Abstract
Significance: The detrimental effects of ionizing radiation (IR) involve a highly orchestrated series of events that are amplified by endogenous signaling and culminating in oxidative damage to DNA, lipids, proteins, and many metabolites. Despite the global impact of IR, the molecular mechanisms underlying tissue damage reveal that many biomolecules are chemoselectively modified by IR. Recent Advances: The development of high-throughput “omics” technologies for mapping DNA and protein modifications have revolutionized the study of IR effects on biological systems. Studies in cells, tissues, and biological fluids are used to identify molecular features or biomarkers of IR exposure and response and the molecular mechanisms that regulate their expression or synthesis. Critical Issues: In this review, chemical mechanisms are described for IR-induced modifications of biomolecules along with methods for their detection. Included with the detection methods are crucial experimental considerations and caveats for their use. Additional factors critical to the cellular response to radiation, including alterations in protein expression, metabolomics, and epigenetic factors, are also discussed. Future Directions: Throughout the review, the synergy of combined “omics” technologies such as genomics and epigenomics, proteomics, and metabolomics is highlighted. These are anticipated to lead to new hypotheses to understand IR effects on biological systems and improve IR-based therapies. Antioxid. Redox Signal. 21: 260–292.
Introduction
Radiation is a phenomenon present in our daily lives, originating from natural and manmade sources. Living organisms are profoundly affected by radiation-induced cellular damage, threatening healthy and diseased tissues alike. In humans, there is a wide range of response to radiation, which is determined by parameters including the radiation source, radiation dosage (amount of radiation energy received), length of exposure, and, importantly, the genetic and epigenetic makeup of the exposed individual. These parameters can range widely, and humans may be exposed to low-dose radiation from commonly used diagnostic tools in medicine such as computed tomography (CT) scanning or high doses of radiation such as those used for radiotherapy and generated by nuclear disasters. The genetic and epigenetic aspects are significant across many conditions and may determine, for example, the likelihood of an individual to develop cancer or to respond to a cancer treatment (e.g., some tumors have either an intrinsic resistance to ionizing radiation (IR) or have acquired this property through cycled accumulation of genetic mutations and selection for increased survival and proliferation).
Though much progress has been made in understanding the basic principles of IR-induced effects on individual components of biological systems, less is known about how localized IR effects on target molecules regulate the cellular networks that contain these modified species, the interactions between networks (e.g., signaling and bioenergy metabolism), and the overall state of a biological system (cell, tissue/tumor, and individual/patient). The availability of high-end technologies and detection methods to monitor single or global radiation-induced modification of biomolecules has increased sharply in recent years, uncovering a much wider context for how radiation impacts the cellular life cycle. In this review, we highlight many recently developed techniques for uncovering radiation targets, though to date, not all have been applied toward further elucidating the unique biological response of radiation. The current review will discuss (i) the impact of IR on biological macromolecules (nucleic acids/DNA, lipids, and proteins); (ii) classical and modern methods of detection of IR-modified species; (iii) cellular processes affected by the interaction of IR with DNA, lipids, and proteins; and (iv) how state-of-the-art “omics” methods and technologies could be applied to decipher the complex interaction networks that exist between the products of IR-induced modifications (e.g., DNA damage, lipid peroxidation, and protein oxidation).
Fundamentals of IR Biochemistry
This chapter discusses the fundamentals of IR in the context of other types of radiation along with the key effector molecules that are responsible for the transforming effects of IR on biomolecules.
Fundamentals
Radiation is classified in two major forms: ionizing and non-ionizing. Environmental radiation is largely the non-ionizing type, including ultraviolet (UV) rays from the sun and electromagnetic radiation associated with radio waves and microwaves. The ability of sources of non-IR, such as UV rays (from the sun or tanning beds), to harm biological tissues is now well-established (168, 382). The interaction of IR with biomolecules is, however, much more aggressive than non-IR due to the ability of IR to induce atom ionization. The source of IR is a class of unstable radionuclides (radioisotopes) that emit high-energy particles which are capable of displacing atomic electrons, facilitating a chain reaction of electron ejection. The major types of IR are alpha particles, beta particles, X-rays, and gamma rays. Since alpha and beta particles can be stopped by physical barriers, such as a sheet of paper or an aluminum plate, while X- and gamma rays are more penetrating, environmental exposure to gamma rays induces a greater degree of biological damage than exposure to alpha or beta particles. However, all four types of radiation are successfully utilized for therapeutic purposes and are capable of causing significant cellular damage (189).
The international unit of measure for absorbed radiation (radiation dose) is the gray (Gy), defined as J/kg of mass. Since equal doses of IR elicit differential effects depending on source and properties of the biological target, the unit of sievert (Sv) is used to express the equivalent dose. Individuals receive an average of 2.4 mSv per year of IR from natural sources, though this figure is increased in more developed nations (173). While natural sources of gamma rays (K-40) exist, gamma rays most widely used in research and therapies are from manmade sources such as Co-60 and Cs-137. The focus of this review will be primarily on the interactions of gamma rays with biological macromolecules, though references to other types of radiation are included where relevant.
Radiation from X-rays, including from CT scans, is a form of IR similar to gamma radiation but of lower energy. The energy level of X-rays enables visualization of dense areas (e.g., bones) in the human body, which cannot be penetrated as efficiently as soft tissue due to the photoelectric effect. CT scanning provides more thorough diagnostic details than X-ray but at the cost of patient exposure to a higher radiation dose, estimated at 15–30 mGy (compared with 0.01–0.15 mGy from X-ray imaging) (47). Cumulative CT-related low-dose IR has been correlated with detrimental biological effects such as DNA damage, bystander effects, tissue injury, and, in some cases, carcinogenesis (218). The biological consequences of low-dose IR have been recently reviewed (218), and illustrate that although CT is a valuable diagnostic tool, excessive exposure, especially in children, may lead to cancer and should be limited (241, 263). On the other hand, exposure to low-dose IR has been reported to increase immunity and induce an adaptive response, defined as a priming that enables an improved protective response to subsequent high-dose IR (261). Since the adaptive response is relevant to both the prevention and treatment of cancer, this intriguing phenomenon is under investigation by a number of groups, with an emphasis on identifying the underlying mechanisms of the adaptive response and the conditions that enable its onset. For example, the exposure of human colon carcinoma cells to a dose similar to that generated in image-guided radiotherapy (<100 mGy) followed by two doses of 2 Gy (24 h apart) resulted in an adaptive response which was further linked to the anti-apoptotic protein survivin (131). In vitro, the adaptive response has also been observed in primary human fibroblasts exposed to 100–500 mGy of X-ray 24 h before a 2 Gy dose (also X-ray) (92). This low-dose IR priming altered the response of the DNA repair protein phosphorylated histone H2AX (γH2AX) and increased secreted cytokine levels relative to non-IR-primed cells. Interestingly, these effects were not replicated by priming the cells with cytokines IL6 or transforming growth factor β (TGF-β) alone, thus ruling out a bystander effect through these signaling cues.
As this exciting area of research continues to develop and the mechanisms involved are further elucidated, new avenues will emerge for the manipulation of low-dose radiation in the prevention and treatment of cancer.
Effectors of IR
Gamma radiation of cellular water rapidly generates the reactive oxygen species (ROS) hydroxyl radical (•OH) and ionized water (H2O+), as well as the less investigated reductants hydrogen radical (H•) and hydrated electrons (eaq−). Within one ps (10−12 s), superoxide (O2•−) and hydrogen peroxide (H2O2) are formed as secondary ROS products of IR (308). Subsequent chemical cascades affect the intracellular stoichiometry of these reactive species and generate additional cell-damaging molecules. For example, metal catalysis by intracellular ferrous and/or cuprous ions converts O2•− and H2O2 to form additional amounts of •OH (64). In a separate, but critically significant process, O2•− couples with endogenous nitric oxide (•NO), forming peroxynitrite anion (ONOO−) (85). Cumulatively, these species, along with peroxynitrous acid (ONOOH), nitrogen dioxide (NO2•), dinitrogen trioxide (N2O3), and others, are referred to as reactive nitrogen species (RNS). The increased formation of RNS and the generation of additional ROS equivalents are particularly harmful to the cell, as the reaction products are in many cases more reactive with biomolecules than their precursors.
Cellular macromolecules are modified by direct ionization and via the reactivity of the high-energy species originating from water radiolysis (indirect effects of ionization), affecting an estimated 2000 primary ionization events (351). The timing attributes of cellular damage inflicted by IR range from chemical reactions occurring as rapidly as 0.01 ps after IR to major cellular effects that occur in the range of minutes to hours (308). Direct radiation damage is initiated in the range of 10−14–10−12 s with the breaking of S–H, O–H, N–H, and C–H bonds. Widespread biomolecular damage induced by radiolytic products of water begins within 1 ps (10−12 s), along with thiol depletion and further bond breaking (e.g., C–C and C–N). By 1 ms after IR exposure, the reactions of nascent •OH, H•, and eaq− are mostly completed and DNA repair processes are initiated. Though the activity of some reactive IR products has diminished, an important event occurring through ∼10 s post-irradiation is the increased intracellular formation of ROS and RNS species through mechanisms described next (section “Endogenous propagation of IR-induced ROS”). The cumulative effects of the early, rapid biochemical processes are manifested in later stages of cellular damage, including the slowing of mitosis, damage to protein signaling networks, and membrane rupture, estimated to occur over the course of minutes to 10 h (308).
Endogenous propagation of IR-induced ROS
The overall amount of ROS generated from primary ionization events is further propagated via the intracellular activation of endogenous ROS-producing systems such as nicotinamide adenine dinucleotide phosphate, reduced form (NADPH) oxidase, and the mitochondrial electron transport chain (ETC) (12, 191, 235, 247, 351). IR exposure has been definitively linked to mitochondria-dependent ROS/RNS generation in tumor cells (95). Increased ROS generation in mitochondria after low-dose IR significantly contributed to radiosensitivity and cell survival (10). Whole body irradiation of rats resulted in the increased activity of cytochrome oxidase and NADH-cytochrome c reductase, decreased antioxidant activity, and increased lipid peroxidation in live mitochondrial fractions (170). Irradiation of A549 cells induced mitochondrial ROS production, increased mitochondrial membrane potential, and promoted respiration and ATP production (367). Similarly, an increased expression of NADPH oxidase was reported after irradiation with 10 Gy in rat brain microvascular endothelial cells, and the inhibition of NADPH oxidase led to a decrease in IR-generated ROS (79). IR-induced chromosomal instability in hematopoietic stem cells (6.5 Gy) was reversed by NADPH oxidase inhibition using diphenylene iodonium (262). The mechanisms of NADPH oxidase activation by IR may involve ceramide signaling, which is discussed later in this review. In addition to NADPH oxidase activation, a 2.5 Gy dosage of IR was shown to induce mitochondrial ROS production that can be blocked by inhibitors of mitochondrial respiration (66).
The temporal propagation of IR effects is also achieved through nitrosative stress mechanisms. A study of murine bone marrow stromal cells showed that irradiation with 2–50 Gy stimulated the expression of nitric oxide synthase (inducible nitric oxide synthase [iNOS]), leading to a dose-dependent increase in •NO levels in vitro along with the increased occurrence of nitrated tyrosine residues in vivo (128). Significant increases in the expression of iNOS and elevated levels of nitrate and nitrite have been associated with radiation-induced epithelial dysfunction in the colon (112). In addition to directly modifying tyrosines, cysteines, and hemes, •NO is the endogenous precursor to ONOO− and other RNS (23). The activation of ROS- and RNS-producing pathways by IR is particularly important, as it illustrates a targeted localized increase in these reactive species as a consequence of global IR-induced ROS production, selectively altering cellular signaling and a host of metabolic pathways.
The complex chemical interplay of ROS/RNS generated directly by IR and through derivative systems such as NADPH oxidase, iNOS, and mitochondrial ETC is summarized in Figure 1. O2•−, H2O2, •OH, and ONOO− are especially reactive, damage a wide range of cellular biomolecules, and react with each other to generate additional ROS/RNS. For example, the powerful oxidant peroxynitrite decays rapidly in acidic conditions (pKa 6.8) and forms the highly potent secondary oxidant NO2• (22, 40). In general, the relatively milder oxidants (e.g., H2O2) target biomolecules in a more selective manner, while ROS with higher reactivity, such as •OH, are promiscuous; selected chemical examples appear throughout the next few sections.
FIG. 1.
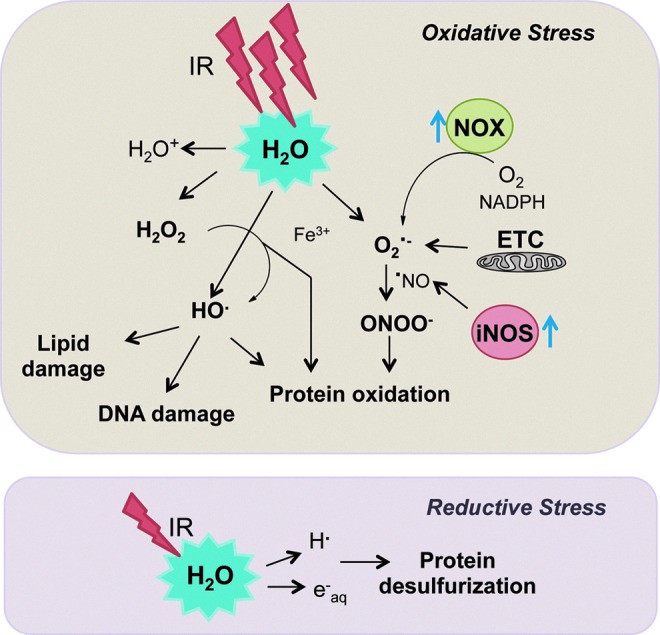
IR generates the potent intracellular oxidants H2O2, O2•−, and •OH, along with reductants H• and eaq−. Endogenous ROS propagation occurs through the mitochondrial ETC and the increased expression of signaling enzymes such as NOX and iNOS. Reductive stress induced by IR leads to loss of sulfur in protein methionine and cysteine residues. •OH, hydroxyl radical; O2•−, superoxide; eaq−, hydrated electron; H•, hydrogen radical; H2O2, hydrogen peroxide; iNOS, inducible nitric oxide synthase; IR, ionizing radiation; NOX, NADPH oxidase; ROS, reactive oxygen species. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Detection of intracellular ROS and RNS is routinely achieved using targeted probes, including fluorescent redox dyes such as dichlorodihydrofluorescein (DCF assay) and chemiluminescent methods (e.g., for •NO detection) (138, 352). Numerous caveats are associated with these methods, particularly with regard to cross-reactivity, and, thus, the development of more selective methods, such as the H2O2-specific peroxylfluor-1, is critically needed (56). Nevertheless, it has been reported that the amount of H2O2 generated by 2 Gy of IR is 10−7 M (351), and the extracellular H2O2 produced immediately after 5 Gy exposure has been measured at 12 μM (3). While seemingly low, these amounts are, at minimum, in the same range as physiological regulatory levels of H2O2, if not 10-fold greater [estimated physiological H2O2 concentration is 10−8–10−7 M (55)]. These concentrations have been shown to affect physiological processes such as proliferation, cell cycle arrest, senescence, and apoptosis (8, 87). We should also point out that a direct correlation between the steady-state levels of physiological or IR-induced ROS and the modification of biomolecules is not straightforward, and one should consider differences in the sensitivity and accuracy of detection methods, ROS compartmentalization, and evolving hypotheses for localized accumulation of ROS such as the “floodgate” hypothesis (360).
Cellular defense against IR-generated ROS
The capability of cells to survive an IR-based insult is dependent on a complex network of ROS-metabolizing molecules and detoxifying enzymes that comprise the cellular oxidative stress defense. O2•−, such as that generated through water radiolysis, is dismutated to H2O2 and O2 by the activity of superoxide dismutases (SODs). Enzymatic detoxification of H2O2 is accomplished through the activity of catalase, peroxiredoxins (Prx), and glutathione peroxidases (GPxs). The roles of these antioxidant enzymes are paramount even under normal cellular conditions to keep ROS levels and pro-oxidant mechanisms in check. Aiding in this cause are low-molecular-weight (MW) endogenous antioxidants such as glutathione (GSH), ascorbate (Vitamin C), melatonin, lipoic acid, ubiquinone (Coenzyme Q10), and Vitamin E, which target water radiolysis products, partially oxidized biomolecules, and peroxynitrite (153, 282, 313, 325, 332).
Almost immediately after cellular exposure to IR, the low MW antioxidant supply becomes compromised, leading, for example, to a rapid decrease in reduced GSH levels [(193), 10 Gy]. In an effort to combat the oxidative burst imparted by IR, cellular transcription factors, including nuclear factor (erythroid-derived 2)-like 2 and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), are activated, resulting in the increased expression of ROS detoxifying enzymes, including catalase and SOD along with GPx, glutathione S-transferase (GST), heme oxygenase-1, and several others (224, 257). In addition, increased levels of many mammalian Prx isoforms have been noted after 10 Gy IR exposure, further enhancing the cellular defense mechanisms (4, 120, 193, 361). Both cumulative and acute IR readily disrupt the cellular redox balance, eventually overwhelming the cellular antioxidant system, an event marked by enzyme inactivation, a low GSH/glutathione disulfide ratio, and a decreased pool of low MW antioxidants. The consequence of such redox imbalance is manifested in the efficient modifications of nucleic acids, lipids, proteins, and other biomolecules (detailed in the next few sections and summarized in Table 1).
Table 1.
Oxidative Modifications of Biomolecules Associated with Exposure to Ionizing Radiation and Their Corresponding Methods of Detection
| Modification | Detection method | References |
|---|---|---|
| Intracellular ROS | DCF assay | Hafer et al. (138) |
| Intracellular RNS | Chemiluminescence | Wardman (352) |
| DNA SSBs and DSBs | TUNEL, Comet, FISH, MS | Weimann et al. (354) |
| 8-oxodG | ELISA, IHC | Rossner and Sram (289) |
| Lipid hydroperoxides (LOOH) | HPLC | Miyazawa et al. (233); Yamamoto (368) |
| Phosphine (DPPP) | Okimoto et al. (252) | |
| Hydrazine with MS | Milic et al. (229, 230) | |
| Hydroxynonenal (HNE) | HPLC/fluorescence | Tanaka et al. (326) |
| HNE, MDA, Acrolein | GC-MS | Kawai et al. (169) |
| Acrolein-DNA adducts | UHPLC-MS/MS | Yin et al. (374) |
| Protein radicals | EPR | Gordy and Miyagawa (129); Symons and Taiwo (320) |
| Protein carbonylation | DNPH antibody | Yan and Forster (369) |
| DNPH with radiolabeling | Lenz et al. (199) | |
| DNPH with MS detection | Guo and Prokai (136); Bernevic et al. (30); Bollineni et al. (43) | |
| GPR with MS detection | Mirzaei and Regnier (232) | |
| Label free MS detection | Rauniyar et al. (276) | |
| Gel fluorescence | Madian and Regnier (219); Tamarit et al. (324) | |
| Biotin-conjugated probes | Chavez et al. (59); Chung et al. (70) | |
| Methionine sulfoxide (MetO) | Infrared spectroscopy | Ravi et al. (277) |
| MetO antibody | Nakaso et al. (246) | |
| Label-free MS detection | Guan et al. (135); Xiang et al. (363) | |
| Reversibly oxidized Cys (SOH, SS, SNO, SN) and tailored for nitrosated Cys | Switch-tag (OxICAT) | Leichert et al. (197); Sethuraman et al. (301) |
| NOxICAT | Lindemann and Leichert (207) | |
| S-nitrosocysteine (CySNO) | Switch-tag | Lu et al. (213); Greco et al. (132); Camerini et al. (52) |
| On-resin (SNO-RAC) | Forrester et al. (111) | |
| Phosphine probes | Wang and Xian (347); Bechtold et al. (21); Zhang et al. (381) | |
| Cysteine sulfenic acid (CySOH) | Switch-tag | Saurin et al. (295) |
| CySOH antibody | Seo and Carroll (300) | |
| 1,3-Dicarbonyls: fluorescent | Poole et al. (267) | |
| 1,3-Dicarbonyls: biotinylated | Charles et al. (57); Poole et al. (267); Klomsiri et al. (178); Nelson et al. (248); Qian et al. (271) | |
| 1,3-Dicarbonyls: click chem. | Reddie et al. (278); Leonard et al. (200); Seo and Carroll (299); Qian et al. (272) |
8-oxodG, 8-oxo-2′-deoxyguanosine; DCF, dichlorofluorescein; DNPH, 2,4-dinitrophenylhydrazine; DPPP, diphenyl-1-pyrenylphosphine; DSB, double-strand break; ELISA, enzyme-linked immunosorbent assay; EPR, electron paramagnetic resonance spectroscopy; FISH, fluorescence in situ hybridization; GC-MS, gas chromatography coupled to mass spectrometry; GPR, Girard's P reagent; HPLC, high-performance liquid chromatography; IHC, immunohistochemistry; MDA, malondialdehyde; MS, mass spectrometry; NOxICAT, isotope-coded affinity tag for detecting nitrosated and oxidized cysteine; OxICAT, isotope-coded affinity tag for detecting oxidized cysteine; RNS, reactive nitrogen species; ROS, reactive oxygen species; SNO-RAC, resin-assisted capture for S-nitrosothiols; SSB, single-strand break; TUNEL, terminal transferase; UHPLC-MS/MS, ultra high-performance LC coupled to tandem MS.
Chronic IR-mediated oxidative effects—evaluation based on in vivo evidence
Without doubt, the fundamental reaction chemistry underlying acute radiation damage involves rapid and widespread oxidation events that play critical functions in tumor cell killing. In addition, indirect contributions from inflammation (240), changes in vasculature (1), growth factor signaling (130), cytokine expression (385), mitochondrial dysfunction (380), and other cellular responses have also been demonstrated to influence the overall outcome of radiation treatment in vivo. Many of these processes have an underlying mechanism of oxidative regulation and/or action.
A remaining question is whether the late effects of radiation on normal tissue (e.g., chronic oxidative stress occurring over weeks to months after treatment) are regulated by oxidative mechanisms (81). For example, when using radiation nephropathy as an indicator of normal tissue damage [see ref. (78) for translational relevance and descriptions of mechanisms involved], there is evidence that both supports and refutes a function of late IR-induced oxidative injury. Supporting evidence includes the identification of chronic oxidative DNA damage (8-oxo-2′-deoxyguanosine [8-oxodG]) in glomerulus and tubules for approximately 24 weeks after IR treatment (288). On the other hand, antioxidant treatment in a rat model of radiation nephropathy using three different reagents (deferiprone, an iron chelator; genistein, an anti-inflammatory isoflavone; and apocynin, an NADPH oxidase inhibitor) did not significantly protect against kidney damage (73). Other studies using microarray analyses for gene expression have found either no or only limited evidence of up-regulation of known antioxidant proteins (72, 185).
In other tissues susceptible to late radiation effects (e.g., skin, lung, and brain), treatment with antioxidants, antioxidant enzymes, or enzyme mimetics has been shown to reduce the late effects of radiation (384). Clinical studies have shown that the administration of Lipsod, a lipid-encapsulated form of antioxidant enzyme SOD, led to a reduction in radiation-induced skin fibrosis (89). Similarly, others have shown that administration of the SOD mimetic EUK-207 along with genistein decreased the extent of radiation damage in normal lung but not skin tissue in a rat model of radiation (147).
A similar prevention or mitigation of radiation late effects was obtained with blockers of the renin–angiotensin system (RAS) and peroxisome proliferator-activated receptor gamma agonists (77). A redox mechanism for their mode of action was proposed for protection against loss of cognitive functions in radiation brain injury (287, 296). Angiotensin II (Ang II), an active peptide of the RAS, binds to angiotensin II type 1 receptor (AT1R) and generates ROS via NADPH oxidase. There is evidence of both increased expression of AT1R in a lung fibrosis model (255) and increased production of Ang II in rat lung at 2 and 6 months post-irradiation with 20 Gy (54). Preclinical studies indicate a role of Ang II blockers in ameliorating radiation-induced tissue damage. Angiotensin-converting-enzyme inhibitors (ACEI) and AT1R antagonists are effective in the treatment and prevention of radiation nephropathy (74–76, 238). Other studies reported that Ang II blockers reduced radiation-induced chronic injury, including lung fibrosis, vascular injury, and inflammation (119, 179, 225, 234). ACEI ramipril, given after stereotactical irradiation with 30 Gy using a single collimated beam, reduced the severity of optic nerve damage and retained nerve function (176, 290). In addition, chronic administration of AT1R antagonist, L-158,809, or ACEI ramipril prevented radiation-induced cognitive impairment in rats assessed at 6 months post-irradiation (196, 286). AT1R antagonist L158,809 blocked radiation-induced expression of heme oxygenase-1, a sensitive indicator of oxidative stress, suggesting a role of Ang II in radiation-mediated chronic oxidative stress (86). In vitro studies further confirmed a role for both Ang II and NADPH oxidase in the generation of ROS after radiation (79). Furthermore, blockade of Ang II with ACEIs or ATR1 antagonists reduced the expression of inflammatory proteins (79, 319) and prevented AP-1 and NF-κB activation (42, 384).
Cumulatively, these results demonstrate tissue- and antioxidant-dependent effects of late radiation injury and its mitigation as a potential explanation for the sometimes opposing reports in this area of research. In the subsequent sections of this review, we reference a number of studies that have identified markers of IR-induced damage (products of IR interaction with nucleic acids, metabolites, and proteins) shown to persist for hours to months after IR exposure. While in vivo evidence linking radiation exposure to both acute and chronic oxidation of biomolecules continues to emerge (11), there is a clear need for more investigations to determine the functional consequences of long-lived products of acute IR on tissue-specific radiation injury.
Interaction of IR and IR Effectors with Nucleic Acids
The interaction of IR with the cell nucleus has for many years been considered the primary mechanism responsible for the genotoxic effects of radiation fueling the central dogma of radiation biology. Among others, investigations by Munro in the early 1970s showed that a significantly higher dosage of radiation is needed to kill cells when the radiation is targeted selectively to cytosol compared with the nucleus (242). This view has evolved in more recent years, and while the interaction of IR with nucleic acids is certainly critical, studies are deciphering new IR-mediated pathways that lead to mutations, carcinogenesis, or cell death. Such mechanisms are discussed in the context of IR interaction with lipids (section “Interaction of IR and IR effectors with lipids”) and proteins (section “Interaction of IR and IR effectors with proteins”).
Chemistry
Nucleic acid damage in response to cellular IR is widely documented and is the classical paradigm for understanding the harmful effects of IR. Given the preponderance of combined therapies, it is important to note that the occurrence of DNA damage is increased when chemotherapeutics such as cisplatin are used in conjunction with radiation (284). DNA damaging events inflicted by IR alone include the deleterious alteration of bases and sugars, cross-link formation, single- and double-strand breaks (SSBs/DSBs), and DNA clustering (96, 331). Of the water radiolysis products, •OH is the most abundant and particularly destructive to nucleic acid molecules.
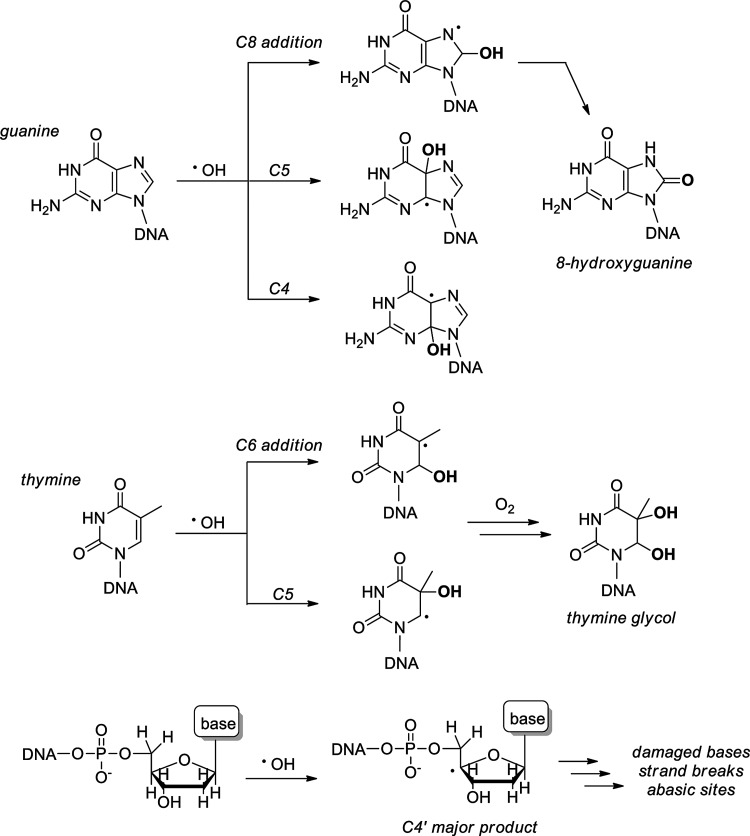
Radiation damage to deoxyriboses is the primary event underlying strand breakages, which occur in a high frequency and randomly along the DNA backbone in response to both direct •OH attack and the activity of nucleic acid-binding enzymes (46). DSBs, in particular, originate through the coordinated reactivity of two •OH radicals at nearby ribose sites, ultimately leading to strand breaks through subsequent radical pathways (14, 24, 350). Both the nucleobases and deoxyribose are targets for •OH -mediated damage. For purine nucleobases, •OH adds at C4, C5, and C8, generating reactive adduct radicals that lead to a variety of products, with the most common being the 8-hydroxypurines, specifically 8-oxodG, which serve as well-known hallmarks for oxidative DNA damage (Fig. 2) (94). The persistence of oxidative DNA damage in vivo has been illustrated by elevated levels of 8-oxodG in the mouse kidney in response to IR (20 Gy maximum) even at 24 weeks after treatment (288). Pyrimidine olefins are also susceptible to •OH addition, particularly at C5 and C6, generating pyrimidine glycols in the presence of O2 (115).
FIG. 2.
Effects of IR on cellular DNA: oxidative damage to nucleobases and riboses mediated by •OH.
Radiation-induced nucleobase lesions include oxidatively modified bases as well as abasic sites, but do not immediately result in strand breakage (344). Both •OH and eaq− react with the nucleobases at diffusion-controlled rates, adding to unsaturated bonds and abstracting H• from methyl and amino substituents (94). These radical products are structurally diverse and are involved in many secondary reactions as oxidants or reductants, depending on the structure and the reactive species in proximity. A common fate for such species is the diffusion-controlled reaction with O2, producing peroxyl radicals, hydroperoxides, ring-opening events, and ring-contraction products. Specific kinetic and thermodynamic details of radiation-induced nucleobase damage, as well as the characterization of downstream oxidation products, have been compiled in a recent review (94).
Without unsaturations, deoxyriboses are modified via .OH-mediated hydrogen abstraction (Fig. 2). Though hydrogen abstraction has been noted at all ribose carbons, radical formation at C4′ dominates (227). Hydrogen abstraction at ribose carbons is the initiation event for reparable damage (e.g., of nucleobases) as well as irreparable DNA strand breaks. The extent of radiation-associated DNA damage is dramatically increased in the presence of bivalent metal ions (Cu2+, Fe2+) along with cellular reductants (GSH) via the Haber–Weiss generation of .OH and H2O2 (9, 270, 279). The capacities of hydrated (eaq−) and prehydrated electrons to facilitate DNA strand breaks have garnered attention only recently but appear to play an important role in the cumulative effects of IR on DNA (44, 307, 346). O2•−, in contrast, has been shown to be significantly less reactive with DNA than with other water radiolysis products (266).
Biological consequences
The immediate response to IR-induced ROS/RNS-mediated DNA damage is the activation of the cell cycle checkpoint response, an intricately controlled network involving sensor, transducer, and effector proteins that respond to the DNA damage signal by initiating a cytoprotective response—the DNA damage response (DDR). The cellular mechanisms for DNA repair are extensive and have been recently reviewed in detail (331). A brief discussion is included here to highlight the interplay between the IR-induced DNA damage, IR-induced protein modifications, and chromatin remodeling that, ultimately, converges to repair nucleic acid damage or signal for the initiation of cell death pathways. Sensor and transducer proteins recognize DNA damage sites, initiate, and amplify a biochemical cascade. The principal proteins implicated as sensors and transducers of DNA damage include the Mre11–Rad50–Nbs1 complex, BRCA1/2 proteins, ATM/ATR, DNA-dependent protein kinase (DNA-PK), checkpoint kinases 1/2 (Chk1/2), and poly(ADP-ribose) polymerase (143, 265, 297, 342, 364, 365). The activities of these proteins are tightly regulated by multiple post-translational modifications (PTMs). For example, ATM, one of the initial enzymes activated in DDR and a key regulator of all three cell cycle checkpoints, is activated by IR in a mechanism involving phosphorylation at Ser1981 (13). ATM is also directly activated by IR-induced ROS, facilitating dimerization through the formation of an intermolecular disulfide (93, 137). In addition to modifying other crucial cell cycle checkpoint proteins (e.g., Chk1/2 and p53), ATM mediates the phosphorylation of Kap1, promoting heterochromatin relaxation and increasing the efficiency of DNA repair (127, 250). The ATM polymorphism was also found to play a function in determining the sensitivity to radiation, as detailed in the section “Genomics and epigenomics.” Another observation pointing to the significance of oxidative PTMs to the DDR comes from studies using freshwater invertebrates, Adineta vaga and Caenorhabditis elegans, revealing that diminished carbonylation maintains the activity of DDR enzymes, enhancing post-IR survival in A. vaga compared with the genomically similar C. elegans (183, 184). Protein carbonylation, an oxidative modification of many amino acids, is discussed in the section “Carbonylation.” Thus, cellular DNA damage after IR exposure is largely dependent on the capacity of the cellular proteins to prevent and repair the DNA modification induced by IR.
Methods of detection
Historically, many techniques and methods have been used to detect the DNA damage. Direct methods include those designed to detect specific SSBs and DSBs such as the well-known terminal transferase, comet, and fluorescence in situ hybridization assays, and those targeted for the analysis of oxidized nucleic acid metabolites [e.g., liquid or gas chromatography (GC) coupled with electrochemical or mass spectrometry (MS) detection] (354). Other methods rely on enzyme-linked immunosorbent assay and immunohistochemistry as commonly applied for detection of 8-oxodG, one of the most abundant oxidized nucleosides (289). Indirect methods for the detection of DNA damage include Western blotting or imaging analysis of proteins involved in the DDR that undergo PTMs after IR-induced DNA damage. Examples include the detection of γH2AX histone that undergoes phosphorylation and binds to sites of DNA DSBs (159, 291). Only the most frequently used methods of detection were highlighted here; for a more in-depth discussion, including the pros and cons of various techniques, the reader is directed to a recent book on this topic (91).
Interaction of IR and IR Effectors with Lipids
Another biomolecule target of radiation-generated ROS is the lipid layer within cell membranes. The lipid component of cell membranes is generally estimated to be ∼5 nm in thickness with significant exposure to the aqueous cellular environment (315). Though radiation is capable of directly damaging lipids, lipid bilayer mimetics have indicated that indirect damage induced by water radiolysis products is a larger contributor toward overall lipid modification by IR (25). Radiation induces lipid peroxidation, particularly the peroxidation of polyunsaturated fatty acids (PUFAs), leading to an increase in membrane permeability, disruption of ion gradients and other transmembrane processes, and altered activity of membrane-associated proteins (80, 359). Studies of IR targeted to the cell membrane revealed the induction of apoptosis at 5–10 Gy via increased ceramide levels (139). This outcome was observed even in cells without a nucleus, revealing another pathway for cellular IR damage independent of nucleic acid damage.
Chemistry
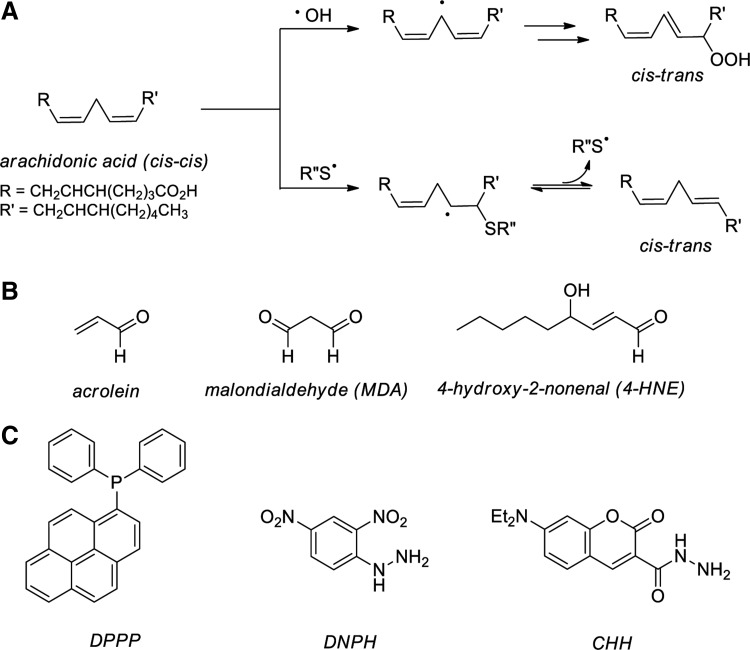
Unsaturated fatty acids are unreactive toward molecular oxygen, but readily oxidized in a number of radical-mediated processes (269). Peroxidation of PUFA, such as linoleic and arachidonic acids, leads to the generation of diene hydroperoxides, isomerization of cis-alkenes, and/or degradation to form small molecule reactive carbonyls such as malondialdehyde (MDA), acrolein, and 4-hydroxy-2-nonenal (HNE) (113). Alkene isomerization is initiated by the abstraction of hydrogen from a PUFA bisallylic site by •OH, thiyl radical (RS•), or other radicals, generating a carbon-centered PUFA radical. Subsequent capture by oxygen results in a cis-trans isomerization and forms a peroxyl radical in a diffusion-controlled reaction (Fig. 3A, top path) (269, 323). Alternatively, cis-trans isomerization may also occur via the direct addition of RS• to a PUFA alkene (Fig. 3A, bottom path). Ejection of the R′′S• radical accompanies isomerization, a process documented even in the presence of oxygen (228). Peroxyl radicals are terminated by the formation of lipid hydroperoxides (LOOH) and PUFA fragmentation to MDA, HNE, and acrolein (Fig. 3B). The reactive aldehyde products of PUFA peroxidation persist intracellularly for approximately 2 min, providing ample time for the modification of proteins, nucleic acids, and other biomolecules (69, 154, 264). In vivo evidence for increased levels of reactive aldehyde MDA in response to IR has been shown in the kidney, lung, and liver of rats exposed to 8 Gy of total body IR (298).
FIG. 3.
Effects of IR on cellular lipids. (A) Oxidative cis-trans isomerization of polyunsaturated fatty acids. (B) Reactive aldehydes generated by lipid peroxidation. (C) Chemical probes for the detection of intracellular ROS (DPPP) and reactive aldehydes (DNPH; CHH). CHH, 7-(diethylamino)coumarin-3-carbohydrazide; DNPH, 2,4-dinitrophenylhydrazine; DPPP, diphenyl-1-pyrenylphosphine.
Biological consequences
A major factor affecting the fate of LOOH and the magnitude of its effects in cells is the extent of lipid peroxidation, correlated directly to cellular redox status. Moderate levels of LOOH activate the oxidative stress response, which, when exceeded, leads to apoptosis; high levels, and widespread LOOH, resulting in global damage to the cell membrane and intracellular content, triggering membrane lysis and necrosis (123). Chemically, the degradation of LOOH proceeds through one-electron and/or two-electron reduction pathways. One-electron reduction of LOOH occurs via aerobic reduction mediated by Fe2+ ions and providing epoxyallylic peroxy radicals (OLOO•) with subsequent radical reactions leading to additional lipid damage. The detoxification of LOOH by two-electron reduction is carried out by selenoperoxidase enzymes such as the GPxs, thioredoxin reductase (TrxR), and phospholipid hydroperoxide glutathione peroxidases (PHGPxs), as well as the seleno-independent GST α (339, 356). These enzymes reduce LOOH to LOH and H2O in the presence of two equivalents of GSH with the exception of TrxR, which instead utilizes NADPH as an electron donor (38). The LOOH-reducing enzymes use similar mechanisms apart from their cofactor differences and vary mainly in terms of size and substrate specificity. To illustrate, though both the GPxs and PHGPx detoxify H2O2, GPx enzymes target polar LOOH such as fatty acid hydroperoxides; whereas PHGPx reduces phospholipid hydroperoxides, cholesterol hydroperoxides, and other hydroperoxides of a lower polarity (330, 339, 340). Apolipoprotein A-I (apoAI) and apolipoprotein A-II (apoAII) have been noted to reduce cholesterol ester hydroperoxides via oxidation of critical Met residues to the corresponding sulfoxides (121). In total, the enzymatic defenses against LOOH cytotoxicity rely heavily on reductant bioavailability (GSH, NADPH) coupled with reduced active site cysteine, selenocysteine, and methionine residues of the repair enzymes. The intense disruption of cellular redox metabolism invoked by IR is sufficient to overwhelm LOOH defense mechanisms in IR-sensitive cells and tissues due to the oxidation and inactivation of detoxifying enzymes, though the timing of such modifications and the lipids most susceptible to radiation damage are not well understood. LOOH are known to persist after IR, as demonstrated in vivo in the mouse hippocampus at 2 weeks after IR exposure (10 Gy) (205).
Sphingolipid metabolism is a key pathway that is altered in response to IR. The sphingolipid ceramide, a product of sphingomyelin hydrolysis catalyzed by acid sphingomyelinase (ASMase) and neutral sphingomyelinase, has, in particular, been closely connected to the cellular IR damage. Cellular exposure to IR leads to ASMase relocalization from the lysosomes to the plasma membrane, where sphingomyelin is hydrolyzed to generate large amounts of ceramide (80). Both mutations and PTMs at Cys629 (and possibly Ser508) are known to regulate the activity and localization of ASMase at the plasma membrane, highlighting the complex interactions among IR-induced protein modifications, lipid raft microdomain rearrangement, and DNA damage repair (273, 378). In addition, DNA damage events activate ceramide synthase, the de novo source of ceramide, contributing to significantly increased levels of intracellular ceramide in response to IR (343). This large in situ generation of ceramide from sphingomyelin within membrane lipid rafts alters the membrane properties, largely because ceramide-containing lipid rafts coalesce and form large, ceramide-enriched membrane platforms. These lipid platforms not only contain membrane receptors and proteins but are also enriched in nuclear enzymes such as DNA-PK that are relocalized on irradiation. In head and neck cancer, in particular, the dynamics of lipid raft microdomains and associated signaling was shown to underlie the response to IR and targeted therapies against epidermal growth factor receptor (EGFR) (33, 101, 157). Membrane rafts have also been associated with the bystander effects of low-dose IR, where cytoplasm-targeted IR of one cell led to an increased micronuclei yield of 36–78% in surrounding cells (302). This outcome was independent of the nucleus and instead was linked to •NO signaling and membrane raft formation. Lipid rafts are also implicated in IR propagation via NADPH oxidase reconstitution within lipid platforms, where it functions as an additional ROS source as discussed in the section “Endogenous propagation of IR-induced ROS” (379).
Methods of detection
Many biochemical approaches have been developed to detect lipid peroxidation. Traditional techniques involve the chemiluminescence detection of LOOH that are first separated by high-performance liquid chromatography (HPLC) and then treated with isoluminol in the presence of a metal ion catalyst (e.g., heme or cytochrome c) with the detection of isoluminol oxidation induced by LOOH (233, 368). A newer approach involves the use of diphenyl-1-pyrenylphosphine (DPPP), which reacts with numerous biological oxidants and forms the highly fluorescent phosphine oxide (Fig. 3C). DPPP localizes to cell membranes, where it is oxidized by LOOH at rates much faster than oxidation by H2O2 or tert-butyl hydroperoxide, likely due to its hydrophobic compatibility with the lipid bilayer (252). This is an especially critical development for the fluorescence imaging of lipids in live cells, as the widely used DCF diacetate (DCF assay reagent) is prohibitively hydrophilic for lipid imaging (252).
Lipid peroxidation events have also been uncovered via the detection of the volatile, lower MW aldehyde end-products. Levels of the lipid-derived aldehyde HNE have been measured using HPLC with pre-column labeling and fluorescence detection (326). Along with HNE, MDA and acrolein have been directly quantified using GC coupled to MS; softer ionization of these species using matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) is inefficient and often requires derivatization (97, 169). 2,4-Dinitrophenylhydrazine (DNPH) readily modifies oxidized phospholipids for MS-based detection in a mechanism similar to that shown in Figure 4, but is not as successful at targeting lower MW peroxidation products in complex samples (Fig. 3C) (229). Recently described derivatization reagent 7-(diethylamino)coumarin-3-carbohydrazide (CHH) provides a means of detecting both high and low MW lipid peroxide products in the same sample (230). Furthermore, the CHH amine improves ionization (by ESI) and offers a unique footprint of reporter ions on MS2 fragmentation by collision-induced dissociation (CID) (230). The biomolecules targeted by these reactive aldehydes, with increased mass and hydrophilicity, are more amenable to analytical separations by liquid chromatography (LC): Acrolein adducts of DNA in human leukocytes have been quantified by ultra high performance LC coupled to tandem MS (374). Ceramide-enriched lipid rafts are detected by imaging in live cells using fluorescent conjugates of cholera toxin subunit B (34). The detection of protein adducts of lipid-derived aldehydes is discussed in the next few sections.
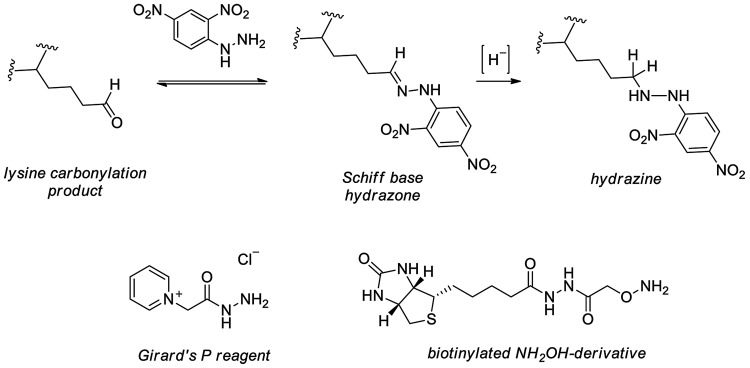
FIG. 4.
Detection of reactive aldehydes and carbonylated proteins. Derivatization of protein carbonyls using DNPH and mechanistically similar Girard's P reagent and a biotinylated hydroxylamine.
Interaction of IR and IR Effectors with Proteins
In addition to nucleic acids and lipids, proteins are highly abundant cellular biomolecules and prominently targeted by IR, inducing changes in their expression and activity as well as oxidative or reductive PTMs. Single cell exposure to microbeam-directed alpha particles in the cytoplasm was shown to be mutagenic with severity of outcome dependent on the intracellular GSH and radicals (362). Importantly, these mutations were shown to occur without much, if any, cell death, illustrating the potential for transforming cell signaling along with cell persistence. In vivo evidence for the oxidative modification of proteins in response to IR has been observed in the mouse brain and rat liver as judged by the increase in protein carbonylation (99, 222).
PTMs critically affect protein structure and function though conformational changes, modulation of enzymatic activity, degradation, and cellular trafficking (268, 275, 295, 349, 376). The paradigm for understanding cellular IR damage in the past several decades has evolved to include the elucidation of protein targets in addition to the classically investigated DNA damage events. Fueling this restructuring are investigations of cytoplasm-directed IR and a wide array of studies that revealed alterations in the cellular proteome at IR doses <10 Gy, once believed to be the threshold for affecting proteins (124, 190). For example, cellular exposure to IR targeted to the cytoplasm led to an increase in levels of RNS, LOOH, cyclooxygenase 2 expression, and activation of extracellular signaling-related kinase signaling, along with increased oxidative DNA damage (151). The modulation of protein expression and protein–protein interactions has been noted in response to doses as low as 1 Gy [see section “Endogenous propagation of IR-induced ROS” and ref. (204)]. The inactivation of redox-sensitive enzymes such as catalase, SOD, GPx, and the protein tyrosine phosphatases has been observed at 2–8 Gy (17, 25). Moreover, IR doses <6 Gy have been shown to disrupt the extent of protein PTMs, including, but certainly not limited to, S-nitrosation and carbonylation (192, 223, 386). Cumulatively, these results demonstrate that the impact of IR on the proteome was long underestimated but is now accepted to profoundly affect cellular processes. The observation that protein signaling is altered at <1 Gy of IR illustrates that the radiosensitivity of proteins is akin to other biomolecules and underscores the need for investigating protein-based IR damage (371). In this chapter, the focus will be on oxidative and reductive PTMs and IR-induced changes in protein expression.
Protein oxidation
More than 35 types of oxidative protein modifications are known currently, including direct amino-acid oxidation (e.g., for Cys and Met), oxidative cleavage of the protein backbone and/or amino acid side chains, carbonylation, and the addition of lipid oxidation products discussed earlier (219). In addition to IR therapy, protein oxidation has been implicated in the progression of many disease states, including diabetes (372), inflammation (312), sepsis (209), Alzheimer's disease (145), multiple sclerosis (37), Parkinson's disease (90), and many cancers [see (283) and references within].
Radical cleavage of the protein backbone and modification of amino-acid side chains
•OH, generated via IR and other processes, initiates cleavage of the protein backbone and reacts with each of the twenty standard amino acids and selenocysteine, with associated rate constants for all such reactions in the 107–1010 M−1 s−1 range (pH 7), reaching diffusion-limited rates for cysteine, methionine, and aromatic amino acids (41, 88). Radicals, including •OH and others, are believed to preferentially react with the protein amide backbone over the amino-acid side chains, readily abstracting hydrogen atoms and forming α-carbon-centered radicals. Similar to DNA-based radicals, protein carbon radicals subsequently react in a diffusion-controlled manner with O2 and may be quenched by other radical species when present in sufficient concentrations. Peroxyl radicals, formed via a reaction with O2, facilitate the formation of additional oxidation products through radical mechanisms, leading to fragmentation of the protein backbone.
Similarly, •OH abstracts hydrogen atoms from aliphatic amino-acid side chains at all carbons, demonstrating that its high degree of reactivity precludes chemoselectivity. Except under anoxic conditions, aliphatic amino-acid radicals are rapidly oxygenated and generate peroxyl radicals or are repaired by cysteine thiols, leading to thiyl radicals, though kinetically this reaction is not believed to compete with oxygenation (31). Aromatic amino acids are significantly more reactive with the dominant reaction pathway being •OH addition to the aromatic ring. In the case of Tyr, •OH addition and subsequent hydrogen abstraction leads to phenoxyl radicals, which in the absence of reductants, form Tyr dimers that are implicated in the formation of intra- and inter-protein linkages (107). Nitrotyrosine products are formed in response to ROS/RNS (e.g., peroxynitrite) and react with eaq− several orders of magnitude faster than tyrosine (k=3.0×1010 M−1 s−1 at pH 7.0 for 3-nitroTyr vs. 2.8×108 M−1 s−1 at pH 6.6 for Tyr) (106, 304). Increased levels of nitrotyrosine have been observed in the mouse hippocampus within 2 h of IR exposure (8 Gy) relative to the control (116). Tryptophan is another target for RNS-initiated attacks to form regioisomeric nitro and hydroxyl derivatives (158, 366).
Methods of detection. Protein backbone fragmentation is classically monitored by the detection of radical products using electron paramagnetic resonance spectroscopy (129, 320).
Carbonylation
Radiation-generated ROS are noted to modify cellular proteins by carbonylation, the post-translational addition of carbonyl moieties to amino-acid side chains, particularly occurring via metal catalysis on Lys, Thr, Pro, Glu, Asp, and Arg residues (220). Along with Lys, Cys and His also undergo carbonylation by reacting with oxidized lipid products such as HNE (337). Sites of carbonylation are formed by the Michael reactivity of Cys, His, and Lys with reactive sugar and other lipid oxidation products. The carbonylation event produces a variety of acyl-modified amino acids and is capable of inducing a lesion in the protein backbone; see (219) for chemical structures. Carbonylation is widely considered an irreversible process and appears to target specific amino acids within a protein rather than many residues indiscriminately; recent evidence suggests that the sites susceptible to carbonylation within a given protein appear to be modulated by the identity of the oxidant as well as structural aspects of the protein (327). Repair processes for proteins modified by carbonylation have not been identified, and, instead, degradation is believed to be the primary mechanism for cellular regulation of proteome carbonylation (201). Increased levels of protein carbonyls have been noted after IR exposure (10 Gy) (310, 317), and in addition, have been implicated in the pathophysiology of many human disease states, including chronic lung disease (357), neurodegenerative diseases (133), diabetes (2), and ischemia-reperfusion injury [(84) and references therein].
Methods of detection. Protein carbonyls, though formed irreversibly, are unstable and readily captured in the Schiff base form by lysine residues, even in frozen samples, making their detection challenging after long-term storage of biological samples (219). Detection of nascent or more stable carbonyls is, however, possible by derivatizing with DNPH (also called Brady's reagent). Nucleophilic carbonyl capture affords hydrazone products (Fig. 4) that are readily detected spectrophotometrically (360 nm) or via immunoblotting using anti-DNP antibodies (369). DNP derivatization is rendered irreversible on a mild reduction of the resulting Schiff base hydrazone product by borohydride reductants; the use of NaB3H4 provides a means for radioactive quantitation of carbonylation (199). Another method to identify and quantify cysteine carbonylation products employs the Raney nickel-catalyzed thioether reduction (338).
Newer techniques for uncovering protein carbonylation have been developed with a focus on compatibility with MS. Sites of His and Cys carbonylation on DNPH-modified peptides have been detected by MS using MS2 with CID for precursor ion fragmentation, though the issue of neutral loss during the acquisition of MS2 spectra complicates the detection of sites carbonylated by HNE (136). A more often utilized approach couples gel electrophoresis and MS, where protein carbonylation is first detected using a DNP-targeted antibody followed by the digestion of selected proteins and MS analysis of their resulting peptides (30). Hydrazine-based detection has been extended using carbonyl-labeling compounds that are appended to biotin for affinity capture or fluorophores such as boron-dipyrromethene, Cy3, and Cy5 for in-gel fluorescence detection (219, 324).
The detection of DNP-derivatized peptide carbonyls has also been achieved by MS using negative ESI with MS2 acquired with pulsed-Q dissociation fragmentation (43). One such carbonyl labeling compound, Girard's P reagent (GPR), utilizes analogous hydrazine chemistry but is equipped with a pyridinium moiety amenable to enrichment using strong cation exchange (SCX) chromatography and is beneficial for peptide ionization (231). Isotope-coded GPR has been used to identify 41 carbonylated peptides in H2O2-treated yeast using SCX followed by MALDI-time-of-flight (TOF)-TOF analysis (232). Alternatively, label-free approaches have been used for the analysis of HNE-modified peptides, using neutral loss-triggered MS3 to pinpoint carbonylated residues (276).
Similar to other PTMs, the detection of carbonylation events is challenged by their low abundance, owing to the relatively low intracellular concentrations of many signaling proteins and the fact that only a small number of amino acids per protein are targeted for oxidation. A complicating factor in distinguishing the two primary types of protein oxidation events, carbonylation and cysteine oxidation, is the recently noted cross-reactivity of DNPH and hydrazine probes with cysteine sulfenic acid (83, 370). Biotin-conjugated hydroxylamines capture protein carbonyl Michael adducts (e.g., those formed via amino-acid modification by HNE) and afford stable oxime adducts at physiological pH (59, 70). Their selectivity for carbonyls over a direct reaction with cysteine sulfenic acid has not been investigated, though such products should be readily distinguished using high-resolution MS techniques. Care should also be taken to avoid nucleic acid contaminants, which generate false positives in DNPH-based assays (215).
Methionine oxidation
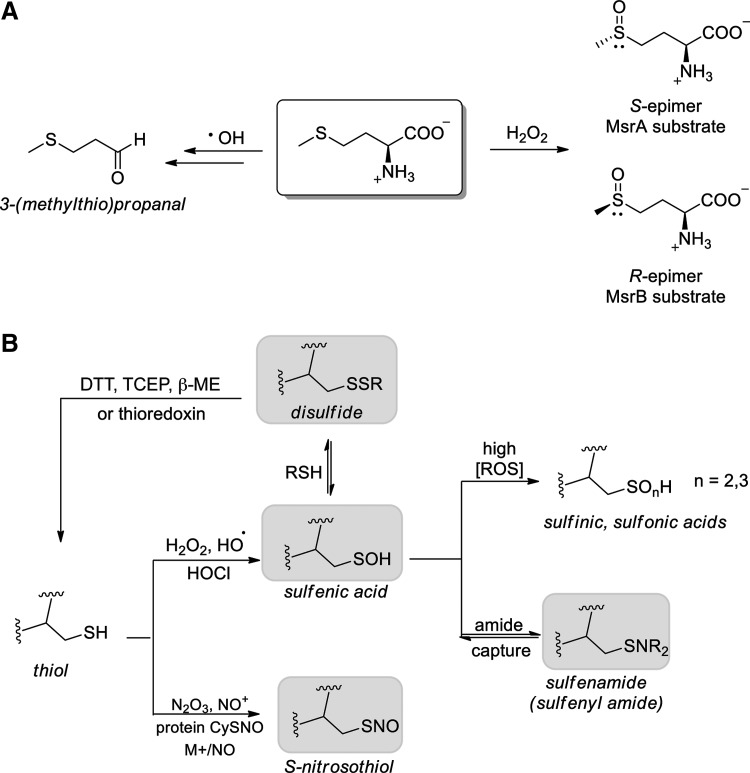
A wide array of one-electron and two-electron oxidants target methionine, including the water radiolysis products. The products of methionine reaction with •OH, H•, eaq−, and H2O2 have been reported in a recent study using aqueous solutions of free methionine (10 mM) that are exposed to a prolonged IR dose (10 h at 6.5 Gy min−1) (15). Characterization of the Met-based products revealed that in the presence of molecular oxygen, (i) the addition of •OH to the sulfur atom of Met did not afford methionine sulfoxide (MetO) but exclusively 3-(methylthio)propanal; (ii) H2O2, derived from water radiolysis or O2•− disproportionation, oxidized Met to MetO (Fig. 5A) (15). Reactions of Met with IR-associated reductants H• and eaq− are discussed in the section “Protein modification by reducing radical species.”
FIG. 5.
Radiation-induced sulfur oxidation in proteins, affecting (A) methionine and (B) cysteine residues. Oxoforms in gray are readily reduced to thiol by DTT and other reductants in vitro.
Other IR-relevant species such as ONOO− and other secondary IR oxidants readily oxidize free methionine and protein methionines to sulfoxide products (Fig. 5A). MetO is significantly more polar than unoxidized Met, and its high degree of thermodynamic stability precludes its reduction by the majority of non-enzymatic intracellular reductants (GSH, ascorbate, and protein thiols) and enables its direct detection. Hyperoxidation of MetO generates methionine sulfone, an irreversible oxidation product identified on irradiation of free methionine (305). Though the fundamental chemical details regarding Met oxidation products are well established, large-scale investigations of IR-induced MetO formation in proteins and the resulting intracellular consequences are still ongoing.
Specific to IR exposure, MetO formation has been noted in vitro only for cytochrome c (329), hemoglobin (363), and bovine α-crystallin (110). MetO has also been observed on ROS treatment of latent TGF-β, an established target of IR (98, 162), providing indirect evidence for the potential formation of MetO in this protein on treatment with IR. Numerous examples have shown, however, that MetO formation occurs in proteins under a range of conditions (non-IR or indirectly linked to IR) with site specificity, suggesting a role of Met oxidation as a regulated mechanism for redox protection and modulation of protein function, though the molecular factors underpinning selectivity are not well established (214, 314). For example, sulfoxide formation at 2 of 9 calmodulin methionine residues was shown to disrupt calmodulin-mediated activation of plasma membrane Ca-ATPase (18, 32). Other examples include MetO formation in the protein Parkinson disease protein 7 (DJ-1), an event that is closely associated with the development of neurodegenerative diseases (65), and in apoAI and apoAII enzymes, in which the reduction of high-density lipoprotein-associated LOOH to LOH by these enzymes was shown to occur with the concomitant formation of MetO at Met112 and Met148 (human apoAI) and Met26 (human apoAII) (121). In total, methionine oxidation selectively modulates numerous cell signaling pathways that are regulated by calcium, phosphorylation, and others (71, 142, 211).
The repair of MetO is catalyzed by the methionine sulfoxide reductases (Msr), which restore reduced methionine from MetO. Chemically, oxidation of the Met thioethers generates the chiral sulfoxide in a racemic mixture of R and S isomers (k=0.006 M−1 s−1 for free Met+H2O2) (285). MsrA selectively reduces the S epimer of MetO [Met(S)O] and MsrB reduces only the R epimer [Met(R)O]. MsrA resides in the mitochondria, cytosol, and nucleus; while three forms of MsrB are known: MsrB1 (cytosol; nucleus), MsrB2 (mitochondria), and MsrB3 (mitochondria; endoplasmic reticulum) (167, 175, 237). MsrA, MsrB2, and MsrB3 have cysteine-based active sites, while MsrB1 contains an active site selenocysteine (Sec) nucleophile, though exceptions have been noted (150). Stereospecific MetO reduction is characterized by oxygen-atom transfer from MetO to Msr, forming an active site with sulfenic acid or selenenic acid (187). Capture by a resolving cysteine with subsequent disulfide (or selenosulfide) reduction by the thioredoxin/TrxR system restores active Msr, reducing MetO at the expense of NADPH (50, 253). Thus, the Msr enzymes help defend cells and tissues from oxidative damage. For example, the expression of MsrA is up-regulated in the epidermis that is exposed to UVA radiation and H2O2, suggesting MetO reduction to be a crucial component of the cellular oxidative stress response (251). To date, the impact of IR on Msr expression and activity has not been elucidated.
Methods of detection. Detection of MetO is accomplished with relatively inexpensive techniques such as Fourier transform infrared spectroscopy with monitoring for vibrational bands at 1044 and 1113 cm−1 or biochemically using a commercial MetO-targeted antibody (246, 277). MS is another major avenue for protein MetO detection, helping identify MetO formation in keratins (195), prion proteins (306), and to correlate MetO with glaucoma (328) and sepsis (122). Both ESI and MALDI were utilized to identify MetO formation in hemoglobin of IR-treated primary human erythrocytes (363). Tandem MS methods are the most widely used for the identification of MetO sites using CID-mediated neutral loss of methanesulfenic acid (loss of 80 Da) or milder precursor ion fragmentation methods such as electron capture or transfer dissociation (135). An essential consideration for all approaches to MetO detection is the possibility for adventitious Met oxidation during sample handling and processing (68, 135). Non-physiological routes to MetO by means of ESI and gel electrophoresis have been uncovered and optimized protocols have been suggested, though many additional avenues likely exist (62, 236, 318, 377).
Cysteine oxidation
Among the amino acids, protein cysteine residues have the lowest redox potential, rendering their thiol groups particularly sensitive to oxidative modification by ROS/RNS. Protein thiols have diverse chemical responses to ROS/RNS, and in a very general sense, cysteine residues with a lower thiol pKa tend to be more readily modified by oxidants and electrophiles and are often termed “redox active” or “redox sensitive” cysteines. However, a range of analyses have uncovered the varying reactivity of cellular thiols with electrophiles, including the endogenous electrophile HNE, and are discussed in greater detail in a recent review (117). It is now well established that decreased thiol pKa does not always correlate with increased reactivity toward H2O2, particularly among redox-regulated enzymes [see ref. (358) for a thorough analysis]. In addition to the cysteine thiol pKa, the differential reactivity of thiols toward ROS/RNS and the products of oxidation are affected by factors, including residue accessibility, polarity and basicity of adjacent amino acids, and identity and concentration of the ROS/RNS source (244, 245, 293). The chemical interplay between thiols and ROS/RNS is further governed by protein subcellular localization, organelle redox potential, and proximity to ROS/RNS sources.
Mechanistically, redox-sensitive cysteine thiols undergo a two-electron oxidation in the presence of H2O2, hydroperoxides (ROOH), peroxynitrite, and other oxidants, forming sulfenic acid (CySOH) as the initial cysteine oxidation product (Fig. 5B). The subsequent capture of CySOH by protein and low MW thiols (e.g., GSH) generates disulfides, which are readily reduced to regenerate the thiols in the presence of cellular reductants or by the activity of disulfide reductases. In addition to its role as a thiol-disulfide intermediate, CySOH may be captured by adjacent lysine residues or an amide nitrogen in the protein backbone and form sulfenamides [also called sulfenyl amides (−SN)] as in the well-defined example of Cys-based protein tyrosine phosphatase 1B (PTP1B) (161, 292). Without sufficient thiol content proximal to the CySOH site, further oxidation results in the formation of sulfinic (CySO2H) and sulfonic (CySO3H) acids, an irreversible process except for the special case of Prx sulfinic acid repair by sulfiredoxin (36, 165). Cysteine thiols react with NO-generated RNS, form S-nitrosothiols, and also undergo transnitrosation, in which the nitroso moiety is passed among cysteine residues in a site-selective manner that is dependent on the local environment and the pKas of thiols involved (49, 149, 309). S-Nitrosation of cysteine occurs through three major pathways: (i) thiol capture of nitrosonium ion (via acidic nitrite) (144); (ii) •NO capture by cysteine thiyl radical; and (iii) directly through N2O3 modification (126). The transition metal-mediated formation of nitrosonium ion from •NO also facilitates S-nitrosation (172). For additional chemical details, including mechanisms of cysteine oxidation and nitrosation, the reader is referred to a recent review on this topic (259).
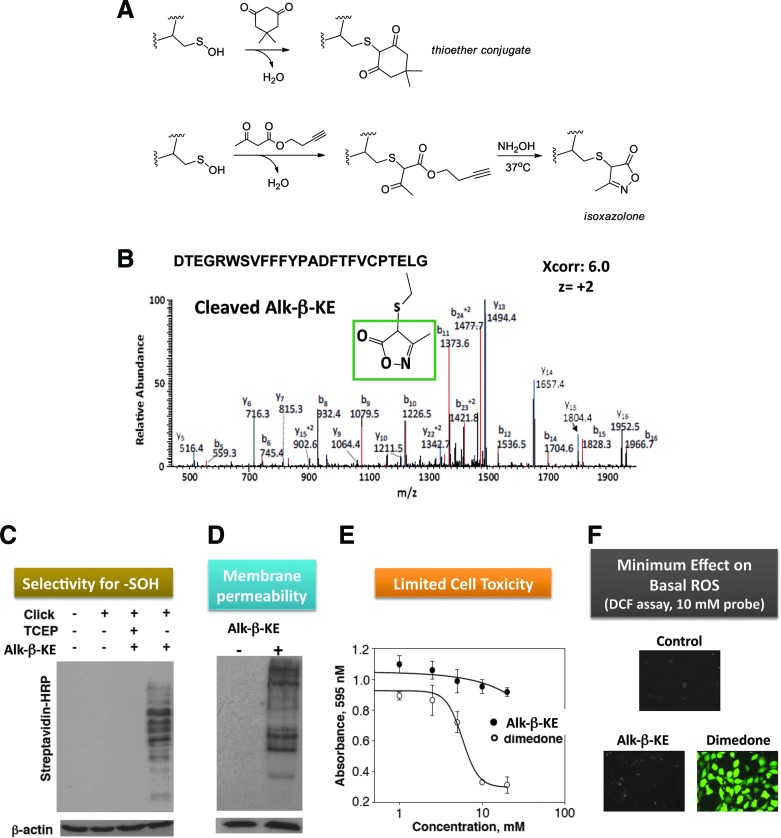
Methods of detection. Despite only a few examples of physiological cysteine sulfinic acid formation, this modification attracts attention, as it indicates a sharply increased intracellular oxidant status and profoundly impacts redox signaling (39, 114, 243, 274). As a largely irreversible cysteine product, CySO2H may be directly detected by MS, though high-resolution detection is required to distinguish this modification from others with a similar mass (e.g., sulfinamide −SONH2; persulfide −SSH). A recent report details the capture of aryl nitroso electrophiles by small-molecule sulfinic acids, leading to stable sulfonamide conjugates, though this chemistry has not yet been applied to protein CySO2H detection (210).
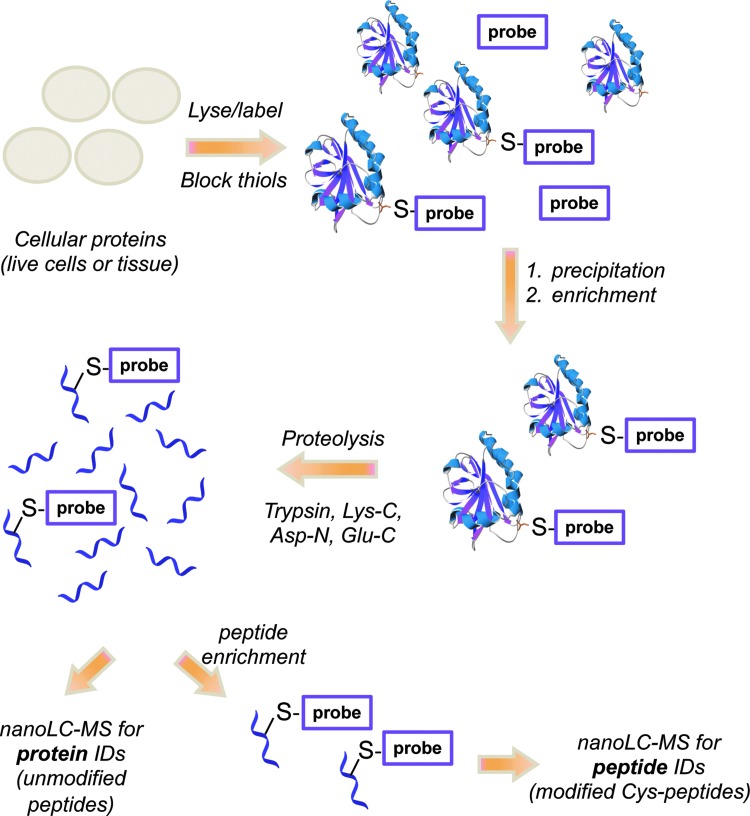
Among the cysteine modifications by ROS/RNS, cysteine nitrosation and sulfenylation are of special interest due to their reversibility and emerging roles in regulating protein function and communication (146, 160, 260, 268, 349). For example, S-nitrosothiol formation on mitogen-activated protein kinase phosphatase-1 is shown to increase phosphatase activity and reduce radiosensitivity in head and neck cancer cells (134). Further evidence linking redox signaling and phosphorylation pathways has emerged with the observations that cysteine oxidation to sulfenic acid (Cys124) inactivates Akt2 kinase (349), while sulfenic acid formation at redox-sensitive Cys797 of EGFR stimulates its kinase activity (260). With the rapid advancements in ESI- and MALDI-based MS methods and newly designed strategies for sub-proteome identification, a host of proteins are now widely accepted ROS/RNS targets (45, 67, 216, 219, 231, 258, 303, 311), though these methods have not yet been applied to investigate the effects of IR. The methods pertaining to the high-throughput identification of the CySOH and CySNO proteins and their cysteine sites of modification are discussed in the section “Redox and phosphoproteomics.” In a research article included in this Forum, we demonstrate the successful application of biotin-tagged probes to monitor the cellular redox status in tumors with various degrees of response to radiation treatment.
Protein modification by reducing radical species
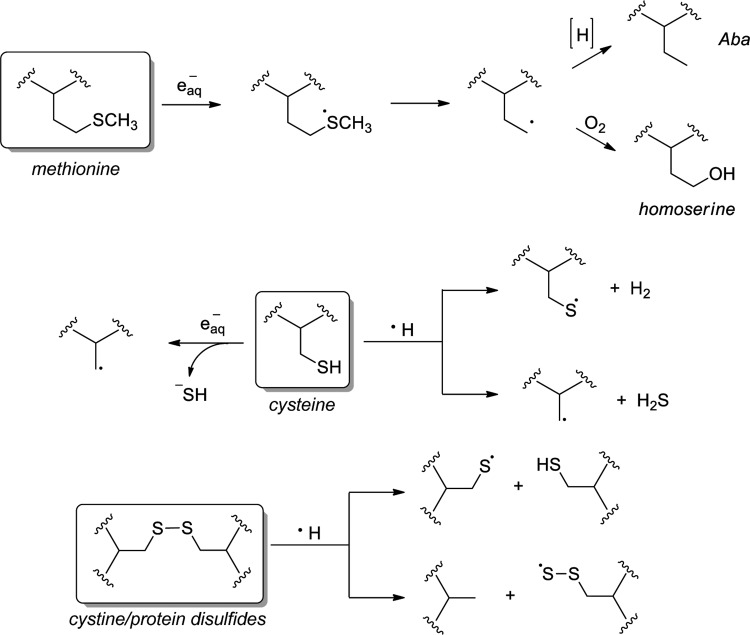
In addition to IR-generated oxidants, the radiolysis of water is a major source of the competent intracellular reductants eaq− and H•. Though reductive stress has been exceedingly understudied in comparison to its oxidative counterpart, the disruption of cellular redox balance toward reduction has been linked to several pathologies, most notably of the cardiovascular system (48). The reductants eaq− and H• readily target the sulfur-containing amino acids, reacting at or near diffusion-controlled rates (51). Methionine serves as a principal target of H•, adding sulfur to generate a sulfuranyl radical (Fig. 6) (15). Subsequent desulfurization forms a carbon-centered radical that may be terminated by a hydrogen atom donor and form α-aminobutyric acid (Aba) or by molecular oxygen to generate homoserine (58). The exposure of methionine-containing peptides to eaq− results in peptide cleavage through a deamination mechanism (316).
FIG. 6.
Impact of IR-induced reductive stress on protein sulfur, including methionine, cysteine, and cystine.
The reaction of eaq− with reduced cysteine generates hydrosulfide ion (HS−) and the corresponding alkyl radical (R•); a reaction with cysteine/protein disulfides (RSSR) provides the thiyl radical (RS•) and the thiolate (RS−) ion (Fig. 6). In contrast, the reactivity of H• is marked by radical abstractions: A reaction with reduced CySH occurs via hydrogen abstraction to generate RS• or through homolytic cleavage to form R• and hydrogen sulfide (H2S). Cystine reactivity with H• induces homolytic disulfide cleavage, forming either RS•+RSH or RSS•+RH, depending on the site of cleavage. Thiyl radicals are also generated by the abstraction of thiol hydrogen by •OH, illustrating how this species, among others, is common to both reductive and oxidative processes (226).
Biochemically, thiyl radicals are gateways to amino-acid conversion and the formation of additional redox-critical species. Thiyl radicals are efficiently captured by thiolate ions (R′S-), yielding a disulfide radical anion (R′SSR•−), which readily reduces O2 to O2•− (Eq. 1) (118, 353).
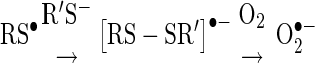 |
Though short lived, RS• reactivity with •NO is a known source of protein S-nitrosothiols, an oxidative cysteine modification whose role in redox signaling has gained increasing attention in recent years (309). Thiyl radicals are quite reactive toward hydrogen abstraction: Cysteine thiyl radical is known to abstract hydrogen from its α-carbon, facilitating the loss of sulfur and cysteine conversion to dehydroalanine (239).
A significant challenge in the investigation of the reductive effects of radiation on cellular proteins is the required quenching of •OH formed in situ, which enables monitoring of reductive processes but may distort the view of the wider cellular implications. A recent study of bovine ribonuclease A (RNase A) using ESI-Q-TOF MS revealed that high-dose IR generated eaq− and H• induced Cys>Ala conversion at three sites along with one Met>Aba desulfurization (108). Interestingly, reductively stressed RNase A affected only three of the eight Cys and one of four Met residues, illustrating a degree of site selectively for reductive protein desulfurizations. The chemoselectivity of these reductive processes, though not well understood, has been observed for the radiation-induced reduction of human serum albumin as well, though at 50–300 Gy doses (294). For instance, perthiyl radical (RSS•) was found to be the major reductive product in lysozyme radiolysis because of the lack of competition reactions for the thiyl radicals (105). It has also been reported that thiyl radicals formed in the Aba production contributed to isomeric formation of trans fatty acid from the cis residues in lipids, which again illustrates the interplay between protein-lipid damage orchestrated by H• atoms (109).
Protein expression
Global profiling of protein expression changes in response to IR is an active area of radiation research. Such investigations include, for example, the comparative proteomics analysis of IR-induced changes in the expression of mouse liver proteins that are associated with specific subcellular fractions. Such findings pointed to the activation of antioxidant and inflammatory responses by IR (206). Another study looked at the alteration of protein expression in different tissues such as the brain, lung, spleen, and intestine in an IR-treated (1 Gy) mouse using MS-based proteomics (204). Transaldolase 1 and phosphoglycerate kinase 1 were assigned as potential tissue-specific biomarkers of IR exposure in the brain and intestine, respectively. Further applications of MS-based proteomics to detect and quantify protein expression and IR-relevant PTMs are reviewed in the section “Redox and phosphoproteomics.”
Emerging Fields and Technologies for Mapping Cellular Responses to IR
Genomics and epigenomics
Differential responses to IR can be partly attributed to the combined effects of genetic and epigenetic variations among individuals, and it is well established that the diversity in “allelic architecture” relates closely to the biological response to IR (6). The field of radiogenomics has thus emerged in recent years to investigate patient-to-patient variability in normal tissue reactions after radiotherapy. Single nucleotide polymorphisms account for 90% of this naturally occurring sequence variation. Over the years, a number of clinical studies have been carried out to test the association of various genetic factors with radiosensitivity with the goal of developing algorithms that predict the normal tissue toxicity after radiotherapy (5). A study of Danish breast cancer patients showed a relationship between the ATM codon Asp1853Asn (5557 G>A) single nucleotide polymorphism and radiosensitivity in which the homozygous (AA) and heterozygous (AG) genotypes of the ATM G5557 G>A variant were more radiosensitive and at an increased risk of radiation-induced subcutaneous fibrosis (7, 63).
Epigenetics, defined as “the structural adaptation of chromosomal regions so as to register, signal, or perpetuate altered activity state,” involves modifications such as DNA methylation, histone methylation and acetylation, ADP-ribosylation, ubiquitination, non-coding RNA-mediated gene silencing, and others (35, 82). Epigenetic modifications are mediators that translate a combination of genetic, metabolic, and environmental factors into a characteristic gene expression pattern. Over the past several years, a number of groups have investigated changes in DNA methylation that were associated with IR and revealed differences which were dependent on the tissue analyzed, gender, and radiation dose (125, 182, 186, 212). Studies using animal models have shown that IR treatment (5 Gy) results in significant DNA hypomethylation in radiation-treated tissues (181). DNA lesions, including strand breakage, deletions, and nucleobase oxidation, are shown to adversely affect the action of DNA methyltransferase enzymes, leading to hypomethylation (345). The presence of 8-oxodG, a prominent oxidation product correlated with IR, reduces the methylation efficiency of nearby cytosines in a proximity-dependent manner, illustrating one example of how IR-generated ROS impacts the epigenome (334, 355). Interestingly, low-dose IR (up to 0.03 Gy in mice) during gestation was linked to an increase in DNA methylation in offspring; this effect was negated with maternal dietary antioxidants, suggesting oxidation as a link between IR and DNA methylation (29).
DNA methylation status was also found to correlate with the response and resistance to IR. For example, the treatment of MCF-7 cells with fractionated IR (2 Gy fractions for a total of 10–20 Gy) decreased DNA methylation at the Forkhead box C1 and Trafficking protein particle complex 9 loci, which was linked to decreased apoptosis and increased post-IR survival (186). In another study, microarray analysis of the CpG methylation profiles of radiosensitive H460 and radioresistant H1299 human non-small-cell lung cancer cell lines identified 1091 differentially methylated genes (174). Interestingly, twice as many hypermethylated genes were identified in the H1299 radioresistant cell line compared with the H460. The function of hypermethylation of the SERPINB5 and S100A6 genes in the radiation resistance phenotype was further validated by down-regulating their expression in H460 cells, which increased their resistance to radiation. Hypomethylation of the gene encoding catalase was observed, consistent with its increased expression in the H1299 cells and a general function of the antioxidant system in regulating the resistance to radiation.
The connection between the central redox metabolism (citric acid cycle and mitochondrial ETC) and epigenetic modulation in cancer was covered in detail in a recent review (148). To the best of our knowledge, there are no studies that present direct evidence of epigenetic modulation as a result of IR-induced modifications of metabolic or signaling proteins, but certainly these could be linked. Many epigenetic enzymes are directly regulated by phosphorylation and oxidative PTMs or use substrates (e.g., S-adenosyl methionine—a methyl donor; acetyl coenzyme A—an acetyl donor) that are products of redox-regulated metabolic pathways. Thus, connective networks that link signaling, metabolism and epigenetics exist and are being discovered using a combination of the emerging technologies discussed here.
Such discoveries would not have been possible without the development of high-throughput technologies such as microarray platforms, ChIP-on-chip, next generation sequencing, and MS. To strengthen these technologies, the development of sophisticated statistical methods is required for an accurate interpretation of the high volume of data generated from such experiments. Often a combination of the aforementioned techniques is used for better results. For example, ChIP from paraffin-embedded patient samples (PAT-ChIP) can be coupled with high-throughput sequencing (PAT-ChIP-seq) for the genome-wide analysis of distinct chromatin modifications (104). Sometimes, ancillary techniques are a prerequisite for the subsequent high-throughput microarray or sequencing analysis. For example, in studies involving the analysis of methylated cytosines, the first step often involves techniques such as Methylated DNA immunoprecipitation, Methylated-CpG island recovery assay, and bisulfite conversion for the enrichment of methylated cytosine DNA followed by an array-based approach (such as tile array, promoter array, or bead array) or a sequence-based approach for epigenetic profiling (180).
Redox and phosphoproteomics
Redox proteomics
Cysteine oxidation, in particular, attracts greater attention in the redox signaling field than other amino acids because of its active site and regulatory roles in numerous signaling and metabolic proteins, including phosphatases, kinases, and transcription factors. Here, we focus on MS-driven proteomic methods for detecting protein sulfenic acids and S-nitrosothiols. Since CySOH and CySNO species are often trapped by proximal thiols and form disulfides, the assessment of cellular thiol-disulfide status is a widely utilized approach for studying cysteine oxidation (140, 198, 280, 373). Isotope-coded affinity tag for detecting oxidized cysteine (OxICAT) is a method for quantifying the cellular thiol and disulfide species in complex samples using an isotopically light electrophile to first capture thiols, followed by global disulfide reduction and nascent thiol capture by an isotopically labeled form of the electrophile (197, 301). An important caveat for OxICAT and related methods is that the reductant employed, tris(2-carboxyethyl)phosphine (TCEP), reduces S-nitrosothiols and sulfenic acids in addition to disulfides. Quantification using this method, therefore, determines the extent of reversibly oxidized cysteines, unless more chemoselective reductants, such as ascorbate or arsenite, are used in lieu of TCEP (52, 295). Slight modifications to the OxICAT approach, such as care to avoid protein acidification, enable the use of this method to detect oxidative and nitrosative protein modifications in the same sample (isotope-coded affinity tag for detecting nitrosated and oxidized cysteine) (207).
Investigations of the CySNO or CySOH proteome are challenged by the low cellular abundance of these PTMs in proteins, necessitating the use of enrichment strategies and therefore requiring the use of biotin-conjugated reagents for labeling the oxidized/nitrosated cysteine in proteins. Unfortunately, the MS signal intensity of a biotin-containing peptide ion is often significantly lower than the ion of the corresponding non-biotinylated peptide. As a result, several groups, including ours, have developed CySOH–, CySNO–, and other oxoform-targeted probes with cleavable biotin tags to facilitate MS compatibility (272, 301, 333). A number of methods developed to selectively detect and quantify the CySNO or CySOH proteomes are discussed next.
Proteomic studies of protein nitrosation (CySNO)
Due to their thermal instability and decomposition in light, nitrosated protein cysteines are not directly detected and require chemical derivatization. A common method for detecting S-nitrosocysteine in biological samples is the biotin-switch technique (BST), a three-step sequence in which (i) free thiols are captured by an electrophile (blocking); (ii) S-nitrosothiols are reduced by ascorbate in the presence of a labeled electrophile; and (iii) labeled thiols are quantified using fluorescence imaging, Western blot, or MS. This approach, though indirect, is widely used and fairly controversial, especially with regard to the chemoselectivity of thiol blocking and ascorbate reduction. Careful controls are paramount to avoid the misinterpretation of BST results. Alternative methods include CySNO decomposition, accompanied by chemiluminesence detection of nascent •NO, and phosphine-based reagents that react directly with the S-nitrosothiol and form chemically unique conjugates (19, 21, 347, 381). Nevertheless, numerous methods have been developed that take advantage of the ascorbate-based reduction of CySNO. Site mapping of an S-nitrosated cysteine in PKBα/Akt1 from rat soleus muscle was determined using trypsin digestion and derivatization of CySNO-containing peptides with isotopically labeled iodoacetic acid (2-12C/13C, 50% ratio) in the presence of ascorbate (213). To globally identify S-nitrosated cysteine sites, the S-nitrosothiol site identification method was adapted from the BST approach but incorporates protein digestion before affinity capture, resulting in an enriched CySNO-containing peptide mixture (132, 141). Alternatively, the His-Tag switch approach uses a highly MS-compatible His-tag linked to iodoacetamide for tagging nascent thiols generated from CySNO reduction by ascorbate (52). A more recent technique is resin-assisted capture (resin-assisted capture for S-nitrosothiols [SNO-RAC]), where a thiol-reactive resin captures ascorbate-generated thiols. SNO-RAC is more streamlined than the classical BST, enables on-resin digestion, is compatible with isotope tagging for relative and absolute quantification (iTRAQ) quantification, and offers higher sensitivity than the BST for high MW proteins (111). These and other CySNO detection methods have been recently reviewed (20).
Proteomic studies of protein sulfenylation (CySOH)
Similar to the S-nitrosothiol, several direct and indirect methods were developed for the detection of CySOH proteome. These are thoroughly reviewed in a recent article (117), and only a brief summary is presented here. The indirect BST approach is used for the detection of CySOH with the difference that arsenite is used instead of ascorbate to achieve selective reduction of CySOH; this is followed by a reaction of the nascent thiol with a biotin (or fluorescein) tagged thiol electrophile (e.g., biotin iodoacetamide) followed by enrichment and detection using MS or imaging technologies (295). A potential caveat is the chemoselectivity of thiol-blocking and oxoform reduction, which are not unequivocally established (281, 348, 383). More direct methods include detection using a CySOH-specific antibody, which relies on the CySOH modification remaining intact during sample processing (300). Another approach uses in vivo labeling by a genetically engineered C-terminal cysteine-rich domain of the transcription factor Yap1 (Yap1-cCRD) (321, 322). The single Cys of the Yap1-cCRD mutant forms mixed disulfides with CySOH in vivo, enabling co-enrichment using nickel affinity columns. Although not yet extensively investigated, this strategy may suffer from false identifications via protein cross-links in vivo.
The lability of sulfenic acids and controversies involving the potential for false positive results with the BST has fueled the shift toward the direct detection of protein sulfenic acids using 1,3-dicarbonyl (often dimedone-based) probes (Fig. 7A). To accommodate the many experimental techniques used for profiling sulfenic acid formation, both in cells and in lysates, a diverse library of 1,3-dicarbonyl probes has been developed with functionalities for affinity capture, MS, and fluorescence imaging. In addition, alkyne- and azide-containing dicarbonyl probes enable their customization using click chemistry (57, 178, 200, 267, 271, 278, 299, 333).
FIG. 7.
Capture of cysteine sulfenic acid by 1,3-dicarbonyl compounds. (A) Dimedone (top), Alk-β-KE (bottom); click chemistry (not shown) installs a biotin affinity tag; the ketoester enables hydroxylamine-mediated ester cleavage, generating MS-friendly isoxazolone. (B) MS2 spectrum for the Alk-β-KE-labeled peptide after Asp-N digestion reveals MS compatibility. (C) Accumulation of intracellular ROS in response to probes is lower for Alk-β-KE compared with dimedone (10 mM each). (D) Click chemistry attaches biotin tag; labeling by Alk-β-KE is specific to CySOH and abolished in the presence of TCEP. (E) Addition of Alk-β-KE to cells, followed by lysis, click to attach the biotin moiety and immunoblotting illustrates membrane permeability. (F) Cellular toxicity of Alk-β-KE is reduced compared with dimedone as measured by the DCF assay. Adapted from Qian et al. (272). DCF, dichlorodihydrofluorescein; MS, mass spectrometry; TCEP, tris(2-carboxyethyl)phosphine. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
MS-based CySOH proteomics studies have been achieved using a biotinylated dimedone analog in heart tissue (57), in HeLa cells using dimedone-based azide DAz-2 subsequently conjugated to biotin via Staudinger ligation (200), and in tumor necrosis factor α-stimulated HEK-293 cells using biotin-containing DCP-Bio1 (248). Despite that fact that a plethora of CySOH-containing proteins have been identified through MS-based proteomics using a diverse set of chemical probes, global mapping of redox-sensitive cysteines has not been successfully completed. As discussed earlier, biotin is essential for enrichment but often problematic for the detection of low-abundance peptides by MS. A recent 1,3-dicarbonyl CySOH probe, Alk-β-KE, incorporates a β-ketoester linkage that is susceptible to chemoselective cleavage by hydroxylamine (NH2OH), enabling on-bead cleavage of labeled proteins or peptides and generating a highly MS-compatible isoxazolone tag (Fig. 7B). This reagent is cell-membrane permeable and has reduced cell toxicity compared with dimedone (Fig. 7C–F) (272). The use of Alk-β-KE and other CySOH probes in large-scale proteomics studies is ongoing with the ultimate goal of uncovering IR-dependent protein oxidation along with the modified cysteine site(s) (Fig. 8). The direct labeling approaches with chemical probes can easily be integrated with relative or absolute quantitative proteomics methods such as stable isotope labeling of amino acids in cell culture (SILAC), iTRAQ, tandem mass tags, absolute quantification, and others, enabling studies of time course or radiation dose-dependence profiles of protein oxidation, phosphorylation, and other PTMs discussed next (103, 203, 254).
FIG. 8.
General workflow for in vitro labeling of CySOH using biotinylated 1,3-dicarbonyl probes. Proteins are precipitated to remove unreacted probe; then, labeled proteins are enriched using avidin-based methods. After proteolytic digestion, protein identifications are obtained by LC-MS/MS analysis of the full peptide mixture, while site mapping requires a second round of enrichment. LC, liquid chromatography. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Phosphoproteomics and other targeted proteomics
SILAC-based quantitative proteomic analyses were recently applied to identify the dynamics of human nuclear protein acetylation and phosphorylation during IR-induced DDR (26, 27). Cumulatively, these two studies have revealed a number of regulatory pathways linking acetylation and phosphorylation signaling to DNA methylation and the regulation of gene expression. In another study, phosphoproteomic methods were employed in conjunction with microarray analyses of mRNA and miRNA expression to quantify changes induced in these species by fractionated radiation versus single-dose radiation. Significant differences were identified in DDR pathways, apoptosis, and immune response (221). Quantitative phosphoproteomics was also applied to identify previously unknown ATM-dependent and independent changes in the phosphorylation of nuclear proteins after DNA damage (28).
Metabolomics
Nuclear magnetic resonance (NMR) and MS-based metabolomics has emerged as a powerful approach for identifying and quantifying biomarkers of IR exposure in both cell culture and in vivo animal studies, including in nonhuman primates. However, the high complexity of the metabolome, the dependency on factors such as gender, diet, age, and genetic background, and the variability of metabolite composition between cell types and tissues make it difficult to employ metabolomics as a stand-alone approach. Nevertheless, this technology is extremely powerful when used in defined systems and in conjunction with genomics, epigenomics, and/or proteomics studies.
Metabolomics studies in radiation biology are divided into two categories: (i) those aimed at the mechanistic understanding of the biological response to IR, and (ii) biodosimetry studies focused on quantifying the damage induced by IR and identifying the respective biomarkers in easily accessible biological fluids (urine, saliva, and serum). For an excellent discussion on the use of radiation metabolomics to identify small-molecule biomarkers that respond to radiation in a dose-dependent manner, the reader is directed to several recent publications (163, 164, 188, 335, 336). These include a description of current and emerging metabolomics technologies, computational methods for data analysis, as well as specific studies that have led to the identification and, ultimately, validation of a number of urinary biomarkers in mice and non-human primates. Analytical methods used to identify metabolite biomarkers and to quantify the extent of metabolite modulation by IR include NMR spectroscopy and MS. One-dimensional and two-dimensional NMR provide structural information for metabolomics studies and are, therefore, critical tools in the characterization of new metabolites. NMR spectroscopy is, however, hampered by low sensitivity, requiring larger amounts of sample than other analytical techniques, but despite this limitation, it has provided valuable information about IR biomarkers (61, 171). MS-based metabolomics methods, in addition to increased sensitivity, also incorporate online analytical separation of metabolites using GC, LC, or capillary electrophoresis.
Examples of radiation biomarkers from studies in non-human primates include 2′-deoxyxanthosine, 2′-deoxyuridine, thymidine, xanthine, and xanthosine (163). Other studies in cell culture showed a good correlation between the changes in small molecule metabolites (e.g., AMP) and the expression of proteins involved in their metabolism as determined by a proteomic analysis (e.g., increase in the adenylate kinase that converts AMP to ADP and a concomitant decrease in the AMP-generating enzymes such as phosphodiesterase (cAMP to AMP) and hypoxanthine-guanine phosphoribosyltransferase, which converts adenine to AMP) (156, 256). In addition, studies of skin exposure to low doses of radiation (3–10 cGy) found significant changes in the metabolites involved in the DNA damage, damage repair, and bioenergy metabolism (155).
In a cell culture, a number of studies have reported metabolomic (small or large scale) changes by IR, and a few of these have applied integrated “omics” analyses (53, 60, 341). Using integrated “omics” analyses, ATM was found to regulate purine, pyrimidine, and urea cycle metabolism through the activation of AMP-activated protein kinase, a crucial sensing enzyme in the regulation of cellular energy pathways (60, 341). In another study and using NMR-based metabolomics, the inhibition of small ubiquitin-like modifier-mediated protein–protein interactions was connected to decreased mitochondrial function and impairment of GSH synthesis after exposure to IR (4 Gy) (53).
GSH content was also shown to undergo dynamic time- and dose-dependent changes in response to IR in human lymphoblast TK6 and human telomerase reverse transcriptase (hTERT)-transformed fibroblast BJ cells (256). In this study, cells were treated with 0.5–8 Gy, and the water-soluble metabolome fraction was analyzed at 1–16 h after IR exposure using ultra performance liquid chromatography-ESI-TOF MS. Extensive metabolomic changes were observed as early as 1 h after IR in a dose-dependent manner. Certain metabolites were depleted at 1 h post-irradiation with 1 Gy, including GSH, AMP, nicotinamide, 5-oxoproline (intermediary metabolite in GSH biosynthesis), phosphocholine (lipid metabolism), uridine monophosphate, proline, NAD+, and spermine (protects against DNA damage). At 4 h, the levels of these metabolites were partially restored to baseline, and by 8 h, there was no significant difference between IR-treated and control. At 16 h (after 4 and 8 Gy only), a reduction of GSH, NAD+, and spermine was only observed in the cells treated with 4 and 8 Gy.
Interestingly, there was a difference in the time- and dose dependence of GSH in the hTERT-transformed fibroblast BJ cells compared with the TK6 cells (256). First, there was an increase in GSH at 1 h after 1 Gy, a depletion of GSH at 4 h and 4 Gy, followed by an increase again at 16 h at the 4 Gy IR dose. The authors attributed these observations to differences between normal and transformed phenotypes, but certainly, additional studies are needed to thoroughly profile crucial metabolites across a range of cell lines, radiation treatments, and time elapsed after radiation treatment.
Within cells, mitochondria are an established target of IR damage (166), and studies have linked the response to IR with changes in specific metabolites. Investigations of IR-induced micronucleus formation have linked the functional status of mitochondrial DNA (mtDNA) to ATP production and radiosensitivity (375). Cells lacking mtDNA showed a decreased sensitivity to radiation and a lower rate of micronucleus formation compared with cells harboring intact mtDNA, which was correlated primarily with decreased ATP production.
Mitochondria-generated ROS were also found to be involved in micronucleus formation (66), and more recent studies have investigated the function of mitochondrial SOD (manganese superoxide dismutase [MnSOD]) in the response to radiation. Increased expression of MnSOD was associated with increased resistance to IR both in vivo (102) and in vitro (152). The function of MnSOD extends beyond protection against IR-induced ROS: A study investigating the function of MnSOD in the adaptive response after low-dose radiation (10 cGy X-ray) using human skin keratinocytes (HK18) found an association of MnSOD with metabolic proteins (100). Thus metabolomic changes associated with altered MnSOD expression are expected but have not been fully elucidated.
IR-induced changes in the serum of rats exposed to approximately 8 Gy of IR showed a decreased prevalence of citric acid cycle intermediates such as isocitrate (3 and 8 Gy) (208), citric acid, and 2-oxoglutaric acid (α-ketoglutarate) (188). Knockdown of the mitochondrial isocitrate dehydrogenase, which catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate, induced G1 cell cycle arrest and autophagy after 5 Gy of IR (177). The inhibition of autophagy using chloroquine in conjunction with IR treatment resulted in apoptotic cell death. Cumulatively, these studies establish a connection between the metabolic intermediates of the citric acid cycle and the biological response to IR. Furthermore, levels of creatine, another mitochondrial metabolite, were increased in human leukocytes exposed to 4 Gy (γ-rays) along with arginine, glutamine, proline, and GSH (measured as reduced GSH) (194). Circulating citrulline, a biomarker of intestinal function and a mitochondrial metabolite, was decreased in mice at day 4 after 8–12 Gy of proton IR (217).
Radiation metabolomics is still an emerging area of research, and efforts to uncover organelle-specific metabolic changes are ongoing. The current chromatography/MS approaches to metabolomics will be enhanced by the growing use of high-resolution mass spectrometers, which offer sharply increased sensitivity and mass-based molecule identification. Thus, metabolomics is expected to become a critical “omics” tool in radiation research and shows significant promise in the discovery of small-molecule biomarkers of IR damage and response that could be exploited for radiation biodosimetry or cancer therapeutics.
Potential caveats of “omics” approaches
Though high-throughput “omics” technologies are providing a wealth of biological information and have proved valuable to identify critical networks affected by radiation, “omics” analyses and the interpretation of their results have a number of limitations. First, the sheer complexity of the human metabolome and inability to predict its composition makes metabolomics and follow-up studies in this area of research highly challenging. Proteomic studies are complicated by the potential for a multitude of protein modifications with varying degrees of contribution to protein activity and as a result, the overall IR effect. Technologies that would enable a high-throughput assessment of protein activity in vivo which could then be used to build computational models of a system (cell, tissue, and body) would be ideal. To date, it is only for selected classes of proteins that these technologies exist (16, 202, 249). “Omics” approaches have traditionally been viewed as “fishing expeditions” and criticized as simply providing snapshots of biological systems. The acquisition of time course and dose-dependence “omics” analysis increased sophistication of data integration and computational analysis, and follow-up studies both in cells and in vivo have made significant strides toward alleviating these concerns and are now a mainstay in the field.
Conclusion and Perspectives
From the studies reviewed here and many others, it has become clear that in order to achieve an understanding of biological systems at the level which would enable a prediction of targets to manipulate the biological outcome, a thorough integration of “omics” and classical technologies through novel computational approaches is needed.
Given the wide range of PTMs induced by IR (Table 1) with their effects on the activity of individual molecules and on the metabolic flux and propagation of signaling networks mostly unknown, future efforts will require the inclusion of protein activity profiles to generate intricate computational models of biological response to IR. Too often in scientific literature, the altered phosphorylation status of a protein is equated with gain or loss of activity. Evidence from our group and others shows that oxidation can easily override the effects of phosphorylation, activating or inhibiting the activity of enzymes (260, 349). This is extremely important in the context of IR-dependent signaling and in studies that integrate metabolomics, proteomics, and epigenetics. While indirect methods for the detection of oxidative PTMs have been applied with various degrees of success, many caveats are associated with these approaches, particularly when a quantitative analysis is attempted. For example, the cross-reactivity of protein sulfenic acids with DNPH and other hydrazines used for detecting protein carbonylation is established, and recently, the cross-reactivity of sulfenic acids with commonly used thiol-blocking electrophiles has been described (281). Therefore, the arsenal of probes that chemically target each of the oxidative and other PTMs should be expanded. As discussed in the current review, the development of robust and selective detection strategies for modified biomolecules is a vibrant area of research both within and outside the radiation biology field. The use of chemical probes in conjunction with high-throughput technologies and multi-scale computational methods opens exciting opportunities for radiation research by facilitating the discovery of new targets and biomarkers of IR and the prediction of long-term effects of IR. Such advances will also enable more detailed investigations of the mechanisms underlying the biological response to IR, such as understanding the mechanistic connections among epigenetic changes (e.g., DNA methylation), DNA repair systems, and metabolism. Further elucidation of the biological mechanisms of IR will impact multiple areas of medicine, from prevention of disease induced by IR exposure to selection of IR-based treatment schedules for cancer.
Abbreviations Used
- γH2AX
phosphorylated histone H2AX
- 8-oxodG
8-oxo-2′-deoxyguanosine
- Aba
α-aminobutyric acid
- ACEI
angiotensin-converting-enzyme inhibitors
- Ang II
angiotensin II
- ApoAI
apolipoprotein A-I
- ApoAII
apolipoprotein A-II
- ASMase
acid sphingomyelinase
- AT1R
angiotensin II type 1 receptor
- BST
biotin-switch technique
- CHH
7-(diethylamino)coumarin-3-carbohydrazide
- Chk1/2
checkpoint kinases 1/2
- CID
collision-induced dissociation
- CT
computed tomography
- DCF
dichlorodihydrofluorescein
- DDR
DNA damage response
- DJ-1
Parkinson disease protein 7
- DNA-PK
DNA-dependent protein kinase
- DNPH
2,4-dinitrophenylhydrazine
- DPPP
diphenyl-1-pyrenylphosphine
- DSB
double-strand break
- eaq−
hydrated electron
- EGFR
epidermal growth factor receptor
- ELISA
enzyme-linked immunosorbent assay
- ESI
electrospray ionization
- ETC
electron transport chain
- FISH
fluorescence in situ hybridization
- GC
gas chromatography
- GC-MS
gas chromatography coupled to mass spectrometry
- GPR
Girard's P reagent
- GPx
glutathione peroxidase
- GSH
glutathione
- GST
glutathione S-transferase
- Gy
gray
- H•
hydrogen radical
- H2O2
hydrogen peroxide
- HNE
4-hydroxy-2-nonenal
- HPLC
high-performance liquid chromatography
- hTERT
human telomerase reverse transcriptase
- IHC
immunohistochemistry
- iNOS
inducible nitric oxide synthase
- IR
ionizing radiation
- iTRAQ
isotope tagging for relative and absolute quantification
- LC
liquid chromatography
- LOOH
lipid hydroperoxide
- MDA
malondialdehyde
- MetO
methionine sulfoxide
- MnSOD
manganese superoxide dismutase
- MS
mass spectrometry
- Msr
methionine sulfoxide reductase
- mtDNA
mitochondrial DNA
- N2O3
dinitrogen trioxide
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced form
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NMR
nuclear magnetic resonance
- •NO
nitric oxide
- NO2•
nitrogen dioxide
- NOX
NADPH oxidase
- NOxICAT
isotope-coded affinity tag for detecting nitrosated and oxidized cysteine
- O2•−
superoxide
- •OH
hydroxyl radical
- ONOO−
peroxynitrite anion
- OxICAT
isotope-coded affinity tag for detecting oxidized cysteine
- PHGPx
phospholipid hydroperoxide glutathione peroxidase
- Prx
peroxiredoxin
- PTM
post-translational modification
- PUFA
polyunsaturated fatty acid
- RAS
rennin–angiotensin system
- RNase A
ribonuclease A
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SCX
strong cation exchange
- SILAC
stable isotope labeling of amino acids in cell culture
- SNO-RAC
resin-assisted capture for S-nitrosothiols
- SOD
superoxide dismutase
- SSB
single-strand break
- Sv
sievert
- TCEP
tris(2-carboxyethyl)phosphine
- TGF-β
transforming growth factor β
- TrxR
thioredoxin reductase
- TUNEL
terminal transferase
- UHPLC-MS/MS
ultra high-performance LC coupled to tandem MS.
- UV
ultraviolet
Acknowledgments
Financial support was provided by funds from NIH R01 CA136810 (C.M.F.) and pilot funds from the Center for Molecular Signaling and Communication at Wake Forest University (C.M.F.). The authors thank Nelmi O. Devarie Baez, PhD, and Jeffrey G. Kuremsky, MD, for helpful feedback on this work.
References
- 1.Ahn GO. and Brown JM. Matrix metalloproteinase-9 is required for tumor vasculogenesis but not for angiogenesis: role of bone marrow-derived myelomonocytic cells. Cancer Cell 13: 193–205, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aljada A, Thusu K, Armstrong D, Nicotera T, and Dandona P. Increased carbonylation of proteins in diabetes mellitus. Diabetes 44: A113, 1995 [Google Scholar]
- 3.Ameziane-El-Hassani R, Boufraqech M, Lagente-Chevallier O, Weyemi U, Talbot M, Métivier D, Courtin F, Bidart J-M, El Mzibri M, Schlumberger M, and Dupuy C. Role of H2O2 in RET/PTC1 chromosomal rearrangement produced by ionizing radiation in human thyroid cells. Cancer Res 70: 4123–4132, 2010 [DOI] [PubMed] [Google Scholar]
- 4.An JH, Kim J, and Seong J. Redox signaling by ionizing radiation in mouse liver. Ann N Y Acad Sci 1030: 86–94, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Andreassen CN. and Alsner J. Genetic variants and normal tissue toxicity after radiotherapy: a systematic review. Radiother Oncol 92: 299–309, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Andreassen CN, Alsner J, and Overgaard J. Does variability in normal tissue reactions after radiotherapy have a genetic basis - where and how to look for it? Radiother Oncol 64: 131–140, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Andreassen CN, Overgaard J, Alsner J, Overgaard M, Herskind C, Cesaretti JA, Atencio DP, Green S, Formenti SC, Stock RG, and Rosenstein BS. ATM sequence variants and risk of radiation-induced subcutaneous fibrosis after postmastectomy radiotherapy. Int J Radiat Oncol Biol Phys 64: 776–783, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Antunes F. and Cadenas E. Cellular titration of apoptosis with steady state concentrations of H2O2: submicromolar levels of H2O2 induce apoptosis through fenton chemistry independent of the cellular thiol state. Free Radic Biol Med 30: 1008–1018, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Ayene IS, Koch CJ, and Krisch RE. DNA strand breakage by bivalent metal ions and ionizing radiation. Int J Radiat Biol 83: 195–210, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Aykin-Burns N, Slane BG, Liu ATY, Owens KM, O'Malley MS, Smith BJ, Domann FE, and Spitz DR. Sensitivity to low-dose/low-LET ionizing radiation in mammalian cells harboring mutations in succinate dehydrogenase subunit C is governed by mitochondria-derived reactive oxygen species. Radiat Res 175: 150–158, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azimzadeh O, Scherthan H, Sarioglu H, Barjaktarovic Z, Conrad M, Vogt A, Calzada-Wack J, Neff F, Aubele M, Buske C, Atkinson MJ, and Tapio S. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 11: 3299–3311, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Azzam EI, Jay-Gerin J-P, and Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett 327: 48–60, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakkenist CJ. and Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506, 2003 [DOI] [PubMed] [Google Scholar]
- 14.Bamatraf MMM, O'Neill P, and Rao BSM. Redox dependence of the rate of interaction of hydroxyl radical adducts of DNA nucleobases with oxidants: consequences for DNA strand breakage. J Am Chem Soc 120: 11852–11857, 1998 [Google Scholar]
- 15.Barata-Vallejo S, Ferreri C, Postigo A, and Chatgilialoglu C. Radiation chemical studies of methionine in aqueous solution:understanding the role of molecular oxygen. Chem Res Toxicol 23: 258–263, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Barglow KT. and Cravatt BF. Activity-based protein profiling for the functional annotation of enzymes. Nat Meth 4: 822–827, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Barrett DM, Black SM, Todor H, Schmidt-Ullrich RK, Dawson KS, and Mikkelsen RB. Inhibition of protein tyrosine phosphatases by mild oxidative stresses is dependent on S-nitrosylation. J Biol Chem 280: 14453–14461, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Bartlett RK, Bieber Urbauer RJ, Anbanandam A, Smallwood HS, Urbauer JL, and Squier TC. Oxidation of Met144 and Met145 in calmodulin blocks calmodulin dependent activation of the plasma membrane Ca-ATPase. Biochemistry 42: 3231–3238, 2003 [DOI] [PubMed] [Google Scholar]
- 19.Basu S, Wang X, Gladwin MT, and Kim-Shapiro DB. Chemiluminescent detection of S-nitrosated proteins: comparison of tri-iodide, copper/CO/cysteine, and modified copper/cysteine methods. Methods Enzymol 440: 137–156, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Bechtold E. and King SB. Chemical methods for the direct detection and labeling of S-nitrosothiols. Antioxid Redox Signal 17: 981–991, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, and King SB. Water-soluble triarylphosphines as biomarkers for protein S-nitrosation. ACS Chem Biol 5: 405–414, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckman JS, Beckman TW, Chen J, Marshall PA, and Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A 87: 1620–1624, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckman JS. and Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol Cell Physiol 271: C1424–C1437, 1996 [DOI] [PubMed] [Google Scholar]
- 24.Beesk F, Dizdaroglu M, Schultefrohlinde D, and Von Sonntag C. Radiation-induced DNA strand breaks in deoxygenated aqueous solutions. The formation of altered sugars as end groups. Int J Radiat Biol 36: 565–576, 1979 [DOI] [PubMed] [Google Scholar]
- 25.Benderitter M, Vincent-Genod L, Pouget JP, and Voisin P. The cell membrane as a biosensor of oxidative stress induced by radiation exposure: a multiparameter investigation. Radiat Res 159: 471–483, 2003 [DOI] [PubMed] [Google Scholar]
- 26.Bennetzen M, Larsen D, Dinant C, Watanabe S, Bartek J, Lukas J, and Andersen J. Acetylation dynamics of human nuclear proteins during the ionizing radiation-induced DNA damage response. Cell Cycle 12: 1688–1695, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, and Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 9: 1314–1323, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang S-Y, Chen DJ, Aebersold R, and Shiloh Y. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 3: rs3, 2010 [DOI] [PubMed] [Google Scholar]
- 29.Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, and Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. FASEB J 27: 665–671, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernevic B, Petre BA, Galetskiy D, Werner C, Wicke M, Schellander K, and Przybylski M. Degradation and oxidation postmortem of myofibrillar proteins in porcine skeleton muscle revealed by high resolution mass spectrometric proteome analysis. Int J Mass Spectrom 305: 217–227, 2011 [Google Scholar]
- 31.Biaglow JE, Varnes ME, Epp ER, Clark EP, Tuttle SW, and Held KD. Role of glutathione in the aerobic radiation response. Int J Radiat Oncol Biol Phys 16: 1311–1314, 1989 [DOI] [PubMed] [Google Scholar]
- 32.Bigelow DJ. and Squier TC. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim Biophys Acta Proteins Proteomics 1703: 121–134, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Bionda C, Athias A, Poncet D, Alphonse G, Guezguez A, Gambert P, Rodriguez-Lafrasse C, and Ardail D. Differential regulation of cell death in head and neck cell carcinoma through alteration of cholesterol levels in lipid rafts microdomains. Biochem Pharmacol 75: 761–772, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Bionda C, Hadchity E, Alphonse G, Chapet O, Rousson R, Rodriguez-Lafrasse C, and Ardail D. Radioresistance of human carcinoma cells is correlated to a defect in raft membrane clustering. Free Radic Biol Med 43: 681–694, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Bird A. Perceptions of epigenetics. Nature 447: 396–398, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Biteau B, Labarre J, and Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425: 980–984, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Bizzozero OA, DeJesus G, Callahan K, and Pastuszyn A. Elevated protein carbonylation in the brain white matter and gray matter of patients with multiple sclerosis. J Neurosci Res 81: 687–695, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Björnstedt M, Hamberg M, Kumar S, Xue J, and Holmgren A. Human thioredoxin reductase directly reduces lipid hydroperoxides by NADPH and selenocystine strongly stimulates the reaction via catalytically generated selenols. J Biol Chem 270: 11761–11764, 1995 [DOI] [PubMed] [Google Scholar]
- 39.Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, and Wilson MA. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the Parkinsonism protein DJ-1. J Biol Chem 284: 6476–6485, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blough NV. and Zafiriou OC. Reaction of superoxide with nitric oxide to form peroxonitrite in alkaline aqueous solution. Inorg Chem 24: 3502–3504, 1985 [Google Scholar]
- 41.Blum J. and Fridovich I. Inactivation of glutathione peroxidase by superoxide radical. Arch Biochem Biophys 240: 500–508, 1985 [DOI] [PubMed] [Google Scholar]
- 42.Blume A, Herdegen T, and Unger T. Angiotensin peptides and inducible transcription factors. J Mol Med 77: 339–357, 1999 [DOI] [PubMed] [Google Scholar]
- 43.Bollineni RC, Fedorova M, and Hoffmann R. Identification of carbonylated peptides by tandem mass spectrometry using a precursor ion-like scan in negative ion mode. J Proteomics 74: 2351–2359, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Boudaïffa B, Cloutier P, Hunting D, Huels MA, and Léon S. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science 287: 1658–1660, 2000 [DOI] [PubMed] [Google Scholar]
- 45.Bregere C, Rebrin I, and Sohal RS. Detection and characterization of in vivo nitration and oxidation of tryptophan residues in proteins. Methods Enzymol 441: 339–349, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breimer LH. Ionizing radiation-induced mutagenesis. Br J Cancer 57: 6–18, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenner DJ. and Hall EJ. Computed tomography—an increasing source of radiation exposure. N Engl J Med 357: 2277–2284, 2007 [DOI] [PubMed] [Google Scholar]
- 48.Brewer A, Mustafi S, Murray T, Rajasekaran N, and Benjamin I. Reductive stress linked to small HSPs, G6PD, and Nrf2 pathways in heart disease. Antioxid Redox Signal 18: 1114–1127, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broniowska KA. and Hogg N. The chemical biology of S-nitrosothiols. Antioxid Redox Signal 17: 969–980, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunell DJ, Lowther T, Sagher D, Brot N, and Weissbach H. A disulfide intermediate is required for the reduction of methionine sulfoxide reductase by thioredoxin. FASEB J 21: A275, 2007 [Google Scholar]
- 51.Buxton GV, Greenstock CL, Helman WP, and Ross AB. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals in aqueous solution. J Phys ChemRef Data 17: 513–886, 1988 [Google Scholar]
- 52.Camerini S, Polci ML, Restuccia U, Usuelli V, Malgaroli A, and Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res 6: 3224–3231, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Cano KE, Li Y-J, and Chen Y. NMR metabolomic profiling reveals new roles of SUMOylation in DNA damage response. J Proteome Res 9: 5382–5388, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao S. and Wu R. Expression of angiotensin II and aldosterone in radiation-induced lung injury. Cancer Biol Med 9: 254–260, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chance B, Sies H, and Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev 59: 527–605, 1979 [DOI] [PubMed] [Google Scholar]
- 56.Chang MC, Pralle A, Isacoff EY, and Chang CJ. A selective, cell-permeable optical probe for hydrogen peroxide in living cells. J Am Chem Soc 126: 15392–15393, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charles RL, Schroder E, May G, Free P, Gaffney PRJ, Wait R, Begum S, Heads RJ, and Eaton P. Protein sulfenation as a redox sensor - proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics 6: 1473–1484, 2007 [DOI] [PubMed] [Google Scholar]
- 58.Chatgilialoglu C, Ferreri C, Torreggiani A, Salzano AM, Renzone G, and Scaloni A. Radiation-induced reductive modifications of sulfur-containing amino acids within peptides and proteins. J Proteomics 74: 2264–2273, 2011 [DOI] [PubMed] [Google Scholar]
- 59.Chavez J, Wu J, Han B, Chung W-G, and Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N′-aminooxymethylcarbonylhydrazino D-biotin. Anal Chem 78: 6847–6854, 2006 [DOI] [PubMed] [Google Scholar]
- 60.Cheema AK, Timofeeva O, Varghese R, Dimtchev A, Shiekh K, Shulaev V, Suy S, Collins S, Ressom H, Jung M, and Dritschilo A. Integrated analysis of ATM mediated gene and protein expression impacting cellular metabolism. J Proteome Res 10: 2651–2657, 2011 [DOI] [PubMed] [Google Scholar]
- 61.Chen C, Brenner DJ, and Brown TR. Identification of urinary biomarkers from X-irradiated mice using NMR spectroscopy. Radiat Res 175: 622–630, 2011 [DOI] [PubMed] [Google Scholar]
- 62.Chen M. and Cook KD. Oxidation artifacts in the electrospray mass spectrometry of Aβ peptide. Anal Chem 79: 2031–2036, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chistiakov DA, Voronova NV, and Chistiakov PA. Genetic variations in DNA repair genes, radiosensitivity to cancer and susceptibility to acute tissue reactions in radiotherapy-treated cancer patients. Acta Oncol 47: 809–824, 2008 [DOI] [PubMed] [Google Scholar]
- 64.Chiu SM, Xue LY, Friedman LR, and Oleinick NL. Copper ion-mediated sensitization of nuclear matrix attachment sites to ionizing radiation. Biochemistry 32: 6214–6219, 1993 [DOI] [PubMed] [Google Scholar]
- 65.Choi J, Sullards MC, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI, Chin L-S, and Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem 281: 10816–10824, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi KM, Kang CM, Cho ES, Kang SM, Lee SB, and Um HD. Ionizing radiation-induced micronucleus formation is mediated by reactive oxygen species that are produced in a manner dependent on mitochondria, Nox1, and JNK. Oncol Rep 17: 1183–1188, 2007 [PubMed] [Google Scholar]
- 67.Chouchani ET, James AM, Fearnley IM, Lilley KS, and Murphy MP. Proteomic approaches to the characterization of protein thiol modification. Curr Opin Chem Biol 15: 120–128, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chowdhury SK, Eshraghi J, Wolfe H, Forde D, Hlavac AG, and Johnston D. Mass spectrometry identification of amino acid transformations during oxidation of peptides and proteins: modifications of methionine and tyrosine. Anal Chem 67: 390–398, 1995 [DOI] [PubMed] [Google Scholar]
- 69.Chung F-L, Chen H-JC, and Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis 17: 2105–2111, 1996 [DOI] [PubMed] [Google Scholar]
- 70.Chung W-G, Miranda CL, and Maier CS. Detection of carbonyl-modified proteins in interfibrillar rat mitochondria using N′-aminooxymethylcarbonylhydrazino-D-biotin as an aldehyde/keto-reactive probe in combination with Western blot analysis and tandem mass spectrometry. Electrophoresis 29: 1317–1324, 2008 [DOI] [PubMed] [Google Scholar]
- 71.Clementi ME, Martorana GE, Pezzotti M, Giardina B, and Misiti F. Methionine 35 oxidation reduces toxic effects of the amyloid beta-protein fragment (31–35) on human red blood cell. Int J Biochem Cell Biol 36: 2066–2076, 2004 [DOI] [PubMed] [Google Scholar]
- 72.Cohen E, Lenarczyk M, Fish B, Jia S, Hessner M, and Moulder J. Evaluation of genomic evidence for oxidative stress in experimental radiation nephropathy. J Genet Disor Genet Rep 2: 1–3, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen EP, Fish BL, Irving AA, Rajapurkar MM, Shah SV, and Moulder JE. Radiation nephropathy is not mitigated by antagonists of oxidative stress. Radiat Res 172: 260–264, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohen EP, Fish BL, and Moulder JE. Treatment of radiation nephropathy with captopril. Radiat Res 132: 346–350, 1992 [PubMed] [Google Scholar]
- 75.Cohen EP, Fish BL, and Moulder JE. Successful brief captopril treatment in experimental radiation nephropathy. J Lab Clin Med 129: 536–547, 1997 [DOI] [PubMed] [Google Scholar]
- 76.Cohen EP, Fish BL, and Moulder JE. Mitigation of radiation injuries via suppression of the renin-angiotensin system: emphasis on radiation nephropathy. Curr Drug Targets 11: 1423–1429, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen EP, Irving AA, Drobyski WR, Klein JP, Passweg J, Talano JA, Juckett MB, and Moulder JE. Captopril to mitigate chronic renal failure after hematopoietic stem cell transplantation: a randomized controlled trial. Int J Radiat Oncol Biol Phys 70: 1546–1551, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen EP. and Robbins ME. Radiation nephropathy. Semin Nephrol 23: 486–499, 2003 [DOI] [PubMed] [Google Scholar]
- 79.Collins-Underwood JR, Zhao WL, Sharpe JG, and Robbins ME. NADPH oxidase mediates radiation-induced oxidative stress in rat brain microvascular endothelial cells. Free Radic Biol Med 45: 929–938, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corre I, Niaudet C, and Paris F. Plasma membrane signaling induced by ionizing radiation. Mutat Res 704: 61–67, 2010 [DOI] [PubMed] [Google Scholar]
- 81.Cruz DN, Perazella MA, and Mahnensmith RL. Bone marrow transplant nephropathy: a case report and review of the literature. J Am Soc Nephrol 8: 166–173, 1997 [DOI] [PubMed] [Google Scholar]
- 82.Cyr AR. and Domann FE. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid Redox Signal 15: 551–589, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dalle-Donne I, Carini M, Orioli M, Vistoli G, Regazzoni L, Colombo G, Rossi R, Milzani A, and Aldini G. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radic Biol Med 46: 1411–1419, 2009 [DOI] [PubMed] [Google Scholar]
- 84.Dalle-Donne I, Giustarini D, Colombo R, Rossi R, and Milzani A. Protein carbonylation in human diseases. Trends Mol Med 9: 169–176, 2003 [DOI] [PubMed] [Google Scholar]
- 85.Darleyusmar VM, Hogg N, Oleary VJ, Wilson MT, and Moncada S. The simultaneous generation of superoxide and nitric oxide can initiate lipid peroxidation in human low density lipoprotein. Free Radic Res Commun 17: 9–20, 1992 [DOI] [PubMed] [Google Scholar]
- 86.Datta PK, Moulder JE, Fish BL, Cohen EP, and Lianos EA. Induction of heme oxygenase 1 in radiation nephropathy: role of angiotensin II. Radiat Res 155: 734–739, 2001 [DOI] [PubMed] [Google Scholar]
- 87.Davies KJA. The broad spectrum of responses to oxidants in proliferating cells: a new paradigm for oxidative stress. IUBMB Life 48: 41–47, 1999 [DOI] [PubMed] [Google Scholar]
- 88.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta 1703: 93–109, 2005 [DOI] [PubMed] [Google Scholar]
- 89.Delanian S, Baillet F, Huart J, Lefaix JL, Maulard C, and Housset M. Successful treatment of radiation-induced fibrosis using liposomal Cu/Zn superoxide dismutase: clinical trial. Radiother Oncol 32: 12–20, 1994 [DOI] [PubMed] [Google Scholar]
- 90.Dexter DT. and Jenner P. Parkinson disease: from pathology to molecular disease mechanisms. Free Radic Biol Med 62: 132–144, 2013 [DOI] [PubMed] [Google Scholar]
- 91.Didenko V. (Ed.). DNA Damage Detection In Situ, Ex Vivo, and In Vivo. New York: Humana Press, 2011 [Google Scholar]
- 92.Dieriks B, De Vos W, Baatout S, and Van Oostveldt P. Repeated exposure of human fibroblasts to ionizing radiation reveals an adaptive response that is not mediated by interleukin-6 or TGF-β. Mutat Res 715: 19–24, 2011 [DOI] [PubMed] [Google Scholar]
- 93.Ditch S. and Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci 37: 15–22, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dizdaroglu M. and Jaruga P. Mechanisms of free radical-induced damage to DNA. Free Radic Res 46: 382–419, 2012 [DOI] [PubMed] [Google Scholar]
- 95.Du C, Gao Z, Venkatesha VA, Kalen AL, Chaudhuri L, Spitz DR, Cullen JJ, Oberley LW, and Goswami PC. Mitochondrial ROS and radiation induced transformation in mouse embryonic fibroblasts. Cancer Biol Ther 8: 1962–1971, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duncan Lyngdoh RH. and Schaefer HF. Elementary lesions in DNA subunits: electron, hydrogen atom, proton, and hydride transfers. Acc Chem Res 42: 563–572, 2009 [DOI] [PubMed] [Google Scholar]
- 97.Eggink M, Wijtmans M, Ekkebus R, Lingeman H, Esch IJPd, Kool J, Niessen WMA, and Irth H. Development of a selective ESI-MS derivatization reagent: synthesis and optimization for the analysis of aldehydes in biological mixtures. Anal Chem 80: 9042–9051, 2008 [DOI] [PubMed] [Google Scholar]
- 98.Ehrhart EJ, Segarini P, Tsang ML, Carroll AG, and Barcellos-Hoff MH. Latent transforming growth factor-β activation in situ: quantitative and functional evidence after low-dose gamma-irradiation. FASEB J 11: 991–1002, 1997 [DOI] [PubMed] [Google Scholar]
- 99.El-Missiry MA, Fayed TA, El-Sawy MR, and El-Sayed AA. Ameliorative effect of melatonin against gamma-irradiation-induced oxidative stress and tissue injury. Ecotoxicol Environ Saf 66: 278–286, 2007 [DOI] [PubMed] [Google Scholar]
- 100.Eldridge A, Fan M, Woloschak G, Grdina DJ, Chromy BA, and Li JJ. Manganese superoxide dismutase interacts with a large scale of cellular and mitochondrial proteins in low-dose radiation-induced adaptive radioprotection. Free Radic Biol Med 53: 1838–1847, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elyassaki W. and Wu SY. Lipid rafts mediate ultraviolet light-induced fas aggregation in M624 melanoma cells. Photochem Photobiol 82: 787–792, 2006 [DOI] [PubMed] [Google Scholar]
- 102.Epperly M, Chaillet J, Kalash R, Shaffer B, Goff J, Franicola D, Zhang X, Dixon T, Houghton F, Wang H, Berhane H, Romero C, Kim J, and Greenberger J. Conditional radioresistance of Tet-inducible manganese superoxide dismutase bone marrow stromal cell lines. Radiat Res 180: 189–204, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Evans C, Noirel J, Ow S-Y, Salim M, Pereira-Medrano A, Couto N, Pandhal J, Smith D, Pham T-K, Karunakaran E, Zou X, Biggs C, and Wright P. An insight into iTRAQ: where do we stand now? Anal Bioanal Chem 404: 1011–1027, 2012 [DOI] [PubMed] [Google Scholar]
- 104.Fanelli M, Amatori S, Barozzi I, Soncini M, Dal Zuffo R, Bucci G, Capra M, Quarto M, Dellino GI, Mercurio C, Alcalay M, Viale G, Pelicci PG, and Minucci S. Pathology tissue-chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples. Proc Natl Acad Sci U S A 107: 21535–21540, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Faucitano A, Buttafava A, Mariani M, and Chatgilialoglu C. The influence of solid-state molecular organization on the reaction paths of thiyl radicals. Chemphyschem 6: 1100–1107, 2005 [DOI] [PubMed] [Google Scholar]
- 106.Feitelson J. and Hayon E. Electron ejection and electron capture by phenolic compounds. J Phys Chem 77: 10–15, 1973 [Google Scholar]
- 107.Ferdinandy P. and Schulz R. Inhibition of peroxynitrite-induced dityrosine formation with oxidized and reduced thiols, nitric oxide donors, and purine derivatives. Antioxid Redox Signal 3: 165–171, 2001 [DOI] [PubMed] [Google Scholar]
- 108.Ferreri C, Chatgilialoglu C, Torreggiani A, Salzano AM, Renzone G, and Scaloni A. The reductive desulfurization of Met and Cys residues in bovine RNase A is associated with trans lipids formation in a mimetic model of biological membranes. J Proteome Res 7: 2007–2015, 2008 [DOI] [PubMed] [Google Scholar]
- 109.Ferreri C, Manco I, Faraone-Mennella MR, Torreggiani A, Tamba M, Manara S, and Chatgilialoglu C. The reaction of hydrogen atoms with methionine residues: a model of reductive radical stress causing tandem protein-lipid damage. Chembiochem 7: 1738–1744, 2006 [DOI] [PubMed] [Google Scholar]
- 110.Finley EL, Dillon J, Crouch RK, and Schey KL. Radiolysis-induced oxidation of bovine α-crystallin. Photochem Photobiol 68: 9–15, 1998 [PubMed] [Google Scholar]
- 111.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, and Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol 27: 557–559, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Freeman SL. and MacNaughton WK. Ionizing radiation induces iNOS-mediated epithelial dysfunction in the absence of an inflammatory response. Am J Physiol Gastrointest Liver Physiol 278: G243–G250, 2000 [DOI] [PubMed] [Google Scholar]
- 113.Fritz KS. and Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol 24: 1411–1419, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fu X, Kassim SY, Parks WC, and Heinecke JW. Hypochlorous acid oxygenates the cysteine switch domain of pro-matrilysin (MMP-7): a mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J Biol Chem 276: 41279–41287, 2001 [DOI] [PubMed] [Google Scholar]
- 115.Fujita S. and Steenken S. Pattern of hydroxyl radical addition to uracil and methyl- and carboxyl-substituted uracils. Electron transfer of hydroxyl adducts with N,N,N',N'-tetramethyl-p-phenylenediamine and tetranitromethane. J Am Chem Soc 103: 2540–2545, 1981 [Google Scholar]
- 116.Fukuda H, Fukuda A, Zhu C, Korhonen L, Swanpalmer J, Hertzman S, Leist M, Lannering B, Lindholm D, Bjork-Eriksson T, Marky I, and Blomgren K. Irradiation-induced progenitor cell death in the developing brain is resistant to erythropoietin treatment and caspase inhibition. Cell Death Differ 11: 1166–1178, 2004 [DOI] [PubMed] [Google Scholar]
- 117.Furdui CM. and Poole LB. Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom Rev [Epub ahead of print]; DOI: 10.1002/mas.21384, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gallogly MM. and Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol 7: 381–391, 2007 [DOI] [PubMed] [Google Scholar]
- 119.Gao F, Fish BL, Moulder JE, Jacobs ER, and Medhora M. Enalapril mitigates radiation-induced pneumonitis and pulmonary fibrosis if started 35 days after whole-thorax irradiation. Radiat Res 180: 546–552, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao M-C, Jia X-D, Wu Q-F, Cheng Y, Chen F-R, and Zhang J. Silencing Prx1 and/or Prx5 sensitizes human esophageal cancer cells to ionizing radiation and increases apoptosis via intracellular ROS accumulation. Acta Pharmacol Sin 32: 528–536, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Garner B, Waldeck AR, Witting PK, Rye K-A, and Stocker R. Oxidation of high density lipoproteins: II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J Biol Chem 273: 6088–6095, 1998 [DOI] [PubMed] [Google Scholar]
- 122.Ghesquiere B, Jonckheere V, Colaert N, Van Durme J, Timmerman E, Goethals M, Schymkowitz J, Rousseau F, Vandekerckhove J, and Gevaert K. Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics 10: M110.006866, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J Lipid Res 39: 1529–1542, 1998 [PubMed] [Google Scholar]
- 124.Giusti AM, Raimondi M, Ravagnan G, Sapora O, and Parasassi T. Human cell membrane oxidative damage induced by single and fractionated doses of ionizing radiation: a fluorescence spectroscopy study. Int J Radiat Biol 74: 595–605, 1998 [DOI] [PubMed] [Google Scholar]
- 125.Goetz W, Morgan MNM, and Baulch JE. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat Res 175: 575–587, 2011 [DOI] [PubMed] [Google Scholar]
- 126.Goldstein S. and Czapski G. Mechanism of the nitrosation of thiols and amines by oxygenated .NO solutions: the nature of the nitrosating intermediates. J Am Chem Soc 118: 3419–3425, 1996 [Google Scholar]
- 127.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, and Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177, 2008 [DOI] [PubMed] [Google Scholar]
- 128.Gorbunov NV, Pogue-Geile KL, Epperly MW, Bigbee WL, Draviam R, Day BW, Wald N, Watkins SC, and Greenberger JS. Activation of the nitric oxide synthase 2 pathway in the response of bone marrow stromal cells to high doses of ionizing radiation. Radiat Res 154: 73–86, 2000 [DOI] [PubMed] [Google Scholar]
- 129.Gordy W. and Miyagawa I. Electron spin resonance studies of mechanisms for chemical protection from ionizing radiation. Radiat Res 12: 211–229, 1960 [PubMed] [Google Scholar]
- 130.Gorski DH, Beckett MA, Jaskowiak NT, Calvin DP, Mauceri HJ, Salloum RM, Seetharam S, Koons A, Hari DM, Kufe DW, and Weichselbaum RR. Blockade of the vascular endothelial growth factor stress response increases the antitumor effects of ionizing radiation. Cancer Res 59: 3374–3378, 1999 [PubMed] [Google Scholar]
- 131.Grdina DJ, Murley JS, Miller RC, Mauceri HJ, Sutton HG, Li JJ, Woloschak GE, and Weichselbaum RR. A survivin-associated adaptive response in radiation therapy. Cancer Res 73: 4418–4428, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, and Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A 103: 7420–7425, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Greilberger J, Koidl C, Greilberger M, Lamprecht M, Schroecksnadel K, Leblhuber F, Fuchs D, and Oettl K. Malondialdehyde, carbonyl proteins and albumin-disulphide as useful oxidative markers in mild cognitive impairment and Alzheimer's disease. Free Radic Res 42: 633–638, 2008 [DOI] [PubMed] [Google Scholar]
- 134.Guan WP, Sha JB, Chen XJ, Xing YL, Yan JQ, and Wang ZQ. S-nitrosylation of mitogen activated protein kinase phosphatase-1 suppresses radiation-induced apoptosis. Cancer Lett 314: 137–146, 2012 [DOI] [PubMed] [Google Scholar]
- 135.Guan Z, Yates NA, and Bakhtiar R. Detection and characterization of methionine oxidation in peptides by collision-induced dissociation and electron capture dissociation. J Am Soc Mass Spectrom 14: 605–613, 2003 [DOI] [PubMed] [Google Scholar]
- 136.Guo J. and Prokai L. To tag or not to tag: a comparative evaluation of immunoaffinity-labeling and tandem mass spectrometry for the identification and localization of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Proteomics 74: 2360–2369, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guo Z, Kozlov S, Lavin MF, Person MD, and Paull TT. ATM activation by oxidative stress. Science 330: 517–521, 2010 [DOI] [PubMed] [Google Scholar]
- 138.Hafer K, Iwamoto KS, and Schiestl RH. Refinement of the dichlorofluorescein assay for flow cytometric measurement of reactive oxygen species in irradiated and bystander cell populations. Radiat Res 169: 460–468, 2008 [DOI] [PubMed] [Google Scholar]
- 139.Haimovitz-Friedman A, Kan C, Ehleiter D, Persaud R, McLoughlin M, Fuks Z, and Kolesnick R. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. J Exp Med 180: 525–535, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hansen RE, Roth D, and Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci U S A 106: 422–427, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hao G, Derakhshan B, Shi L, Campagne F, and Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A 103: 1012–1017, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hardin SC, Larue CT, Oh MH, Jain V, and Huber SC. Coupling oxidative signals to protein phosphorylation via methionine oxidation in Arabidopsis. Biochem J 422: 305–312, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harper JW. and Elledge SJ. The DNA damage response: ten years after. Mol Cell 28: 739–745, 2007 [DOI] [PubMed] [Google Scholar]
- 144.Hart TW. Some observations concerning the S-nitroso and S-phenylsulfonyl derivatives of L-cysteine and glutathione. Tetrahedron Lett 26: 2013–2016, 1985 [Google Scholar]
- 145.Hensley K, Butterfieldld DA, Hall N, Cole P, Subramaniam R, Mark R, Mattson MP, Markesbery WR, Harris ME, Aksenov M, Aksenova M, Wu JF, and Carney JM. Reactive oxygen species as causal agents in the neurotoxicity of the Alzheimer's disease-associated amyloid beta peptide. Ann N Y Acad Sci 786: 120–134, 1996 [DOI] [PubMed] [Google Scholar]
- 146.Hess DT, Matsumoto A, Kim SO, Marshall HE, and Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166, 2005 [DOI] [PubMed] [Google Scholar]
- 147.Hill RP, Zaidi A, Mahmood J, and Jelveh S. Investigations into the role of inflammation in normal tissue response to irradiation. Radiother Oncol 101: 73–79, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hitchler MJ. and Domann FE. Redox regulation of the epigenetic landscape in cancer: a role for metabolic reprogramming in remodeling the epigenome. Free Radic Biol Med 53: 2178–2187, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hogg N. The kinetics of S-transnitrosation-a reversible second-order reaction. Anal Biochem 272: 257–262, 1999 [DOI] [PubMed] [Google Scholar]
- 150.Hondal R, Marino S, and Gladyshev V. Selenocysteine in thiol/disulfide-like exchange reactions Antioxid Redox Signal 18: 1675–1689, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hong M, Xu A, Zhou H, Wu L, Randers-Pehrson G, Santella RM, Yu Z, and Hei TK. Mechanism of genotoxicity induced by targeted cytoplasmic irradiation. Br J Cancer 103: 1263–1268, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hosoki A, Yonekura S, Zhao QL, Wei ZL, Takasaki I, Tabuchi Y, Wang LL, Hasuike S, Nomura T, Tachibana A, Hashiguchi K, Yonei S, Kondo T, and Zhang-Akiyama QM. Mitochondria-targeted superoxide dismutase (SOD2) regulates radiation resistance and radiation stress response in HeLa cells. J Radiat Res 53: 58–71, 2012 [DOI] [PubMed] [Google Scholar]
- 153.Houée-Levin C. and Bobrowski K. The use of the methods of radiolysis to explore the mechanisms of free radical modifications in proteins. J Proteomics 92: 51–62, 2013 [DOI] [PubMed] [Google Scholar]
- 154.Hu W, Feng Z, Eveleigh J, Iyer G, Pan J, Amin S, Chung F-L, and Tang M-S. The major lipid peroxidation product, trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at codon 249 of human p53 gene, a unique mutational hotspot in hepatocellular carcinoma. Carcinogenesis 23: 1781–1789, 2002 [DOI] [PubMed] [Google Scholar]
- 155.Hu Z-P, Kim Y-M, Sowa MB, Robinson RJ, Gao X, Metz TO, Morgan WF, and Zhang Q. Metabolomic response of human skin tissue to low dose ionizing radiation. Mol Biosyst 8: 1979–1986, 2012 [DOI] [PubMed] [Google Scholar]
- 156.Hu ZZ, Huang H, Cheema A, Jung M, Dritschilo A, and Wu CH. Integrated bioinformatics for radiation-induced pathway analysis from proteomics and microarray data. J Proteomics Bioinform 1: 47–60, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Irwin ME, Mueller KL, Bohin N, Ge Y, and Boerner JL. Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol 226: 2316–2328, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Itakura K, Uchida K, and Kawakishi S. Selective formation of oxindole- and formylkynurenine-type products from tryptophan and its peptides treated with a superoxide-generating system in the presence of iron(III)-EDTA: a possible involvement with iron-oxygen complex. Chem Res Toxicol 7: 185–190, 1994 [DOI] [PubMed] [Google Scholar]
- 159.Jakob B. and Durante M. Radiation dose detection by imaging response in biological targets. Radiat Res 177: 524–532, 2012 [DOI] [PubMed] [Google Scholar]
- 160.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der and Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med 45: 1–17, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jeong JH, Jung YS, Na SJ, Lee ES, Kim MS, Choi S, Shin DH, Paek E, Lee HY, and Lee KJ. Novel oxidative modifications in redox-active cysteine residues. Mol Cell Proteomics 10: M110.000513, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jobling ME, Mott JD, Finnegan MT, Jurukovski V, Erickson AC, Walian PJ, Taylor SE, Ledbetter S, Lawrence CM, Rifkin DB, and Barcellos-Hoff MH. Isoform-specific activation of latent transforming growth factor β (LTGF-β) by reactive oxygen species. Radiat Res 166: 839–848, 2006 [DOI] [PubMed] [Google Scholar]
- 163.Johnson C, Patterson A, Krausz K, Kalinich J, Tyburski J, Kang D, Luecke H, Gonzalez F, Blakely W, and Idle J. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res 178: 328–340, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Johnson C, Patterson A, Krausz K, Lanz C, Kang D, Luecke H, Gonzalez F, and Idle J. Radiation metabolomics. 4. UPLC-ESI-QTOF MS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res 175: 473–484, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jonsson TJ, Tsang AW, Lowther WT, and Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J Biol Chem 283: 22890–22894, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kam WW-Y. and Banati RB. Effects of ionizing radiation on mitochondria. Free Radic Biol Med 65: 607–619, 2013 [DOI] [PubMed] [Google Scholar]
- 167.Kantorow M, Lee W, and Chauss D. Focus on molecules: methionine sulfoxide reductase A. Exp Eye Res 100: 110–111, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Karagas MR, Stannard VA, Mott LA, Slattery MJ, Spencer SK, and Weinstock MA. Use of tanning devices and risk of basal cell and squamous cell skin cancers. J Natl Cancer Inst 94: 224–226, 2002 [DOI] [PubMed] [Google Scholar]
- 169.Kawai Y, Takeda S, and Terao J. Lipidomic analysis for lipid peroxidation-derived aldehydes using gas chromatography-mass spectrometry. Chem Res Toxicol 20: 99–107, 2007 [DOI] [PubMed] [Google Scholar]
- 170.Kergonou JF, Braquet M, and Rocquet G. Influence of whole-body gamma irradiation upon rat liver mitochondrial fractions. Radiat Res 88: 377–384, 1981 [PubMed] [Google Scholar]
- 171.Khan A, Rana P, Devi M, Chaturvedi S, Javed S, Tripathi R, and Khushu S. Nuclear magnetic resonance spectroscopy-based metabonomic investigation of biochemical effects in serum of γ-irradiated mice. Int J Radiat Biol 87: 91–97, 2011 [DOI] [PubMed] [Google Scholar]
- 172.Kharitonov VG, Sundquist AR, and Sharma VS. Kinetics of nitrosation of thiols by nitric oxide in the presence of oxygen. J Biol Chem 270: 28158–28164, 1995 [DOI] [PubMed] [Google Scholar]
- 173.Khong PL, Ringertz H, Donoghue V, Frush D, Rehani M, Appelgate K, and Sanchez R. Radiological protection in paediatric diagnostic and interventional radiology. Ann ICRP 42: 1–63, 2013 [DOI] [PubMed] [Google Scholar]
- 174.Kim EH, Park AK, Dong SM, Ahn JH, and Park WY. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene 29: 4725–4731, 2010 [DOI] [PubMed] [Google Scholar]
- 175.Kim H-Y. and Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell 15: 1055–1064, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim JH, Brown SL, Kolozsvary A, Jenrow KA, Ryu S, Rosenblum ML, and Carretero OA. Modification of radiation injury by ramipril, inhibitor of angiotensin-converting enzyme, on optic neuropathy in the rat. Radiat Res 161: 137–142, 2004 [DOI] [PubMed] [Google Scholar]
- 177.Kim SY, Yoo YH, and Park JW. Silencing of mitochondrial NADP(+)-dependent isocitrate dehydrogenase gene enhances glioma radiosensitivity. Biochem Biophys Res Commun 433: 260–265, 2013 [DOI] [PubMed] [Google Scholar]
- 178.Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu SE, King SB, Furdui CM, and Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection: evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol 473: 77–94, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kma L, Gao F, Fish BL, Moulder JE, Jacobs ER, and Medhora M. Angiotensin converting enzyme inhibitors mitigate collagen synthesis induced by a single dose of radiation to the whole thorax. J Radiat Res 53: 10–17, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kondo Y. and Issa JPJ. DNA methylation profiling in cancer. Expert Rev Mol Med 12: 1–14, 2010 [DOI] [PubMed] [Google Scholar]
- 181.Koturbash I, Pogribny I, and Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue—a possible mechanism contributing to radiation carcinogenesis? Biochem Biophys Res Commun 337: 526–533, 2005 [DOI] [PubMed] [Google Scholar]
- 182.Krakowczyk L, Blamek S, Strzelczyk J, Plachetka A, Maciejewski A, Poltorak S, and Wiczkowski A. Effects of X-ray irradiation on methylation levels of p16, MGMT and APC genes in sporadic colorectal carcinoma and corresponding normal colonic mucosa. Med Sci Monit 16: CR469–CR474, 2010 [PubMed] [Google Scholar]
- 183.Krisko A, Leroy M, Radman M, and Meselson M. Extreme anti-oxidant protection against ionizing radiation in bdelloid rotifers. Proc Natl Acad Sci U S A 109: 2354–2357, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Krisko A. and Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci U S A 107: 14373–14377, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kruse JJ, te Poele JA, Velds A, Kerkhoven RM, Boersma LJ, Russell NS, and Stewart FA. Identification of differentially expressed genes in mouse kidney after irradiation using microarray analysis. Radiat Res 161: 28–38, 2004 [DOI] [PubMed] [Google Scholar]
- 186.Kuhmann C, Weichenhan D, Rehli M, Plass C, Schmezer P, and Popanda O. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiother Oncol 101: 116–121, 2011 [DOI] [PubMed] [Google Scholar]
- 187.Kumar RA, Koc A, Cerny RL, and Gladyshev VN. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J Biol Chem 277: 37527–37535, 2002 [DOI] [PubMed] [Google Scholar]
- 188.Lanz C, Patterson A, Slavik J, Krausz K, Ledermann M, Gonzalez F, and Idle J. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res 172: 198–212, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lawrence T. and Rosenberg S. (Eds.). Cancer: Principles and Practice of Oncology. Philadelphia, PA: Lippincott Williams and Wilkins, 2008 [Google Scholar]
- 190.Le Maire M, Thauvette L, de Foresta B, Viel A, Beauregard G, and Potier M. Effects of ionizing radiation on proteins: evidence of non-random fragmentations and a caution in the use of the method for determination of molecular mass. Biochem J 267: 431–439, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Leach JK, Van Tuyle G, Lin PS, Schmidt-Ullrich R, and Mikkelsen RB. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res 61: 3894–3901, 2001 [PubMed] [Google Scholar]
- 192.Lee JH. and Park J-W. Oxalomalate regulates ionizing radiation-induced apoptosis in mice. Free Radic Biol Med 42: 44–51, 2007 [DOI] [PubMed] [Google Scholar]
- 193.Lee K, Park J-S, Kim Y-J, Soo Lee Y, Sook Hwang T, Kim D-J, Park E-M, and Park Y-M. Differential expression of Prx I and II in mouse testis and their up-regulation by radiation. Biochem Biophys Res Commun 296: 337–342, 2002 [DOI] [PubMed] [Google Scholar]
- 194.Lee R. and Britz-McKibbin P. Metabolomic studies of radiation-induced apoptosis of human leukocytes by capillary electrophoresis-mass spectrometry and flow cytometry: adaptive cellular responses to ionizing radiation. Electrophoresis 31: 2328–2337, 2010 [DOI] [PubMed] [Google Scholar]
- 195.Lee SH, Miyamoto K, Goto T, and Oe T. Non-invasive proteomic analysis of human skin keratins: screening of methionine oxidation in keratins by mass spectrometry. J Proteomics 75: 435–449, 2011 [DOI] [PubMed] [Google Scholar]
- 196.Lee TC, Greene-Schloesser D, Payne V, Diz DI, Hsu FC, Kooshki M, Mustafa R, Riddle DR, Zhao W, Chan MD, and Robbins ME. Chronic administration of the angiotensin-converting enzyme inhibitor, ramipril, prevents fractionated whole-brain irradiation-induced perirhinal cortex-dependent cognitive impairment. Radiat Res 178: 46–56, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, and Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 105: 8197–8202, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Leichert LI. and Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal 8: 763–772, 2006 [DOI] [PubMed] [Google Scholar]
- 199.Lenz A-G, Costabel U, Shaltiel S, and Levine RL. Determination of carbonyl groups in oxidatively modified proteins by reduction with tritiated sodium borohydride. Anal Biochem 177: 419–425, 1989 [DOI] [PubMed] [Google Scholar]
- 200.Leonard SE, Reddie KG, and Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol 4: 783–799, 2009 [DOI] [PubMed] [Google Scholar]
- 201.Levine RL. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med 32: 790–796, 2002 [DOI] [PubMed] [Google Scholar]
- 202.Li N, Kuo C-L, Paniagua G, van den Elst H, Verdoes M, Willems LI, van der Linden WA, Ruben M, van Genderen E, Gubbens J, van Wezel GP, Overkleeft HS, and Florea BI. Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat Protoc 8: 1155–1168, 2013 [DOI] [PubMed] [Google Scholar]
- 203.Liang S, Xu Z, Xu X, Zhao X, Huang C, and Wei Y. Quantitative proteomics for cancer biomarker discovery. Comb Chem High Throughput Screen 15: 221–231, 2012 [DOI] [PubMed] [Google Scholar]
- 204.Lim YB, Pyun BJ, Lee HJ, Jeon SR, Jin YB, and Lee YS. Proteomic identification of radiation response markers in mouse intestine and brain. Proteomics 11: 1254–1263, 2011 [DOI] [PubMed] [Google Scholar]
- 205.Limoli CL, Rola R, Giedzinski E, Mantha S, Huang TT, and Fike JR. Cell-density-dependent regulation of neural precursor cell function. Proc Natl Acad Sci U S A 101: 16052–16057, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lin RX, Zhao HB, Li CR, Sun YN, Qian XH, and Wang SQ. Proteomic analysis of ionizing radiation-induced proteins at the subcellular level. J Proteome Res 8: 390–399, 2009 [DOI] [PubMed] [Google Scholar]
- 207.Lindemann C. and Leichert L. Quantitative redox proteomics: The NOxICAT method. In: Quantitative Methods in Proteomics, edited by Marcus K. New York: Humana Press, 2012, pp. 387–403 [DOI] [PubMed] [Google Scholar]
- 208.Liu H, Wang Z, Zhang X, Qiao Y, Wu S, Dong F, and Chen Y. Selection of candidate radiation biomarkers in the serum of rats exposed to gamma-rays by GC/TOFMS-based metabolomics. Radiat Prot Dosimetry 154: 9–17, 2013 [DOI] [PubMed] [Google Scholar]
- 209.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, and Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116: 617–628, 2004 [DOI] [PubMed] [Google Scholar]
- 210.Lo Conte M. and Carroll KS. Chemoselective ligation of sulfinic acids with aryl-nitroso compounds. Angew Chem Int Ed Engl 51: 6502–6505, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Long LH, Liu J, Liu RL, Wang F, Hu ZL, Xie N, Fu H, and Chen JG. Differential effects of methionine and cysteine oxidation on Ca2+ in cultured hippocampal neurons. Cell Mol Neurobiol 29: 7–15, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Loree J, Koturbash I, Kutanzi K, Baker M, Pogribny I, and Kovalchuk O. Radiation-induced molecular changes in rat mammary tissue: possible implications for radiation-induced carcinogenesis. Int J Radiat Biol 82: 805–815, 2006 [DOI] [PubMed] [Google Scholar]
- 213.Lu XM, Lu M, Tompkins RG, and Fischman AJ. Site-specific detection of S-nitrosylated PKBα/Akt1 from rat soleus muscle using CapLC-Q-TOF(micro) mass spectrometry. J Mass Spectrom 40: 1140–1148, 2005 [DOI] [PubMed] [Google Scholar]
- 214.Luo S. and Levine RL. Methionine in proteins defends against oxidative stress. FASEB J 23: 464–472, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Luo S. and Wehr NB. Protein carbonylation: avoiding pitfalls in the 2,4-dinitrophenylhydrazine assay. Redox Rep 14: 159–166, 2009 [DOI] [PubMed] [Google Scholar]
- 216.Lushchak VI. Free radical oxidation of proteins and its relationship with functional state of organisms. Biochemistry (Mosc) 72: 809–827, 2007 [DOI] [PubMed] [Google Scholar]
- 217.Lutgens LC, Deutz NE, Gueulette J, Cleutjens JP, Berger MP, Wouters BG, von Meyenfeldt MF, and Lambin P. Citrulline: a physiologic marker enabling quantitation and monitoring of epithelial radiation-induced small bowel damage. Int J Radiat Oncol Biol Phys 57: 1067–1074, 2003 [DOI] [PubMed] [Google Scholar]
- 218.Ma S, Kong B, Liu B, and Liu X. Biological effects of low-dose radiation from computed tomography scanning. Int J Radiat Biol 89: 326–333, 2013 [DOI] [PubMed] [Google Scholar]
- 219.Madian AG. and Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res 9: 3766–3780, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E, and Dukan S. Rules governing selective protein carbonylation. PLoS One 4: 1–12, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Makinde AY, John-Aryankalayil M, Palayoor ST, Cerna D, and Coleman CN. Radiation survivors: understanding and exploiting the phenotype following fractionated radiation therapy. Mol Cancer Res 11: 5–12, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Manda K, Ueno M, Moritake T, and Anzai K. Radiation-induced cognitive dysfunction and cerebellar oxidative stress in mice: protective effect of alpha-lipoic acid. Behav Brain Res 177: 7–14, 2007 [DOI] [PubMed] [Google Scholar]
- 223.Manda K, Ueno M, Moritake T, and Anzai K. α-Lipoic acid attenuates x-irradiation-induced oxidative stress in mice. Cell Biol Toxicol 23: 129–137, 2007 [DOI] [PubMed] [Google Scholar]
- 224.McDonald JT, Kim K, Norris AJ, Vlashi E, Phillips TM, Lagadec C, Della Donna L, Ratikan J, Szelag H, Hlatky L, and McBride WH. Ionizing radiation activates the Nrf2 antioxidant response. Cancer Res 70: 8886–8895, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Medhora M, Gao F, Fish BL, Jacobs ER, Moulder JE, and Szabo A. Dose-modifying factor for captopril for mitigation of radiation injury to normal lung. J Radiat Res 53: 633–640, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mezyk SP. Rate constant determination for the reaction of hydroxyl and glutathione thiyl radicals with glutathione in aqueous solution. J Phys Chem 100: 8861–8866, 1996 [Google Scholar]
- 227.Miaskiewicz K. and Osman R. Theoretical study on the deoxyribose radicals formed by hydrogen abstraction. J Am Chem Soc 116: 232–238, 1994 [Google Scholar]
- 228.Mihaljevic B, Tartaro I, Ferreri C, and Chatgilialoglu C. Linoleic acid peroxidation vs. isomerization: a biomimetic model of free radical reactivity in the presence of thiols. Org Biomol Chem 9: 3541–3548, 2011 [DOI] [PubMed] [Google Scholar]
- 229.Milic I, Fedorova M, Teuber K, Schiller J, and Hoffmann R. Characterization of oxidation products from 1-palmitoyl-2-linoleoyl-sn-glycerophosphatidylcholine in aqueous solutions and their reactions with cysteine, histidine and lysine residues. Chem Phys Lipids 165: 186–196, 2012 [DOI] [PubMed] [Google Scholar]
- 230.Milic I, Hoffmann R, and Fedorova M. Simultaneous detection of low and high molecular weight carbonylated compounds derived from lipid peroxidation by electrospray ionization-tandem mass spectrometry. Anal Chem 85: 156–162, 2012 [DOI] [PubMed] [Google Scholar]
- 231.Mirzaei H. and Regnier F. Enrichment of carbonylated peptides using Girard P reagent and strong cation exchange chromatography. Anal Chem 78: 770–778, 2005 [DOI] [PubMed] [Google Scholar]
- 232.Mirzaei H. and Regnier F. Identification and quantification of protein carbonylation using light and heavy isotope labeled Girard's P reagent. J Chromatogr A 1134: 122–133, 2006 [DOI] [PubMed] [Google Scholar]
- 233.Miyazawa T, Fujimoto K, Suzuki T, and Yasuda K. Determination of phospholipid hydroperoxides using luminol chemiluminescence - high-performance liquid chromatography. Methods Enzymol 233: 324–332, 1994 [DOI] [PubMed] [Google Scholar]
- 234.Molthen RC, Wu Q, Fish BL, Moulder JE, Jacobs ER, and Medhora MM. Mitigation of radiation induced pulmonary vascular injury by delayed treatment with captopril. Respirology 17: 1261–1268, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Morales A, Miranda M, Sanchez-Reyes A, Biete A, and Fernandez-Checa JC. Oxidative damage of mitochondrial and nuclear DNA induced by ionizing radiation in human hepatoblastoma cells. Int J Radiat Oncol Biol Phys 42: 191–203, 1998 [DOI] [PubMed] [Google Scholar]
- 236.Morand K, Talbo G, and Mann M. Oxidation of peptides during electrospray ionization. Rapid Commun Mass Spectrom 7: 738–743, 1993 [DOI] [PubMed] [Google Scholar]
- 237.Moskovitz J, Singh VK, Requena J, Wilkinson BJ, Jayaswal RK, and Stadtman ER. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem Biophys Res Commun 290: 62–65, 2002 [DOI] [PubMed] [Google Scholar]
- 238.Moulder JE, Fish BL, and Cohen EP. Radiation nephropathy is treatable with an angiotensin converting enzyme inhibitor or an angiotensin II type-1 (AT1) receptor antagonist. Radiother Oncol 46: 307–315, 1998 [DOI] [PubMed] [Google Scholar]
- 239.Mozziconacci O, Kerwin BA, and Schöneich C. Reversible hydrogen transfer reactions of cysteine thiyl radicals in peptides: the conversion of cysteine into dehydroalanine and alanine, and of alanine into dehydroalanine. J Phys Chem B 115: 12287–12305, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Multhoff G. and Radons J. Radiation, inflammation and immune responses in cancer. Front Oncol 2: 58, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Munley MT, Moore JE, Walb MC, Isom SP, Olson JD, Zora JG, Kock ND, Wheeler KT, and Miller MS. Cancer-prone mice expressing the Ki-rasG12C gene show increased lung carcinogenesis after CT screening exposures. Radiat Res 176: 842–848, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Munro T. The site of the target region for radiation-induced mitotic delay in cultured mammalian cells. Radiat Res 44: 747–757, 1970 [PubMed] [Google Scholar]
- 243.Nagashima S, Nakasako M, Dohmae N, Tsujimura M, Takio K, Odaka M, Yohda M, Kamiya N, and Endo I. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. Nat Struct Mol Biol 5: 347–351, 1998 [DOI] [PubMed] [Google Scholar]
- 244.Nagy P. Kinetics and mechanisms of thiol-disulfide exchange covering direct substitution and thiol oxidation-mediated pathways. Antioxid Redox Signal 18: 1623–1641, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nagy P, Karton A, Betz A, Peskin AV, Pace P, O'Reilly RJ, Hampton MB, Radom L, and Winterbourn CC. Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide: a kinetic and computational study. J Biol Chem 286: 18048–18055, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Nakaso K, Tajima N, Ito S, Teraoka M, Yamashita A, Horikoshi Y, Kikuchi D, Mochida S, Nakashima K, and Matsura T. Dopamine-mediated oxidation of methionine 127 in α-synuclein causes cytotoxicity and oligomerization of α-synuclein. PLoS One 8: e55068, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Narayanan PK, Goodwin EH, and Lehnert BE. Alpha particles initiate biological production of superoxide anions and hydrogen peroxide in human cells. Cancer Res 57: 3963–3971, 1997 [PubMed] [Google Scholar]
- 248.Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, and Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods Enzymol 473: 95–115, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Nodwell M. and Sieber S. ABPP methodology: introduction and overview. In: Activity-Based Protein Profiling, edited by Sieber SA. Berlin Heidelberg: Springer, 2012. pp. 1–41 [DOI] [PubMed] [Google Scholar]
- 250.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, and Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184, 2010 [DOI] [PubMed] [Google Scholar]
- 251.Ogawa F, Sander CS, Hansel A, Oehrl W, Kasperczyk H, Elsner P, Shimizu K, Heinemann SH, and Thiele JJ. The repair enzyme peptide methionine-S-sulfoxide reductase is expressed in human epidermis and upregulated by UVA radiation. J Invest Dermatol 126: 1128–1134, 2006 [DOI] [PubMed] [Google Scholar]
- 252.Okimoto Y, Watanabe A, Niki E, Yamashita T, and Noguchi N. A novel fluorescent probe diphenyl-1-pyrenylphosphine to follow lipid peroxidation in cell membranes. FEBS Lett 474: 137–140, 2000 [DOI] [PubMed] [Google Scholar]
- 253.Olry A, Boschi-Muller S, and Branlant G. Kinetic characterization of the catalytic mechanism of methionine sulfoxide reductase B from Neisseria meningitidis. Biochemistry 43: 11616–11622, 2004 [DOI] [PubMed] [Google Scholar]
- 254.Ong S-E. The expanding field of SILAC. Anal Bioanal Chem 404: 967–976, 2012 [DOI] [PubMed] [Google Scholar]
- 255.Otsuka M, Takahashi H, Shiratori M, Chiba H, and Abe S. Reduction of bleomycin induced lung fibrosis by candesartan cilexetil, an angiotensin II type 1 receptor antagonist. Thorax 59: 31–38, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Gonzalez FJ, and Idle JR. UPLC-ESI-TOF MS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem 80: 665–674, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Patwardhan RS, Checker R, Sharma D, Sandur SK, and Sainis KB. Involvement of ERK-Nrf-2 signaling in ionizing radiation induced cell death in normal and tumor cells. PLoS One 8: 1–12, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Paulsen CE. and Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol 5: 47–62, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Paulsen CE. and Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev 113: 4633–4679, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, and Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol 8: 57–64, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pauwels EKJ. and Bourguignon M. Cancer induction caused by radiation due to computed tomography: a critical note. Acta Radiol 52: 767–773, 2011 [DOI] [PubMed] [Google Scholar]
- 262.Pazhanisamy SK, Li HL, Wang Y, Batinic-Haberle I, and Zhou DH. NADPH oxidase inhibition attenuates total body irradiation-induced haematopoietic genomic instability. Mutagenesis 26: 431–435, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pearce MS, Salotti JA, Little MP, McHugh K, Lee C, Kim KP, Howe NL, Ronckers CM, Rajaraman P, Craft AW, Parker L, and Berrington de González A. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 380: 499–505, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Petersen DR. and Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med 37: 937–945, 2004 [DOI] [PubMed] [Google Scholar]
- 265.Petrini JHJ. and Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol 13: 458–462, 2003 [DOI] [PubMed] [Google Scholar]
- 266.Pogozelski WK, McNeese TJ, and Tullius TD. What species is responsible for strand scission in the reaction of [FeIIEDTA]2- and H2O2 with DNA? J Am Chem Soc 117: 6428–6433, 1995 [Google Scholar]
- 267.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, and King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem 18: 2004–2017, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Poole LB. and Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol 12: 18–24, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Porter NA, Caldwell SE, and Mills KA. Mechanisms of free radical oxidation of unsaturated lipids. Lipids 30: 277–290, 1995 [DOI] [PubMed] [Google Scholar]
- 270.Prutz WA. and Monig H. On the effect of oxygen or copper(II) in radiation-induced degradation of DNA in the presence of thiols. Int J Radiat Biol 52: 677–682, 1987 [DOI] [PubMed] [Google Scholar]
- 271.Qian J, Klomsiri C, Wright MW, King SB, Tsang AW, Poole LB, and Furdui CM. Simple synthesis of 1,3-cyclopentanedione derived probes for labeling sulfenic acid proteins. Chem Commun 47: 9203–9205, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, and Furdui CM. A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of β-ketoesters as cleavable probes. Chem Commun 48: 4091–4093, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Qiu HW, Edmunds T, Baker-Malcolm J, Karey KP, Estes S, Schwarz C, Hughes H, and Van Patten SM. Activation of human acid sphingomyelinase through modification or deletion of C-terminal cysteine. J Biol Chem 278: 32744–32752, 2003 [DOI] [PubMed] [Google Scholar]
- 274.Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, and Lunardi J. Proteomics analysis of cellular response to oxidative stress: evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem 277: 19396–19401, 2002 [DOI] [PubMed] [Google Scholar]
- 275.Radi R, Beckman JS, Bush KM, and Freeman BA. Peroxynitrite oxidation of sulfhydryls: the cytotoxic potential of superoxide and nitric oxide. J Biol Chem 266: 4244–4250, 1991 [PubMed] [Google Scholar]
- 276.Rauniyar N, Stevens SM, Prokai-Tatrai K, and Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography−tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal Chem 81: 782–789, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ravi J, Hills A, Cerasoli E, Rakowska P, and Ryadnov M. FTIR markers of methionine oxidation for early detection of oxidized protein therapeutics. Eur Biophys J 40: 339–345, 2011 [DOI] [PubMed] [Google Scholar]
- 278.Reddie KG, Seo YH, Muse WB, Leonard SE, and Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst 4: 521–531, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Reed CJ. and Douglas KT. Chemical cleavage of plasmid DNA by glutathione in the presence of Cu(II) ions - the Cu(II)-thiol system for DNA strand scission. Biochem J 275: 601–608, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rehder DS. and Borges CR. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 49: 7748–7755, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Reisz JA, Bechtold E, King SB, Poole LB, and Furdui CM. Thiol-blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J 280: 6150–6161, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Reiter RJ, Korkmaz A, Ma S, Rosales-Corral S, and Tan D-X. Melatonin protection from chronic, low-level ionizing radiation. Mutat Res 751: 7–14, 2012 [DOI] [PubMed] [Google Scholar]
- 283.Reuter S, Gupta SC, Chaturvedi MM, and Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 49: 1603–1616, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rezaee M, Sanche L, and Hunting DJ. Cisplatin enhances the formation of DNA single- and double-strand breaks by hydrated electrons and hydroxyl radicals. Radiat Res 179: 323–331, 2013 [DOI] [PubMed] [Google Scholar]
- 285.Richardson DE, Regino CAS, Yao H, and Johnson JV. Methionine oxidation by peroxymonocarbonate, a reactive oxygen species formed from CO2/bicarbonate and hydrogen peroxide. Free Radic Biol Med 35: 1538–1550, 2003 [DOI] [PubMed] [Google Scholar]
- 286.Robbins ME, Payne V, Tommasi E, Diz DI, Hsu FC, Brown WR, Wheeler KT, Olson J, and Zhao W. The AT1 receptor antagonist, L-158,809, prevents or ameliorates fractionated whole-brain irradiation-induced cognitive impairment. Int J Radiat Oncol Biol Phys 73: 499–505, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Robbins ME, Zhao W, Garcia-Espinosa MA, and Diz DI. Renin-angiotensin system blockers and modulation of radiation-induced brain injury. Curr Drug Targets 11: 1413–1422, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Robbins MEC, Zhao WL, Davis CS, Toyokuni S, and Bonsib SM. Radiation-induced kidney injury: a role for chronic oxidative stress? Micron 33: 133–141, 2002 [DOI] [PubMed] [Google Scholar]
- 289.Rossner P. and Sram RJ. Immunochemical detection of oxidatively damaged DNA. Free Radic Res 46: 492–522, 2012 [DOI] [PubMed] [Google Scholar]
- 290.Ryu S, Kolozsvary A, Jenrow KA, Brown SL, and Kim JH. Mitigation of radiation-induced optic neuropathy in rats by ACE inhibitor ramipril: importance of ramipril dose and treatment time. J Neurooncol 82: 119–124, 2007 [DOI] [PubMed] [Google Scholar]
- 291.Sak A. and Stuschke M. Use of γH2AX and other biomarkers of double-strand breaks during radiotherapy. Semin Radiat Oncol 20: 223–231, 2010 [DOI] [PubMed] [Google Scholar]
- 292.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, and Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423: 769–773, 2003 [DOI] [PubMed] [Google Scholar]
- 293.Salsbury FR, Knutson ST, Poole LB, and Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci 17: 299–312, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Salzano AM, Renzone G, Scaloni A, Torreggiani A, Ferreri C, and Chatgilialoglu C. Human serum albumin modifications associated with reductive radical stress. Mol Biosyst 7: 889–898, 2011 [DOI] [PubMed] [Google Scholar]
- 295.Saurin AT, Neubert H, Brennan JP, and Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A 101: 17982–17987, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Schnegg CI. and Robbins ME. Neuroprotective mechanisms of PPARδ: modulation of oxidative stress and inflammatory processes. PPAR Res 373560: 29, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Scully R, Puget N, and Vlasakova K. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene 19: 6176–6183, 2000 [DOI] [PubMed] [Google Scholar]
- 298.Sener G, Kabasakal L, Atasoy BM, Erzik C, Velioglu-Ogunc A, Cetinel S, Contuk G, Gedik N. and Yegen BC. Propylthiouracil-induced hypothyroidism protects ionizing radiation-induced multiple organ damage in rats. J Endocrinol 189: 257–269, 2006 [DOI] [PubMed] [Google Scholar]
- 299.Seo YH. and Carroll KS. Facile synthesis and biological evaluation of a cell-permeable probe to detect redox-regulated proteins. Bioorg Med Chem Lett 19: 356–359, 2009 [DOI] [PubMed] [Google Scholar]
- 300.Seo YH. and Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A 106: 16163–16168, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sethuraman M, McComb ME, Heibeck T, Costello CE, and Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics 3: 273–278, 2004 [DOI] [PubMed] [Google Scholar]
- 302.Shao C, Folkard M, Michael BD, and Prise KM. Targeted cytoplasmic irradiation induces bystander responses. Proc Natl Acad Sci U S A 101: 13495–13500, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sharov VS, Dremina ES, Galeva NA, Gerstenecker GS, Li X, Dobrowsky RT, Stobaugh JF, and Schoneich C. Fluorogenic tagging of peptide and protein 3-nitrotyrosine with 4-(aminomethyl)-benzenesulfonic acid for quantitative analysis of protein tyrosine nitration. Chromatographia 71: 37–53, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Shi W-Q, Fu H-Y, Bounds PL, Muroya Y, Lin M-Z, Katsumura Y, Zhao Y-L, and Chai Z-F. Nitration activates tyrosine toward reaction with the hydrated electron. Radiat Res 176: 128–133, 2011 [DOI] [PubMed] [Google Scholar]
- 305.Shimazu F, Kumta US, and Tappel AL. Radiation damage to methionine and its derivatives. Radiat Res 22: 276–287, 1964 [PubMed] [Google Scholar]
- 306.Silva CJ, Dynin I, Erickson ML, Requena JR, Balachandran A, Hui C, Onisko BC, and Carter JM. Oxidation of methionine 216 in sheep and elk prion protein is highly dependent upon the amino acid at position 218 but is not important for prion propagation. Biochemistry 52: 2139–2147, 2013 [DOI] [PubMed] [Google Scholar]
- 307.Simons J. How do low energy (0.1−2 eV) electrons cause DNA strand breaks? Acc Chem Res 39: 772–779, 2006 [DOI] [PubMed] [Google Scholar]
- 308.Singh A. and Singh H. Time-scale and nature of radiation-biological damage: approaches to radiation protection and post-irradiation therapy. Prog Biophys Mol Biol 39: 69–107, 1983 [DOI] [PubMed] [Google Scholar]
- 309.Smith BC. and Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol 16: 498–506, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Smutna M, Benova K, Dvorak P, Nekvapil T, Kopriva V, and Mate D. Protein carbonyls and traditional biomarkers in pigs exposed to low-dose gamma-radiation. Res Vet Sci 94: 214–218, 2013 [DOI] [PubMed] [Google Scholar]
- 311.Soreghan BA, Yang F, Thomas SN, Hsu J, and Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm Res 20: 1713–1720, 2003 [DOI] [PubMed] [Google Scholar]
- 312.Spychalowicz A, Wilk G, Sliwa T, Ludew DJ, and Guzik T. Novel therapeutic approaches in limiting oxidative stress and inflammation. Curr Pharm Biotechnol 13: 2456–2466, 2012 [PubMed] [Google Scholar]
- 313.Sram RJ, Binkova B, and Rossner P, Jr., Vitamin C for DNA damage prevention. Mutat Res 733: 39–49, 2012 [DOI] [PubMed] [Google Scholar]
- 314.Stadtman E, Moskovitz J, and Levine R. Oxidation of methionine residues of proteins: biological consequences. Antioxid Redox Signal 5: 577–582, 2003 [DOI] [PubMed] [Google Scholar]
- 315.Stark G. The effect of ionizing radiation on lipid membranes. Biochim Biophys Acta 1071: 103–122, 1991 [DOI] [PubMed] [Google Scholar]
- 316.Strzelczak G, Krzysztof B, and Holcman J. Reduction of selected oligopeptides containing methionine induced by hydrated electrons. Radiat Res 150: 688–694, 1998 [PubMed] [Google Scholar]
- 317.Sukharev SA, Pleshakova OV, Moshnikova AB, Sadovnikov VB, and Gaziev AI. Age- and radiation-dependent changes in carbonyl content, susceptibility to proteolysis, and antigenicity of soluble rat liver proteins. Comp Biochem Physiol Biochem Mol Biol 116: 333–338, 1997 [DOI] [PubMed] [Google Scholar]
- 318.Sun G. and Anderson VE. Prevention of artifactual protein oxidation generated during sodium dodecyl sulfate-gel electrophoresis. Electrophoresis 25: 959–965, 2004 [DOI] [PubMed] [Google Scholar]
- 319.Suzuki Y, Ruiz-Ortega M, Lorenzo O, Ruperez M, Esteban V, and Egido J. Inflammation and angiotensin II. Int J Biochem Cell Biol 35: 881–900, 2003 [DOI] [PubMed] [Google Scholar]
- 320.Symons M. and Taiwo F. Radiation damage to proteins: an electron paramagnetic resonance study. J Chem Soc Perkin Trans 2 2: 1413–1415, 1992 [Google Scholar]
- 321.Takanishi CL, Ma LH, and Wood MJ. A genetically encoded probe for cysteine sulfenic acid protein modification in vivo. Biochemistry 46: 14725–14732, 2007 [DOI] [PubMed] [Google Scholar]
- 322.Takanishi CL. and Wood MJ. A genetically encoded probe for the identification of proteins that form sulfenic acid in response to H2O2 in Saccharomyces cerevisiae. J Proteome Res 10: 2715–2724, 2011 [DOI] [PubMed] [Google Scholar]
- 323.Tallman KA, Roschek B, and Porter NA. Factors influencing the autoxidation of fatty acids: effect of olefin geometry of the nonconjugated diene. J Am Chem Soc 126: 9240–9247, 2004 [DOI] [PubMed] [Google Scholar]
- 324.Tamarit J, de Hoogh A, Obis E, Alsina D, Cabiscol E, and Ros J. Analysis of oxidative stress-induced protein carbonylation using fluorescent hydrazides. J Proteomics 75: 3778–3788, 2012 [DOI] [PubMed] [Google Scholar]
- 325.Tan D-X, Manchester LC, Terron MP, Flores LJ, and Reiter RJ. One molecule, many derivatives: a never-ending interaction of melatonin with reactive oxygen and nitrogen species? J Pineal Res 42: 28–42, 2007 [DOI] [PubMed] [Google Scholar]
- 326.Tanaka R, Sugiura Y, and Matsushita T. Simultaneous identification of 4-hydroxy-2-nonenal in foods by pre-column fluorigenic labeling with 1,3-cyclohexanedione and reversed-phase high-performance liquid chromatography with fluorescence detection. J Liq Chromatogr Relat Technol 36: 881–896, 2013 [Google Scholar]
- 327.Temple A, Yen T-Y, and Gronert S. Identification of specific protein carbonylation sites in model oxidations of human serum albumin. J Am Soc Mass Spectrom 17: 1172–1180, 2006 [DOI] [PubMed] [Google Scholar]
- 328.Tezel G, Thornton IL, Tong MG, Luo C, Yang X, Cai J, Powell DW, Soltau JB, Liebmann JM, and Ritch R. Immunoproteomic analysis of potential serum biomarker candidates in human glaucoma. Invest Ophthalmol Vis Sci 53: 8222–8231, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Thariat J, Collin F, Marchetti C, Ahmed-Adrar NS, Vitrac H, Jore D, and Gardes- Albert M. Marked difference in cytochrome c oxidation mediated by HO· and/or O2•− free radicals in vitro. Biochimie 90: 1442–1451, 2008 [DOI] [PubMed] [Google Scholar]
- 330.Thomas JP, Maiorino M, Ursini F, and Girotti AW. Protective action of phospholipid hydroperoxide glutathione peroxidase against membrane-damaging lipid peroxidation: in situ reduction of phospholipid and cholesterol hydroperoxides. J Biol Chem 265: 454–461, 1990 [PubMed] [Google Scholar]
- 331.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res 751: 158–246, 2012 [DOI] [PubMed] [Google Scholar]
- 332.Traber MG. and Atkinson J. Vitamin E, antioxidant and nothing more. Free Radic Biol Med 43: 4–15, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Truong TH, Garcia FJ, Seo YH, and Carroll KS. Isotope-coded chemical reporter and acid-cleavable affinity reagents for monitoring protein sulfenic acids. Bioorg Med Chem Lett 21: 5015–5020, 2011 [DOI] [PubMed] [Google Scholar]
- 334.Turk PW, Laayoun A, Steven SS, and Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis 16: 1253–1255, 1995 [DOI] [PubMed] [Google Scholar]
- 335.Tyburski J, Patterson A, Krausz K, Slavik J, Fornace AJ, Gonzalez F, and Idle J. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res 170: 1–14, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Tyburski J, Patterson A, Krausz K, Slavik J, Fornace AJ, Gonzalez F, and Idle J. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res 172: 42–57, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Uchida K. and Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc Natl Acad Sci U S A 89: 4544–4548, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Uchida K. and Stadtman ER. Selective cleavage of thioether linkage in proteins modified with 4-hydroxynonenal. Proc Natl Acad Sci U S A 89: 5611–5615, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ursini F. and Bindoli A. The role of selenium peroxidases in the protection against oxidative damage of membranes. Chem Phys Lipids 44: 255–276, 1987 [DOI] [PubMed] [Google Scholar]
- 340.Vankuijk F, Sevanian A, Handelman GJ, and Dratz EA. A new role for phospholipase-A2: protection of membranes from lipid peroxidation damage. Trends Biochem Sci 12: 31–34, 1987 [Google Scholar]
- 341.Varghese RS, Cheema A, Cheema P, Bourbeau M, Tuli L, Zhou B, Jung M, Dritschilo A, and Ressom HW. Analysis of LC-MS data for characterizing the metabolic changes in response to radiation. J Proteome Res 9: 2786–2793, 2010 [DOI] [PubMed] [Google Scholar]
- 342.Venkitaraman AR. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci 114: 3591–3598, 2001 [DOI] [PubMed] [Google Scholar]
- 343.Vit JP. and Rosselli F. Role of the ceramide-signaling pathways in ionizing radiation-induced apoptosis. Oncogene 22: 8645–8652, 2003 [DOI] [PubMed] [Google Scholar]
- 344.von Sonntag C. The chemistry of free-radical-mediated DNA damage. Basic Life Sci 58: 287–317; discussion 317–321, 1991 [DOI] [PubMed] [Google Scholar]
- 345.Wachsman JT. DNA methylation and the association between genetic and epigenetic changes: relation to carcinogenesis. Mutat Res 375: 1–8, 1997 [DOI] [PubMed] [Google Scholar]
- 346.Wang C-R, Nguyen J, and Lu Q-B. Bond breaks of nucleotides by dissociative electron transfer of nonequilibrium prehydrated electrons: a new molecular mechanism for reductive DNA damage. J Am Chem Soc 131: 11320–11322, 2009 [DOI] [PubMed] [Google Scholar]
- 347.Wang H. and Xian M. Fast reductive ligation of S-nitrosothiols. Angew Chem Int Ed Engl 47: 6598–6601, 2008 [DOI] [PubMed] [Google Scholar]
- 348.Wang X, Kettenhofen NJ, Shiva S, Hogg N, and Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med 44: 1362–1372, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, and Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc Natl Acad Sci U S A 108: 10550–10555, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat Res 86: 185–195, 1981 [PubMed] [Google Scholar]
- 351.Ward JF. DNA damage as the cause of ionizing radiation-induced gene activation. Radiat Res 138: S85–S88, 1994 [PubMed] [Google Scholar]
- 352.Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic Biol Med 43: 995–1022, 2007 [DOI] [PubMed] [Google Scholar]
- 353.Wardman P. and von Sonntag C. Kinetic factors that control the fate of thiyl radicals in cells. Methods Enzymol 251: 31–45, 1995 [DOI] [PubMed] [Google Scholar]
- 354.Weimann A, Broedbaek K, Henriksen T, Stovgaard ES, and Poulsen HE. Assays for urinary biomarkers of oxidatively damaged nucleic acids. Free Radic Res 46: 531–540, 2012 [DOI] [PubMed] [Google Scholar]
- 355.Weitzman SA, Turk PW, Milkowski DH, and Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci U S A 91: 1261–1264, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Wenk J, Schuller J, Hinrichs C, Syrovets T, Azoitei N, Podda M, Wlaschek M, Brenneisen P, Schneider LA, Sabiwalsky A, Peters T, Sulyok S, Dissemond J, Schauen M, Krieg T, Wirth T, Simmet T, and Scharffetter-Kochanek K. Overexpression of phospholipid-hydroperoxide glutathione peroxidase in human dermal fibroblasts abrogates UVA irradiation-induced expression of interstitial collagenase/matrix metalloproteinase-1 by suppression of phosphatidylcholine hydroperoxide-mediated NFκB activation and interleukin-6 release. J Biol Chem 279: 45634–45642, 2004 [DOI] [PubMed] [Google Scholar]
- 357.Winterbourn CC, Chan T, Buss IH, Inder TE, Mogridge N, and Darlow BA. Protein carbonyls and lipid peroxidation products as oxidation markers in pretern infant plasma: associations with chronic lung disease and retinopathy and effects of selenium supplementation. Pediatr Res 48: 84–90, 2000 [DOI] [PubMed] [Google Scholar]
- 358.Winterbourn CC. and Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med 45: 549–561, 2008 [DOI] [PubMed] [Google Scholar]
- 359.Wong-ekkabut J, Xu Z, Triampo W, Tang IM, Peter Tieleman D, and Monticelli L. Effect of lipid peroxidation on the properties of lipid bilayers: a molecular dynamics study. Biophys J 93: 4225–4236, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Wood ZA, Poole LB, and Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653, 2003 [DOI] [PubMed] [Google Scholar]
- 361.Woolston CM, Storr SJ, Ellis IO, Morgan DAL, and Martin SG. Expression of thioredoxin system and related peroxiredoxin proteins is associated with clinical outcome in radiotherapy treated early stage breast cancer. Radiother Oncol 100: 308–313, 2011 [DOI] [PubMed] [Google Scholar]
- 362.Wu L-J, Randers-Pehrson G, Xu A, Waldren CA, Geard CR, Yu Z, and Hei TK. Targeted cytoplasmic irradiation with alpha particles induces mutations in mammalian cells. Proc Natl Acad Sci U S A 96: 4959–4964, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Xiang W, Weisbach V, Sticht H, Seebahn A, Bussmann J, Zimmermann R, and Becker C-M. Oxidative stress-induced posttranslational modifications of human hemoglobin in erythrocytes. Arch Biochem Biophys 529: 34–44, 2013 [DOI] [PubMed] [Google Scholar]
- 364.Xu B, Kim ST, Lim DS, and Kastan MB. Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol Cell Biol 22: 1049–1059, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Xu XZ, Tsvetkov LA, and Stern DF. Chk2 activation and phosphorylation-dependent oligomerization. Mol Cell Biol 22: 4419–4432, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Yamakura F. and Ikeda K. Modification of tryptophan and tryptophan residues in proteins by reactive nitrogen species. Nitric Oxide 14: 152–161, 2006 [DOI] [PubMed] [Google Scholar]
- 367.Yamamori T, Yasui H, Yamazumi M, Wada Y, Nakamura Y, Nakamura H, and Inanami O. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med 53: 260–270, 2012 [DOI] [PubMed] [Google Scholar]
- 368.Yamamoto Y. Chemiluminescence-based high-performance liquid chromatography assay of lipid hydroperoxides. Methods Enzymol 233: 319–324, 1994 [DOI] [PubMed] [Google Scholar]
- 369.Yan L-J. and Forster MJ. Chemical probes for analysis of carbonylated proteins: a review. J Chromatogr B 879: 1308–1315, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Yang CY, Gu ZW, Yang ML, Lin SN, Garcia-Prats AJ, Rogers LK, Welty SE, and Smith CV. Selective modification of apoB-100 in the oxidation of low density lipoproteins by myeloperoxidase in vitro. J Lipid Res 40: 686–698, 1999 [PubMed] [Google Scholar]
- 371.Yang F, Waters KM, Miller JH, Gritsenko MA, Rui Z, Xiuxia D, Livesay EA, Purvine SO, Monroe ME, Yingchun W, Camp DG, II, Smith RD, and Stenoien DL. Phosphoproteomics profiling of human skin fibroblast cells reveals pathways and proteins affected by low doses of ionizing radiation. PLoS One 5: 1–11, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Yang H, Jin X, Kei Lam CW, and Yan S-K. Oxidative stress and diabetes mellitus. Clin Chem Lab Med 49: 1773–1782, 2011 [DOI] [PubMed] [Google Scholar]
- 373.Yang Y, Song Y, and Loscalzo J. Regulation of the protein disulfide proteome by mitochondria in mammalian cells. Proc Natl Acad Sci U S A 104: 10813–10817, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Yin RC, Liu SQ, Zhao C, Lu ML, Tang MS, and Wang HL. An ammonium bicarbonate-enhanced stable isotope dilution UHPLC-MS/MS method for sensitive and accurate quantification of acrolein-DNA adducts in human leukocytes. Anal Chem 85: 3190–3197, 2013 [DOI] [PubMed] [Google Scholar]
- 375.Yoshida K, Yamazaki H, Ozeki S, Inoue T, Yoshioka Y, Yoneda M, Fujiwara Y, and Inoue T. Mitochondrial genotypes and radiation-induced micronucleus formation in human osteosarcoma cells in vitro. Oncol Rep 8: 615–619, 2001 [DOI] [PubMed] [Google Scholar]
- 376.You K, Benitez LV, McConachie WA, and Allison WS. The conversion of glyceraldehyde-3-phosphate dehydrogenase to an acylphosphatase by trinitroglycerin and inactivation of this activity by azide and ascorbate. Biochim Biophys Acta Enzymol 384: 317–330, 1975 [DOI] [PubMed] [Google Scholar]
- 377.Zang L, Carlage T, Murphy D, Frenkel R, Bryngelson P, Madsen M, and Lyubarskaya Y. Residual metals cause variability in methionine oxidation measurements in protein pharmaceuticals using LC-UV/MS peptide mapping. J Chromatogr B 895–896: 71–76, 2012 [DOI] [PubMed] [Google Scholar]
- 378.Zeidan YH, Wu BX, Jenkins RW, Obeid LM, and Hannun YA. A novel role for protein kinase Cδ-mediated phosphorylation of acid sphingomyelinase in UV light-induced mitochondrial injury. FASEB J 22: 183–193, 2008 [DOI] [PubMed] [Google Scholar]
- 379.Zhang AY, Yi F, Zhang G, Gulbins E, and Li PL. Lipid raft clustering and redox signaling platform formation in coronary arterial endothelial cells. Hypertension 47: 74–80, 2006 [DOI] [PubMed] [Google Scholar]
- 380.Zhang B, Davidson MM, Zhou H, Wang C, Walker WF, and Hei TK. Cytoplasmic irradiation results in mitochondrial dysfunction and DRP1-dependent mitochondrial fission. Cancer Res 73: 6700–6710, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Zhang J, Li S, Zhang D, Wang H, Whorton AR, and Xian M. Reductive ligation mediated one-step disulfide formation of S-nitrosothiols. Org Lett 12: 4208–4211, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Zhang M, Qureshi AA, Geller AC, Frazier L, Hunter DJ, and Han J. Use of tanning beds and incidence of skin cancer. J Clin Oncol 30: 1588–1593, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Zhang Y, Keszler A, Broniowska KA, and Hogg N. Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins. Free Radic Biol Med 38: 874–881, 2005 [DOI] [PubMed] [Google Scholar]
- 384.Zhao W. and Robbins ME. Inflammation and chronic oxidative stress in radiation-induced late normal tissue injury: therapeutic implications. Curr Med Chem 16: 130–143, 2009 [DOI] [PubMed] [Google Scholar]
- 385.Zhou DH, Yu T, Chen G, Brown SA, Yu ZF, Mattson MP, and Thompson JS. Effects of NF-κB1 (p50) targeted gene disruption on ionizing radiation-induced NF-κB activation and TNF-α, IL-1 alpha, IL-1 beta and IL-6 mRNA expression in vivo. Int J Radiat Biol 77: 763–772, 2001 [DOI] [PubMed] [Google Scholar]
- 386.Zhu H, Hunter TC, Pan S, Yau PM, Bradbury EM, and Chen X. Residue-specific mass signatures for the efficient detection of protein modifications by mass spectrometry. Anal Chem 74: 1687–1694, 2002 [DOI] [PubMed] [Google Scholar]