Abstract
Hematopoietic stem cells (HSCs) sustain blood formation throughout life and are the functional units of bone marrow transplantation. We show that transient expression of six transcription factors RUNX1T1, HLF, LMO2, PRDM5, PBX1, and ZFP37 imparts multi-lineage transplantation potential onto otherwise committed lymphoid and myeloid progenitors, and myeloid effector cells. Inclusion of MYC-N and MEIS1, and use of polycistronic viruses increase reprogramming efficacy. The reprogrammed cells, designated induced-HSCs (iHSCs), possess clonal multi-lineage differentiation potential, reconstitute stem/progenitor compartments, and are serially transplantable. Single-cell analysis revealed that iHSCs derived under optimal conditions exhibit a gene expression profile that is highly similar to endogenous HSCs. These findings demonstrate that expression of a set of defined factors is sufficient to activate the gene networks governing HSC functional identity in committed blood cells. Our results raise the prospect that blood cell reprogramming may be a strategy for derivation of transplantable stem cells for clinical application.
INTRODUCTION
Within the hematopoietic system, HSCs are the only cells with the functional capacity to differentiate to all blood lineages, and to self-renew for life. These properties, in combination with the ability of HSCs to engraft conditioned recipients upon transplantation, have established the paradigm for stem cell use in regenerative medicine. Allogeneic and autologous HSC transplantation is used in the treatment of ~50,000 patients/year for congenital and acquired hematopoietic diseases and other malignancies (Gratwohl et al., 2010). Despite wide clinical use, HSC transplantation has inherent risks with transplantation outcomes impacted by multiple factors including relapse of primary disease, the numbers of HSCs transplanted, graft failure, and opportunistic infection. Moreover, allogeneic transplantation often leads to graft versus host disease (GVHD), a devastating T-cell mediated condition resulting from minor histoincompatibility differences between donor and recipient. In spite of advances in HLA-typing to identify histocompatible donors, GVHD remains a significant cause of morbidity and mortality for ~60–80% of patients receiving grafts from unrelated donors (Petersdorf, 2013). De novo generation of isogenic HSCs from patient derived cells would obviate these issues, and extend transplantation to patients for whom a histocompatible donor cannot be identified. Furthermore, deriving HSCs from patients with hematological diseases would be invaluable for gaining insights into disease etiology through in vitro and in vivo disease modeling, as well as providing a cell-based platform for therapeutic screening. Deriving HSCs from alternative cell types has thus been a long sought goal in regenerative medicine.
Considerable effort has been mounted towards developing strategies for generating transplantable HSCs from alternative cell types, such as pluripotent (ES/iPS) stem cells (Choi et al., 2009; Kennedy et al., 2012). The advantages of using pluripotent cells to derive HSCs are many and include the ease by which iPS cells can be derived from patient cells, thereby putting autologous cell-based therapies within reach if HSCs can be successfully generated. However, despite considerable progress in defining the developmental pathways leading to HSCs from pluripotent cells (Sturgeon et al., 2013), the generation of robustly transplantable definitive HSCs from pluripotent cells remains elusive. The developmental plasticity of fibroblasts and success in converting them to other cell types has prompted efforts to generate HSCs from these cells as an alternative strategy to pluripotent stem cell based methodologies. In one study, ectopic expression of OCT4 combined with the instructive signals of hematopoietic cytokines led to the generation of blood cell progenitors from human fibroblasts, though the resulting cells showed limited self-renewal potential and were unable to give rise to all blood cell lineages (Szabo et al., 2010). More recently, expression of Gata2, Gfi1b, cFos, and Etv6 in murine fibroblasts led to the production of hematopoietic progenitors through an endothelial-like cell intermediate, though the resulting cells ultimately did not possess HSC potential (Pereira et al., 2013). In another study, expression of 5 transcription factors HOXA9, RORA, ERG, SOX4, and MYB imparted transient myeloerythroid engraftment potential onto iPS-derived blood cell progenitors, but were unable to instill the multi-lineage differentiation and self-renewal potential characteristic of HSCs (Doulatov et al., 2013).
The reasons underlying the current inability to generate transplantable HSCs from fibroblasts or pluripotent stem cells may be many but likely include the failure of current ex vivo conditions to support maintenance and propagation of HSCs. Moreover, the epigenetic landscape underlying HSC functional identity may be difficult to establish from divergent lineages such as fibroblasts or pluripotent stem cells. An alternative strategy that has the potential to surmount such challenges would be to reprogram differentiated blood cells back to HSCs. Striking examples in which hematopoietic cells have been experimentally reprogrammed to alternative blood cell fates by forced expression or ablation of lineage-affiliated transcription factors (Choi et al., 1990; Hanna et al., 2008; Iwasaki et al., 2006; Laiosa et al., 2006; Rolink et al., 1999; Taghon et al., 2007; Xie et al., 2004) demonstrate that cells of the hematopoietic system are amenable to reprogramming to alternative fates. Indeed, studies by Busslinger and colleagues have shown that ablation of a single transcription factor, Pax5, in early B-cell progenitors (Nutt et al., 1999; Rolink et al., 1999), or terminally differentiated B-cells (Cobaleda et al., 2007) was sufficient to dedifferentiate these cells to a primitive progenitor state that possessed multi-lineage myeloid and T-cell potential, but ultimately did not possess HSC activity. Pax5 ablation in peripheral B-cells also led to the formation of highly penetrant and aggressive lymphomas in vivo (Cobaleda et al., 2007) thus limiting prospects for translating Pax5 ablation as a strategy to derive engraftable multi-lineage progenitors.
HSCs rely on complex gene regulatory networks to enable their full functional potential (Orkin and Zon, 2008). Cell fate reprogramming studies including those mentioned above have however demonstrated that ectopic expression of surprisingly few regulators is often sufficient to rewrite the gene networks regulating cell identity for many cell types. We hypothesized that the regulatory networks governing HSC identity might similarly be accessed through the action of a small number of genes, and if so, that the functional identity of HSCs might be endowed to downstream blood cells upon ectopic expression of such factors. In this study, we have used the approach of transcription factor mediated cellular reprogramming within the hematopoietic system to derive transplantable HSC-like cells that possess the functional and molecular properties of HSCs.
RESULTS
Identification of factors capable of imparting alternative lineage potential in vitro and multi-lineage engraftment potential on committed progenitors in vivo
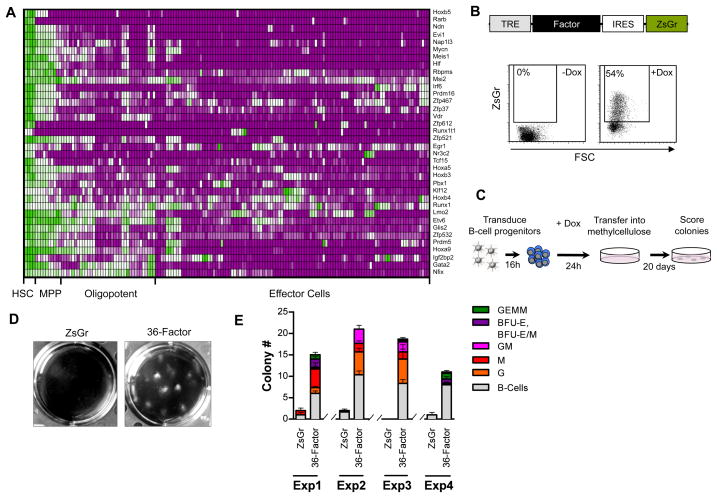
Experimental strategies for reprogramming diverse cell types generally rely on the action of one or more genes to impart the cellular and molecular properties of one cell type onto a different cell type. We reasoned that regulatory factors with relatively restricted expression in HSCs in relation to their downstream hematopoietic progeny are likely to be involved in defining the functional identity of HSCs through regulation of the gene networks underlying their fundamental properties. We therefore hypothesized that transient ectopic expression of such factors in committed blood cells might therefore endow them with the functional potential of HSCs and potentially stably reprogram them back to an HSC-like state. To identify such factors in an unbiased manner we analyzed microarray data of 40 different hematopoietic cell types that others and we have generated comprising the majority of hematopoietic progenitor and effector cells in addition to HSCs. These datasets (142 arrays in total) were normalized into a single database through which we identified 36 regulatory factors with relatively restricted expression in HSCs (Figure 1A, Table S1). These included 33 genes encoding transcription factors, and 3 genes encoding translational regulators many of which have been previously identified in studies profiling different hematopoietic cell types to define HSC transcriptional signatures (Chambers et al., 2007; Gazit et al., 2013). In addition to genes with known roles in regulating HSCs such Ndn, Evi1, Meis1, and Egr1, we also identified several genes that remain unstudied in HSC biology. The 36 factors were then cloned into doxycycline-inducible lentiviruses bearing a reporter cassette (Zs-Green) and high-titer viruses produced (Figure 1B).
Figure 1. Identification of factors capable of imparting alternative lineage potential in vitro.
(A) Heat map showing relative expression (green;high, to purple;low) of 36 regulatory genes identified as HSC-specific in the indicated cell types (see also Table S1).
(B) Schematic representation of lentivirus transgene expression cassette (top), and flow cytometry plots showing reporter cassette (ZsGr) expression in Pro/Pre B-cells +/− doxycycline induction (48 hours post).
(C) Schematic representation of in vitro screening strategy for cell fate conversion.
(D) Representative images of wells showing colonies arising in methylcellulose from Pro/Pre B cells transduced with ZsGr or 36-factor cocktail.
(E) Colony number and type arising in methylcellulose from Pro/Pre B cells transduced with ZsGr or 36-factor cocktail. Four independent experiments are shown and each condition performed in triplicate.
See Also Table S1.
It has been recognized that one of the challenges to reprogramming mature cells is that they are inherently stable. This is not necessarily true of oligo-potent and lineage-committed hematopoietic progenitors, which are in the process of differentiation. Moreover, since progenitor cells proximal to HSCs are more epigenetically related to HSCs (Bock et al., 2012), we reasoned that such proximity might be leveraged to lower the epigenetic barriers to HSC derivation. We first sought to determine if we could impart myeloid lineage potential onto otherwise B-cell restricted progenitors by expression of the 36-factors. Towards this we purified Pro/Pre B-cells (CD19+B220+AA4.1+IgM−) from mice expressing the reverse tetracycline-controlled transactivator (rtTA) from the Rosa26 locus (Rosa26rtTA) (Figure S1), and transduced them with control virus (Zs-green), or the 36-factor viral cocktail. Transduced cells were then cultured in the presence of doxycycline for 2 days followed by plating into methylcellulose in the presence of myeloid promoting cytokines, as well Flt3L and IL7 to promote B-cell survival (Figure 1C). These experiments showed that whereas control-transduced Pro/Pre B-cells were unable to form myeloid colonies as expected, cells transduced with the 36-factor cocktail readily gave rise to colonies containing diverse myeloid lineages including granulocytes, erythrocytes, megakaryocytes and macrophages (Figure 1D–E).
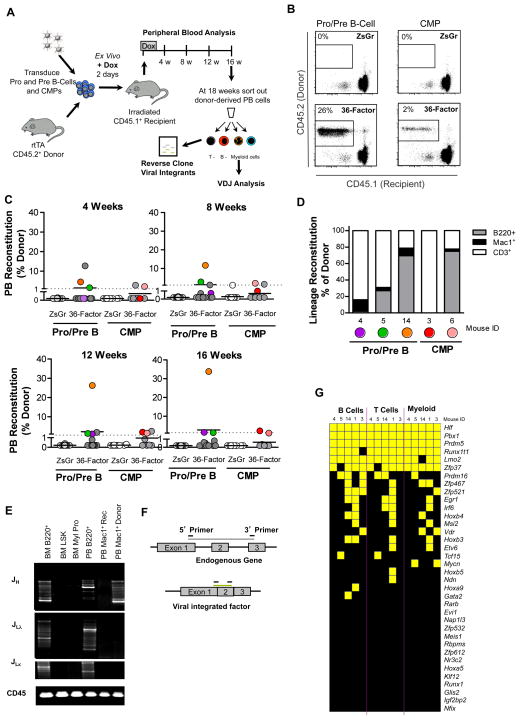
We next determined if transient ectopic expression of the 36-factor cocktail imparted HSC-like potential onto lineage-restricted lymphoid or myeloid progenitors in vivo. We took advantage of the fact that HSCs are the only hematopoietic cells capable of long-term multi-lineage reconstitution in myeloablated recipients upon transplantation, and progenitors transduced with a combination of factors able to instill them with long-term reconstitution potential would be readily detected in these assays (Figure 2A). Moreover, the sensitivity of transplantation is such that the activity of even a single HSC can read out, suggesting that even rare reprogramming events might be evidenced. Moreover, we reasoned that an in vivo setting could provide access to cues present in the microenvironment of the hematopoietic system that might facilitate or even be required for reprogramming. For these experiments we purified Pro/Pre B-cells or common myeloid progenitors (CMPs: lin−c-kit+Sca1−FcγrlowCD34+) from Rosa26rtTA mice (CD45.2) and following a 2-day transduction protocol with control (Zs-green) or viruses bearing the 36-factors in the presence of doxycycline, we competitively transplanted them along with whole bone marrow cells (CD45.1) into lethally irradiated congenic recipients (CD45.1) (Figure 2A). Doxycycline was maintained in the drinking water for 2 weeks post-transplant to maintain ectopic expression of the introduced factors, followed by doxycycline withdrawal. Peripheral blood analysis of the reconstituted mice over the 16-week course of the experiment revealed that, as expected, control-transduced Pro/Pre B-cells or CMPs did not give rise to long-term engraftment (Figure 2B–C). By contrast, a few of the recipients transplanted with the 36-factor transduced B-cell progenitors (3/15) or CMPs (2/8) exhibited long-term donor-derived reconstitution (Figures 2B–C, S2A). All but one of the reconstituted mice showed multi-lineage engraftment of B-, T- and myeloid cells though the degree of engraftment of each lineage varied amongst the different recipients (Figure 2D). Donor-derived cells were found to be negative for expression of the reporter gene present in all viruses indicating an absence of continued factor expression post-dox-induction (Figure S2A). Analysis of V(D)J recombination in sorted donor-derived myeloid cells from the Pro/Pre B-cell arm of the experiment confirmed the B-lineage origin of the reconstituting cells as evidenced by recombination of the heavy chain of the Ig locus (Figure 2E). The observation of multiple bands in the gel indicated that the reconstituting cells were polyclonal.
Figure 2. Identification of factors capable of imparting multi-lineage engraftment potential onto committed progenitors in vivo.
(A) Schematic of experimental strategy to identify factors capable of imparting multi-lineage engraftment potential on committed progenitors in vivo.
(B) Representative flow cytometry plots showing donor (CD45.2) reconstitution of mice transplanted with control (ZsGr) or 36-factor transduced Pro/Pre B cells or CMPs 16-weeks post-transplant.
(C) Donor reconstitution of mice transplanted with ZsGr or 36-factor transduced Pro/Pre B cells or CMPs at indicated time points post-transplantation. Only mice with >1% donor chimerism (dotted line) were considered reconstituted. Recipients transplanted; Pro/PreB;ZsGr n=15, Pro/PreB;36-factor n=15, CMP;ZsGr n=8, and CMP;36-factor n=8.
(D) Reconstitution of indicated peripheral blood cell lineages of individual recipients showing >1% donor chimerism presented as % of donor.
(E) PCR analysis of immunoglobulin rearrangement showing heavy (JH), and light chain (JLλ, JLκ) in bone marrow (BM) cells including B-cells (B220+), stem/progenitor (LSK) cells, myeloid progenitors (Myl Pro), and peripheral blood (PB) cells including B-cells (B220+), recipient myeloid cells (Mac1+ Rec), and donor myeloid cells (Mac1+ Donor) originating from Pro/Pre B cell;36-factor experiment. Loading control; genomic PCR for CD45.
(F) PCR-based strategy to identify virally integrated factors and discriminate from endogenous genes.
(G) Summary of data showing presence (yellow) or absence (black) of each of the indicated factors in donor B-, T-, and myeloid cells in each of the reconstituted mice shown in (C).
See also Figure S1–S2.
These experiments indicated that one or more factors from the 36-factor cocktail could imbue multi-lineage reconstituting potential onto otherwise committed lymphoid and myeloid progenitors. To determine which factors might be involved in conferring this potential, we sorted donor-derived myeloid, B- and T-cells to test for the presence of each of the 36 factors using a PCR-based strategy (Figure 2F, Table S2). This analysis revealed that whereas multiple factors could be identified in the donor-derived cells from each of the reconstituted mice, 6 transcription factors, Hlf, Runx1t1, Pbx1, Lmo2, Zfp37, and Prdm5 were consistently detected in all of the reconstituted recipients in multiple lineages (Figure 2G).
Transient ectopic expression of six transcription factors in committed progenitors is sufficient to alter lineage potential in vitro, and impart long-term multi-lineage engraftment potential in vivo
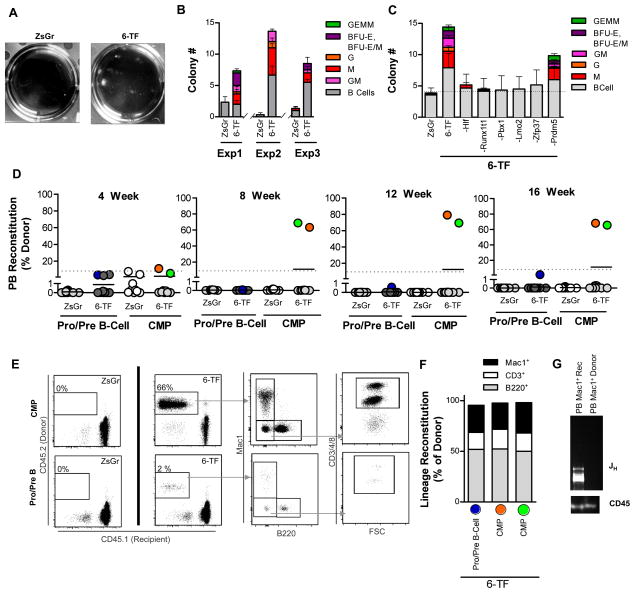
We next assessed if the 6 transcription factors we had identified in our in vivo screen were sufficient to confer myeloid colony forming potential onto Pro/Pre B-cells in methylcellulose. As we had observed with the 36-factor cocktail (Figure 1D–E), transduction with the viral combination of Hlf, Runx1t1, Pbx1, Lmo2, Zfp37, and Prdm5 was able to imbue lineage-restricted B-cell progenitors with myeloid lineage potential in these assays while also augmenting the ability of the cells to give rise to B-cell colonies (Figure 3A–B). To test the requirement for each of the 6 transcription factors (6-TF) we performed “N minus 1” experiments in which each of the factors was sequentially omitted from the transduction cocktail (Figure 3C). These experiments revealed that whereas Hlf, Runx1t1, Pbx1, Lmo2, and Zfp37 were all required for instilling myeloid colony forming potential onto Pro/Pre B-cells in vitro, the 5-factor cocktail minus Prdm5 gave rise to myeloid colonies albeit at lower numbers than the 6-TF combination (Figure 3C).
Figure 3. Transient ectopic expression of six transcription factors in committed progenitors is sufficient to alter lineage potential in vitro and impart long-term engraftment potential on committed progenitors in vivo.
(A) Representative images of wells showing colonies arising in methylcellulose from Pro/Pre B cells transduced with ZsGr or 6-TF cocktail.
(B) Colony number and indicated colony type arising in methylcellulose from Pro/Pre B cells transduced with ZsGr or 6-TF cocktail. 3 independent experiments are shown with each condition performed in triplicate.
(C) Colony number and type arising in methylcellulose from Pro/Pre B cells transduced with ZsGr, 6-TF cocktail, or 6-TF minus the indicated factor. Each condition performed in triplicate.
(D) Donor reconstitution of mice transplanted with ZsGr or 6-TF transduced Pro/Pre B cells or CMPs at indicated time points post-transplantation. Only mice with >1% donor chimerism (dotted line) were considered reconstituted. Recipients transplanted; Pro/PreB;ZsGr n=10, Pro/PreB;6-TF n=12, CMP;ZsGr n=9, and CMP;6-TF n=9.
(E) Representative flow cytometry plots showing donor reconstitution and lineage composition of mice transplanted with control (ZsGr) or 6-TF transduced Pro/Pre B cells or CMPs 16-weeks post-transplant. Lineage contribution to Mac1+ myeloid cells, B220+ B-cells, and CD3/4/8+ T-cells is shown.
(F) Reconstitution of indicated peripheral blood cell lineages of individual recipients showing >1% donor chimerism presented as % of donor.
(G) PCR analysis of immunoglobulin heavy (JH) chain rearrangement in recipient myeloid cells (Mac1+ Rec), and donor myeloid cells (Mac1+ Donor) originating from Pro/Pre B cell;6-TF experiment. Loading control; genomic PCR for CD45.
See also Figure S2.
We next tested whether transient expression of these 6 transcription factors was sufficient to impart long-term reconstituting potential onto committed myeloid or B-cell progenitors in competitive transplantation assays. Purified Pro/Pre B-cells or CMPs were transduced with control (Zs-Green) virus or the 6-TF cocktail followed by transplantation into congenic recipients (CD45.1). In contrast to control-transduced cells, long-term multi-lineage reconstitution was observed in 1/13 and 2/12 recipients transplanted with 6-TF transduced Pro/Pre B-cells or CMPs cells, respectively (Figures 3D). Peripheral blood analysis of recipient mice throughout the course of the experiment revealed that in all cases, donor-derived cells from the reconstituted recipients showed multi-lineage engraftment (Figures 3D–F, S2B). Ig heavy chain rearrangement was observed in donor-derived myeloid cells sorted from the Pro/Pre B-cell reconstituted mouse confirming the B-cell origin of the reconstituting cells (Figure 3G). These results indicate that transient ectopic expression of Hlf, Runx1t1, Pbx1, Lmo2, and Zfp37, and Prdm5 is sufficient to impart long-term, multi-lineage transplantation potential onto otherwise committed myeloid and lymphoid progenitors.
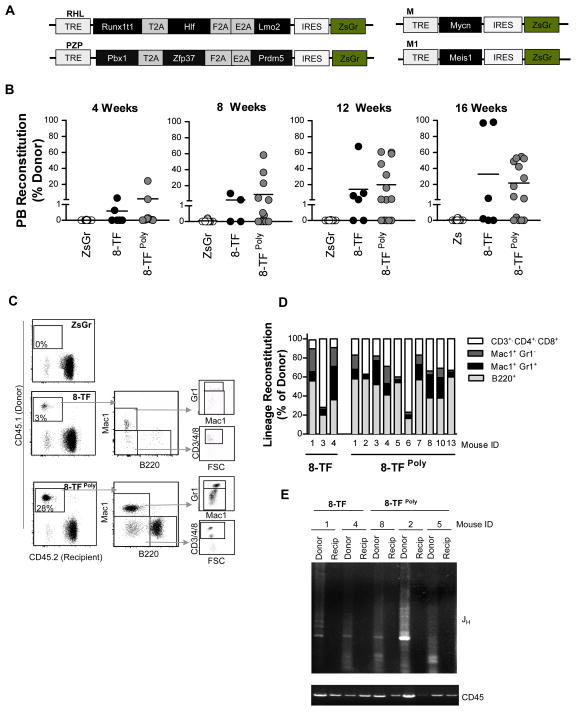
Inclusion of Meis1 and Mycn and use of polycistronic viruses improves in vivo reprogramming efficiency
The absence of donor-derived reconstitution in many of the recipient mice in our 6-TF transplantation experiments (Figure 3D) suggested that the efficiency of conferring long-term multi-lineage potential onto committed progenitors was low. In order to increase the probability that target cells would be co-transduced with all the factors we developed polycistronic doxycycline-inducible lentiviruses bearing three transcription factors each separated by 2A peptide sequences (Runx1T1•Hlf•Lmo2 (RHL), Pbx1•Zfp37•Prdm5 (PZP)). We also included two additional transcription factors (Mycn and Meis1) that we had repeatedly identified from primitive colonies generated in in vitro colony forming experiments (Figures 4A, S3, and data not shown). To test the utility of these strategies we transduced purified Pro/Pre B-cells (CD45.1) with control virus, or the 8-transcription factor cocktail as individual viruses (8-TF), or using the RHL and PZP polycistronic viruses along with viruses bearing Mycn, and Meis1 (8-TFPoly), and transplanted them along with Sca1-depleted (CD45.2) marrow into irradiated congenic recipients (CD45.2). Peripheral blood analysis of transplanted mice over the course of 16 weeks revealed that in contrast to the control-transduced cells that showed no donor-derived chimerism (0/12), multiple recipients transplanted with either the 8-TF (3/6) or the 8-TFPoly (9/14) transduced cells exhibited donor-derived chimerism (Figure 4B). All recipients showed multi-lineage reconstitution 18–22 weeks post-transplant though again the degree of B-cell, T-cell and myeloid chimerism varied amongst recipients (Figures 4C–D, S2C). The B-cell origin of the reconstituting cells was confirmed through evidence of Ig heavy chain rearrangement in donor-derived myeloid cells (Figure 4E).
Figure 4. Inclusion of Meis1 and Mycn and use of polycistronic viruses improves in vivo reprogramming efficiency.
(A) Schematic representation of RHL (Runxt1t1, Hlf, Lmo2) and PZP (Pbx1, Zfp37, Prdm5) polycistronic, and Meis1 and Mycn single factor viral constructs.
(B) Donor reconstitution of mice transplanted with ZsGr, 8-TF (8 single factor viruses), or 8-TFPoly (RHL, PZP polycistronic viruses plus Meis1 and Mycn viruses), transduced Pro/Pre B cells at indicated time points post-transplantation. Only mice with >1% donor chimerism were considered reconstituted. Recipients transplanted; ZsGr; n=12, 8-TF; n=6, 8TFPoly; n=14.
(C) Representative flow cytometry plots showing donor reconstitution and lineage contribution of mice transplanted with control (ZsGr), 8-TF, or 8TFPoly transduced Pro/Pre B cells 16-weeks post-transplant. Lineage contribution to Mac1+GR1- myeloid cells, Mac+GR1+ granulocytes, B220+ B-cells, and CD3/4/8+ T-cells is shown.
(D) Reconstitution of indicated peripheral blood cell lineages of individual recipients showing >1% donor chimerism presented as % of donor.
(E) PCR analysis of immunoglobulin heavy (JH) chain rearrangement in recipient (Recip), and donor (Donor) myeloid cells. Loading control; genomic PCR for CD45.
See also Figures S2, S3, Table S2.
Reprogrammed cells engraft bone marrow progenitor compartments and can reconstitute secondary recipients
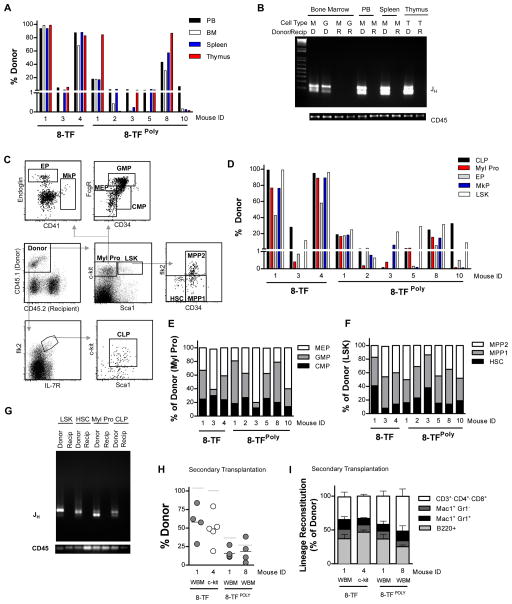
In addition to reconstituting the peripheral blood, HSCs efficiently repopulate secondary hematopoietic organs and bone marrow progenitor cell compartments upon transplantation. To determine if the B-cell progenitors transduced with the 8-TF or 8-TFPoly cocktails possessed this ability, reconstituted mice were sacrificed and analyzed 18–20 weeks post-transplant, showing that all the mice had donor-derived chimerism of the bone marrow, spleen and thymus though the level of chimerism varied between recipients as we had observed in the peripheral blood (Figures 5A). The Pro/Pre B-cell origin of the engrafting cells was confirmed through analysis of Ig rearrangement from DNA isolated from granulocytes and myeloid cells purified from the bone marrow and spleen, and T-cells derived from the thymus (Figure 5B). Immunophenotyping of bone marrow cells revealed donor contribution to common lymphoid progenitors (CLPs: lin−Flk2+IL7Rα+ckitlowSca1low), CMPs, granulocyte/monocyte progenitors (GMPs: lin−ckit+Sca1−FcγrhighCD34+), megarkaryocyte/erythrocyte progenitors (MEPs: lin−ckit+Sca1−Fcγr−CD34−), and primitive LSK progenitors (lin−Sca1+ckit+) (Figure 5C–F), which were comparable to host-derived progenitors analyzed from the same mice (Figure S4A–B). Importantly, we also observed donor contribution to megakaryocyte progenitors (MkPs: lin−c-kit+Sca1−CD41+), and erythroid progenitors (EPs: lin−ckit+Sca1−Endoglin+) demonstrating that the reconstituting cells were able of give rise to precursor cells of platelets and erythrocytes. Radioprotection transplantation assays performed using donor-derived MEPs (Na Nakorn et al., 2002) confirmed that the reprogrammed cells possessed a robust ability to generate platelets and red blood cells in vivo (Figure S4C–D). Importantly, subfractionation of the LSK compartment revealed donor-derived reconstitution of the multi-potent progenitor (MPP1: lin−ckit+Sca1+CD34+Flk2−, MPP2: lin−c-kit+Sca1+CD34+Flk2+) and HSC (lin−c-kit+Sca1+CD34−Flk2−) compartments (Figure 5C–D, F). Donor-marked progenitors and HSCs were subsequently analyzed for V(D)J recombination, which revealed Ig heavy chain rearrangement confirming their B-cell origin (Figure 5G).
Figure 5. Reprogrammed cells engraft secondary hematopoietic organs, bone marrow progenitor compartments and reconstitute secondary recipients.
(A) Donor reconstitution of peripheral blood (PB), bone marrow (BM), spleen, and thymus of mice transplanted with 8-TF, or 8-TFPoly transduced Pro/Pre B cells 18–20 weeks post-transplantation.
(B) PCR analysis of immunoglobulin heavy (JH) chain rearrangement in recipient (R), and donor (D) cells. Cell types analyzed include Mac1+ myeloid cells (M), Mac1+GR1+ granulocytes (G), and T-cells (T). Loading control; genomic PCR for CD45.
(C) Representative bone marrow stem and progenitor analysis of a recipient transplanted with 8-TFPoly transduced Pro/Pre B cells 18-weeks post-transplantation showing donor-reconstitution of myeloid progenitors (Myl Pro), megarkaryocyte/erythrocyte progenitors (MEP), granulocyte/monocyte progenitors (GMP), common myeloid progenitors (CMP), megakaryocyte progenitors (MkP), erythroid progenitors (EP), common lymphoid progenitors (CLP), Lineage-negative Sca1+ckit+ multipotent progenitors (LSK), multipotent progenitors (MPP1, MPP2), and hematopoietic stem cells (HSC). All cells were pre-gated through doublet-discriminated, live (propidium iodide negative), and lineage negative cells.
(D) Total donor reconstitution of the indicated populations in mice analyzed in (A).
(E–F) Reconstitution of the indicated myeloid progenitor (E) and primitive multi-potent and stem cell (F) populations in mice analyzed in (A) presented as percentage of donor.
(G) PCR analysis of immunoglobulin heavy (JH) chain rearrangement in the indicated recipient and donor populations. Loading control; genomic PCR for CD45.
(H) Donor reconstitution of secondary recipient mice transplanted with whole bone marrow (WBM) or c-Kit positive bone marrow cells derived from primary transplants of 8-TF transduced Pro/Pre B cells analyzed at 16–22 weeks. Number of recipients transplanted; WBM; n=5, c-Kit+; n=4. Grey bar indicates donor reconstitution level of primary recipient.
(I) Reconstitution of indicated peripheral blood cell lineages of individual recipients presented as % of donor.
See also Figures S2, S4, S5.
A defining property of HSCs is their ability to self-renew, a potential that can be evidenced by an ability to reconstitute secondary recipients upon serial transplantation. To test if the cells generated in our experiments possessed this potential we sacrificed primary recipient mice 18 weeks post-transplant and transplanted unsorted whole bone marrow or c-kit+ cells into irradiated secondary congenic recipients. Peripheral blood analysis at long-term time points post-transplant (16–22 weeks), revealed donor reconstitution of B-, T- and myeloid cells in all secondary recipients (Figure 5H–I).
In addition to sustained self-renewal potential, a hallmark property of HSCs is their ability to give rise to multi-lineage differentiation at the clonal level. Although we had observed clonal multi-lineage differentiation potential in vitro after induction of our factors (Figures 3B–C), our in vivo transplantation experiments, which were done at the population level, precluded us from concluding clonal differentiation potential in vivo. We reasoned that Ig heavy chain rearrangements arising in Pro/Pre B-cells could be used as a bar code that could be used to trace the clonal origin of donor-derived cells. We therefore isolated DNA from sorted donor-derived B- and T-cells, granulocytes, and macrophage/monocytes from primary recipients transplanted with Pro/Pre B-cells transduced with either 8-TF single or polycistronic viral cocktails (8-TF, 8-TFPoly). Ig heavy chain-specific PCR spanning the V(D)J junction was then performed and products common in size to all lineages analyzed were gel purified, cloned and sequenced. Analysis of these sequences revealed unique V(D)J junction sequences common to all four donor-derived lineages analyzed from three independent donors (Figure S5A). These results provide strong evidence that a single reprogrammed cell possessed multi-lineage differentiation potential, though there is an extremely remote possibility that these results could reflect the contribution of cells bearing identical V(D)J junctions. Using the same approach, we next analyzed DNA isolated from FACS purified donor-derived Mac1+ myeloid cells, B-cells and T-cells from secondary recipients that had been transplanted with the bone marrow of one of the mice we had analyzed during primary transplant. We identified the same V(D)J junction in the donor-derived myeloid, B- and T- cells of the secondary recipient, that had been previously identified in the primary recipient (Figure S5B). From these experiments we conclude that reprogrammed cells possess self-renewal and multi-lineage differentiation potential at the clonal level.
Taken together, these results demonstrate that transient ectopic expression of 8 transcription factors in lineage-restricted B-cell progenitors imparts them with the functional hallmarks of HSCs including clonal multi-lineage reconstituting potential, capacity to reconstitute bone marrow stem and progenitor cell compartments, and long-term self-renewal potential.
Transient expression of defined transcription factors in myeloid effector cells is sufficient to instill them long-term multi-lineage transplantation potential in vivo
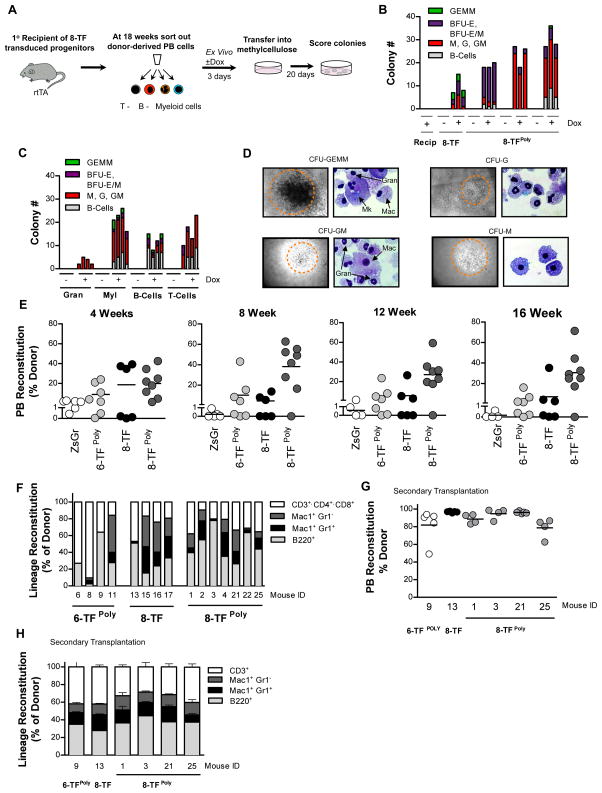
Eventual clinical translation of blood cell reprogramming to derive HSCs would benefit from an ability to reprogram cell types that can be readily and non-invasively obtained from the peripheral blood. We therefore sought to determine if multi-lineage progenitor activity could be conferred onto terminally differentiated blood cells using the transcription factors we identified. To test this, recipient and donor-derived peripheral blood was sorted from mice engrafted with Pro/Pre B-cells transduced with the 8-factor cocktail (8-TF or 8TFPoly) 16–22 weeks post-transplant. Sorted cells were then cultured in the absence or presence of doxycycline – with the latter condition expected to re-induce expression of the transduced factors – followed by plating the cells in methylcellulose (Figure 6A). As expected, neither the recipient-marked cells, nor the donor-derived cells cultured and plated in the absence of doxycycline gave rise to colonies (Figure 6B). By contrast, plates seeded with donor cells that had seen reactivation of the 8 transcription factors by exposure to doxycycline gave rise to mixed myeloid lineage colonies (Figure 6B). To determine which lineage(s) in the peripheral blood had the potential to give rise to these colonies upon re-expression of the transduced transcription factors, we purified B-cells, T-cells, myeloid cells and granulocytes from 8-TFPoly reconstituted mice, and tested their colony forming potential following culturing and plating in the absence or presence of doxycycline. Strikingly, plates seeded with each of the four peripheral blood cell types all developed colonies in the presence of dox, indicating that progenitor activity had been instilled into single cells upon reinduction of the factors. Of these, granulocytes gave rise to the fewest colonies whereas Mac1+ macrophages/monocytes yielded the largest number of colonies and the greatest number of primitive GEMM colonies (Figure 6C–D).
Figure 6. Transient expression of defined transcription factors in myeloid effector cells is sufficient instill them with progenitor activity in vitro, and long-term multi-lineage transplantation potential in vivo.
(A) Schematic representation of experimental strategy for assaying the colony forming potential of 8-TF transduced peripheral blood cells.
(B) Colony number and type arising in methylcellulose from peripheral blood cells from recipient (left-most lanes) or donor-derived cells from a recipient transplanted with Pro/Pre B cells transduced with 8-TF or 8-TFPoly cocktail, plus (+) or minus (−) exposure to doxycycline. Results from individual mouse performed in triplicate are shown.
(C) Colony number and type arising in methylcellulose from plated granulocytes, macrophages/monocytes (Myl), B-cells, and T-cells purified from the peripheral blood of cells pooled recipients transplanted with Pro/Pre B cells transduced with 8-TFPoly cocktail plus (+) or minus (−) exposure to doxycycline. n=4 biological replicates per cell type, per condition.
(D) Representative colony types and cytospins stained with May Grunwald of colonies derived in (C).
(E) Donor reconstitution of mice transplanted with ZsGr, 6-TFPoly, 8-TF or 8-TFPoly transduced Mac1+cKit- myeloid effector cells at indicated time points post-transplantation. Only mice with >1% donor chimerism were considered reconstituted. Recipients transplanted; ZsGr; n=6, 6-TFPoly; n=7, 8-TF; n=6, and 8-TFPoly; n=8.
(F) Reconstitution of indicated peripheral blood cell lineages of mice showing >1% donor chimerism presented as % of donor.
(G) Donor reconstitution 16 weeks post-transplant of secondary recipient mice transplanted non-competitively with 5x106 donor-derived (CD45.2+) bone marrow cells derived from primary recipients of 6-TFPoly, 8-TF or 8-TFPoly transduced Mac1+cKit- myeloid effector cells. Cells from individual primary donor mice (indicated by ID) were transplanted into N=5 secondary recipients each.
(H) Average reconstitution of indicated peripheral blood cell lineages presented as % of donor. n=5 recipients per group.
See also Figures S2, S6.
Encouraged by these results we next sought to determine if the transcription factors we identified could impart multi-lineage reconstituting potential onto terminally differentiated cells in transplantation assays. We focused on differentiated myeloid cells because unlike differentiated lymphoid cells that have rearranged TCR (T-cells) or Ig (B-cells) loci, multi-lineage reconstituting cells derived via reprogramming of myeloid cells would be expected to have the potential to give rise to full repertoires of lymphoid effector cells upon differentiation. We therefore sorted Mac1+c-kit− myeloid effector cells, which include monocytes, macrophages and granulocytes, from Rosa26rtTA mice and transduced them with either 6-factor (6-TFPoly), or 8-factor cocktails (8-TF and 8-TFPoly) and transplanted them into irradiated congenic recipients. Peripheral blood analysis at monthly intervals revealed that, whereas none of mice transplanted with cells transduced with control virus were reconstituted, multiple recipients transplanted with cells transduced with 6-TFPoly (4/7), 8-TF (3/6), and 8-TFPoly (7/8) exhibited long-term donor-derived engraftment (Figure 6F, S6A). Lineage analysis of the reconstituted mice showed donor-derived contribution to B-cell, T-cell, myeloid, and granulocyte lineages with contribution to each lineage varying between recipients (Figures 6F, S2D). Donor-derived contribution to secondary hematopoietic organs, and bone marrow progenitor cell compartments was also observed in mice sacrificed and analyzed 20 weeks post-transplant (Figure S6B–C). Serial transplantation of sorted donor-derived bone marrow cells demonstrated that the 6-TF or 8-TF transduced myeloid effectors could engraft secondary recipients in all lineages to 16 weeks post-transplant (Figure 6G–H).
Based on the functional data presented in Figures 1–6, we conclude that transient ectopic expression of 6 (Hlf, Runx1t1, Pbx1, Lmo2, Zfp37, and Prdm5) or 8 (Hlf, Runx1t1, Pbx1, Lmo2, and Zfp37, Prdm5, Mycn, and Meis1) transcription factors is sufficient to reprogram differentiated hematopoietic progenitors and effector cells to cells that possess the functional properties of HSCs. We term these reprogrammed cells induced-HSCs (iHSCs).
Single cell expression profiling of iHSCs reveals evidence of partial and full reprogramming
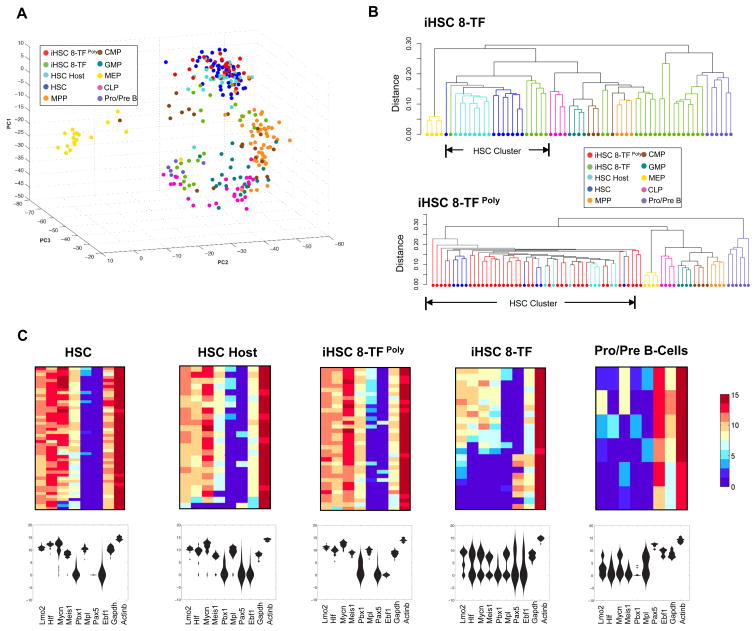
To assess the extent to which reprogrammed iHSCs recapitulate the molecular properties of endogenous HSCs, we employed a recently developed single cell gene expression profiling methodology that accurately defines hematopoietic stem and progenitor identity through the simultaneous quantification of expression of 151 lineage-specific transcription factors, epigenetic modifiers, cell surface molecules, and cell-cycle regulators (Guo et al., 2013). We sorted and analyzed donor-derived iHSCs by immunophenotype (CD45.2+lineage−ckit+Sca1+Fk2−CD34−/lowCD150+) 18 weeks post-transplantation from two different experiments in which Pro/Pre B cells had been transduced with the 8-TF cocktail as single viruses (8-TF), or with polycistronic viruses (8-TFPoly) (Figure 4). We also sorted and analyzed host-derived HSCs (CD45.1+lineage−ckit+Sca1+Fk2−CD34−/lowCD150+) from the same mice to serve as controls. Single cell expression data generated from iHSCs and host HSCs was then analyzed in comparison to data generated from Pro/Pre B-cells (the starting cell type), and also to data previously generated from HSCs, MPPs, CLPs, CMPs, GMPs, and MEPs purified at steady-state (Guo et al., 2013). Analysis of the raw data revealed high correlation between gene expression for the vast majority of control and test cell types (Figure S7, Table S3). To interrogate the transcriptional relationships amongst all the cell types analyzed, we performed principal component analysis (PCA) to define the transcriptional distances between the cells. As expected, steady-state HSCs and progenitor cells were largely positioned in agreement with established lineal relationships with HSCs forming a clearly defined cluster, and with MPPs positioned proximal, and oligopotent progenitors (MEPs, GMPs, CLPs) positioned more distal to HSCs (Figure 7A). Pro/Pre B-cells positioned closely to CLPs consistent with the lineal relationship between these cell types, while the host-derived HSCs were positioned within the steady-state HSC cluster as expected (Figure 7A). Interestingly, iHSCs derived from the two experiments (8-TF or 8-TFPoly) exhibited very distinct patterns of expression with the iHSCs derived from the 8-TF single virus experiment being more heterogeneous than the iHSCs derived from the 8-TFPoly transduced cells (Figures 7A, S7, Table S3). Whereas some of the iHSCs 8-TF positioned closely or within the HSC cluster, others mapped closer to MPPs while others positioned closely to the Pro/Pre B cluster (Figure 7A). By contrast, all of the iHSCs derived using the polycistronic virsues (iHSC 8-TFPoly) clustered within the HSC node (Figure 7A). Unsupervised hierarchical clustering analysis showed that whereas approximately equal numbers of iHSCs derived using single 8-TF viruses mapped closely to HSCs (6/23), others mapped closely to MPPs (7/23), while the remainder mapped more closely to Pro/Pre B cells (10/23) (Figure 7B). In contrast, all of the iHSCs derived using polycistronic viruses clustered very closely to host and control HSCs (35/35).
Figure 7. Single cell expression profiling of iHSCs reveals evidence of partial and full reprogramming.
(A) Principal component analysis of single cell expression data of iHSCs and the indicated control cell types. iHSCs were derived from experiments in which Pro/Pre B cells were transduced with the 8 identified transcription factors via single (iHSC 8-TF) or polycistronic viruses (iHSC 8-TFPoly). Data for individual cells of given type indicated in the legend.
(B) Dendrograms showing unsupervised hierarchical clustering of single cell expression data of representative control cells, and all iHSCs generated using single viruses (top panel), or polycistronic viruses (lower panel) as described in (A). Dendrogram branches are color-coded according to cell types indicated in the legend.
(C) Violin plots and the correlation heatmaps showing single cell expression data of the indicated genes. Expression levels are shown with high expression in red, and low expression in blue.
The inclusion of five (Mycn, Hlf, Lmo2, Meis1 and Pbx1) of the eight reprogramming factors amongst the 151 genes analyzed in these experiments allowed us to address how endogenous levels of these factors was re-established in iHSCs post-reprogramming. Consistent with their known roles in regulating HSCs, high levels of each of MycN, Hlf, Lmo2, and Meis1 were observed in steady-state HSCs, which contrasted the low levels observed in Pro/Pre B cells (Figure 7C). Pbx1 expression was lower in the majority of HSCs and absent in Pro/Pre B cells. Conversely, Ebf1 and Pax5, which are critical transcription factors for B-cell development were expressed at high levels in Pro/Pre B cells and negligible levels in HSCs. Analysis of the expression of these genes in iHSCs again revealed distinct differences depending upon whether or not single or polycistronic viruses were used for their derivation. Whereas high levels of endogenous MycN, Hlf, Lmo2, Meis1 and moderate levels of Pbx1 was reestablished in many of the iHSCs derived using single viruses, low levels of these genes and high levels of Ebf1 and Pax5 were still observed in a significant fraction of the cells (Figure 7C). By contrast, the expression of each of these genes in iHSCs derived using the polycistronic viruses closely recapitulated the expression patterns observed in the control HSCs (Figure 7C), as did the expression of all other genes analyzed known to be critical for HSCs function including the transcription factors Gfi1b, Gata2, and Ndn, and the cytokine receptors Mpl, and c-kit (Figure 7C, Table S3). Taken together, these results demonstrate that 8-TF reprogramming of Pro/Pre B using single viruses generates iHSCs with transcriptional properties consistent with either full or partial reprogramming, whereas iHSCs derived under optimal polycistronic viral conditions exhibit an expression profile highly similar with HSCs.
DISCUSSION
The de novo generation of transplantable HSCs has been a long sought goal in regenerative medicine. Here we report the generation of induced-HSCs via reprogramming from committed hematopoietic progenitor and effector cells. Through functional screening of 36 HSC-enriched factors, we identified 6 transcription factors Hlf, Runx1t1, Pbx1, Lmo2, Zfp37, and Prdm5 whose transient ectopic expression was sufficient to impart HSC functional potential onto committed blood cells in vivo. Inclusion of two additional transcription factors, Mycn, and Meis1, and the use of polycistronic viruses increased reprogramming efficiency. Reprogrammed cells showed multi-lineage differentiation and extensive self-renewal at the clonal level, and continued ectopic expression of the virally-transduced reprogramming factors was not required to sustain the functional potential of the reprogrammed cells in vivo (Figure S2). These findings demonstrate that ectopic expression of a set of defined transcription factors in committed blood cells is sufficient to stably activate the gene regulatory networks governing HSC functional identity.
HSCs are the most well characterized tissue-specific stem cells yet surprisingly little is known about the molecular mechanisms involved in regulating their central properties. The identification of a defined set of transcription factors capable of stably imparting HSC potential onto otherwise non-self-renewing, lineage-restricted cells, implies that these factors may be critically involved in regulating the transcriptional networks underlying HSC functional identity. Consistent with this, several of the factors that we identified have previously been established to regulate diverse aspects of HSC biology. For example, PBX1 and MEIS1, which interact and can form heterodimeric and heterotrimeric complexes with HOX proteins, regulate HSC self-renewal by maintaining HSC quiescence (Ficara et al., 2008; Kocabas et al., 2012; Unnisa et al., 2012). LMO2 is required for hematopoiesis and in its absence, neither primitive or definitive blood cells form (Warren et al., 1994; Yamada et al., 1998). And while MycN is dispensable for HSC activity due to functional redundancy with Myc, combined ablation of both severely disrupts HSC self-renewal and differentiation potential (Laurenti et al., 2008). In contrast to these well-characterized genes, Prdm5 and Zfp37 remain unstudied in HSC biology; and though the role of RUNX1T1 (as known as ETO) as a fusion partner with RUNX1 in acute myeloid leukemia is well established, its role in normal hematopoiesis remains unclear. Defining the roles that each of the reprogramming factors play in normal HSC biology will be critical for understanding their function in blood cell reprogramming.
Going forward it will also be important to elucidate how the reprogramming factors activate and maintain HSC transcriptional networks. Given that 6 of the 8 factors we identified (Hlf, Meis1, Lmo2, Mycn, Pbx1, and Runx1t1), are proto-oncogenes, suggests that iHSC reprogramming likely involves the activation and/or repression of gene networks common to stem cells and cancer cells (Reya et al., 2001). This is also consistent with the finding that virtually all the transcription factors required for HSC formation, maintenance, or lineage commitment are targeted by somatic mutation or translocation in hematologic malignancy (Orkin and Zon, 2008). Insights into how individual reprogramming factors may mediate their activity has been provided by recent studies. In one study, LMO2 overexpression in T-cell progenitors led to a pre-leukemic state with sustained self-renewal yet without blocking T-cell differentiation potential (McCormack et al., 2010). Similarly, ectopic expression of HLF in committed progenitors imbued them with sustained self-renewal activity ex vivo without blocking differentiation potential (Gazit et al., 2013), though this activity was insufficient to impart HSC potential onto downstream progenitors in vivo (RG, BG, DJR unpublished). These studies show that while expression of HLF or LMO2 can instill some of the functional and molecular properties of HSCs onto committed blood cells, by themselves they cannot activate the full repertoire of programs needed to establish HSC identity. In these regards, it is interesting that whereas iHSCs generated using polycistronic viruses exhibited expression profiles that were indistinguishable from control HSCs, iHSCs generated using monocistronic viruses were heterogeneous at the molecular level, possibly as a result of not receiving the full complement of reprogramming factors.
Although the transcriptional properties of iHSCs derived under optimal 8-TF polycistronic conditions were indistinguishable from endogenous HSCs, further analysis will be required to determine if the epigenetic landscape of these cells is fully reset. In this regard, it was interesting that the donor reconstitution observed in our experiments sometimes, though not always, evolved over time post-transplantation, with donor-derived chimerism and lineage potential changing over time. These results suggest that some reprogrammed iHSCs may need time to fully reset their epigenetic landscape to achieve balanced HSC potential, in a manner similar to the erasure of epigenetic memory observed with continued passage of iPS cells (Polo et al., 2010). Whether or not cell passage influences epigenetic resetting during iHSC derivation is at this point unclear. It is plausible that iHSCs may require a period of “maturation” in the stem cell niche to achieve full HSC potential. In these regards it is noteworthy that some of the partially reprogrammed iHSCs we identified had failed to appropriately upregulate the Mpl or cKit receptors suggesting that an inability to transduce signals in response to TPO or SCF emanating from the niche, may have been a factor in the incomplete resetting of their transcriptional state.
Transcription factors play a critical role in the specification of different lineages during development, and as such the discovery of a set of transcription factors capable of activating the gene regulatory networks underlying HSC functional identity suggests that it may be possible to use these factors on cells derived from pluripotent stem cells to facilitate the generation of definitive HSCs. It will also be interesting to test if the reprogramming factors we identified can be used to convert cell types outside of the hematopoietic system to an iHSC fate in a manner similar to the ability of the Yamanaka factors to bestow pluripotency onto cells of diverse lineages. It remains possible however that iHSCs derivation using the factors we defined will be limited to the blood system. In these regards it is important to note that although our approach relied on delivery of the reprogramming factors to the target cells ex vivo, it is possible that the reintroduction of the target cells into the hematopoietic microenvironment via transplantation may have provided cues that were critical for iHSC generation. Whether or not derivation of iHSCs from blood cells or other cell types can be achieved ex vivo without input from the hematopoietic microenvironment is unknown, though clearly the lack of suitable culture conditions for ex vivo HSC maintenance may preclude this. Nonetheless, the generation of iHSCs via blood cell reprogramming represents a powerful new experimental paradigm for studying the fundamental mechanisms underlying HSC identity that might eventually be lead to the derivation of transplantable stem cells with clinical potential.
Experimental Procedures
Identification of HSC-enriched factors
HSC-enriched factors were identified using microarray data (Affymetrix 430 2.0) of 40 FACs purified populations curated from the Gene Expression Omnibus. For complete list of populations used and accession numbers see Table S1. All datasets were subjected to quality control (QC) measures provided in the ArrayQualityMetrics package of R/Bioconductor. Datasets were normalized (gcRMA) using R. We applied a filter in which the ratio of expression in HSCs to all others was greater than 2.5-fold to define a list of potential HSC regulators that was further defined by cross-referencing the literature.
Colony forming assays
Pro/Pre B cells and CMPs sorted from Rosa26rtTA mice were plated in 200uL of S-clone SF-03 media (IWAI North America) supplemented with 10 ng/mL SCF, IL-12, and TPO, with 5 ng/mL Flk-3, and 5 ng/mL IL-7 included for Pro/Pre B-cells. Viruses were added for 16 hours, followed by addition of 1.0 mg/ml doxycycline for 24–48 hours. Cells were seeded at 10,000 cells/well (Pro/Pre B-cells) and 1000 cells/well (CMPs), into 6-well culture plates, by transferring the contents of the transduction wells including doxycycline into 1.75 mL/well of M3434 methylcellulose (Stem Cell Technologies). For experiments using Pro/Pre b-cells, M3434 was supplemented with 5 ng/mL Flk-3, and 5 ng/mL IL-7. For CFC assays involving peripheral blood of transplanted mice, donor-derived blood cells were sorted and cultured at 10,000 cells/well in F12 media supplemented with 10 ng/mL SCF, IL-12, and TPO, and 5 ng/mL Flk-3, and IL-7 in the presence or absence of 1.0 mg/ml doxycycline for 3 days followed by transferring the contents of the transduction wells including doxycycline into 1.75 mL/well of M3434 methylcellulose supplemented with IL7 and Flt3L as above. Colonies arising in these assays emerged later than typically observed (10–12 days) when primary HSPCs are plated (Gazit et al., 2013), and were thus scored at 20–22 days post-plating.
Transplantation assays
For experiments with 36-factor and 6-factor single viruses, Pro/Pre B-cells or CMPS were transduced with viruses for 2 days with doxycycline added on day 2. On day 3, transduced cells (ZsGr+) were sorted and transplanted at 1x104 cells/recipient into irradiated congenic recipients along with 2x105 marrow cells. For experiments in which polycistronic viruses were used, sorted Pro/Pre B-cells or Mac1+ckit- myeloid effector cells were transduced and transplanted without re-sorting at 2x106 or 5x106 cells per recipient into irradiated recipients along with 2x105 Sca1-depleted radio-protective bone marrow cells. Due to markedly reduced transduction efficiency of the polycistronic viruses, recipients in experiments involving polycistronic viruses were transplanted at greater cell numbers without pre-sorting transduced cells. Doxycycline was maintained in the drinking water for 2 weeks post-transplant. Serial transplantation from Pro/Pre B-cell experiments were performed by transplanting 1x107 unfractionated bone marrow cells, or 1x104 c-kit enriched bone marrow along with 2x105 Sca1-depleted radio-protective bone marrow cells. Serial transplantation in myeloid effector experiments was performed by transplanting 5x106 sorted donor-derived cells per recipient. In all transplant experiments, peripheral blood analysis was performed at four-week intervals with antibodies against TER-119, B220, Mac1, Gr1, CD3, CD45.1, and CD45.2 and propidium iodide to discriminate dead cells.
Supplementary Material
Highlights.
Six defined factors impart HSC potential onto committed blood cells.
Adding MYC-N and MEIS1 and polycistronic viruses improves reprogramming efficacy.
Induced-HSCs possess differentiation and self-renewal potential at clonal level.
Transcriptional properties of induced-HSCs recapitulate true HSCs at clonal level.
Acknowledgments
We wish to thank Isabel Beerman, Lydia Fang, Mike Bamberg, Ana Zguro, Rab Prinjha and Duane Wesemann for expertise, input and assistance. This work was supported by the National Institutes of Health RO1HL107630 (DJR), R00AG029760 (DJR), UO1DK072473-01 (DJR), and U01HL100001 (SHO), and grants from GlaxoSmithKline (DJR), The Leona M. and Harry B. Helmsley Charitable Trust (DJR), The New York Stem Cell Foundation (DJR), and The Harvard Stem Cell Institute (DJR, SHO, GC-Y). SHO is an Investigator of the Howard Hughes Medical Institute. DJR is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci U S A. 1990;87:7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KD, Vodyanik MA, Slukvin II. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J Clin Invest. 2009;119:2818–2829. doi: 10.1172/JCI38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R, Garrison BS, Rao TN, Shay T, Costello JF, Erikson J, Collins JJ, Regev A, Wagers A, Rossi DJ. Transcriptome Analysis Identifies Regulators of Hematopoietic Stem and Progenitor Cells. Stem Cell Reports. 2013;1:266–280. doi: 10.1016/j.stemcr.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratwohl A, Baldomero H, Aljurf M, Pasquini MC, Bouzas LF, Yoshimi A, Szer J, Lipton J, Schwendener A, Gratwohl M, et al. Hematopoietic stem cell transplantation: a global perspective. Jama. 2010;303:1617–1624. doi: 10.1001/jama.2010.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Luc S, Marco E, Lin TW, Peng C, Kerenyi MA, Beyaz S, Kim W, Xu J, Das PP, et al. Mapping Cellular Hierarchy by Single-Cell Analysis of the Cell Surface Repertoire. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zuniga-Pflucker JC, Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell reports. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, Deberardinis RJ, Zhang C, Sadek HA. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood. 2012 doi: 10.1182/blood-2012-05-432260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky G, Kotton DN, Fabian AJ, Gray JT, Lee JS, Mulligan RC. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol Ther. 2005;11:932–940. doi: 10.1016/j.ymthe.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Na Nakorn T, Traver D, Weissman IL, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J Clin Invest. 2002;109:1579–1585. doi: 10.1172/JCI15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma’ayan A, et al. Induction of a Hemogenic Program in Mouse Fibroblasts. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersdorf EW. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood. 2013;122:1863–1872. doi: 10.1182/blood-2013-05-355982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–606. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat Biotechnol. 2013;31:416–418. doi: 10.1038/nbt.2571. [DOI] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG, Jenkins NA, Grimes HL, Kumar AR. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood. 2012;120:4973–4981. doi: 10.1182/blood-2012-06-435800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.