Abstract
Acyl-Coenzyme A:cholesterol Acyltransferases (ACATs), members of the membrane bound O-acyltransferase (MBOAT) family, catalyze the conversion of cholesterol to cholesteryl esters. Mammals contain two isoenzymes ACAT1 and ACAT2. Both enzymes are drug targets for treating human diseases. ACAT1 is present ubiquitously in various cell types. It contains 9 transmembrane domains (TMDs) with the active site H460 locating within TMD #7, and another active site N421 locating within the 4th large cytoplasmic loop. In human ACAT1, a single nucleotide polymorphism (SNP) exists for residue 526: the codon is either CAG for glutamine (Q), or CGG for arginine (R). Q/R 526 is present within the Cterminal loop. Its biochemical significance is unknown. In addition, within the Cterminal half of ACAT1, numerous residues conserved with those of ACAT2 are present; the functions of these conserved residues are largely unknown. Here we performed single substitution mutagenesis experiments to investigate the roles of individual residues present in the C-terminal loop including Q/R526, and the 8 conserved prolines (P) located near/at various TMDs. The results show that the enzyme activity of ACAT1 Q526 is less active than that of ACAT1 R526 by 40%. In addition, several residues in the C-terminal loop are important to maintain proper ACAT1 protein stability. Other results show that the P347 plays important role in modulating enzyme catalysis. Overall, our results implicate that the CAG/CGG polymorphism can be utilized to perform ACAT1 activity/human disease susceptibility studies, and that P347 located near TMD #5 plays an important role in modulating enzyme catalysis.
Keywords: Single nucleotide polymorphism, Cholesterol metabolism, Atherosclerosis, Alzheimer’s disease, Site-specific mutagenesis
Introduction
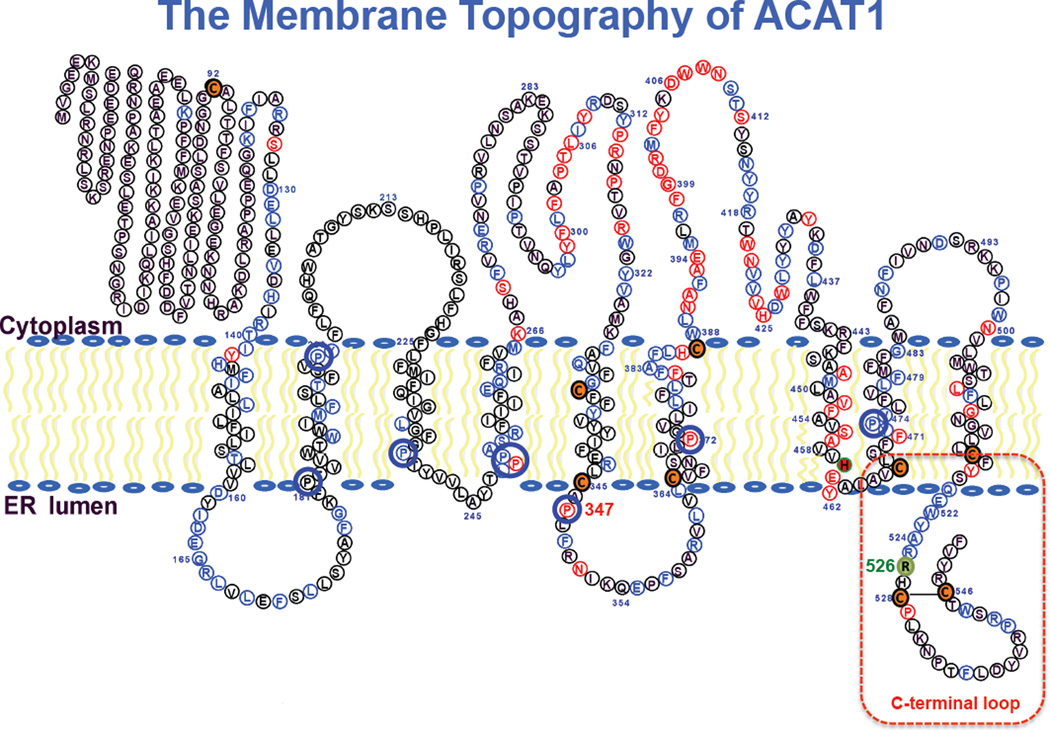
Cholesterol is an essential lipid for the growth and maintenance of all mammalian cells. Cholesterol tends to interact with sphingolipids and form lipid raft domains in cell membranes [1]. Large increases in membrane cholesterol content can be harmful to cells [2, 3]. To prevent excess (unesterified) cholesterol from building up in cell membranes, a major role of the enzyme acyl-CoA:cholesterol acyltransferases (ACAT) (also known as sterol O-acyltransferase; SOAT) is to catalyze the conversion of cholesterol to its storage form cholesteryl esters, by forming an ester linkage between the 3-beta OH moiety in cholesterol, and the carboxyl group of a long chain fatty acid donated from the long chain fatty acyl coenzyme A. Unlike cholesterol, cholesteryl esters do not partition well in lipid bilayers; they coalesce together to form cytoplasmic lipid droplets. Cholesteryl esters can also be packaged as part of the neutral lipid core within plasma lipoproteins for lipid transport purposes [4]. In mammals, there are two ACAT isoenzymes, ACAT1 [5] and ACAT2 [6–8]. Both enzymes are integral membrane proteins with multiple transmembrane domains (TMD). ACAT1 is present ubiquitously in many cell types; ACAT2 is enriched mainly in intestinal enterocytes. Neither enzyme is under the direct control of the key transcription factor sterol response element binding protein 2 (SREBP2), which controls gene expressions of many enzymes involved in lipid metabolism [9]. Instead, both enzymes are under allosteric control by its own substrate cholesterol [10, 11]. Both ACAT1 and ACAT2 are potential drug targets for atherosclerosis [12]; recent results suggest that ACAT1 is also a potential target for Alzheimer’s Disease [13–15] as well as for patients with low white blood cell count [16]. ACAT1 and ACAT2 are members of the membrane bound acyltransferase (MBOAT) family [17]. MBOAT enzymes contain multiple TMDs and share the first active site histidine located in a long hydrophobic region, and the second active site asparagine in a long hydrophilic region. Based on biochemical reactions, the MBOAT family contains 3 subgroups, as reviewed in: the first group (comprises of ACAT1, ACAT2, and diacylglycerol acyltransferase 1 (DGAT1)) acylates the -OH moiety of cholesterol or diacylglycerol; the second acylates an amino acid (residue) within a protein or a peptide hormone[18, 19]; the third acylates a lysophopholipid to reform a phospholipid [20]. Currently, high-resolution structural models are not available for any MBOAT member. Knowledge on MBOAT is mainly based on biochemical studies. ACAT1 polypeptide contains 550 amino acids, and is located mainly at the endoplasmic reticulum (ER). It is a homo-tetramer in vitro and in intact cells [21]. The enzyme contains 9 trans-membrane domains (TMD), with 5 loops located at the cytoplasmic side and 3 loops located at the luminal side of the ER. The first large loop, not conserved in ACAT2 or DGAT1, is not needed for enzyme activity, but plays a key role in forming a dimerization domain [22]. The active site histidine (H460 in human ACAT1) is located within TMD #7, while the other active site asparagine (N421 in human ACAT1) is located within the 4th large cytoplasmic loop [23, 24] (Fig.1). The helical coil rich domains present in TMD #7 and TMD #8 can be dissected into two distinct functional sides: one side is involved in substrate binding and catalysis, while the other side is involved in subunit interaction [25]. Key residues in TMD #7 in ACAT1 are conserved in ACAT2, and DGAT1, while key residues in TMD #8 in ACAT1 are conserved in ACAT2, but not in DGAT1. In addition, within the C-terminal half of ACAT1, numerous residues conserved with those of ACAT2 are present; the functions of these conserved residues are largely unknown at present. Of note, within the coding region of human ACAT1 cDNA, a single nucleotide polymorphism (SNP) exists at residue 526: the codon is either CAG for glutamine (Q), or CGG for arginine (R). Based on the various sources of the database available online (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=13306731), about 13% Japanese and 14% Chinese populations are homozygous with R526, while about 75 to 88% European and 100% African American are homozygous with Q526. The significance of this SNP at the biochemical level remains unknown. Q/R526 is located in the C-terminal loop of ACAT1. Within this region, we had previously shown that C528 and C546 form a disulfide bond; the lack of disulfide bond formation significantly decreases ACAT1 protein content [26]. Here we perform site-specific mutagenesis experiments to analyze the role of individual residues in the C-terminal loop, including residue Q/R526. We also note that there are 8 prolines located within or nearby TMDs #2,3,4,5,6, and 8; 5 of them are conserved between ACAT1 and ACAT2, while the other 3 are conserved between ACAT1, ACAT2, and DGAT1 (Fig. 1). The functionality of these proline residues remained unknown. Prolines within the TMD can act as molecular hinges or swivels [27, 28]. Here we perform site-specific mutagenesis experiments to analyze the functions of the 8 prolines present in various TMDs. The results of our analyses reveal biochemical significance of the SNP at residue 526, and provide new information regarding the role of proline 347 near the 5th transmembrane domain in modulating enzyme catalysis.
Figure 1. The ACAT1 transmembrane topology model.
The C-terminal loop is marked with a red rectangle. Conserved proline residues within TMDs are shown in blue. Additional residues in blue are conserved between ACAT1 and ACAT2; residues in red are conserved between ACAT and diacylglycerol:acyltransferase 1 (DGAT1). Native cysteines are in orange; the active site His460 within TMD #7 is in red. Human SNP R526 in C-terminal loop is in green. Modified Adapted from [12].
Result and Discussion
SNP at residue 526 affects ACAT1 enzyme activity
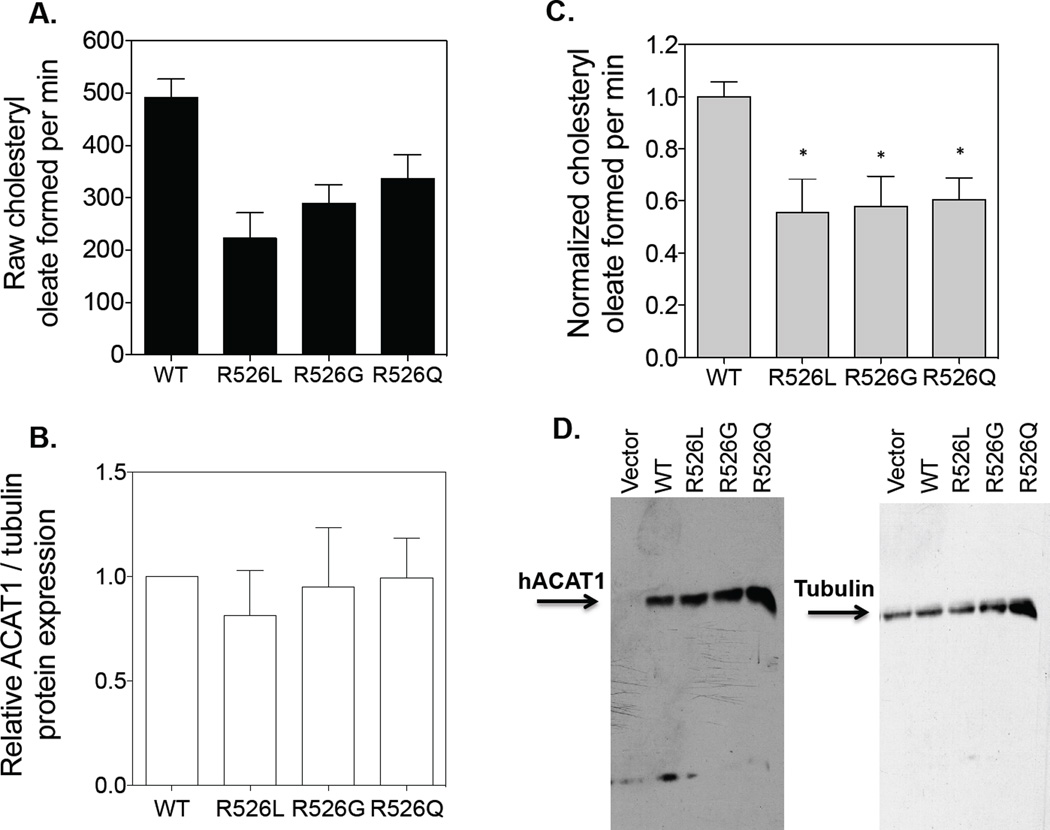
Ohta and colleagues [29] had previously reported the presence of the human ACAT1 SNP (missense variant, R526G) in a group of hyperlipidemia subjects; the R526G variant did not affect plasma concentrations of lipids or apolipoproteins. However, it was not known whether the R526G variation affects ACAT1 enzyme activity. To address this issue, we used the WT hACAT1 as the template and produced three mutant hACAT1 constructs by site-specific mutagenesis, with single mutation at residue 526: R526R (WT), R526Q, R526G, and R526L. These constructs were individually transiently transfected into ACAT deficient mutant CHO cells (AC29). The ACAT activity expressed by each of these constructs was monitored by standard procedures described in Methods. Fig.2A shows the amount of radioactive cholesteryl oleate formed by each construct. Fig. 2B shows the amount of hACAT1 protein expression of each construct normalized with tubulin expression. Fig. 2C shows the normalized ACAT1 enzyme activity (obtained by dividing values in A by values in B). Western blot analysis shown in Fig. 2D demonstrates the specificity of the ACAT1 antibodies used, the protein expression of hACAT1 of each construct as indicated, and the expression of tubulin used as the loading control. The results show that the enzyme activity of ACAT1 Q526 is less active than that of ACAT1 R526 by 40%. ACAT1 is a potential drug target for treating human diseases. Our result here suggests that in the future, the CAG/CGG polymorphism at ACAT1 residue 526 can be utilized to perform ACAT1 activity/human disease susceptibility studies.
Figure 2. Enzymatic activities of WT and mutant R526 hACAT1s.
The ACAT deficient mutant CHO cells (AC29) were transiently transfected with various hACAT1 plasmids as indicated. The procedures for transfection and for measuring ACAT1 enzyme activity in intact cells are described in Materials and Methods. For A, B, and C, results reported are means ± SEM and are averages of two separate experiments. Values in C were obtained from dividing values in A by the values in B with the WT hACAT1 activity set as 1.0. For D, the ACAT1 and tubulin protein expression by western blot analysis were from one experiment representative of two separate experiments. The 1st lane is the negative control with cells transfected with empty vector. The p values were obtained by comparing values of individual mutant hACAT1s vs value of WT hACAT1. * P<0.05.
Mutations within C-terminal loop affect ACAT1 protein content and/or ACAT1 enzyme activity
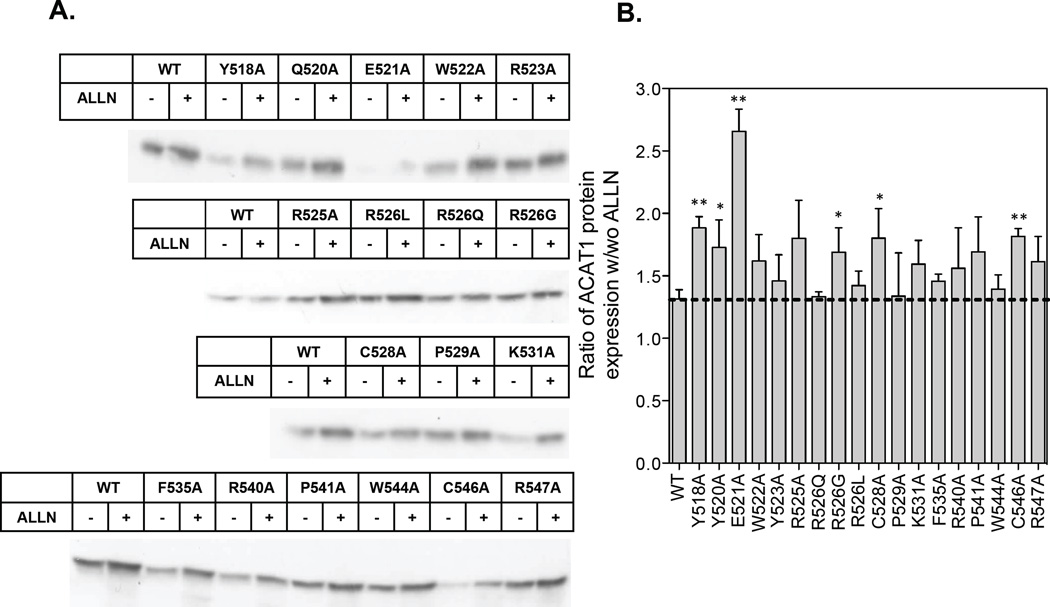
R526 is located in C-terminal loop of ACAT1. The result described above raises the possibility that certain residues within C-terminal loop may modulate ACAT1 enzyme activity and/or ACAT1 protein stability in intact cells. To test this possibility, we performed alanine scanning mutagenesis experiments aiming at 13 conserved amino acids (Y518, Q520, E521, W522, Y523, R525, R526, C528, P529, F535, P541, C546, and W544), and 3 non-conserved amino acids with positive charge side chain (K531, R540 and R547). After transfecting into mutant CHO cells lacking ACAT activity, we monitored ACAT1 protein content expressed by each mutant construct (Fig. 3). We found that mutation to Ala at several positions (i.e., Y518, Q520, E521, C528, and C546) significantly decrease ACAT1 protein content (Fig. 4A without ALLN treatment; ALLN, a proteasome specific inhibitor, stands for acetyl-leucyl-leucyl-norleucinal). These results suggest that certain residues at the C-terminal loop maybe involved in maintaining the structural integrity of ACAT1; mutation in any of these residues may cause instability/misfolding of the protein. Misfolded proteins at the ER often undergo proteasome-mediated degradation. We employed the proteasome specific inhibitor ALLN to test this possibility. We treated individually transfected cells with or without ALLN for 16 hr. Afterwards, cells were harvested for ACAT1 protein measurement by western blot analysis. The effect of ALLN is reported as the ratio of ACAT1 protein expression in cells with ALLN treatment versus that of cells without ALLN treatment. The results (Fig. 3 A,B) show that, in the WT hACAT1 group, the ratio is 1.3. In the first mutant group (Y518A, Q520A, E521A, R526G, C528A, C546A) (Fig. 3B), the value of this ratio is significantly higher than 1.3. In the second mutant group (W522A, R525A, K531A, R540A, P541A, and R547A) (Fig. 3B), the values of the ratios all had trends toward being higher than 1.3. These results suggest that mutations in these first two groups may cause significant structural alteration(s) and lead to destabilization of the ACAT1 protein in intact cells. In the third mutant group (W522A, Y523A, R525A, P529A, K531A, F535A, R540A, P541A, W544A, and R547A) (Fig. 3B), the ratios of ACAT1 protein expression w/wo ALLN were comparable to that of WT ACAT1 (1.3), suggesting that mutations in these residues do not cause significant alteration in structural integrity of the ACAT1 protein.
Figure 3. Effect of ALLN on hACAT1 protein expression levels.
The cell preparations prepared in the same manner as described in Fig. 2 were treated without or with ALLN to monitor ACAT protein expression by western analysis. For each well, the entire cell lysates with and without ALLN treatment were loaded. A. Relative WT or mutant hACAT1 protein expression levels with or without 100uM ALLN treatment for 16 hr. B. Quantification of results from A; shown as ratios of band intensity with ALLN versus band intensity without ALLN. Results reported are means ± SEM; for A, results are from one single experiment representative of three separate experiments. For B, results are averages of three separate experiments. The p values were obtained by comparing the ratio values of individual mutant hACAT1s protein expression vs the ratio value of WT hACAT1 protein expression. * P<0.05; ** P<0.01
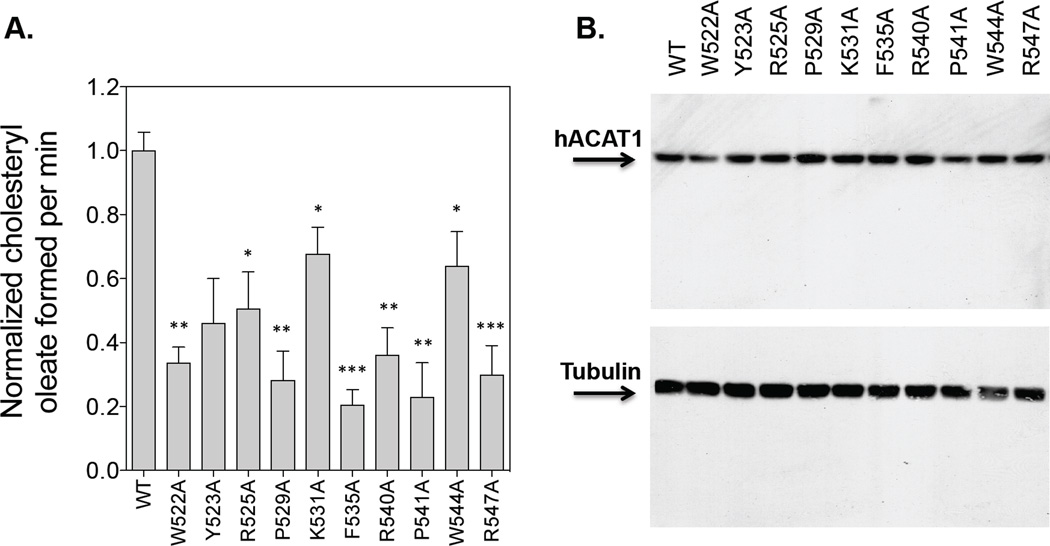
Figure 4. Enzyme activities of WT hACAT1 or hACAT1s with mutations in the C-terminal loop.
The hACAT1 is identified as a single band at 50 kDa. For A, the hACAT1 mutant activities are measured in the same manner as described in Fig. 2. The WT hACAT1 activity is set as 1.0. The results reported are means ± SEM and are average of two separate experiments. For B, the ACAT1 and tubulin protein expression by western blot analysis were from one single experiment representative of two separate experiments. The p values were obtained by comparing values of individual mutant hACAT1s vs value of WT hACAT1. * P<0.05; ** P<0.01; * P<0.001.
We next focused on testing the effects of the third mutant group on ACAT1 enzyme activity. After transfecting each mutant construct into mutant CHO cells lacking ACAT activity, we monitored ACAT enzyme activity and ACAT1 protein content expressed (Fig. 4A, B). Tubulin was used as the loading control. The results show that for each of these amino acids tested, mutation to alanine decreases enzyme activity by at least 35% (Fig. 4A). This analysis supports the interpretation that the main function of the C-terminal loop is to provide structural integrity of the ACAT1 polypeptide and ACAT1 enzyme activity in intact cells.
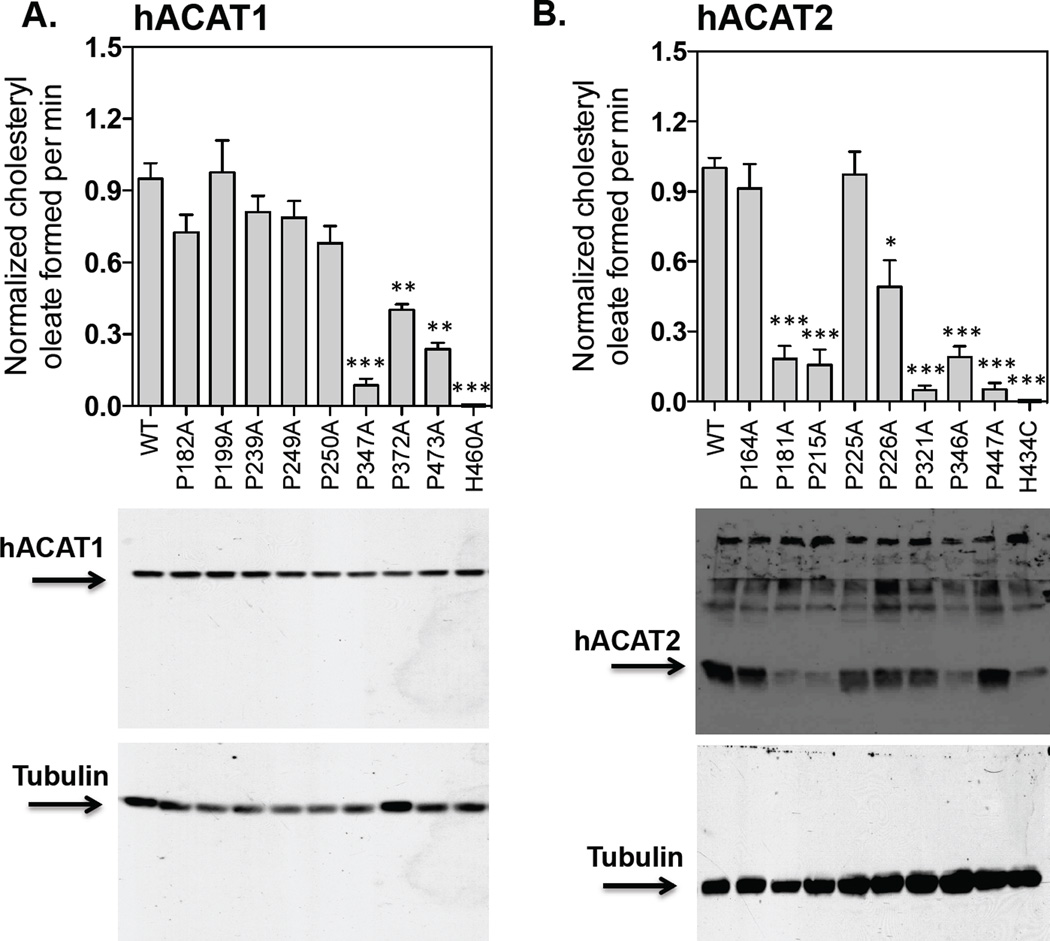
Mutations in conserved P347 in hACAT1 decreases ACAT1 enzyme activity without affecting ACAT1 protein content
Based on the ACAT1 membrane topography (Fig. 1), there are eight prolines within or near TMDs #2, 3, 4, 5, 6 and 8, of which five are conserved between ACAT1 and ACAT2, while the other three are conserved between ACAT1, ACAT2, and DGAT1. To probe their functionalities, for each proline, we mutated to alanine, and examined the effects of mutation in ACAT1 enzyme activity and ACAT1 protein content in intact cell; the amount of hACAT1 protein expression of each construct was normalized with tubulin expression (Fig. 5A). The results show that the P347A mutation results in a decrease in enzyme activity by about 95% without significantly affecting the ACAT1 protein content. The P372A and the P473A mutations also caused decreases in enzyme activity. Other P to A mutations did not significantly affect the ACAT1 enzyme activity. To test whether the effect of the P347A mutation on ACAT1 is conserved in ACAT2, we also tested the effect of the P to A mutation on each corresponding Ps in ACAT2. The amount of hACAT2 protein expression of each construct was normalized with tubulin expression. The result (Fig. 5B) shows that the P321A mutation in ACAT2, corresponding to the P347A mutation in ACAT1, also leads to decrease in enzyme activity by about 95%. The P181A, P215A, and P346A mutations cause decrease in both enzyme activity and in ACAT2 protein expression.
Figure 5. Protein expression and enzymatic activities of various hACAT1 or hACAT2 proline mutants within the TMDs.
The ACAT deficient mutant CHO cells (AC29) were transiently transfected with various hACAT1 plasmids (A) or various hACAT2 plasmids (B) as indicated. The procedures for transfection and for measuring ACAT enzyme activity in intact cells are described in Materials and Methods, as demonstrated in Fig. 2. By western analysis, hACAT1 is identified as a single band at 50 kDa; hACAT2 is identified as a single band at 46 kDa. The anti-ACAT1 antibodies used here were high-titer antibodies. Instead, the anti-ACAT2 antibodies used here were not high-titer antibodies; in addition to the ACAT2 protein band, these antibodies also detected several non-specific bands located at regions above the ACAT2 protein band. Values for the WT hACAT1 or WT hACAT2 activity were set 1.0. The western blot results are from one single experiment representative of two separate experiments. The enzyme activity data are reported as means ± SEM and are averages of two separate experiments. The p values were obtained by comparing values of individual mutant hACATs vs value of WT hACAT. ** P<0.01; *** P<0.001.
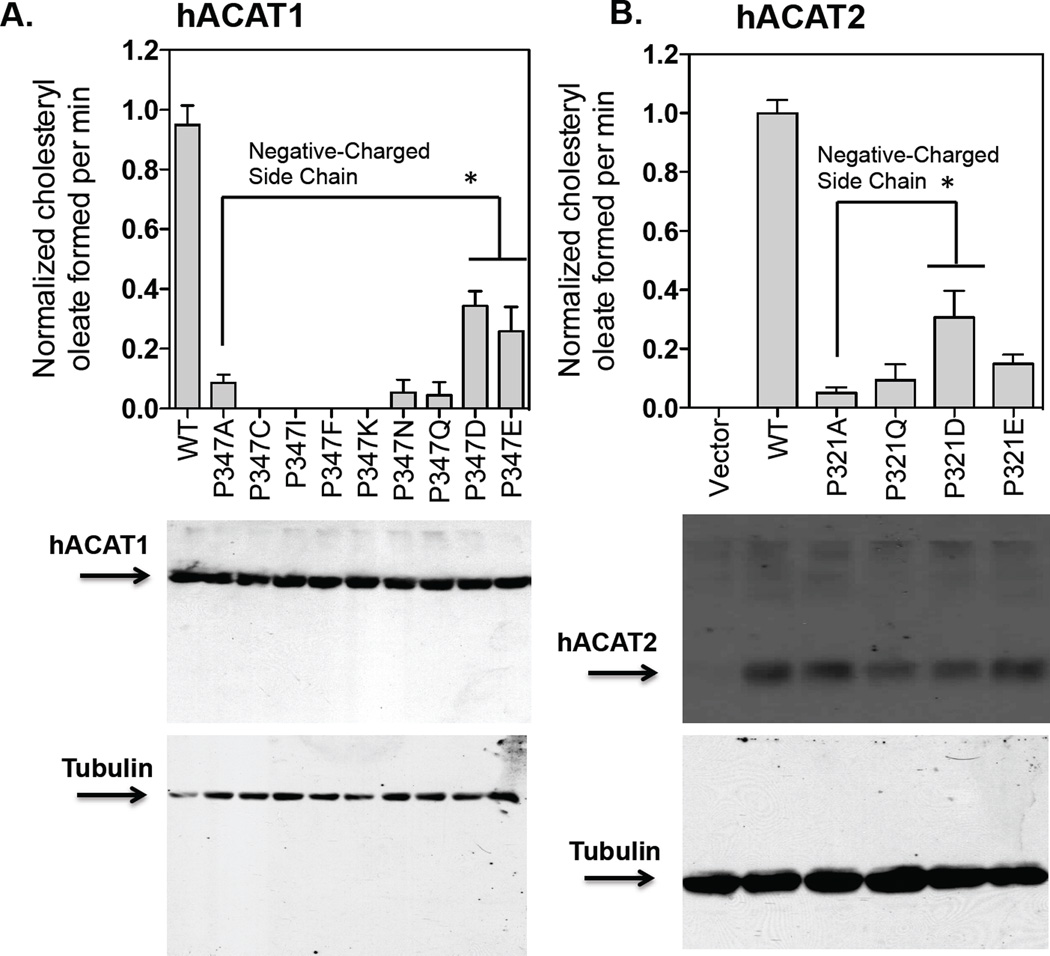
Mutating P347 in hACAT1 to D or to E restores partial enzyme activity
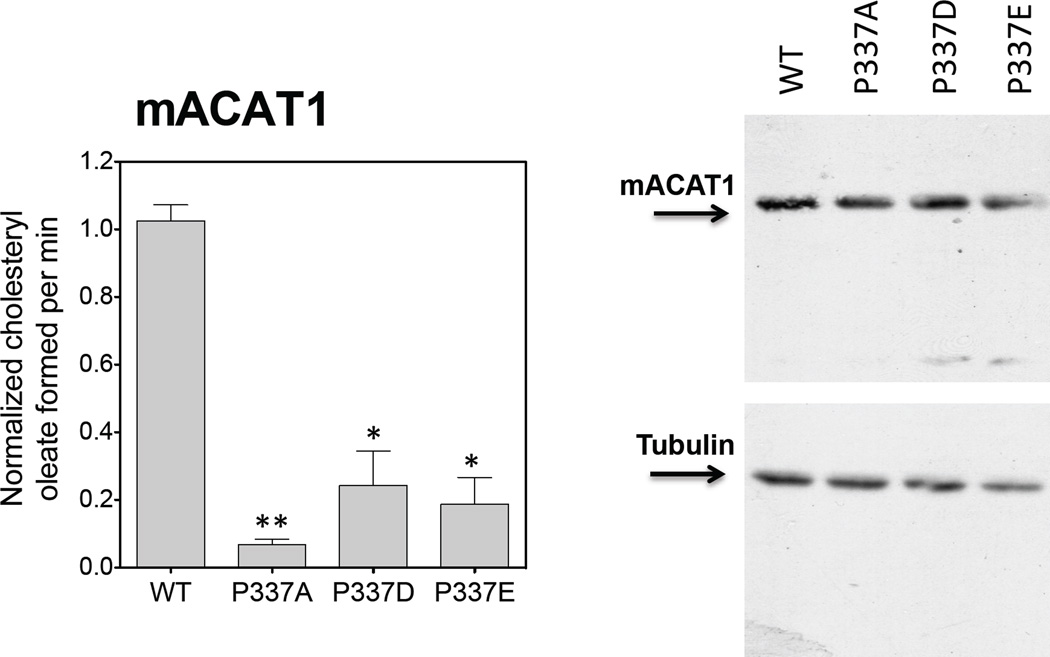
The results described above show that replacing P347 with A in ACAT1 significantly decreases its enzyme activity without significantly affecting the ACAT1 protein expression level. We wanted to test whether replacement of P347 with other residues may provide partial enzyme activity, and performed additional substitution mutagenesis experiments to address this issue. The result (Fig. 6A) shows that substituting P347 with any of several residues as indicated did not significantly affect the ACAT1 protein expression level; however, only substituting with either glutamate (D) or aspartate (E), but not with any other residues as indicated, could cause partial regain in enzyme activity. This result suggests that at position 347, replacing P with residue that contains negative charged side chain can provide partial enzyme activity. To validate this conclusion, we performed similar substitution experiments using hACAT2 as the template, and showed that consistent with the results shown in ACAT1, the P321D mutant clearly provided partial enzyme activity in ACAT2 (Fig. 6B). In addition, we used the mouse ACAT1 as the template, performed similar substitution experiment at the equivalent P position (P337 in mouse ACAT1), and obtained a result (Fig. 7) very similar to those obtained by using hACAT1 as the template. Taken together, these results show that P347 in hACAT1 plays an important role to modulate enzyme activity.
Figure 6. Relative protein expression and enzyme activity of P347 mutants in hACAT1, and P321 mutants in hACAT2.
The experiments were carried out in the same manner as described in Fig. 5 with P347 mutants shown in A. and with P321 mutants as shown in B. The hACAT1 and hACAT2 western blots as shown are from a single experiment representative of two separate experiments. For enzyme activity measurement, results reported as means ± SEM and are averages of two separate experiments. The p values were obtained by comparing values of individual mutant hACAT1 or hACAT2 vs value of hACAT1(P347A) or value of hACAT2(P321A). * P<0.05.
Figure 7. Relative protein expressions and enzyme activities of various mouse ACAT1 (mACAT1) P337 mutants.
The experiments were carried out in the same manner as described in Fig. 5. The mACAT1 western blots as shown are from one single experiment representative of two separate experiments. For enzyme activity measurement, results reported are means ± SEM and are averages of two separate experiments. The p values were obtained by comparing values of individual mutant mACAT1s vs value of WT mACAT1. * P<0.05; ** P<0.01
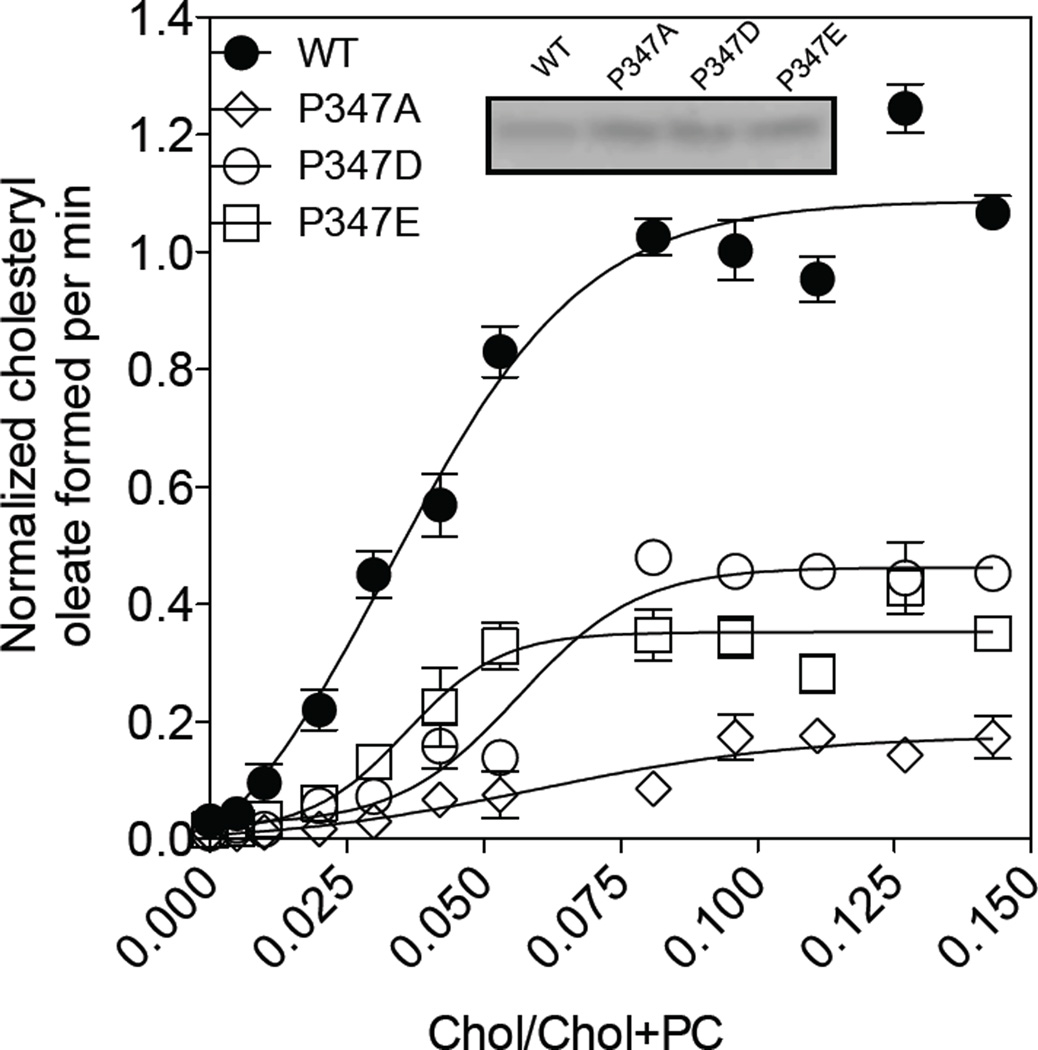
Both ACAT1 and ACAT2 are allosteric enzymes regulated by their own substrate cholesterol. When assayed in mixed micelles, both enzymes respond to increasing concentrations of cholesterol in sigmoidal manner. Since both the P347D mutant and the P347E mutant exhibits partial enzyme activity, we ask whether the D or E mutation affects the ability of the enzyme to respond to cholesterol. To address this issue, we expressed the WT enzyme or the individual mutant P347 enzymes in CHO cells, prepared membrane fractions from the transfected cells, and used them as the enzyme source to perform cholesterol substrate saturation curve in mixed micelles. The result (Fig. 8) shows that similar to the WT enzyme, both the 347E enzyme and the 347D enzyme responds to cholesterol in sigmoidal manner, suggesting that the D mutation or the E mutation does not alter the ability of the enzyme to respond to cholesterol in significant manner. The result also shows that when compared to the WT enzyme, the P347A mutant enzyme contains less than ~10% of the activity, while both the P347E mutant and the P347D mutant contain 30 to 40% of the activity of the normal enzyme. This result recapitulates what we observed when the enzyme activities were measured in intact cells, supporting the conclusion that P347 plays important role(s) in modulating the ACAT enzyme activity. It is possible that P347 located at the luminal end of TMD #5 maybe in close proximity with key amino acids located in TMDs #7 and #8 to facilitate enzyme catalysis. It is also possible that P347 plays important roles by maintaining optimal configuration/conformation during substrate binding and/or product release; etc.
Figure 8. Cholesterol substrate saturation curves of various hACAT1 P347 mutants.
The ACAT enzyme assay in vitro, described in Materials and methods, was performed in mixed micelles with varying concentrations of cholesterol as indicated. Activity of WT hACAT1 was set at 1.0. Western blot result as shown is from one single experiment representative of three separate experiments; the activity data as shown are averages of 3 separate experiments.
Conclusion
The current results reveal that the A/G single nucleotide polymorphism present within hACAT1 bears functional significance at the biochemical level: ACAT1 with Q526 is catalytically less active than ACAT1 R526 by 40%. This result raises the possibility that the CAG/CGG polymorphism present in ACAT1 can be utilized to perform ACAT1 SNP/disease susceptibility studies in humans. It will also be interesting to express normal hACAT1 (with R526) in mouse that lacks endogenous Acat1, and monitor the effect of replacing R 526 with Q in mouse models for atherosclerosis or for Alzheimer’s disease. We also show that the main function of the C-terminal loop is to maintain proper ACAT1 protein configuration. This work also shows that, in addition to H460 located within TMD #7 and N421 located in the 4th hydrophilic loop, P347, located near the luminal end of TMD #5, also participates in enzyme catalysis. The P337A mutation in mouse still possesses 10% residual ACAT1 enzyme activity. In the future, one can generate a P337A knock-in mouse line and use this line to evaluate the effects of partial ACAT1 inhibition on mouse models for atherosclerosis and for Alzheimer’s disease.
Materials and Methods
Materials
FuGENE 6 Transfection Reagent was from Roche Molecular Biology, Nutley, NJ, USA. [9,10-3H]Oleic acid was from Amersham Biosciences, Piscataway, NJ, USA. The rabbit polyclonal antibodies (DM10) generated against the N-terminal fragment (1-131) of hACAT1 were described previously [30]. β-tubulin antibody (2G7D4) was from GenScript (A01717). Goat anti-rabbit IgG(L+H)-HRP conjugate and Goat anti-mouse IgG(L+H)-HRP conjugate were from Bio-Rad, Hercules, CA, USA. The SuperSignal West Pico Chemiluminescent Substrate was from Pierce, Rockford, Ill, USA. CHAPS, taurocholate, oleoyl CoA, egg PC, cholesteryl oleate, cholesterol, fatty acid-free bovine serum albumin, the proteosome specific inhibitor ALLN, and the protease inhibitor cocktails were all from Sigma-Aldrich, St. Louis, Mo, USA. [3H] oleoyl-coenzyme A was synthesized as described [31]
Cell culture
The CHO cells were cultured in F-12/Dulbecco's modified Eagle's medium (50:50) supplemented with 10% fetal bovine serum in a 5% CO2 incubator at 37 °C. The ACAT1-deficient CHO cell line AC29 [32] was used to express the Nterminal His6-tagged hACAT1(C92A)(His-hACAT1(C92A)) or its various engineered mutants as described. For transfections, the AC29 cells were cultured in 6-well plates to 70–80% confluence and transfected with 2 μg of pcDNA3 vectors encoding His-hACAT1 or its mutants, using FuGENE 6 transfection Reagent according to the manufacturer's protocols. On the second day, the cells were trypsinized, divided equally into three wells, and grown for 1 day in G-418 containing medium for intact cell ACAT enzyme activity and for protein expression studies as described in [22, 25].
Recombinant DNA technology
All of the ACAT1 mutant constructs were prepared by using the QuikChange mutagenesis kit from Stratagene (Grand Island, NY, USA) according to the manufacturer's manual. The His-hACAT1(C92A) [26] was used as the mutagenesis template, and all the mutations were confirmed by DNA sequencing.
ACAT activity assay in transiently transfected cells
This method measures the rate of [3H]cholesteryl oleate synthesis in intact cells [33]. The transiently transfected AC29 cells were cultured in 6-well plates at 37 °C as described above. 20 μl of 10 mM [3H]oleate in 10% bovine serum albumin was added to the media at 37 °C for 30 min. The amount of [3H]cholesteryl oleate formed in intact cells from [3H]oleate was quantitated by scintillation counting after lipid extraction and separation by thin-layer chromatography. The ACAT enzyme activity of AC29 cells expressing the mutant H460A hACAT1, or the mutant H434C hACAT2, which are devoid of ACAT enzyme activity are used as the blank values [26, 34].
ACAT protein content analysis and determination of normalized hACAT enzyme activity in transiently transfected cells
The transiently transfected cells in 6-well plates were washed with 2 ml of phosphate-buffered saline and lysed by 120 μl of 10% SDS per sample. The protein concentrations of the cell lysates were quantified by using Lowry method. Aliquots of lysates that contained 50 μg protein per sample were transferred to Eppendorf tubes, and 30 μl/tube of SDS-PAGE Loading Buffer (10% SDS, 20% glycerol, 0.05% bromophenol blue, 50 mm Tris-Cl, pH 6.8) was added. The samples were mixed well by vigorous vortexing, and loaded onto a 10% SDS-gel. After electrophoresis, the proteins were transferred to a PVDF membrane, and the ACAT1 or ACAT2 protein bands were visualized by western blot using DM10 or DM 54 as the primary antibodies. For each lane, the ACAT1 or ACAT2 protein expression was normalized by the tubulin expression in the same blot. The normalized ACAT1 or ACAT2 enzyme activity was obtained by dividing the enzyme activity present in each well by the normalized ACAT1 or ACAT2 protein content. The normalized WT ACAT1 or WTACAT2 enzyme activity is set as 1.0.
ACAT enzyme assay in mixed micelles
The hACAT1 (C92A) and other mutant plasmids were individually transfected in AC29 cells in one 150cm2 dish of each genotype when cells reach to 70–80% confluence. On the second day, the cells were trypsinized, divided equally into three 150cm2 plates, and grown for 2 days in G-418 containing medium. Afterwards, cells were harvested by hypotonic shock and scraping [35]; the protein concentration of the cell homogenates was kept at 2–4 mg/ml in Buffer A (50 mM Tris, 1 mM EDTA at pH 7.8 with protease inhibitors). In order to prepare the subcellular components from tissue culture cells, whole cell extract was homogenized with a stainless steel tissue grinder (from Wheaton: Dura-Grind). After centrifugation (800 × g, 5 min) to remove the unbroken cells and nuclei, the whole cell homogenates underwent ultracentrifugation (100,000 × g, 30 min) at 4 °C. After ultracentrifugation the pellets were gently re-suspended in the 300 uL cold buffer containing a final concentration of 1 M KCl and 2.5% CHAPS in buffer A, with total protein concentration of ~4–6mg/mL, followed by gently stirred for 5 mins under ice-cold water bath to solubilize the enzyme. To assay the enzyme in bile salt/cholesterol/PC mixed micelles, the membrane prep (20 μl) was diluted into 100 μl of bile salt/cholesterol/PC mixed micelles. The bile salt/cholesterol/PC mixed micelles were prepared as follows: the cholesterol/PC mixture was prepared as described previously [36] and lyophilized to remove residual organic solvents. After lyophilization, sodium taurocholate in Buffer A was added to reach a final concentration of 10 mg/ml. The mixture was purged with nitrogen, followed by sonication until this mixture was clear. ACAT assay was initiated by adding 20 μl of solution containing 10 nmol of [3H]oleoyl-CoA and 10 nmol of fatty acid-free bovine serum albumin to the enzyme-vesicle mixture or the enzyme-mixed micelle mixture prepared on ice. The reaction mixture was incubated at 37°C for up to 30 min, during which the activity was found to be linear. The formation of [3H]cholesteryl oleate was quantitated by scintillation counting after lipid extraction and separation by thin-layer chromatography. 5uL of membrane prep of each genotype was used to measure ACAT1 protein expression by western blotting as described earlier.
Statistical analysis
Results are reported as mean ± SEM. Two-tailed parametric student’s test was used to evaluate the statistical significance among various study groups. Analysis was performed and plotted by using GraphPad Prism 5.0. The difference between two sets of values was considered significant when the p value was < 0.05 (*p<0.05, **p<0.01, ***p<0.001).
Acknowledgements
We thank Dr. John Hwa for initiating the idea of examining SNP in human ACAT1 coding region. We also thank Dr. Dean Madden at Geisel School of Medicine at Dartmouth, and thank Dr. Yasuomi Urano and other members of the Chang lab for stimulating discussions. This work is supported by NIH grant HL 060306 to TYC and CCYC.
Abbreviations
- ACAT
Acyl-Coenzyme A:cholesterol Acyltransferases
- TMD
transmembrane domains
- AC29
the ACAT deficient mutant CHO cells
- CHO
Chinese hamster ovary
- ER
endoplasmic reticulum
References
- 1.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Warner GJ, Stoudt G, Bamberger M, Johnson WJ, Rothblat GH. Cell toxicity induced by inhibition of acyl coenzyme A:cholesterol acyltransferase and accumulation of unesterified cholesterol. J Biol Chem. 1995;270:5772–5778. doi: 10.1074/jbc.270.11.5772. [DOI] [PubMed] [Google Scholar]
- 3.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. The Journal of clinical investigation. 2002;110:905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang TY, Chang CC, Cheng D. Acyl-coenzyme A:cholesterol acyltransferase. Annual review of biochemistry. 1997;66:613–638. doi: 10.1146/annurev.biochem.66.1.613. [DOI] [PubMed] [Google Scholar]
- 5.Chang CC, Huh HY, Cadigan KM, Chang TY. Molecular cloning and functional expression of human acyl-coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells. The Journal of biological chemistry. 1993;268:20747–20755. [PubMed] [Google Scholar]
- 6.Buhman KF, Accad M, Farese RV. Mammalian acyl- CoA:cholesterol acyltransferases. Biochimica et biophysica acta. 2000;1529:142–154. doi: 10.1016/s1388-1981(00)00144-x. [DOI] [PubMed] [Google Scholar]
- 7.Oelkers P, Behari A, Cromley D, Billheimer JT, Sturley SL. Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase-related enzymes. The Journal of biological chemistry. 1998;273:26765–26771. doi: 10.1074/jbc.273.41.26765. [DOI] [PubMed] [Google Scholar]
- 8.Anderson RA, Joyce C, Davis M, Reagan JW, Clark M, Shelness GS, Rudel LL. Identification of a form of acyl-CoA:cholesterol acyltransferase specific to liver and intestine in nonhuman primates. The Journal of biological chemistry. 1998;273:26747–26754. doi: 10.1074/jbc.273.41.26747. [DOI] [PubMed] [Google Scholar]
- 9.Brown AJ, Jessup W. Oxysterols: Sources, cellular storage and metabolism, and new insights into their roles in cholesterol homeostasis. Mol Aspects Med. 2009;30:111–122. doi: 10.1016/j.mam.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Bolick DT, Srinivasan S, Kim KW, Hatley ME, Clemens JJ, Whetzel A, Ferger N, Macdonald TL, Davis MD, Tsao PS, Lynch KR, Hedrick CC. Sphingosine-1-phosphate prevents tumor necrosis factor-{alpha}-mediated monocyte adhesion to aortic endothelium in mice. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:976–981. doi: 10.1161/01.ATV.0000162171.30089.f6. [DOI] [PubMed] [Google Scholar]
- 11.Maue RA, Burgess RW, Wang B, Wooley CM, Seburn KL, Vanier MT, Rogers MA, Chang CC, Chang TY, Harris BT, Graber DJ, Penatti CA, Porter DM, Szwergold BS, Henderson LP, Totenhagen JW, Trouard TP, Borbon IA, Erickson RP. A novel mouse model of Niemann-Pick type C disease carrying a D1005G-Npc1 mutation comparable to commonly observed human mutations. Human molecular genetics. 2012;21:730–750. doi: 10.1093/hmg/ddr505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang TY, Li BL, Chang CC, Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. American journal of physiology Endocrinology and metabolism. 2009;297:E1–E9. doi: 10.1152/ajpendo.90926.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryleva EY, Rogers MA, Chang CC, Buen F, Harris BT, Rousselet E, Seidah NG, Oddo S, LaFerla FM, Spencer TA, Hickey WF, Chang TY. ACAT1 gene ablation increases 24(S)- hydroxycholesterol content in the brain and ameliorates amyloid pathology in mice with AD. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3081–3086. doi: 10.1073/pnas.0913828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy SR, Chang CC, Dogbevia G, Bryleva EY, Bowen Z, Hasan MT, Chang TY. Acat1 Knockdown Gene Therapy Decreases Amyloid-beta in a Mouse Model of Alzheimer's Disease. Molecular therapy : the journal of the American Society of Gene Therapy. 2013 doi: 10.1038/mt.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huttunen HJ, Havas D, Peach C, Barren C, Duller S, Xia W, Frosch MP, Hutter-Paier B, Windisch M, Kovacs DM. The acyl-coenzyme A: cholesterol acyltransferase inhibitor CI-1011 reverses diffuse brain amyloid pathology in aged amyloid precursor protein transgenic mice. Journal of neuropathology and experimental neurology. 2010;69:777–788. doi: 10.1097/NEN.0b013e3181e77ed9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang LH, Gui J, Artinger E, Craig R, Berwin BL, Ernst PA, Chang CCY, Chang TY. Acat1 gene ablation in mice increases hematopoietic progenitor cell proliferation in bone marrow and causes leukocytosis. Arterioscler Thromb Vasc Biol. 2013;33 doi: 10.1161/ATVBAHA.112.301080. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CC, Sun J, Chang TY. Membrane bound Oacyltransferases (MBOAT) Frontiers in Biology. 2011;6:177–182. [Google Scholar]
- 18.Chen J, Zhao XN, Yang L, Hu GJ, Lu M, Xiong Y, Yang XY, Chang CC, Song BL, Chang TY, Li BL. RNA secondary structures located in the interchromosomal region of human ACAT1 chimeric mRNA are required to produce the 56-kDa isoform. Cell research. 2008;18:921–936. doi: 10.1038/cr.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. The Journal of biological chemistry. 2008;283:22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shindou H, Eto M, Morimoto R, Shimizu T. Identification of membrane O-acyltransferase family motifs. Biochemical and biophysical research communications. 2009;383:320–325. doi: 10.1016/j.bbrc.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Yu C, Chen J, Lin S, Liu J, Chang CC, Chang TY. Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro. The Journal of biological chemistry. 1999;274:36139–36145. doi: 10.1074/jbc.274.51.36139. [DOI] [PubMed] [Google Scholar]
- 22.Yu C, Zhang Y, Lu X, Chen J, Chang CC, Chang TY. Role of the N-terminal hydrophilic domain of acyl-coenzyme A:cholesterol acyltransferase 1 on the enzyme's quaternary structure and catalytic efficiency. Biochemistry. 2002;41:3762–3769. doi: 10.1021/bi0120188. [DOI] [PubMed] [Google Scholar]
- 23.Guo ZY, Lin S, Heinen JA, Chang CC, Chang TY. The active site His-460 of human acyl-coenzyme A:cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain. The Journal of biological chemistry. 2005;280:37814–37826. doi: 10.1074/jbc.M508384200. [DOI] [PubMed] [Google Scholar]
- 24.Guo Z-Y, Lin S, Heinen JA, Chang CCY, Chang T-Y. The Active Site His-460 of Human Acyl-coenzyme A:Cholesterol Acyltransferase 1 Resides in a Hitherto Undisclosed Transmembrane Domain. Journal of Biological Chemistry. 2005;280:37814–37826. doi: 10.1074/jbc.M508384200. [DOI] [PubMed] [Google Scholar]
- 25.Guo ZY, Chang CC, Chang TY. Functionality of the seventh and eighth transmembrane domains of acyl-coenzyme A:cholesterol acyltransferase 1. Biochemistry. 2007;46:10063–10071. doi: 10.1021/bi7011367. [DOI] [PubMed] [Google Scholar]
- 26.Guo ZY, Chang CC, Lu X, Chen J, Li BL, Chang TY. The disulfide linkage and the free sulfhydryl accessibility of acyl-coenzyme A:cholesterol acyltransferase 1 as studied by using mPEG5000-maleimide. Biochemistry. 2005;44:6537–6546. doi: 10.1021/bi047409b. [DOI] [PubMed] [Google Scholar]
- 27.Stitham J, Martin KA, Hwa J. The critical role of transmembrane prolines in human prostacyclin receptor activation. Molecular pharmacology. 2002;61:1202–1210. doi: 10.1124/mol.61.5.1202. [DOI] [PubMed] [Google Scholar]
- 28.Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 29.Ohta T, Takata K, Katsuren K, Fukuyama S. The influence of the acyl-CoA:cholesterol acyltransferase-1 gene (-77G-->A) polymorphisms on plasma lipid and apolipoprotein levels in normolipidemic and hyperlipidemic subjects. Biochimica et biophysica acta. 2004;1682:56–62. doi: 10.1016/j.bbalip.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Chang CC, Chen J, Thomas MA, Cheng D, Del Priore VA, Newton RS, Pape ME, Chang TY. Regulation and immunolocalization of acyl-coenzyme A: cholesterol acyltransferase in mammalian cells as studied with specific antibodies. The Journal of biological chemistry. 1995;270:29532–29540. doi: 10.1074/jbc.270.49.29532. [DOI] [PubMed] [Google Scholar]
- 31.Bishop JE, Hajra AK. A method for the chemical synthesis of 14C-labeled fatty acyl coenzyme A's of high specific activity. Anal Biochem. 1980;106:344–350. doi: 10.1016/0003-2697(80)90531-x. [DOI] [PubMed] [Google Scholar]
- 32.Cadigan KM, Chang TY. A simple method for reconstitution of CHO cell and human fibroblast acyl coenzyme A: cholesterol acyltransferase activity into liposomes. J Lipid Res. 1988;29:1683–1692. [PubMed] [Google Scholar]
- 33.Chang CC, Doolittle GM, Chang TY. Cycloheximide sensitivity in regulation of acyl coenzyme A:cholesterol acyltransferase activity in Chinese hamster ovary cells. 1. Effect of exogenous sterols. Biochemistry. 1986;25:1693–1699. doi: 10.1021/bi00355a038. [DOI] [PubMed] [Google Scholar]
- 34.Lin S, Lu X, Chang CC, Chang TY. Human acyl-coenzyme A:cholesterol acyltransferase expressed in chinese hamster ovary cells: membrane topology and active site location. Molecular biology of the cell. 2003;14:2447–2460. doi: 10.1091/mbc.E02-11-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang TY, Limanek JS, Chang CC. Evidence indicating that inactivation of 3-hydroxy-3-methylglutaryl coenzyme A reductase by low density lipoprotein or by 25-hydroxycholesterol requires mediator protein(s) with rapid turnover rate. The Journal of biological chemistry. 1981;256:6174–6180. [PubMed] [Google Scholar]
- 36.Chang CC, Lee CY, Chang ET, Cruz JC, Levesque MC, Chang TY. Recombinant acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) purified to essential homogeneity utilizes cholesterol in mixed micelles or in vesicles in a highly cooperative manner. The Journal of biological chemistry. 1998;273:35132–35141. doi: 10.1074/jbc.273.52.35132. [DOI] [PubMed] [Google Scholar]