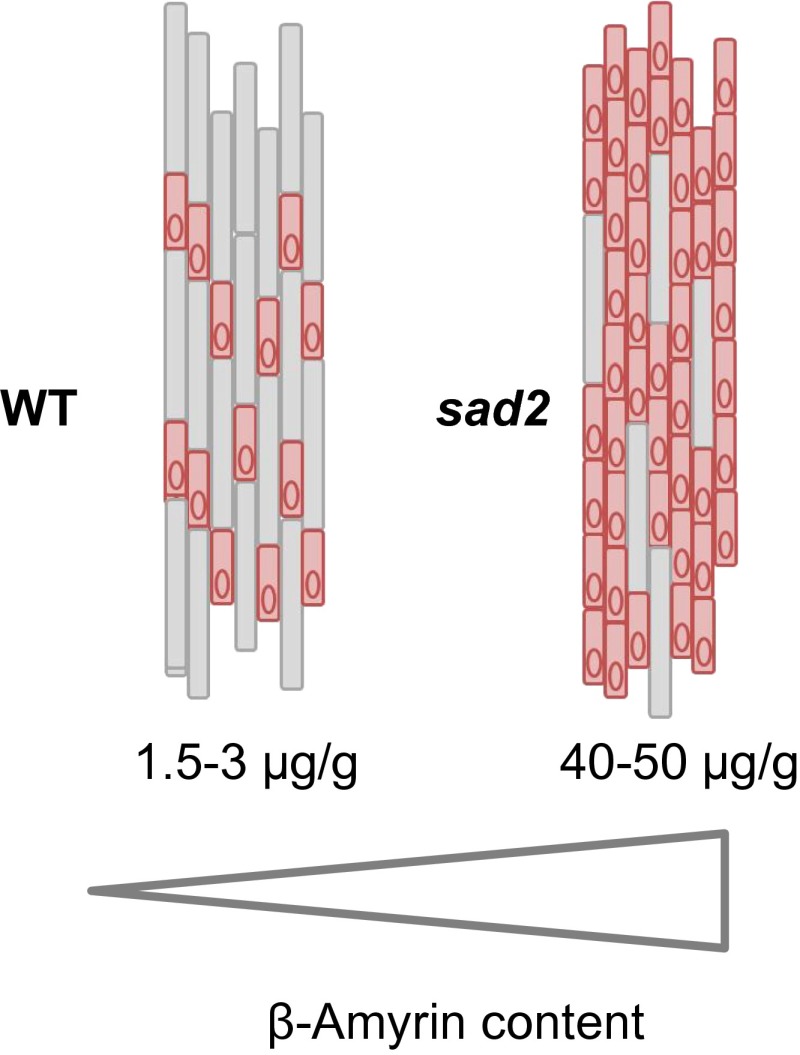
Fig. 5.
Increased β-amyrin content alters epidermal cell patterning in oat roots. The levels of β-amyrin in root extracts of wild-type and sad2 mutant lines are shown (9, 35). sad2 mutants are unable to metabolize β-amyrin to avenacin A-1 and therefore accumulate considerably elevated levels of this simple triterpene. The effects of the accumulation of elevated levels of β-amyrin are seen early in the establishment of the root epidermis, causing a greater proportion of epidermal cells to be specified as trichoblasts (specialized shorter cells that give rise to root hairs) (indicated in pink).
Interestingly, our earlier mRNA FISH experiments using highly sensitive fluorophore detection methods detected low levels of Sad1 transcript in 50% of the cells of the subepidermal cell layer, but the Sad2 transcript was not detectable in this cell layer (16). The subepidermal cell layer is known to influence cell specification in the root epidermis through as yet unknown signaling processes (18–21). Therefore it is tempting to speculate that β-amyrin may play a role in such processes, perhaps by influencing asymmetric cell division and hence specification of trichoblasts and atrichoblasts as the epidermal cells leave the meristematic zone. Application of exogenous β-amyrin to wild-type and sad1 mutant A. strigosa lines did not result in an increase in root hair cells (Fig. S7). The failure to phenocopy the sad2 root phenotype was not caused by the conversion of β-amyrin to avenacins, as evidenced by LC-MS analysis (Fig. S7). Likewise transgenic rice lines expressing SAD1 under the control of the maize ubiquitin promoter did not show a short root or root hair phenotype (Fig. S8). This finding is not surprising, however, given that the effect of β-amyrin on cell fate is likely to depend on local concentrations of the triterpene in particular cell types at a specific stage in development.
Discussion
Previously we reported the discovery and characterization of a gene cluster for avenacin synthesis in oat and showed that Sad1 and Sad2, the first two genes in the pathway, have been recruited from sterol biosynthesis (8, 9, 15). These genes have arisen by gene duplication and neofunctionalization, coupled with a change in expression pattern (from being widely expressed to being highly tissue specific). Here we show that the promoters of these genes retain their characteristic primary and lateral root meristem expression patterns when introduced into other plant species, indicating that the promoters are appropriately regulated in diverse heterologous plant backgrounds, including both monocots and dicots. Other genes within the avenacin cluster have expression patterns very similar to those of Sad1 and Sad2, with expression being tightly restricted to the epidermal cells of the root meristems (11–13). Thus, the genes for avenacin synthesis have acquired a common and specific expression pattern during pathway evolution. Our deletion analysis of the Sad1 promoter identified a classical L1-box motif with a pseudopalindromic border sequence that is essential for promoter function, implicating the HD-ZIP IV family of transcription factors in the regulation of triterpene synthesis. A future challenge will be to determine the function of AsHDZ1 in the regulation of triterpene synthesis and to establish the roles of lipid and sterol ligands in pathway regulation.
2,3-Oxidosqualene can be converted to a diverse array of triterpene products in plants. Of these, the most common cyclization products β-amyrin, α-amyrin, and lupeol are widespread in plants and have been reported in unconjugated form in numerous species, including A. thaliana (1–5). Little is known about the physiological functions of these compounds in planta. However, an increasing body of evidence suggests that they may have important roles. For example, they accumulate in the epicuticular layers of stem and leaf surfaces and have been implicated in protection against dehydration and potentially also against herbivores (reviewed in ref. 5). They are also involved in signaling. Previously we reported that lupeol is synthesized in the nodules of the legume Lotus japonicas and is involved in nodule development (36). Interestingly, accumulation of the simple triterpene thalianol in A. thaliana leads to enhanced root length (37). Our discovery that the accumulation of elevated levels of β-amyrin in oat roots causes a change in epidermal cell patterning provides further intriguing evidence suggesting that simple triterpenes may have widespread and as yet largely unrecognized functions in plant growth and development. At present we cannot distinguish between direct effects (e.g., specific binding to a receptor) and indirect effects (e.g., perturbation of membranes). Nevertheless, elucidation of the mechanistic basis of these effects is likely to provide new insights into the factors that govern plant growth and development.
Materials and Methods
Plant Material.
Wild-type and mutant lines of A. strigosa are as described (7, 9). The methods used to measure root length and metabolite analysis after β-amyrin feeding are described in SI Materials and Methods. Col-O was used as the wild-type A. thaliana accession for this work. Mutant A. thaliana lines used were tir1-1 (N3798) (27) and pin1-7 (SALK_047613) (26), mp12G (28), pid9 (25), and hdg7 (SALK_132114), all from Nottingham Arabidopsis Stock Center. Crosses with the pSad1-GUS reporter line were genotyped as described (38). Transgenic rice lines expressing oat SAD1 were analyzed as described in SI Materials and Methods.
Accession Numbers.
Sequence data for the genes and promoters referred to in this article can be found in the GenBank/European Molecular Biology Laboratory database libraries under accession nos. DQ680849 (Sad1 and Sad2, including promoter regions); KF857483 (CAS1 promoter sequence); KF857482 (AsCYP51G1 promoter sequence); AY618693 (CAS1 coding sequence); DQ680850 (AsCYP51G1 coding sequence); and KF857484 (AsHDZ1 coding sequence). The oat root tip transcriptome data have been deposited in the European Bioinformatics Institute database, accession no. ERA148431.
Transgenic Plants.
The promoter regions of the Sad1 and Sad2 genes were amplified from BAC clone 460D15 (9) using the primers listed in Table S2. For construct pK1848aGUS a BamHI/EcoRI fragment harboring 1,848 bp of the Sad1 promoter, the GUS gene, and the NOS-terminator were cloned into the respective sites of the linearized pBI101 vector. Inserts for all other constructs were amplified by PCR, cloned into Gateway-compatible vectors, and introduced into the binary vector pHGWFS7.0 (39). The maize ubiquitin promoter sequence (+ intron) was amplified from pAL156 (40).
Details of plant transformation can be found in SI Materials and Methods.
Generation of Oat Root Transcriptome Database, Phylogenetic Analysis, and Microscopy.
Details are provided in SI Materials and Methods.
Quantitative RT-PCR.
Total RNA was extracted from roots of 3-d-old oat seedlings. Details of RNA extraction are described in SI Materials and Methods. Quantitative RT-PCR (qRT-PCR) and relative quantification were performed as previously described (41). Gene-specific primers for Sad1, CAS1, Sad2, CYP51G1, and AsHDZ1 were designed around introns within the coding sequences (Table S3). Oat elongation factor1-α (EF1-α) was used as an internal standard. The method described by Livak and Schmittgen (42) was used to quantify gene expression.
Supplementary Material
Acknowledgments
We thank Lionel Hill for metabolite analysis, the John Innes Centre horticulture staff for plant care, Andrew Davis for assistance with photography of plants, the Genome Analysis Centre for generation of the oat root transcriptome resource, and Brenda Blacklock for useful discussions. This work was supported by Deutsche Forschungsgemeinschaft Fellowship KE 1519/1-1 (to A.C.K.); a joint Engineering and Physical Sciences Research Council/National Science Foundation (NSF/EPSRC) award (to A.O. and S.J.R.) as part of the Syntegron consortium NSF/EPSRC Grant EP/K03459/1; the University of Glasgow Work Placement Programme (S.H.); Plant Bioscience Limited; the Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant BB/J004561/1 “Understanding and Exploiting Plant and Microbial Secondary Metabolism”; and the John Innes Foundation.
Footnotes
Conflict of interest statement: A.O., K. Haralampidis, and R.E.M. are named as inventors on US patent 7,982,096 (awarded July 19, 2011) relating to the oat promoters described in this paper.
This article is a PNAS Direct Submission. J.C. is a guest editor invited by the Editorial Board.
Data deposition: Sequence data for the genes and promoters referred to in this article have been deposited in the GenBank/European Molecular Biology Laboratory database libraries [accession nos. KF857483 (CAS1 promoter sequence), KF857482 (AsCYP51G1 promoter sequence), and KF857484 (AsHDZ1)]. The oat root tip transcriptome data have been deposited in the European Bioinformatics Institute database (accession no. ERA148431).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401553111/-/DCSupplemental.
References
- 1.Chappell J. The genetics and molecular genetics of terpene and sterol origami. Curr Opin Plant Biol. 2002;5(2):151–157. doi: 10.1016/s1369-5266(02)00241-8. [DOI] [PubMed] [Google Scholar]
- 2.Augustin JM, Kuzina V, Andersen SB, Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72(6):435–457. doi: 10.1016/j.phytochem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Moses T, Pollier J, Thevelein JM, Goossens A. Bioengineering of plant (tri)terpenoids: From metabolic engineering of plants to synthetic biology in vivo and in vitro. New Phytol. 2013;200(1):27–43. doi: 10.1111/nph.12325. [DOI] [PubMed] [Google Scholar]
- 4.Sawai S, Saito K. Triterpenoid biosynthesis and engineering in plants. Front Plant Sci. 2011;2:25. doi: 10.3389/fpls.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thimmappa R, Geyser K, Louveau T, O’Maille P, Osbourn A. Triterpene biosynthesis in plants. Ann Rev Plant Biol. 2014;65:16.1–16.33. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 6.Turner EM. The nature of resistance of oats to the take-all fungus. III. Distribution of the inhibitor in oat seedlings. J Exp Bot. 1960;11:403–412. [Google Scholar]
- 7.Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA. 1999;96(22):12923–12928. doi: 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haralampidis K, et al. A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc Natl Acad Sci USA. 2001;98(23):13431–13436. doi: 10.1073/pnas.231324698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi X, et al. A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc Natl Acad Sci USA. 2006;103(49):18848–18853. doi: 10.1073/pnas.0607849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler K, et al. Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc Natl Acad Sci USA. 2013;110(35):E3360–E3367. doi: 10.1073/pnas.1309157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mugford ST, et al. A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell. 2009;21(8):2473–2484. doi: 10.1105/tpc.109.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mugford ST, et al. Modularity of plant metabolic gene clusters: A trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell. 2013;25(3):1078–1092. doi: 10.1105/tpc.113.110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owatworakit A, et al. Glycosyltransferases from oat (Avena) implicated in the acylation of avenacins. J Biol Chem. 2013;288(6):3696–3704. doi: 10.1074/jbc.M112.426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mylona P, et al. Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell. 2008;20(1):201–212. doi: 10.1105/tpc.107.056531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi X, et al. A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA. 2004;101(21):8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegel E, Koumproglou R, Shaw P, Osbourn A. Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell. 2009;21(12):3926–3936. doi: 10.1105/tpc.109.072124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5(6):R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masucci JD, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122(4):1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 19.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119(1):71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 20.Datta S, et al. Root hairs: Development, growth and evolution at the plant-soil interface. Plant Soil. 2011;346:1–14. [Google Scholar]
- 21.Grebe M. The patterning of epidermal hairs in Arabidopsis—updated. Curr Opin Plant Biol. 2012;15(1):31–37. doi: 10.1016/j.pbi.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Osbourn A, et al. An oat species lacking avenacin is susceptible to infection by Gaeumannomyces graminis var. tritici. Physiol Mol Plant Pathol. 1994;45(6):457–467. [Google Scholar]
- 23.Abe M, Takahashi T, Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;26(5):487–494. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura M, et al. Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 2006;141(4):1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100(4):469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 26.Smith RS, et al. A plausible model of phyllotaxis. Proc Natl Acad Sci USA. 2006;103(5):1301–1306. doi: 10.1073/pnas.0510457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 28.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17(5):1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu R, et al. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa) PLoS ONE. 2012;7(2):e31149. doi: 10.1371/journal.pone.0031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Depège-Fargeix N, et al. Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J Exp Bot. 2011;62(1):293–305. doi: 10.1093/jxb/erq267. [DOI] [PubMed] [Google Scholar]
- 31.Zou L-P, et al. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 2011;156(3):1589–1602. doi: 10.1104/pp.111.176016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubo H, Peeters AJ, Aarts MG, Pereira A, Koornneef M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell. 1999;11(7):1217–1226. doi: 10.1105/tpc.11.7.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Cristina M, et al. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10(3):393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- 34.McCallum CM, Comai L, Greene EA, Henikoff S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123(2):439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin B, et al. High throughput screening of mutants of oat that are defective in triterpene synthesis. Phytochemistry. 2010;71(11-12):1245–1252. doi: 10.1016/j.phytochem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Delis C, et al. Role of lupeol synthase in Lotus japonicus nodule formation. New Phytol. 2011;189(1):335–346. doi: 10.1111/j.1469-8137.2010.03463.x. [DOI] [PubMed] [Google Scholar]
- 37.Field B, Osbourn AE. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science. 2008;320(5875):543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- 38.Thomas CL, Schmidt D, Bayer EM, Dreos R, Maule AJ. Arabidopsis plant homeodomain finger proteins operate downstream of auxin accumulation in specifying the vasculature and primary root meristem. Plant J. 2009;59(3):426–436. doi: 10.1111/j.1365-313X.2009.03874.x. [DOI] [PubMed] [Google Scholar]
- 39.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 40.Amoah BK, Wu H, Sparks C, Jones HD. Factors influencing Agrobacterium-mediated transient expression of uidA in wheat inflorescence tissue. J Exp Bot. 2001;52(358):1135–1142. doi: 10.1093/jexbot/52.358.1135. [DOI] [PubMed] [Google Scholar]
- 41.Horst I, et al. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 2007;144(2):806–820. doi: 10.1104/pp.107.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.