Significance
Human induced pluripotent stem cells (hiPSCs) could be used as an unlimited source of retinal cells for the treatment of retinal degenerative diseases. The production of retinal cells from hiPSCs for personalized therapeutic approaches must comply with certain criteria, such as safety, efficiency, reproducibility, and low production cost. Here, we report a simple and scalable retinal differentiation process for the generation of retinal pigmented epithelial cells and neural retinal tissues containing retinal progenitor cells. These progenitors can be differentiated into all retinal cell types, including retinal ganglion cells and precursors of photoreceptors, which could find important applications in regenerative medicine. This method also provides an accessible in vitro model to investigate mechanisms involved in human retinogenesis and retinal diseases.
Keywords: retinal ganglion cells, rods, cones
Abstract
Progress in retinal-cell therapy derived from human pluripotent stem cells currently faces technical challenges that require the development of easy and standardized protocols. Here, we developed a simple retinal differentiation method, based on confluent human induced pluripotent stem cells (hiPSC), bypassing embryoid body formation and the use of exogenous molecules, coating, or Matrigel. In 2 wk, we generated both retinal pigmented epithelial cells and self-forming neural retina (NR)-like structures containing retinal progenitor cells (RPCs). We report sequential differentiation from RPCs to the seven neuroretinal cell types in maturated NR-like structures as floating cultures, thereby revealing the multipotency of RPCs generated from integration-free hiPSCs. Furthermore, Notch pathway inhibition boosted the generation of photoreceptor precursor cells, crucial in establishing cell therapy strategies. This innovative process proposed here provides a readily efficient and scalable approach to produce retinal cells for regenerative medicine and for drug-screening purposes, as well as an in vitro model of human retinal development and disease.
Irreversible blindness caused by retinal diseases, such as inherited retinopathies, age-related macular degeneration (AMD), or glaucoma, is mainly due to the impairment or loss of function of photoreceptor cells, supporting retinal pigmented epithelium (RPE) or retinal ganglion cells (RGCs). Rescuing the degenerated retina is a major challenge for which specific cell replacement is one of the most promising approaches (1, 2). Pluripotent stem cells, like human embryonic stem cells (hESCs) or induced pluripotent stem cells (hiPSCs), have the ability to be expanded indefinitely in culture and could be used as an unlimited source of retinal cells for the treatment of retinal degenerative diseases (3, 4). Several publications have indicated that hESCs and hiPSCs can be differentiated into RPE cells spontaneously after fibroblast growth factor (FGF) 2 removal (5–7) or by different floating aggregate methods (8–11). Concerning neural retinal cells, a growing body of convergent data has demonstrated the ability of hESCs or hiPSCs to be committed into the neural retinal lineage and further differentiated into cells expressing photoreceptor markers (12–15). Recent innovative approaches using 3D cultures from embryoid bodies (EBs) of hESCs or hiPSCs allowed the self-formation of optic cup (OC) structures (16) or the generation of optic vesicle (OV)-like structures (17), depending on the addition of exogenous molecules and different substrates used. These protocols require multiple steps and trained handling, which are not always compatible with the manufacturing process for therapeutic approach or drug screening that need a large-scale production of cells of interest. Therefore, very simple and reliable approaches minimizing the use of exogenous molecules should be developed to generate hESCs or hiPSC-derived retinal cells.
In the present study, we report a new retinal differentiation process using confluent hiPSCs, without cell clumps or EB formation and in the absence of Matrigel or serum. We demonstrate that integration-free hiPSCs derived from adult human dermal fibroblasts (AHDFs) cultured in proneural medium can simultaneously generate RPE cells and self-forming neural retinal (NR)-like structures within 2 wk and that, when switched to floating cultures, structures containing retinal progenitor cells (RPCs) can differentiate into all retinal cell types, including RGCs and precursors of photoreceptors, needed for therapeutic applications.
Results
Generation and Characterization of Integration-Free hiPSCs.
AHDFs were cotransfected with three plasmids coding for the transcription factors OCT4, NANOG, SOX2, LIN28, KLF4, and C-MYC, corresponding to plasmid OriP/EBNA vectors (18). Transfected AHDF were primarily cultured in fibroblast medium followed by iPS medium, in the presence of small molecules (Fig. S1A) previously described to improve and accelerate the reprogramming process (19). Between day 30 and 40, almost all tightly packed dome-like structures were picked up and replated onto mouse embryonic fibroblasts (MEFs), noted as passage 0 (P0). Emerging hiPSC colonies were detectable in less than 1 wk (Fig. S1B, arrow). Full characterization of hiPSC line 2 (hiPSC-2) is depicted in Fig. S1. Expanded hiPSC colonies developed alkaline phosphatase (AP) activity along with expression of the pluripotency markers NANOG, TRA1-81, OCT4 (also known as POU Class 5 homeobox 1), and SSEA4 (Fig. S1 C–I). Quantitative (q) RT-PCR revealed that the expression of pluripotency genes markedly increased over the respective fibroblast population and was comparable with that seen in hESCs (Fig. S1J). PCR analysis on the OriP site on genomic DNA or episomal fractions demonstrated the absence of genomic integration of the transgenes and the complete loss of episomal vectors after 15 passages (Fig. S1K). hiPSC-2 could be differentiated in vitro and in vivo into derivatives of all three germ layers, as shown by qRT-PCR (Fig. S1L), immunohistochemistry on EBs (Fig. S1 M–O) and teratoma formation in NSG mouse (Fig. S1 P–U). hiPSC-2 exhibited a normal karyotype (Fig. S1V).
Differentiation of Confluent hiPSCs into RPE Cells and Self-Forming Neural Retina-Like Structures.
Because a prerequisite for hiPSC differentiation is the shutdown of the self-renewal machinery, FGF2 was removed from the iPS medium during 2 d to encourage the spontaneous differentiation of confluent hiPSCs. FGF2 withdrawal from the culture medium may also promote neuroectoderm induction as demonstrated in hESCs (20). To favor this differentiation of hiPSCs into a neuroectoderm lineage, colonies were cultured in a serum-free proneural medium containing N2 supplement (Fig. 1A). This process led to the appearance of pigmented colonies within 4 d, and, after 7 d, bright-phase structures started to appear close to over half of patches of pigmented cells (Fig. 1B). Within 2 wk, all these structures formed a neuroepithelial tissue partly surrounded with a patch of pigmented cells (Fig. 1C), corresponding to 1–2 structures per cm2 of culture dish. The formation of these structures was rarely observed in nonpigmented areas, and the pigmented patches that did not develop neuroepithelial-like structures were used for RPE cell culture and expansion.
Fig. 1.
Efficient generation of RPE cells and NR-like structures from confluent hiPSCs. (A) Schematic diagram showing the stages of the differentiation protocol. (B and C) Neuroepithelial-like structure derived from hiPSC-2 at D7 and D14. (D) qRT-PCR analysis of NOGGIN and DKK1 in hiPSC-2 at D0, D7, and D14. Data are normalized on hiPSC-2 single colonies. (E) qRT-PCR analysis of eye-field transcription factors NRL, CRX, NEUROD1, and pluripotency marker OCT4 in NR-like structures at D14. Data are normalized to hiPSC-2 at D0. (F–P) Immunofluorescence staining of cryosectioned NR-like structures at D14 for PAX6 and RAX (F–H), Ki67 and LHX2 (I–K), or MITF and VSX2 (L–P). (Scale bars: B, C, F, I, and L, 100 µm; G, H, J, and M–P, 50 µm.)
Gene-expression analysis revealed endogenous expression of Wnt and BMP antagonists, DKK1 and NOGGIN, in confluent hiPSC cultures, and both genes were up-regulated during the formation of the neuroepithelial-like structures (Fig. 1D). By 14 d (D14), isolated neuroepithelial-like structures had lost the expression of the pluripotency-related gene OCT4 and acquired expression of transcription factors associated with eye-field specification, such as LHX2, RAX, PAX6, and SIX3 (Fig. 1E). Immunostaining of the neuroepithelial-like structures demonstrated that all cells coexpressed PAX6 and RAX (Fig. 1 F–H), a characteristic of eye-field cells (21). Nearly all of the cells were LHX2+, and their progenitor status was confirmed using the cell proliferation marker Ki67 (Fig. 1 I–K). The expression of both MITF and VSX2 (also known as CHX10), two transcription factors involved in retinal specification during OV and OC formation (22), was increased by 10- and 100-fold, respectively, at D14 (Fig. 1E). Immunohistochemistry experiments revealed an opposite gradient of VSX2 and MITF expression, with the most intense staining for VSX2 in the neuroepithelial-like structures whereas the strongest MITF expression was found in the distal pigmented part (Fig. 1 L–P). Together, these findings demonstrate that neuroepithelial-like structures have a marker expression profile typical of neural retinal progenitor cells (RPCs) and can be renamed neural retina-like (NR-like) structures. Interestingly, the expression of transcription factors typical to photoreceptor precursors such as NRL and CRX is increased by fivefold as early as after 14 d in culture (Fig. 1E and Fig. S2A). The presence of CRX+ cells in the NR-like structures at D14 (Fig. S2B) confirms that some RPCs are already committed to the photoreceptor lineage.
Isolation and Amplification of hiPSC-Derived RPE Cells.
Given the fast appearance of RPE cells from confluent hiPSCs cultured in proneural medium, pigmented patches of cells were mechanically isolated at D14 and spread onto gelatin-coated plates for expansion (Fig. 2A). After 1 mo, they formed a confluent cell monolayer that displayed the classical cobblestone morphology (Fig. 2 B and C). The cells were immunoreactive for a key RPE-specific transcription factor, MITF, and the tight junction marker ZO-1 (Fig. 2D). qRT-PCR analysis, normalized to adult human RPE cells, demonstrated that hiPSC-derived RPE (hiRPE) cells retained the expression of mature RPE-associated markers such as MERTK, RPE65, BEST1, and PEDF, after several passages (Fig. 2E). To determine whether hiRPE cells were functional, we tested their ability to carry out phagocytosis of FITC-labeled photoreceptor outer segments (POSs). We detected a pronounced phagocytosis activity as efficient as for the control RPE-J cell line with an average of 30% internalized POSs within 3 h (Fig. 2F).
Fig. 2.
Amplification and characterization of hiRPE cells. (A) Schematic illustration outlining the differentiation protocol to amplify hiRPE (arrow 1) or to maturate NR-like structures (arrow 2). (B and C) Phase-contrast images of hiRPE cell monolayer after 45 d. (D) ZO-1 and MITF immunostaining of hiRPE cell monolayer after 45 d. (E) qRT-PCR analysis of mature RPE markers in hiRPE cells at passage 0 (P0), P1, and P2. Data are normalized to control RNA isolated from human adult RPE cells. (F) Evaluation of RPE cell phagocytic activity; ratio of FITC/DAPI fluorescence in hiRPE cell cultures at P1 and in control RPE-J cell line after 3 h incubation with FITC-labeled POS. Binding and uptake of POS were assayed as described in SI Materials and Methods. (Scale bars: B and C, 100 µm; D, 50 µm.)
Differentiation of RPCs from NR-Like Structures into All Retinal Cell Types in Floating Cultures.
NR-like structures with the surrounding pigmented patch of cells (Fig. 1C) were mechanically isolated at D14 and further cultured as floating structures in the presence of FGF2 during 1 wk (Fig. 2A), to favor neural retinal differentiation rather than differentiation into the RPE lineage (23). The isolated NR-like structure formed a hollow sphere, which continued to increase in size (Fig. 3 A–C and Fig. S3). Quantitative analysis showed an increase in thickness of the neuroepithelium from 139 ± 19 µm to 251 ± 41 µm between D17 and D24 (Fig. S3). We analyzed the time course and the acquisition of specific retinal phenotypes from growing spheres, using both immunohistochemistry and qRT-PCR. Throughout the process from D14 to D42, transcription factors involved in retinal specification and differentiation were still expressed (Fig. 3 D and E). At D21, VSX2+ cells were located in the developing neuroepithelium, and MITF+ cells were found mainly in the RPE cells in the distal part of the NR-like structures (Fig. 3F, arrow). VSX2+ cells were predominantly located along the outer part of the neuroepithelium and also expressed PAX6 (Fig. 3G). The population of PAX6+/VSX2− cells congregated at the inner part of the neuroepithelium, which correspond to the retinal neurons that differentiated first and did not carry the proliferation marker Ki67 (Fig. 3I).
Fig. 3.
Characterization of early differentiating cells in floating NR-like structures. (A–C) Morphology of representative isolated floating NR-like structures at D15, D21, and D28. (D and E) qRT-PCR analysis of eye-field and photoreceptor-specific transcription factors in NR-like structures from D14 to D42. Data are normalized to NR-like structures at D14. (F–N) Immunohistochemistry analysis of cryosectioned NR-like structures at D21 or D28 for MITF (F), VSX2 (F and G), PAX6 (G, I, and N), OTX2 (H and N), BRN3A (H), CRX (L and M), CALRETININ (J), LIM1 (K), and Ki67 (I). (Scale bars: A–C and F–H, 100 µm; I–N, 50 µm.)
Indeed, as early as D21, RGCs and amacrine cells were identified by immunohistochemistry at the same location as the one shown by BRN3A, also known as POU class 4 homeobox 1 (Fig. 3H) or CALRETININ (Fig. 3J) staining. Lim homeobox 1 (LIM1)+ cells corresponding to differentiating horizontal cells were also found in the neuroepithelium (Fig. 3K). The expression of OTX2 and NEUROD1, two transcription factors involved in the differentiation of RPCs into photoreceptors (24), increased during the floating culture step (Fig. 3E). The photoreceptor cell fate commitment was confirmed by the large increase in NRL and CRX expression from D14 to D42 (Fig. 3E and Fig. S2A). Immunohistochemical analysis showed the expression of OTX2 in hiPSC-derived RPE cells in the distal part of the structures (Fig. 3H, arrow), as well as the presence of OTX2+ cells in the neuroepithelium (Fig. 3H). CRX+ cells were also clearly identifiable in the neuroepithelium at D21 (Fig. 3L) and coexpressed OTX2 (Fig. S2 C–H). At D28, CRX is essentially expressed in postmitotic Ki67− cells (Fig. 3M). As expected (25), OTX2+ committed photoreceptor precursors did not express PAX6 (Fig. 3N). These data demonstrated that our culture conditions allowed for the rapid differentiation of hiPSCs into RGCs, amacrine cells, or horizontal cells, and precursors of photoreceptors in 3 wk. Moreover, the protocol developed here showed a good reproducibility when two distinct nonintegrative hiPSC lines (hiPSC-1 and hiPSC-2) were compared (Fig. S4).
Prolonged maintenance of isolated NR-like structures in floating culture allowed further differentiation of the RPCs into the late-born retinal cell types as demonstrated by qRT-PCR (Fig. 4 A–E). Indeed, after the first expression of early-born retinal markers of maturing RGCs (BRN3A and B), amacrine (CALRETININ and GAD2), and horizontal (LIM) cells (Fig. 4 A and B), the emergence of markers of late-born retinal cell types was observed later, corresponding to cone (R/G OPSIN, BLUE OPSIN, and CONE ARRESTIN) and rod photoreceptors (RHODOPSIN and RECOVERIN) (Fig. 4 C and D), bipolar [protein kinase C alpha (PKCα)] cells, and Müller glial cells (the glutamate/aspartate transporter GLAST1) (Fig. 4E). Transient expression of BRN3A and BRN3B (Fig. 4A) was confirmed by immunohistochemical data, showing a gradual decrease in the BRN3A+ cell population in NR-like structures as the culture was extended (Fig. S5). Between D21 (Fig. 4F) and D42 (Fig. 4G), most of the NR-like structures lost their laminar appearance and developed internal rosettes that contained OTX2+, CRX+, and RECOVERIN+ cells corresponding to the differentiating photoreceptors (Fig. 4 G and J–L and Fig. S2 F–H), surrounded by cells that expressed different markers of RGCs (BRN3A and CALRETININ), amacrine (CALRETININ and AP2) cells, and horizontal (LIM) cells (Fig. 4 G–J). Interestingly, at D42, RECOVERIN+ cells expressed the cell surface marker CD73 (Fig. 4L), a marker used for cell sorting of photoreceptor precursors for transplantation (26). At D77, PAX6 was present only outside the rosettes in postmitotic cells (KI67−), consistent with its expression in RGCs, amacrine cells, and horizontal cells (Fig. 4M). By D112, RHODOPSIN and R/G OPSIN appeared in NR-like structures, reflecting the maturation of both rods and cones (Fig. 4 N and O). RECOVERIN+ and RHODOPSIN+ cells were commonly localized at the most inner part of the residual rosettes at D112 (Fig. S6 A and B). Interestingly, immunohistochemistry using the connecting cilium marker acetylated TUBULIN revealed the existence of very thin structures in the luminal zone of rosettes juxtaposed to RECOVERIN+ cells, suggesting the formation of potential cilia and photoreceptor outer segments (Fig. S6 C and D). The differentiation of the two other late-born retinal cell types, bipolar and Müller glial cells, also required a longer time in culture (D112) to be detected, respectively, by PKCα staining (Fig. 4P) and by coexpression of glutamine synthetase (GS) and SOX9 (Fig. 4Q). Thereby, our cell-culture conditions allowed the generation of all retinal-cell types from the RPCs present in the NR-like structures in a sequential manner.
Fig. 4.
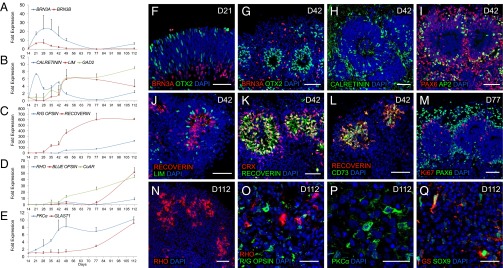
Differentiation of all retinal cell types from floating NR-like structures. (A–E) qRT-PCR analysis of selected neural retinal cell types in NR-like structures at different times. Data are normalized to NR-like structures at D14 and at D35 for both R/G and BLUE OPSIN. (F–Q) Immunohistochemical analysis of cryosectioned NR-like structure at different stages of differentiation using markers for RGCs (BRN3A, PAX6, and CALRETININ), amacrine cells (PAX6, AP2, and CALRETININ), horizontal cells (LIM1, PAX6, and CALRETININ), photoreceptors (OTX2, RECOVERIN, CRX, CD73, RHODOPSIN, and R/G OPSIN), bipolar cells (PKCα), Müller glial cells (GS and SOX9), and for mitotic progenitors (Ki67). (Scale bars: F–N, 50 µm; O–Q, 25 µm.)
Acceleration of Photoreceptor Precursor Generation by Notch Inhibition.
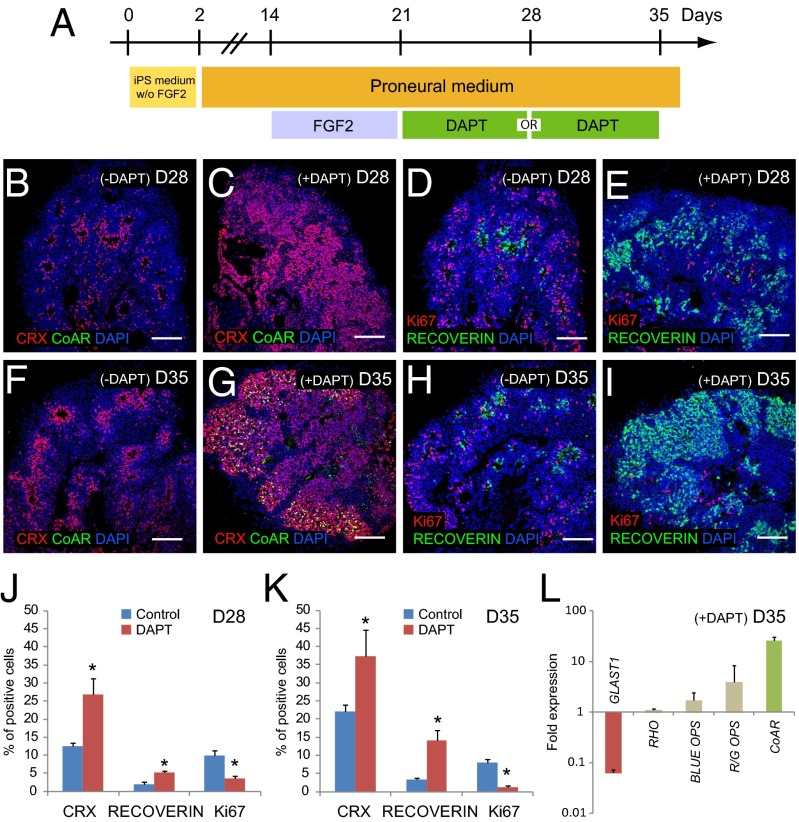
Based on the role of Notch in retinal progenitor cell fate (16, 27), we next investigated the effect of the Notch pathway inhibition during floating cultures of NR-like structures on the generation of the precursors of photoreceptors. Because, in NR-like structures between D21 and D35, most RPCs are committed to the photoreceptor lineage (corresponding to the increased expression of the two photoreceptor-specific transcription factors, CRX and NRL) (Fig. 4E and Fig. S2A), the Notch inhibitor DAPT was added during this time window. A 1-wk treatment with DAPT between D21 and D28 is sufficient to increase both CRX and RECOVERIN staining (Fig. 5 B–E). This treatment enables enhanced generation of photoreceptor precursors because, at D28, the number of CRX+ and RECOVERIN+ cells increased 2.2- and 2.6-fold, respectively, compared with the control (Fig. 5J). Concomitantly, the population of mitotic progenitors evaluated at D28 by Ki67 staining largely decreased (threefold) after treatment at D28 (Fig. 5J). We further chose to evaluate the effect of Notch inhibition between D28 and D35 rather than prolonged exposure to DAPT because few RPCs remained at D28 after DAPT treatment. Under these conditions, Notch inhibition also led to an increase in the number of photoreceptor precursors within the NR-like structures, namely a 1.7- and 4.1-fold increase at D35 in the number of CRX+ and RECOVERIN+ cells, respectively (Fig. 5 F–I and K). Interestingly, CONE-ARRESTIN+ cells that were not normally detected at D35 could be clearly identified at that time after DAPT treatment (Fig. 5G). Moreover, qRT-PCR analysis confirmed the increase in CONE-ARRESTIN expression at D35 after DAPT treatment whereas no significant changes in gene expression of RHODOPSIN, BLUE OPSIN, or R/G OPSIN were observed (Fig. 5L). GLAST1 expression was decreased after DAPT treatment (Fig. 5L). All together, these data suggest that inhibition of Notch signaling accelerated the generation of photoreceptor precursors from multipotent RPCs.
Fig. 5.
Acceleration of photoreceptor precursor generation from floating NR-like structures by Notch inhibition. (A) Schematic illustration of the experiment with DAPT treatment either from D21 to D28 or from D28 to D35. (B–I) Immunohistochemical analysis of cryosectioned NR-like structures at D28 or D35, with (C, E, G, and I) or without (B, D, F, and H) DAPT treatment. (J and K) Quantification of photoreceptor precursors (CRX and RECOVERIN) and mitotic progenitors (Ki67) at D28 (J) and D35 (K) with or without (control) DAPT. Values represent the mean percentage of positive cells ± SEM (n = 4, *P < 0.05). (L) qRT-PCR analysis of maturing photoreceptor markers and GLAST1 (marker for Müller glial cells) in D35 NR-like structures treated with DAPT. Data are normalized to NR-like structures at D35 without DAPT treatment. (Scale bars: 100 µm.)
Discussion
In this study, we report the finding that a simple culture of confluent hiPSCs in a serum-free proneural medium is sufficient to generate NR-like structures and RPE cells in 2 wk. The process we describe avoids the steps of EB formation and selection and addition of inductive molecules and/or Matrigel, as well as EB coating on adherent substrates. Early generated structures present an OV phenotype revealed by coexpression of PAX6 and RAX, and an opposite gradient of VSX2 and MITF between the neuroepithelium and the RPE. This efficiency is likely due in part to the increasing endogenous production by confluent hiPSCs of DKK1 and NOGGIN, two inducers of neural and retinal specification, generally added for retinal differentiation of hESCS or hiPSCs (17, 28). Nevertheless, previous studies reported that IGF-I added to the culture medium or present in the Matrigel can direct hESCs to a retinal progenitor identity (12, 29), suggesting that insulin, already present in the N2 supplement, could be sufficient to play a similar role in our conditions.
Floating cultures of isolated hiPSC-derived NR-like structures allowed the differentiation of the RPCs into all of the retinal cell types, in a sequential manner consistent with the in vivo vertebrate retinogenesis, demonstrating the multipotency of hiPSC-derived RPCs. In terms of timing, differentiation of NR-like structures is quite similar to the ones reported for the in vitro self-formed OC and OV-like structures (16, 17), with the detection of RGCs between D21 and D35 and photoreceptor precursors around D30–D40 whereas more mature photoreceptors, stained for the presence of OPSIN or RHODOPSIN, were not clearly identified before D77. The use of different molecules during the culture, such as taurine, sonic hedgehog, retinoic acid, FGF1, and FGF2, could accelerate photoreceptor maturation, as recently reported for hESC aggregates in the presence of Matrigel (28). Interestingly, we report that inhibition of the Notch pathway when RPCs are committed to the photoreceptor lineage clearly enhances the proportion of photoreceptor precursors in the NR-like structures, with a twofold increase in CRX+ cells, presumably achieved by forcing RPCs to exit early from the cell cycle, as previously described (16). A 1-wk treatment with the Notch inhibitor, DAPT, is indeed sufficient to induce cell-cycle exit for a large majority of the RPCs, allowing the generation after 35 d of about 40% of CRX+ photoreceptor precursors, also expressing cone precursor markers. This strategy could be considered for the efficient generation of cells with therapeutic applications. NR-like structures did not invaginate to form bilayered cups as elegantly reported in an EBs/Matrigel-dependent protocol using hESCs by Nakano et al. (16). Instead, the hiPSC-derived structures maintained a laminar organization until D21 and subsequently developed rosettes containing photoreceptor-like cells in the central region, surrounded by both cells with a retinal inner nuclear layer-specific identity and RGCs. This kind of inverted retina, which has also been reported in OV-like structures (30), could be due to the absence of RPE cells in close contact with the differentiating neuroepithelium in the NR-like structures. Nevertheless, generating mature and stratified NR tissue may not be requisite for future cell-therapy strategies based on purified photoreceptor precursors or other retinal-derived cells. In this context, our protocol allows, in 42 d, the generation of promising candidates for transplantation: i.e., CD73+ photoreceptor precursors that have previously been purified and successfully transplanted in mouse retina (26). The possibility of combining NOTCH inhibition and CD73 selection should enable the isolation of a large number of transplantable cells, holding great promise for the replacement of degenerated photoreceptors in retinal dystrophies. The ability to produce RGCs from the NR-like structures could have important implications for the treatment of glaucoma. Nevertheless, optimization of culture conditions will be required to prolong RGC survival in floating NR-like structures. In addition to the generation of retinal neurons, our protocol concomitantly allows the generation of RPE cells (hiRPE) that can be easily passaged and amplified while retaining their phenotype, close to their in vivo state. The present protocol hereby holds great potential to rapidly generate banks of hiRPE cells intended for the future treatment of AMD and other RPE-related diseases.
With the goal of maintaining a clinical grade, we generated hiPSCs by episomal reprogramming because the use of lentiviral vectors bears a risk of genotoxicity. Reactivation of or residual transgene expression during hiPSC differentiation could indeed affect lineage choice and the functionality of hiPSC derivatives, as recently demonstrated for the RPE cell lineage (31). Although autologous feeders may be used for the maintenance of hiPSCs (32), developments toward a xeno-free and feeder-free system will be required for regenerative therapy. Switching to a nonxenogeneic system should not be a brake for the generation of NR-like structures and hiRPE cells as recently reported in others differentiation approaches (6, 33, 34). From a pharmacological perspective, hiPSCs offer valuable potential to profile new compounds in the first process of drug discovery (35). The proliferative capacity of hiPS-derived RPCs and RPE cells should ensure the development of new cellular tools for phenotype- and target-based high-throughput screening with the goal of identifying specific active compounds for future treatments of retinal dystrophies (36).
This new protocol, which eliminates the need for the time-consuming and labor-intensive manual steps usually required to differentiate hiPSCs into a specific retinal lineage, provides a readily scalable approach to generate large numbers of both RPE cells and multipotent RPCs. Thus, in a relatively short period, the methods described here produce a source of photoreceptor precursors or RGCs holding the promise for a novel approach to regenerative medicine and pharmaceutical testing/drug screening. Our strategy using hiPSCs also provides an opportunity to study the molecular and cellular mechanisms underlying human retinal development and should advance the development of in vitro models of human retinal degenerative diseases.
Materials and Methods
Reprogramming of Human Fibroblasts.
The reprogramming was done with an episomal approach as described (18). Detailed protocols are available in the SI Materials and Methods.
Retinal Differentiation and hiPSC-Derived Retinal Cell Cultures.
hiPSCs were expanded to confluence in 3-cm MEF-seeded dishes in iPS medium. At this point, defined as day 0 (D0), confluent hiPSCs were cultured in iPS medium without FGF2. After 2 d, the medium was switched to a “proneural medium” composed of DMEM:Nutrient Mixture F-12 (DMEM/F12, 1:1, l-Glutamine), 1% MEM nonessential amino acids, and 1% N2 supplement (Life Technologies). The medium was changed every 2–3 d. On D14, identified NR-like structures were isolated using a needle with the surrounding pigmented cells and were individually cultured in 24-well-plates as floating structures in the proneural medium supplemented with 10 ng/mL FGF2. Plates were mounted on a 3D Nutator shaker (VWR) during the two first days to avoid the adhesion of NR-like structures to the culture plate bottom, and the medium was changed every 2–3 d. At D21, FGF2 was removed, and half of the proneural medium was changed every 2–3 d for the next several weeks. For the DAPT experiment, structures were treated with 10 µM DAPT (Selleck) during 7 d between either D21 and D28 or D28 and D35.
For hiPSC-derived RPE (hiRPE) cell cultures, identified pigmented patches were cut at D14 without the nonpigmented budding structures and transferred onto 0.1% gelatin-coated plates, noted as passage 0 (P0). hiRPE cells were expanded in the proneural medium, and the medium was changed every 2–3 d. At confluency, cells were dissociated and replated onto 24-well gelatin-coated plates for amplification or onto 96-well gelatin-coated plates for phagocytosis assays.
More information is provided in SI Materials and Methods and Tables S1 and S2.
Supplementary Material
Acknowledgments
We thank M. Peschanski, Y. Laabi, and A. Bayot for helpful discussions and S. Fouquet of the Institut de la Vision Imaging Facility. We acknowledge T. Léveillard for RNA isolated from human adult RPE cells, C. Varela for performing karyotype analysis, and O. Féraud for Teratoma analysis. This work was financed by Institut National de la Santé et de la Recherche Médicale, Université Pierre-et-Marie-Curie-Université Paris 6. This work was supported by the Agence Nationale de la Recherche (ANR) (GPiPS: ANR-2010-RFCS005) and by French state funds managed by the ANR within the Investissements d'Avenir programme (ANR-11-IDEX-0004-02) in the frame of the LABEX LIFESENSES (ANR-10-LABX-65). I-Stem is part of the Biotherapies Institute for Rare Diseases supported by the Association Française contre les Myopathies-Téléthon. S.R. and A.T. were supported by the Regional Council of Ile-de-France.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324212111/-/DCSupplemental.
References
- 1.Barber AC, et al. Repair of the degenerate retina by photoreceptor transplantation. Proc Natl Acad Sci USA. 2013;110(1):354–359. doi: 10.1073/pnas.1212677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson RA, et al. Restoration of vision after transplantation of photoreceptors. Nature. 2012;485(7396):99–103. doi: 10.1038/nature10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucherie C, Sowden JC, Ali RR. Induced pluripotent stem cell technology for generating photoreceptors. Regen Med. 2011;6(4):469–479. doi: 10.2217/rme.11.37. [DOI] [PubMed] [Google Scholar]
- 4.Rowland TJ, Buchholz DE, Clegg DO. Pluripotent human stem cells for the treatment of retinal disease. J Cell Physiol. 2012;227(2):457–466. doi: 10.1002/jcp.22814. [DOI] [PubMed] [Google Scholar]
- 5.Buchholz DE, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27(10):2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- 6.Vaajasaari H, et al. Toward the defined and xeno-free differentiation of functional human pluripotent stem cell-derived retinal pigment epithelial cells. Mol Vis. 2011;17:558–575. [PMC free article] [PubMed] [Google Scholar]
- 7.Zahabi A, et al. A new efficient protocol for directed differentiation of retinal pigmented epithelial cells from normal and retinal disease induced pluripotent stem cells. Stem Cells Dev. 2012;21(12):2262–2272. doi: 10.1089/scd.2011.0599. [DOI] [PubMed] [Google Scholar]
- 8.Idelson M, et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5(4):396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Kokkinaki M, Sahibzada N, Golestaneh N. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29(5):825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu B, et al. Long-term safety and function of RPE from human embryonic stem cells in preclinical models of macular degeneration. Stem Cells. 2009;27(9):2126–2135. doi: 10.1002/stem.149. [DOI] [PubMed] [Google Scholar]
- 11.Maruotti J, et al. A simple and scalable process for the differentiation of retinal pigment epithelium from human pluripotent stem cells. Stem Cells Transl Med. 2013;2(5):341–354. doi: 10.5966/sctm.2012-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamba DA, Karl MO, Ware CB, Reh TA. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci USA. 2006;103(34):12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellough CB, Sernagor E, Moreno-Gimeno I, Steel DH, Lako M. Efficient stage-specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells. 2012;30(4):673–686. doi: 10.1002/stem.1037. [DOI] [PubMed] [Google Scholar]
- 14.Meyer JS, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106(39):16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osakada F, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26(2):215–224. doi: 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 16.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Meyer JS, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29(8):1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Li W, Laurent T, Ding S. Small molecules, big roles: The chemical manipulation of stem cell fate and somatic cell reprogramming. J Cell Sci. 2012;125(Pt 23):5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greber B, et al. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 2011;30(24):4874–4884. doi: 10.1038/emboj.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathers PH, Jamrich M. Regulation of eye formation by the Rx and pax6 homeobox genes. Cell Mol Life Sci. 2000;57(2):186–194. doi: 10.1007/PL00000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horsford DJ, et al. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132(1):177–187. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- 23.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35(9):565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Nishida A, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6(12):1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 26.Eberle D, Schubert S, Postel K, Corbeil D, Ader M. Increased integration of transplanted CD73-positive photoreceptor precursors into adult mouse retina. Invest Ophthalmol Vis Sci. 2011;52(9):6462–6471. doi: 10.1167/iovs.11-7399. [DOI] [PubMed] [Google Scholar]
- 27.Jadhav AP, Cho SH, Cepko CL. Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc Natl Acad Sci USA. 2006;103(50):18998–19003. doi: 10.1073/pnas.0608155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boucherie C, et al. Brief report: Self-organizing neuroepithelium from human pluripotent stem cells facilitates derivation of photoreceptors. Stem Cells. 2013;31(2):408–414. doi: 10.1002/stem.1268. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y, et al. Three-dimensional neuroepithelial culture from human embryonic stem cells and its use for quantitative conversion to retinal pigment epithelium. PLoS ONE. 2013;8(1):e54552. doi: 10.1371/journal.pone.0054552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips MJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53(4):2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toivonen S, et al. Comparative analysis of targeted differentiation of human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells reveals variability associated with incomplete transgene silencing in retrovirally derived hiPSC lines. Stem Cells Transl Med. 2013;2(2):83–93. doi: 10.5966/sctm.2012-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K, Narita M, Yokura M, Ichisaka T, Yamanaka S. Human induced pluripotent stem cells on autologous feeders. PLoS ONE. 2009;4(12):e8067. doi: 10.1371/journal.pone.0008067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sridhar A, Steward MM, Meyer JS. Nonxenogeneic growth and retinal differentiation of human induced pluripotent stem cells. Stem Cells Transl Med. 2013;2(4):255–264. doi: 10.5966/sctm.2012-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker BA, Anfinson KR, Mullins RF, Stone EM, Young MJ. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Transl Med. 2013;2(1):16–24. doi: 10.5966/sctm.2012-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charbord J, et al. High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells. 2013;31(9):1816–1828. doi: 10.1002/stem.1430. [DOI] [PubMed] [Google Scholar]
- 36.Swoboda JG, et al. Small molecule mediated proliferation of primary retinal pigment epithelial cells. ACS Chem Biol. 2013;8(7):1407–1411. doi: 10.1021/cb4001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.