Abstract
Nucleoside analogues have long been recognized as prospects for the discovery of direct acting antivirals (DAAs) to treat hepatitis C virus because they have generally exhibited cross-genotype activity and a high barrier to resistance. C-Nucleosides have the potential for improved metabolism and pharmacokinetic properties over their N-nucleoside counterparts due to the presence of a strong carbon–carbon glycosidic bond and a non-natural heterocyclic base. Three 2′CMe-C-adenosine analogues and two 2′CMe-guanosine analogues were synthesized and evaluated for their anti-HCV efficacy. The nucleotide triphosphates of four of these analogues were found to inhibit the NS5B polymerase, and adenosine analogue 1 was discovered to have excellent pharmacokinetic properties demonstrating the potential of this drug class.
Keywords: C-Nucleoside, HCV, NS5B polymerase
Hepatitis C virus (HCV) is a major cause of chronic viral hepatitis that often leads to liver cirrhosis and hepatocellular carcinoma.1,2 In 2011 the protease inhibitors boceprevir and telaprevir became available to treat HCV infection with genotype 1 in combination with ribavirin and pegylated interferon.3,4 The recent approval of two new direct acting antivirals (DAAs), simeprevir and sofosbuvir,5 will significantly enhance the available armory, but there remains a need for new DAAs for use in safe and effective combination therapies.
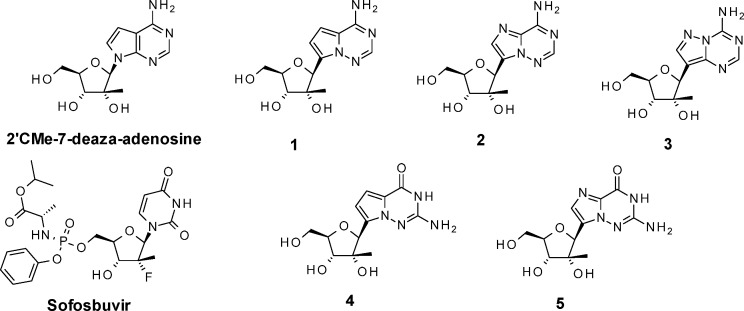
Nucleoside and nucleotide inhibitors of HCV (NIs) are a class of DAA that tend to demonstrate broad activity across HCV genotypes and a high barrier to the emergence of viral resistance. Structural modifications have been made to both the carbohydrate (“sugar”) and heterocyclic base components of natural nucleosides to develop potent and selective antiviral analogues.6 The early promise of 2′CMe-7-deaza-adenosine (Figure 1) and, more recently, the excellent clinical success of sofosbuvir have validated the use of NIs as anti-HCV DAAs.5,7,8
Figure 1.
Structures of sofosbuvir, 2′CMe-7-deaza-adenosine, and target C-nucleoside analogues.
Structural analogues that have attracted relatively little attention to date for application to HCV are C-nucleosides,9−14 which comprise a sugar moiety and non-natural heterocylic base connected by a carbon–carbon bond. This strong glycosidic bond makes C-nucleosides more resistant to enzymatic and hydrolytic cleavage than their N-nucleoside counterparts. This feature, combined with the broad structural variation possible in the heterocyclic base, offers the potential for improved drug metabolism and pharmacokinetic properties. As part of an effort to investigate new nucleoside agents for HCV, C-nucleoside adenosine analogues 1–3 and guanosine analogues 4 and 5 were targeted. Reported herein are the synthesis and biological characterization of these compounds (Figure 1).
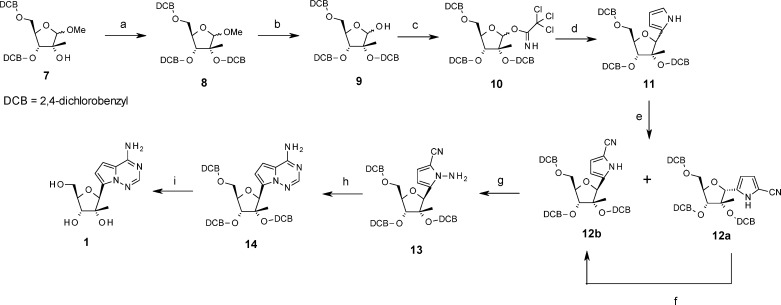
Pyrrolo[2,1-f][1,2,4]triazine-4-amine adenosine analogue, 1, was first synthesized using the linear route shown in Scheme 1. This is a highly modified approach to that used by Patil et al., for the synthesis of the unsubstituted ribo-analogue (4-aza-7,9-dideazaadenosine).15 The 3,5-bis-dichlorobenzyl protected 2′CMe-riboside 7 was protected at the 2′-position to afford 8, then hydrolyzed under acidic conditions to afford 9. The key glycosylation reaction was performed using a procedure adapted from a method reported for the reaction of pyrroles with O-glycosylimidates.16 Riboside 9 was converted to the activated trichloroacetimidate 10. Slow addition of pyrrole to 10 at low temperature in the presence of BF3·Et2O afforded an anomeric mixture of the pyrrolo-nucleoside, 11. Modest improvement in β-selectivity was achieved at higher temperature (−50 °C, α/β = 1:2, 60% yield), but at the cost of yield (−78 °C, α/β = 1:1, 80% yield). Because pyrrolo-nucleoside 11 decomposed slowly on standing, it was immediately converted to the more stable pyrrolonitrile nucleoside 12 by reaction with chlorosulfonyl isocyanate in DMF. The α/β mixture was separated at this stage by flash chromatography, and more of the desired β-anomer (12b) recovered after BF3·Et2O catalyzed anomerization of the purified α-anomer (12a). The β-stereochemistry was established at this stage by the observation of strong NOE between the H4′ and the H1′ protons. Electrophilic amination of the pyrrole nitrogen, ring closure with formamidine actetate, followed by buffered hydrogenolysis afforded the free adenosine analogue 1.
Scheme 1. Synthesis of 1.
Reagents and conditions: (a) NaH, 2,4-dichlorobenzyl chloride; (b) TFA/water 9:1; (c) Cl3CCN, Cs2CO3; (d) (i) 4 Å mol. sieves, DCM, 2 h, rt, (ii) pyrrole, BF3·OEt2, −50 °C (60% yield, β/α 2:1) or −78 °C (80% yield, β/α 1:1), (iii) NH3 in MeOH, −78 °C; (e) ClSO2NCO, CH3CN/DMF, 0 °C; (f) BF3·OEt2, DCM, reflux; (g) (i) NaH, THF, 0 °C, (ii) Ph2P(O)ONH2, THF, 0 °C; (h) formamidine acetate, DMA, 140 °C, 1–2 h; (i) H2, Pd–C (10%), 45 °C, 18 h, NaOAc, MeOH, HOAc.
An analogous linear route toward the pyrrolo[2,1-f][1,2,4]triazine-4-amine guanosine analogue 4 was attempted. Evidence was obtained that this may be achieved via hydrolysis of the 2,6-diamino analogue of 1 under strongly basic conditions at high temperature. However, this approach was abandoned in favor of the convergent synthesis described below.
Imidazo[2,1-f][1,2,4]triazine-4-amine adenosine analogue, 2, was synthesized using a convergent approach, which utilized the direct coupling of a bicyclic heterocycle, as shown in Scheme 2. After selective bromination of 15, the bromo-base 16 was coupled with ribolactone 17. Amine displacement of the thiomethyl group followed by selective lactol reduction yielded the desired β-nucleoside 20 and subsequently the free nucleoside 2 after hydrogenolysis. A similar convergent synthesis of the pyrrolo analogue 1 was developed for large scale synthesis. Convergent methods for 1 and 2 have also been reported by others since we completed this work.12
Scheme 2. Convergent Synthesis of A-Analogue 2.
Reagents and conditions: (a) NBS, DMF, 86 °C, 1 h; (b)(i) nBuLi, −78 °C, THF, (ii) 17, −78 °C; (c) NH3 in MeOH, RT to reflux; (d) BF3·OEt2, Et3SiH, DCM, −78 °C to RT, 3 h; (e) H2, Pd–C (10%), 60 °C, 18 h, NaOAc, MeOH/DCM (9:1), AcOH.
Guanosine analogues 4 and 5 were prepared using an analogous approach, as shown in Scheme 3. Compounds 21 and 26 were chosen as appropriate starting heterocycles. Under standard conditions for Boc-protection, N-Boc protection was achieved for both starting materials, together with tBu-protection of the 4-oxygen, a surprising transformation that has been observed for other guanine analogues.17 After selective bromination, the direct glycosylation of the bromo-bases 23 and 28 with the lactone 17 was achieved at very low temperature (−100 °C). It was noted that the anions of these guanosine base analogues were much less stable than the anions of the corresponding adenosine base analogues. The coupling thus required careful, slow addition of the lactone to the organolithium base to maintain a low temperature and minimize anion quenching. The protecting groups on the base were removed in the subsequent stereoselective lactol reduction to afford DCB-protected β-nucleosides 25 and 30. The β-stereochemistry of 25 was established at this stage by the observation of an interaction between H1′ and H4′ in the NOESY spectrum. Such unambiguous signals were not observed for 30; so, the formamidine analogue 31 was prepared under mild conditions. The observation of a strong interaction between H1′ and H4′ in the NOESY spectrum of this compound provided evidence of β-stereochemistry. The free guanosine nucleoside analogues 4 and 5 were obtained upon hydrogenolysis of the 2,4-dichlorobenzyl groups under buffered conditions.
Scheme 3. Convergent Synthesis of G-Analogues 4 and 5.
Reagents and conditions: (a) TEA, DMAP, Boc2O, MeCN, RT, 46 h; (b) NBS, DCE, −10 to 0 °C, 1 h; (c)(i) nBuLi, THF −100 °C, 23 or 28 (ii) 17; (d) BF3·OEt2, Et3SiH, MeCN, −78 °C to RT, 3 h; (e) H2 (60 psi) Pd–C (10%), 60 °C, 18 h, NH4OAc, MeOH/EtOAc (13:2) or BBr3, −78 °C to −30 °C (30 to 5); (f) DMF-dimethylacetal, DCM/MeCN, RT to 60 °C.
Because of the anion instability of the intermediate organolithium base, formed from 23, the synthesis of 4 was only possible in small, low milligram quantities using the method outlined in Scheme 3. A more robust procedure was therefore developed, which is shown in Scheme 4. The heterocycle 35 was chosen for glycosidic coupling and subsequent transformation to nucleoside 4 based on observations made during the initial synthesis of 4. These included the relative instability of anions on heterocycles bearing a protected nitrogen at the 2-position, the requirement for O4 protection to enable good metalation, and the susceptibility of the 2,4-dihalides to uncontrolled hydrolysis. Heterocycle 35 was prepared from the bis-chloro analogue 32 by displacement of the 4-chloro with BnOLi followed by bromination with NBS in DCE to yield an inseparable mixture of 35 along with its C7 isomer (3:1). As the C7 isomer was found to be inert in the glycosylation reaction, the mixture was used without separation. Glycosylation of the per-benzylated lactone 34 was achieved in 60% yield based on 35. After selective reduction of the lactol 36 to afford β-nucleoside 37, the 2-amino was installed by applying cross-coupling chemistry,18 which also resulted in partial deprotection of the 4-oxygen to afford a mixture of 38 and 39. The benzyl groups were cleaved under hydrogenolysis and the acetamide group cleaved under basic conditions to afford the free guanosine nucleoside 4.
Scheme 4. Improved Synthesis of 4.
Reagents and conditions: (a) nBuLi, BnOH, −10 °C to RT, THF; (b) NBS, DCM, −10 °C to RT; (c)(i) nBuLi, −100 °C, 2-Me-THF, (ii) 34 containing C7 isomer, −100 °C to RT; (d) BF3·OEt2, Et3SiH, DCM, −78 °C, 15 min; (e) acetamide, Pd2(dba)2, xantphos, Cs2CO3, 130 °C, 1 h; (f) H2, MeOH, Pd–C (10%), RT to 50 °C, 70 h; (g) NaOMe, RT to 80 °C.
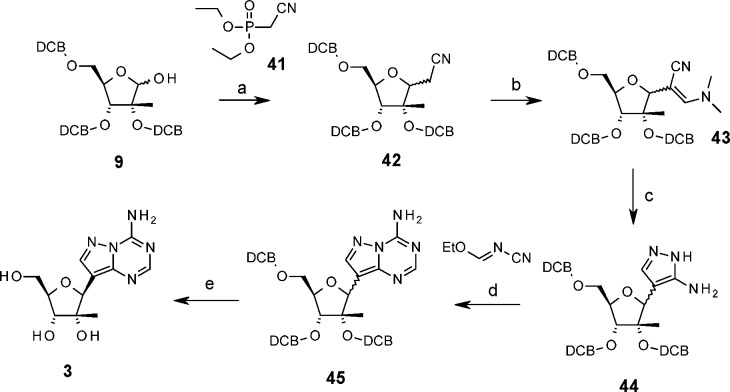
Application of the convergent chemistry used for the pyrrolo[2,1-f][1,2,4]triazine and imidazo[2,1-f][1,2,4]triazine bases was unsuccessful for the synthesis of pyrazolo[1,5-a]-1,3,5-triazine adenosine 3. Scheme 5 shows its successful linear synthesis, which adopts similar methodology reported by Klein et al. for the synthesis of the unsubstituted riboside analogue.19,20 Condensation of 2-diethoxyphosphorylacetonitrile, 41, with the tris-benzyl riboside 9 afforded the C-nucleoside intermediate 42. Separation of the anomers was possible at this stage but unnecessary, as interchange between anomers was observed under reaction conditions for the dimethylamine adduct 43 and for the amino-pyrazole intermediate 44. In situ condensation of 44 with ethyl cyanoimidoformate afforded the protected nucleoside 45. The anomers were separated, and the β-stereochemistry was established by the observation of an interaction between H1′ and H4′ in the NOESY spectrum. The pyrazolotriazine adenosine analogue 3 was obtained after hydrogenolysis.
Scheme 5. Synthesis of Pyrazolo-Nucleoside 3.
Reagents and conditions: (a) NaH, DME, 41, 0 °C to RT; (b) (CH3)3COCH[N(CH3)2]2, DMF, 60 °C, 15 h; (c) NH2NH2·HCl, EtOH, 105 °C, 2 h; (d) ethyl N-cyanoformimidate, 85 °C, toluene; (e) H2, MeOH, Pd–C (10%), 45 °C, 17 h.
The in vitro anti-HCV results for the parent C-nucleosides 1–5 are shown in Table 1. The low IC50 values observed for the pyrrolotriazine and imidazotriazine adenosine and guanosine nucleoside triphosphates show they are well recognized by the NS5B enzyme. The adenosine analogues 1 and 2 were furthermore active in the replicon assay, although the imidazotriazine analogue, 2, also showed some cytotoxicity. The corresponding guanosine analogues 4 and 5 were inactive in the cell-based replicon assay, a result consistent with the relatively poor activity reported for N-nucleoside guanosine analogues 2′CMe-G (EC50 = 2–4 μM) and 7-deaza-2′CMe-G (EC50 > 50 μM).7,21 The activity of the corresponding NTP suggests they were not efficiently phosphorylated by host kinases. In contrast, the poor enzyme activity observed for the pyrazolotriazine adenosine analogue, 3, is consistent with its poor activity in the replicon assay. On the basis of these initial results, 1 was selected for further investigation as a potential HCV DAA.
Table 1. Inhibitory Potency (EC50) of Nucleosides on HCV RNA Replication in Replicon Assay; Cytotoxicity (CC50); and Inhibitory Potency (IC50) of Corresponding Nucleoside-Triphosphate (NTP) on NS5B Polymerase Enzyme (WT GT1b(Con1)).
| compd | GT1b EC50 (μM)a | CC50 (μM)b | NTP IC50 (μM)c |
|---|---|---|---|
| 2′CMe-7-deaza-A | 0.20, 0.3 (lit.)21 | >100, >100 (lit.)21 | 0.20, 0.07 (lit.)21 |
| pyrrolo-A (1) | 1.6, 1.98 (lit.)12 | >100, 85 (lit.)12 | 0.37, 0.31 (lit.)12 |
| pyrrolo-G (4) | >100 | >100 | 0.21 |
| imidazo-A (2) | 1.3, 1.28 (lit.)12 | 34, >89 (lit.)12 | 0.55, 2.1 (lit.)12 |
| imidazo-G (5) | >100, >89 (lit.)12 | >100, >89 (lit.)12 | 0.31, 0.19 (lit.)12 |
| pyrazolo-A (3) | >100 | >100 | 33 |
Measured using subgenomic replicon cell lines described by Blight et al.22 See Supporting Information for methods.
Measured using Huh-7 derived replicon cell lines with MTT as the vital dye indicator.
See Supporting Information for methods.
Compound 1 displayed excellent activity against HCV genotypes 1a, 2a, 2b, 3a, 4a, 5a, 6a, and 7a. EC50 and IC50 values ranged from 0.39 to 1.0 μM (see Supporting Information for details) demonstrating its potential suitability as part of a pan-genotypic HCV therapy.
Because the nucleotide triphosphate is the compound responsible for anti-HCV activity, it is important to measure the ability of relevant cells to anabolize the parent nucleoside. The in vitro anabolism of 1 was studied in adherent Huh-7 cells and plated primary human hepatocytes. Following incubation of Huh-7 cells with 10 μM initial concentration of 1 for 24 h, the intracellular concentration of 1-NTP reached 9.3 pmol/million cells. Surprisingly, primary human hepatocytes generated much higher (∼50-fold) intracellular levels of 1-NTP (487 pmol/million cells) after 24 h with the same starting concentration, offering the promise of enhanced HCV antiviral activity in humans. Intracellular levels of 1 were found to be comparable in Huh-7 cells and hepatocytes, but the mono- and diphosphate species were about 10-fold lower in the replicon cell line suggesting that the first and second phosphorylation steps are more efficient in hepatocytes. The decay half-life (t1/2) of 1-NTP displays a biphasic profile in both replicon cells and primary human hepatocytes: a rapid initial decay t1/2 (0.7 and 3.3 h, respectively), followed by a prolonged elimination phase (t1/2 ≥ 35 h).
The pharmacokinetic properties of 1 were determined in male Sprague–Dawley rats, beagle dogs, and cynomolgus monkeys. Compound 1 displayed excellent cross-species pharmacokinetics with elimination t1/2 ranging from 4 to 7.6 h and high bioavailabilities (33–96%). On the basis of these data, the drug may have sufficient PK properties for a once daily dosing regimen in humans taking into account the long intracellular half-life of its triphosphate in hepatocytes.
Compound 1 was evaluated in a range of in vitro safety assays to assess its suitability for progression into animal toxicity studies. In a mitochondrial toxicity study over 14 days in HepG2 cells, 1 showed no increase in lactate production or decrease in mtDNA up to 10 μM. Low levels of cytotoxicity were observed in rat cells (CC50 = 190 μM for liver epithelial cells and CC50 > 250 μM for kidney fibroblasts and heart myoblasts). Compound 1 did not inhibit erythroid and myeloid progenitor proliferation in human bone marrow cells up to 100 μM and was negative in an AMES mutagenicity test. In addition, compound 1 triphosphate did not inhibit human DNA polymerases (IC50 >100 μM for α, β, and γ enzymes). Despite this promising in vitro profile, unacceptable adverse effects, including death, were observed during single oral dose toxicity studies in rats (dosed up to 400 mg/kg). This result caused us to focus development efforts on derivatives of 1 with a potentially improved safety margin. Since the completion of this work, the results of safety pharmacology and toxicology studies of 1 have been published.11
In summary, the synthesis and anti-HCV activity of a series of C-nucleoside purine analogue inhibitors of HCV NS5B polymerase have been investigated. Four of the five nucleosides showed excellent enzyme activity as their triphosphates, with two adenosine analogues showing activity in the cell-based replicon assay. The enzyme active guanosine analogues were, however, inactive in the whole cell assay, presumably due to inefficient anabolism to their nucleotide triphosphates. The pyrrolotriazine adenosine analogue 1 was chosen for further profiling because it showed excellent cross-genotype activity, low in vitro toxicity, good anabolism, and excellent pharmacokinetic properties in three species. Compound 1 incorporates the key structural features of a 2′CMe-ribose moiety with a pyrrolotriazine 7-deazaadenine base mimic, connected by a highly stable C–C bond. Unfortunately, the observation of adverse effects in rat safety studies caused us to re-evaluate our pursuit of 1 as a drug candidate. Despite this, these results demonstrated the potential for C-nucleosides to be used in new antiviral HCV therapies provided a sufficient therapeutic margin can be established.
Supporting Information Available
Experimental procedures, analytical data, and the description and results of in vitro assays and in vivo pharmacokinetics. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Centers for Disease Control and Prevention. Hepatitis C Information for Health Professionals.http://www.cdc.gov/hepatitis/HCV/HCVfaq.htm (accessed October 11, 2013).
- Lavanchy D. The Global Burden of Hepatitis C. Liver Int. 2009, 74–81. [DOI] [PubMed] [Google Scholar]
- Poordad F.; Dieterich D. Treating Hepatitis C: Current Standard of Care and Emerging Direct-Acting Antiviral Agents. J. Viral Hepatitis 2012, 19, 449–524. [DOI] [PubMed] [Google Scholar]
- Pawlotsky J.-M. Hepatitis C Virus: Standard-of-Care Treatment. Adv. Pharmacol. 2013, 67, 169–215. [DOI] [PubMed] [Google Scholar]
- US Food and Drug Administration. Viral Hepatitis Therapies. http://www.fda.gov/ForConsumers/ByAudience/ForPatientAdvocates/ucm151494.htm (accessed December 23, 2013).
- Sofia M. J.; Chang W.; Furman P. A.; Mosley R. T.; Ross B. S. Nucleoside, Nucleotide, and Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B RNA-Dependent RNA-Polymerase. J. Med. Chem. 2012, 55, 2481–2531. [DOI] [PubMed] [Google Scholar]
- Olsen D. B.; Eldrup A. B.; Bartholomew L.; Bhat B.; Bosserman M. R.; Ceccacci A.; Colwell L. F.; Fay J. F.; Flores O. A.; Getty K. L.; Grobler J. A.; LaFemina R. L.; Markel E. J.; Migliaccio G.; Prhavc M.; Stahlhut M. W.; Tomassini J. E.; MacCoss M.; Hazuda D. J.; Carroll S. S. A 7-Deaza-Adenosine Analog Is a Potent and Selective Inhibitor of Hepatitis C Virus Replication with Excellent Pharmacokinetic Properties. Antimicrob. Agents Chemother. 2004, 48103944–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinusi A.; Meissner E. G.; Lee Y.-J.; Bon D.; Heytens L.; Nelson A.; Sneller M.; Kohli A.; Barrett L.; Proschan M.; Herrmann E.; Shivakumar B.; Gu W.; Kwan R.; Teferi G.; Talwani R.; Silk R.; Kotb C.; Wroblewski S.; Fishbein D.; Dewar R.; Highbarger H.; Zhang X. X.; Kleiner D.; Wood B. J.; Chavez J.; Symonds W. T.; Subramanian M.; McHutchison J.; Polis M. A.; Fauci A. S.; Masur H.; Kottilil S. Sofosbuvir and Ribavirin for Hepatitis C Genotype 1 in Patients With Unfavorable Treatment Characteristics: A Randomized Clinical Trial. JAMA 2013, 3108804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biota Scientific Management & Boehringer Ingelheim International GMBH. Bicyclic Nucleosides and Nucleotides as Therapeutic Agents. WO2010/002877, January 7, 2010.
- Butora G.; Olsen D. B.; Caroll S. S.; McMasters D. R.; Schmitt C.; Leone J. F.; Stahlhut M.; Burlein C.; MacCoss M. Synthesis and HCV Inhibitory Properties of 9-Deaza-and 7,9-Dideaza-7-oxa-2′-C-methyladenosine. Bioorg. Med. Chem. 2007, 15, 5219–5229. [DOI] [PubMed] [Google Scholar]
- Cho A.; Zhang L.; Xu J.; Lee R.; Butler T.; Metobo S.; Aktoudianakis V.; Lew W.; Ye H.; Clarke M.; Doerffler E.; Byun D.; Wang T.; Babusis D.; Carey A. C.; German P.; Sauer D.; Zhong W.; Rossi S.; Fenaux M.; McHutchison J. G.; Perry J.; Feng J.; Ray A. S.; Kim C. U. Discovery of the First C-Nucleoside HCV Polymerase Inhibitor (GS-6620) with Demonstrated Antiviral Response in HCV Infected Patients. J. Med. Chem. 2014, 57, 1812–1825. [DOI] [PubMed] [Google Scholar]
- Cho A.; Zhang L.; Xu J.; Babusis D.; Butler T.; Lee R.; Saunders O. L.; Wang T.; Parrish J.; Perry J.; Feng J. Y.; Ray A. S.; Kim C. U. Synthesis and Characterization of 2′-C-Me Branched C-Nucleosides as HCV. Bioorg. Med. Chem. Lett. 2012, 22, 4127–4132. [DOI] [PubMed] [Google Scholar]
- Feng J. Y.; Cheng G.; Perry J.; Barauskas O.; Xu Y.; Fenaux M.; Eng S.; Tirunagari N.; Peng B.; Yu M.; Tian Y.; Lee Y.-J.; Stepan G.; Lagpacan L. L.; Jin D.; Hung M.; Ku K. S.; Han B. Inhibition of Hepatitis C Virus Replication by GS-6620, A Potent C-Nucleoside Monophosphate Prodrug. Antimicrob. Agents Chemother. 2014, 58, 1930–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E.; Wang T.; Babusis D.; Lepist E.-I.; Sauer D.; Park Y.; Vela J. E.; Shih R.; Birkus G.; Stefanidis D.; Kim C. U.; Cho A.; Ray A. S. Metabolism and Pharmacokinetics of the Anti-HCV Nucleotide Prodrug GS-6620. Antimicrob. Agents Chemother. 2014, 10.1128/AAC.02350-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S. A.; Otter B. A.; Klein R. S. 4-Aza-7,9-Dideazaadenosine, a New Cytotoxic Synthetic C-Nucleoside Analogue of Adenosine. Tetrahedron Lett. 1994, 35, 5339–5342. [Google Scholar]
- Hungerford N. L.; Armitt D. J.; Banwell M. G. Syntheses of Showdomycin and Its Anomer Using N-(Triisopropylsilyl)pyrrole as a Synthetic Equivalent for the Maleimide C3-Anion. Synthesis 2003, 12, 1837–1843. [Google Scholar]
- Wang R.-W.; Gold B. A Facile Synthetic Approach to 7-Deazaguanine Nucleosides via a Boc Protection Strategy. Org. Lett. 2009, 11, 2465–2468. [DOI] [PubMed] [Google Scholar]
- Bosch L.; Cialîcu I.; Caner J.; Ariza X.; Costa A. M.; Terrazas M.; Vilarrasa J. Pd-Catalysed Amidation of 2,6-Dihalopurine Nucleosides. Replacement of Iodine at 0°C. Tetrahedron Lett. 2012, 53, 1358–1362. [Google Scholar]
- Tam S. Y.-K.; Hwang J.-S.; de Las Heras F. G.; Klein R. S.; Fox J. J. Nucleosides CV. Synthesis of the 8-(β-d-ribofuranosyl)pyrazolo[1,5-a]-1,3,5-triazine Isosteres of Adenosine and Inosine. J. Heterocycl. Chem. 1976, 13, 1305–1308. [Google Scholar]
- Tam S. Y. K.; Klein R. S.; Wempen I.; Fox J. J. Nucleosides. 112. Synthesis of Some New Pyrazolo[1,5-a]-1,3,5-triazines. J. Org. Chem. 1979, 44, 4547–4553. [Google Scholar]
- Eldrup A. B.; Allerson C. R.; Bennett C. F.; Bera S.; Bhat B.; Bhat N.; Bosserman M. R.; Brooks J.; Burlein C.; Carroll S. S.; Cook P. D.; Getty K. L.; MacCoss M.; McMasters D. R.; Olsen D. B.; Prakash T. P.; Prhavc M.; Song Q.; Tomassini J. E.; Xia J. Structure–Activity Relationship of Purine Ribonucleosides for Inhibition of Hepatitis C Virus RNA-Dependent RNA Polymerase. J. Med. Chem. 2004, 47, 2283–2295. [DOI] [PubMed] [Google Scholar]
- Blight K. J.; Kolykhalov A. A.; Rice C. M. Efficient Initiation of HCV RNA Replication in Cell Culture. Science 2000, 290, 1972–1974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.