Abstract
PR/SET domain containing 9 (Prdm9) mediates histone modifications such as H3K4me3 and marks hotspots of meiotic recombination. In many mammalian species, the Prdm9 gene is highly polymorphic. Prdm9 polymorphism is assumed to play two critical roles in evolution: to diversify the spectrum of meiotic recombination hotspots and to cause male hybrid sterility, leading to reproductive isolation and speciation. Nevertheless, information about Prdm9 sequences in natural populations is very limited. In this study, we conducted a comprehensive population survey on Prdm9 polymorphism in the house mouse, Mus musculus. Overall M. musculus Prdm9 displays an extraordinarily high level of polymorphism, particularly in regions encoding zinc finger repeats, which recognize recombination hotspots. Prdm9 alleles specific to various M. musculus subspecies dominate in subspecies territories. Moreover, introgression into other subspecies territories was found for highly divergent Prdm9 alleles associated with t-haplotype. The results of our phylogeographical analysis suggest that the requirement for hotspot diversity depends on geographical range and time span in mouse evolution, and that Prdm9 polymorphism has not been maintained by a simple balanced selection in the population of each subspecies.
Keywords: Prdm9, mouse, polymorphism, evolution
1. Introduction
Meiotic recombination enhances the genetic diversity in natural populations and contributes to genome evolution. In organisms as diverse as yeasts and mammals, meiotic recombination events do not take place at random but are clustered at specific genomic regions, referred to as recombination hotspots.1,2 Nevertheless, until recently, the molecular basis underlying determination of the hotspots has been elusive.
We previously reported that wm7, a wild mouse-derived haplotype of the major histocompatibility complex (MHC) on chromosome 17, enhances meiotic recombination at a hotspot within the MHC.3 Subsequently, we found that a factor genetically linked to this hotspot determines its recombination rate, and that the wm7 haplotype carries a recombination-enhancing factor.4 Recently, the factor was reported to be a histone methyltransferase, PR/SET domain containing 9 (Prdm9).5–7 Prdm9 mediates histone modifications, such as H3K4me3, and is thought to mark recombination hotspots. Since its identification, many reports have shown that Prdm9 polymorphisms correlate well with site variation in hotspots in different mammalian species, including humans, chimpanzees, and mice.8–12 Genome-wide ChIP analysis with antibodies that recognize DMC1 and RAD51 performed for two different Prdm9 alleles in a common genetic background revealed that Prdm9 variation can account for the site specificity of almost all of the DNA double-strand breaks that initiate meiotic recombination.11 Thus, Prdm9 appears to be a major trans-acting factor for determining the spectrum of hotspots in mice and humans, and perhaps in other mammalian species as well.8–11
A Prdm9 knockout mouse shows meiosis arrest, indicating that Prdm9 histone methyltransferase activity is involved in progression of meiosis.13 This function has also been implicated in reproductive isolation, a process that prevents free exchange of genes between two genetically divergent populations, leading to speciation. In crosses between the mouse subspecies Mus musculus domesticus and M. m. musculus, the F1 male hybrids are sometimes sterile. The locus responsible for this hybrid sterility was named Hybrid sterility 1 (Hst1), and mapped to chromosome 17. Recent results revealed that Hst1 is identical to Prdm9, such that Prdm9 became the first speciation gene to be reported in mammalian species.14,15
Comprehensive information on Prdm9 polymorphisms in natural populations should provide an important insight into Prdm9 functions, especially in evolution. Past studies have focused on Prdm9 polymorphism in human populations. The results revealed that the worldwide population is quite diverse, with differences including repeat number variations of the zinc finger (ZF) DNA-binding repeat and amino acid substitutions in the zinc finger array (ZFA) of the Prdm9 C-terminal domain.7,10,16,17 Importantly, non-synonymous substitutions preferentially occur at three amino acids along the α-helix domain of the ZF, which are involved in recognition of the hotspot nucleotide motifs.18 Currently, it is thought that hotspot diversity in natural human populations can be attributed to polymorphism of the Prdm9 ZFA.19
There are marked advantages to studying Prdm9 polymorphism in natural populations of M. musculus. First, the phylogenetics of M. musculus is well established.20,21 Mus musculus is a complex species, and comprises distinct ‘phylogroups’ or subspecies. The results of extensive phylogenetic analysis of Eurasian wild mice revealed that these subspecies diverged roughly 0.5–1.0 million years ago.20 Their habitats are demarcated throughout the Eurasian continent.21,22 Secondly, whereas subspecies of M. musculus are thought to be in an early stage of speciation, neighbouring species, including M. spretus, M. macedonicus, and M. spicilegus, inhabit areas overlapping those of M. musculus.22 Thus, M. musculus and its neighbouring species provide an ideal model system to study phylogeography and speciation. Thus far, mouse Prdm9 polymorphisms has been investigated in commonly used laboratory inbred strains and inbred strains derived from wild mice.5,7,23 However, in these studies, sample collection was limited, as laboratory inbred strains originate predominantly from a single western European subspecies, M. m. domesticus.24,25 Moreover, only a limited number of inbred strains derived from wild-captured mice were included in previous studies.5,23
In this study, we extended the population survey to wild mice collected in natural populations of M. musculus subspecies and neighbouring species, as well as inbred strains derived from wild mice. We also investigated Prdm9 polymorphism in mice with the t-haplotype chromosome variant, which is characterized by long inversions on chromosome 17 and is linked to the Prdm9 locus. Genes on the t-haplotype are highly divergent from those on wild-type chromosome 17.26 The results of our study confirm that Prdm9 polymorphisms are concentrated in ZFA with extensive variation of the ZF repeat number and hyper-variation of amino acids at the three DNA recognition sites within the ZF. Our survey of 79 wild-captured mice and 37 inbred strains revealed as many as 57 different Prdm9 alleles in M. musculus. In contrast, some alleles were predominant in two subspecies, M. m. domesticus and M. m. musculus. The overall phylogeography of mouse Prdm9 reflects evolutionary episodes of this species. More importantly, Prdm9 alleles that predominate in one subspecies are often found in territories of other subspecies. Likewise, highly divergent Prdm9 alleles associated with the t-haplotype were found to introgress into all subspecies of M. musculus.
2. Materials and methods
2.1. Mice
Nine mouse strains (M. m. molossinus, MSM/Ms; M. m. musculus, NJL/Ms, KJR/Ms, BLG2/Ms, SWN/Ms, CHD/Ms; M. m. castaneus, HMI/Ms; M. m. domesticus, PGN2/Ms, BFM/Ms), one Japanese fancy mouse-derived strain (M. m. molossinus, JF1/Ms), and two MHC congenic mouse strains (B10.R209, B10D2.TCH/+) were maintained at the Genetic Strains Research Center, National Institute of Genetics (NIG). A classical laboratory mouse strain, C57BL/10Snf, was purchased from the Jackson Laboratory and maintained at the NIG. The inbred strain SPR2/Rbrc, derived from M. spretus, was provided by the RIKEN BioResource Center (BRC) through the National BioResource Project, which is funded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Inbred strains used in this study are listed in Supplementary Table S1. All animal experiments were performed in accordance with protocols approved by the Animal Care and Use Committee of NIG.
2.2. Prdm9 cDNA synthesis
Total testes RNA from each mouse strain was isolated with Isogen (Nippon Gene). Complementary DNA (cDNA) was synthesized using the Primescript RT reagent kit (TAKARA) according to the manufacturer's instructions.
2.3. Mouse genomic DNA samples and PCR conditions
Most genomic DNA samples, including classical inbred and wild-captured mice, were prepared by the Genetic Strains Research Center at the NIG. Some were prepared at Hokkaido University. Several genomic DNA samples were purchased from the Jackson Laboratory. Genomic DNA samples from t-haplotype mice were kindly provided by Joe Nadeau or by the RIKEN BRC. All the genomic DNA samples are listed in Supplementary Table S2. To prevent errors in PCR, we used high-fidelity DNA polymerases (KAPA HiFi from Kapa Biosystems or KOD Neo FX from Toyobo). In addition, we repeated independent amplification at least three times for each sample. PCR primer sets and conditions are shown in Supplementary Table S3, and the amplified sites are shown in Supplementary Fig. S6.
2.4. Sequence analysis
To determine the sequences of cDNA, ZFA, high SNP region 1 (HSR1) of Prdm9, and intron of T-complex protein 1 (Tcp1), we sequenced PCR products directly or after subcloning. For subcloning, PCR products were extracted with a QIAquick Gel Extraction Kit (QIAGEN) after electrophoresis. Then, the purified PCR products were subcloned into pCR-Blunt II-TOPO (Invitrogen). For sequencing, we used the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and a 3130XL DNA Analyser (Applied Biosystems). Primers used for sequence analyses are listed in Supplementary Table S4. We analysed at least six clones per sample and carried out multiple independent experiments for each allele. All sequence data from this study were submitted to the DDBJ Sequence Read Archive (Accession numbers AB843858 to AB844116 and AB846828).
2.5. Coding of ZF repeats
The Prdm9 ZFA nucleotide sequence was conceptually translated (Fig. 1B), and then the sequence of 28 amino acids from 512S to the last residue, which corresponds to the ZFA, was extracted from all ZF repeats. We assigned a one-letter box with a given colour to every ZF repeat that had a given amino acid triad at the most variable positions of the α-helix domain (−1, 3, and 6). When amino acid variation was found for a ZF repeat at less variable positions (−5, −2, and 1), the right bottom of the box was labelled with the variant amino acids.
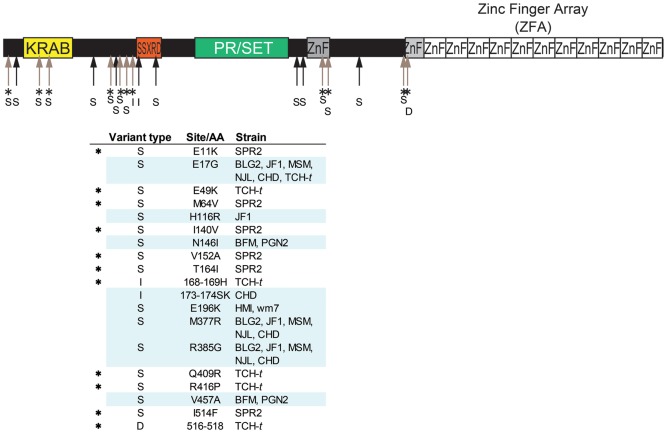
Figure 1.
Polymorphisms in Prdm9 cDNA sequences from inbred mouse strains. Amino acid polymorphism of Prdm9 excluding ZFA is summarized. Full-length cDNA of Prdm9 was sequenced for 12 inbred strains derived from M. musculus and one inbred strain derived from a neighbouring species, M. spretus. Nucleotide sequences of the inbred strains were compared with the C57BL/6J reference sequence (NCBI mm9). The C57BL/10J sequence was identical to the reference. Comparison revealed nucleotide substitutions that lead to 16 amino acid substitutions in total, as well as two insertions and one deletion of an amino acid relative to the reference. No amino acid substitution was observed in the PR/SET domains. Upper arrows indicate positions of amino acid variation. An asterisk (*) indicates an interspecific variation (or variation in B10.D2-TCH/+). All amino acid variations are listed in the table below the diagram of Prdm9 protein. The letters S, I, and D in variant types indicate substitution, insertion, and deletion of an amino acid, respectively.
2.6. Phylogenetic analysis
To construct a phylogenetic tree of ZFA, we aligned ZFA repeat units using a progressive multiple sequence alignment algorithm implemented in ClustalW.27 Briefly, each repeat unit was converted to a one-letter code according to the amino acid residues at five signature sites, i.e. the three most variable and two less variable among ZF repeats. Mismatch scores between repeat units were given by the number of different signature sites between two units. The gap open penalty was set to 0.5 and the gap extension penalty we used was 0.1. The algorithm aligns ZF repeat unit sequences by maximizing the alignment score, as is typical for nucleotide and protein sequence alignment. After alignment, the repeat unit sequences were transformed into nucleotide sequences and nucleotide distance was measured using Kimura's two-parameter method.28 A phylogenetic tree was constructed using the neighbour-joining method29 and implemented using the MEGA 5 software.30 A phylogenetic tree of HSR1 and Tcp1 was similarly constructed using neighbour-joining with Kimura's two-parameter distances. Bootstrap resampling tests were conducted with 1000 iterations.
3. Results
3.1. Polymorphism in the cDNA sequence of Prdm9
To determine the overall nature of mouse Prdm9 polymorphism, we first cloned Prdm9 cDNAs by PCR from total RNAs prepared from the testes of 12 inbred strains (Supplementary Table S1). These comprised seven strains derived from wild-captured mice belonging to different subspecies: two congenic strains harbouring wild-derived Prdm9 alleles, one inbred strain of the Japanese fancy mouse (JF1/Ms), and one inbred strain derived from a different species, M. spretus. The PCR products showed marked variation in size. For each sample, the major band was cloned and then sequenced using a capillary sequencer.
Comparison between these 12 sequences yielded a total of 28 nucleotide changes in the 1565-bp Prdm9 ORF, excluding the ZFA (Fig. 1). Of these, 19 cause amino acid changes. When compared with the C57BL/6 reference sequence (NCBI mm9) and our C57BL/10 sequence (this study), insertions and deletions (in/dels) without frame shifts were observed in two strains, CHD/Ms and B10.D2-TCH/+ (Supplementary Fig. S1). No difference was found in the PR/SET domains of the 12 strains examined. In contrast, the repeat numbers of ZF were largely variable among these strains, consistent with changes in the electrophoretic mobility of the PCR products (Supplementary Fig. S2A). In addition, we found numerous nucleotide changes in ZFA. Almost all of these were associated with amino acid changes.
3.2. ZFA polymorphism of wild-captured mice
We next extended our survey of Prdm9 ZFA polymorphisms to wild-captured mice. The entire ZFA region is encoded by the last exon. We directly amplified this exon by PCR from genomic DNA from total of 79 wild mice collected at different locations (Supplementary Table S3). The samples include populations belonging to three major M. musculus subspecies and a neighbouring species. To avoid artificial PCR products, we used high-fidelity DNA polymerase and highly stringent conditions. Moreover, PCR primer sets were designed for regions 200 bp apart from both ends of the ZFA to prevent mis-annealing of PCR intermediates. PCR products were separated on an agarose gel and the major band, which displays extensive variation in size among samples (Supplementary Fig. S2B), was subjected to subcloning. When two major bands existed, the both bands were subcloned, as they are likely to represent heterozygous alleles. Reproducibility was confirmed by repeated independent PCR amplifications. More than six clones were sequenced for each sample with two different primer sets. If two types of ZFA sequences were reproducibly obtained, they were judged as heterozygous alleles.
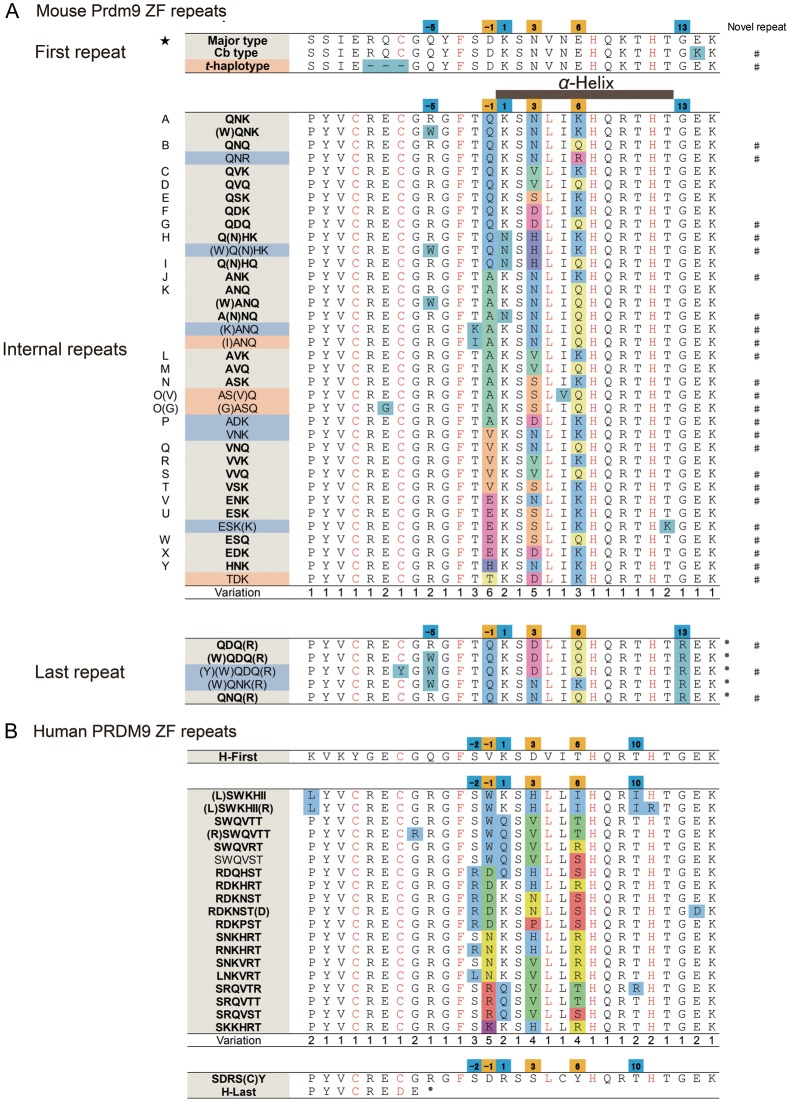
We aligned all ZF repeats identified in this study (Fig. 2A). The first ZF repeat appears to be uniform, but the internal ZF repeats were highly variable. In particular, DNA recognition positions −1, 3, and 6 were highly variable (Fig. 2A), consistent with a previous report.17 A lower level of variation was found at positions, −5, −2, and 1. The last repeat tends to be less variable than the internal repeats. In total, we identified 36 unique ZF repeats in ZFA. Of these, 24 are newly identified in this study (Fig. 2A).
Figure 2.
Amino acid variation in Prdm9 ZF repeats. (A) Multiple alignment of ZF repeat sequences from inbred strains and wild-captured mice. The alignments were made separately for the first repeat, the internal repeats of the ZFA, and the last repeat, as their degrees of variation are different (see text). The α-helix domain of the ZF is shown at top of the internal repeat. The one-letter code is shown to the left of the repeats. A novel repeat found in this study is indicated by # to the right of the repeat. Amino acid variation at specific positions along aligned ZF repeats are shown in colour. The star on the left side in the first repeat corresponds to those shown in Fig. 3. (B) Multiple alignment of ZF repeats of human PRDM9 based on the publically available data (http://www.ncbi.nlm.nih.gov/nuccore). The format of the alignment is the same as that used for mouse Prdm9.
We further compared variation in the mouse ZF repeats with those in human PRDM9, as obtained from a public database (http://www.ncbi.nlm.nih.gov/nuccore) (Fig. 2B). Amino acids in the backbone of C2H2-class ZFs are well conserved between the two species; however, some species-specific amino acid variations are found. For example, the amino acid at position −2 of the α-helix domain is mostly tyrosine (T) in the mouse, but serine (S) or arginine (R) in humans. The amino acid at position 5 is mostly isoleucine (I) in mouse, but exclusively leucine (L) in human. These positions reside in the α-helix domain, close to the most variable DNA recognition positions, i.e. positions −1 and 6. Amino acid variation is also found within each species. It is conceivable that this variation results in changes to DNA recognition patterns.
3.3. Phylogeography of polymorphisms in the ZFA of Prdm9 in natural populations
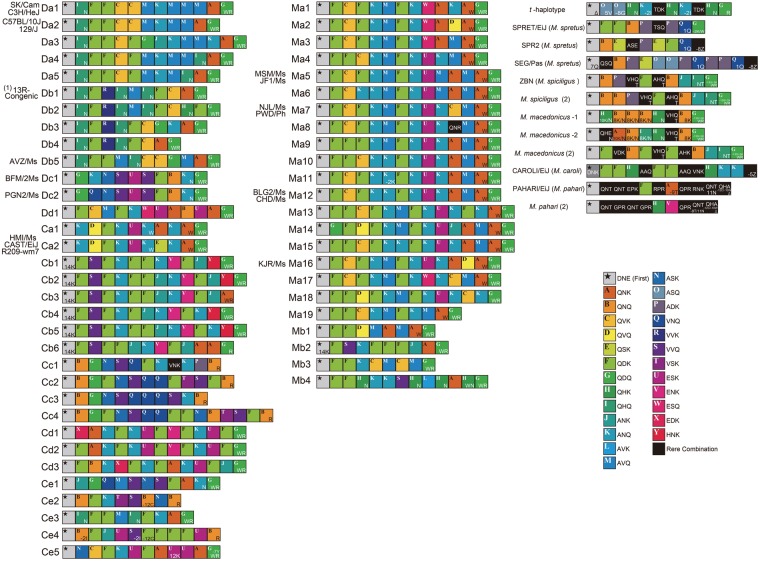
To simplify annotation of Prdm9 ZFAs in subsequent analyses, we used a one-letter code to identify each ZF repeat depending on the amino acid triads present at the most variable three positions, −1, 3, and 6, as well as additional alterations (Fig. 2A) (see Materials and Methods for detail). Figure 3 presents an alignment of all ZFA diagrams of wild-captured mice, inbred strains derived from wild mice, and commonly used laboratory inbred strains. The data indicate that wild-captured mice have ZFAs of various lengths. The ZF repeat numbers were between 9 and 16. We could identify as many as 57 ZFA variants within a single species, M. musculus. In addition, we found that neighbouring species of M. musculus have ZFA variant types different from those of M. musculus.
Figure 3.
ZFA alignment for inbred and wild-captured mice. We first assigned a one-letter coding system to ZFAs (see text), then aligned the ZFA diagrams based on the sequences of classical laboratory strains, wild-derived inbred strains, wild-captured mice, neighbouring species of M. musculus, and t-haplotype bearing mice. Using this code, we easily classified all variant types of ZFA. ZFA boxes are aligned from the N-terminal (left of the diagram) to the C-terminal end of the protein (right of the diagram). The diagrams are categorized into DOM- and CAS-related ZFA variant types (left block), MUS-related ZFA variant type (centre block), and other ZFA types of neighbouring species and t-haplotype (right block). The identification (ID) code for each ZFA diagram is indicated onto the left of the diagram. The names of inbred strains and taxonomy are indicated to the left of the ID codes.
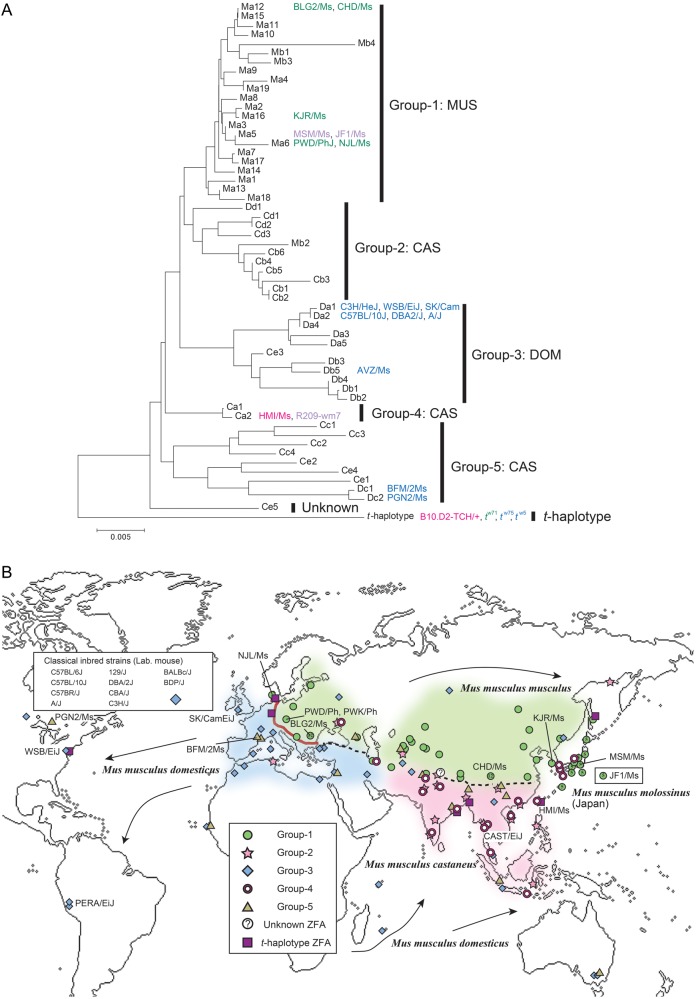
To elucidate the phylogenetic relationships among ZFA variants in M. musculus, multiple alignment of the ZFA was performed using the one-letter code. After alignment, the phylogenetic tree was constructed using the neighbour-joining method.29 This phylogenetic analysis revealed that the ZFA variants could be divided into five major groups (Fig. 4A). The localities where the wild mice were collected are then plotted on the territories of three major subspecies of M. musculus (Fig. 4B).
Figure 4.
Phylogeny and phylogeography of Prdm9 ZFAs. (A) Phylogenic tree constructed from 57 Prdm9 ZFAs. The identification codes of ZFA variant types are labelled according to the ZFA diagrams (Fig. 3). The 57 ZFA variant types can be divided into five major groups. The names of inbred strains carrying a given ZFA variant type are shown. (B) Points of collection of wild-captured mice are shown on a world map. Background colours indicate the ranges (territories) of each M. musculus subspecies as follows. Blue, DOM; green, MUS; purple, CAS. The red line in Europe indicates a DOM–MUS hybrid zone, where the two subspecies come into contact. Dashed lines indicate borders between other subspecies. For some species, the border regions are not clear. Japanese wild mice, M. m. molossinus, are a hybrid of two subspecies, MUS and CAS.
Group 1 exclusively includes mice collected in the territory of subspecies M. m. musculus (hereafter, MUS), which extends on the North Eurasian continent from Eastern Europe to the Far East. Groups 2, 4, and 5 mainly include mice collected in the territory of subspecies M. m. castaneus (CAS), which includes a wide range, from Southwestern Asia to India, and extends to Southeast Asia, South China, Indonesia, and the southern and northern edges of Japanese islands. Group 3 includes mice collected in the territory of subspecies M. m. domesticus (DOM), which extends from western and southern Europe to the Middle East and the shores around the Mediterranean Sea. Group 3 territory also includes the New World, North and South America, coincident with human migration. This group also includes commonly used classical inbred strains, which are overwhelmingly derived from the DOM lineage.31 Groups 2 and 5 also include a small number of mice collected in the MUS and DOM territories, respectively. Likewise, Group 3 includes one mouse collected in the CAS territory. In the Japanese population, 12 of 13 mice (including two inbred strains) showed only a single ZFA variant type (Ma5), classified as Group 1. The remaining strain has the variant type of Group 4 (Ca2).
Altogether, although M. musculus as a whole holds huge variations in ZFA, the mice collected in the MUS and DOM territories exhibit mutually exclusive ZFA variant patterns (Group 1 for MUS and Group 3 for DOM, respectively). The existence of such predominant variant types is not seen in the case of the mice collected in the CAS territory. In this region, ZFA variant types can be further divided into at least the three major groups.
We calculated the frequency of wild-captured mice heterozygous for ZFA variant types in different territories of subspecies (Supplementary Fig. S3). The value for the total M. musculus population is 35% (26/74); for DOM, 10% (2/20); for MUS, 44% (11/25); for CAS, 50% (12/24); and for Japanese wild mice (M. m. molossinus), 10% (1/10). These results are consistent with the degree of ZFA diversity observed in the three subspecies populations.
3.4. Phylogeny of Prdm9 ZFA in t-haplotype mice
B10.D2-TCH/+ is a heterozygous MHC haplotype mouse stock that harbours a recessive lethal mutation. In this stock, the wild-derived MHC haplotype is transmitted to the next generation at a highly distorted ratio (unpublished data). Therefore, we inferred that B10.D2-TCH/+ has t-haplotype (Supplementary Fig. S4). Prdm9 cDNA from this strain did not have polymorphisms observed for other strains in the PR/SET domain; however, unique substitutions were found outside the ZFA, as for M. spretus, in addition to a DOM-type sequence. The result suggests that this stock is heterozygous for DOM-type and more divergent Prdm9 alleles (Fig. 1). Its ZFA has 11 repeats of ZF with a rare type of the amino acid triad. To analyse other strains carrying the t-haplotype, we amplified the ZFA from seven t-haplotype samples, tw5/+, tw71/+, tw75/+, t12/+, tw12/+, t0/+, and tw2/ tw2. With the exception of for tw2, all samples were heterozygous for a DOM-type chromosome. Sequencing of ZFA from all strains showed a rare variant type in addition to a DOM-type ZFA. Notably, the sequence of the rare variant is identical among samples carrying the t-haplotype and B10.D2-TCH/+. Phylogenetic analysis showed that the ZFA sequence associated with the t-haplotype is more divergent from those of the five major groups in M. musculus (Figs 3 and 4A).
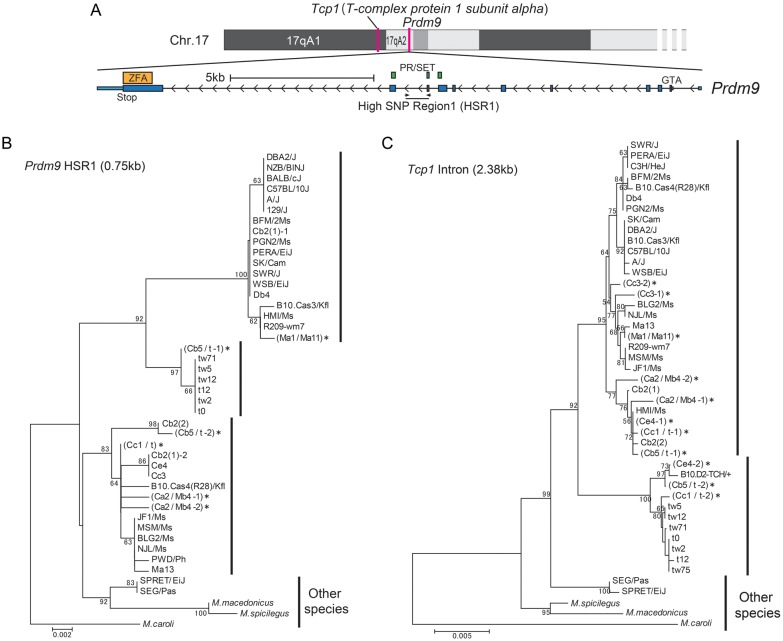
We compared Prdm9 genome sequences among three inbred strains, C57BL/6J, CAST/EiJ, and PWK/PhJ, which are derived from three subspecies, DOM, CAS, and MUS, respectively. We found nucleotide sequence polymorphism in a 750-bp intronic region of Prdm9 residing in the interval between two exons that encode the PR/SET domain (Supplementary Fig. S5). We named this HSR1 (Fig. 5A). To clarify the phylogeny of regions outside the Prdm9 ZFA, we sequenced HSR1 from different subspecies of wild-captured M. musculus and neighbouring species. We also sequenced a 2.38-kb region that includes introns 8–11 of Tcp1, a t-haplotype marker,26 to analyse the phylogeny of a gene linked to Prdm9. We constructed phylogenetic trees from these two sequences, HSR1 and Tcp1, using the neighbour-joining method.29 For tree construction, M. caroli, which is more divergent from M. musculus than other neighbouring species,20 was used as an outgroup. The overall topology of the Tcp1 tree is similar to the tree based on Prdm9 ZFA (Fig. 5C). In particular, Tcp1 in the t-haplotype has diverged from those in the wild-type chromosome, consistent with a previous report.26 However, the sequence is similar to that found in some mice from the CAS territory. In contrast, the phylogenetic tree of the HSR1 sequence shows that Prdm9 in the t-haplotype is similar to that found in mice from the CAS and DOM territories, but distant from those of mice from the MUS territory (Fig. 5B). These results indicate that the single Prdm9 allele in t-haplotype introgressed into all subspecies of M. musculus, and suggest that intragenic recombination somewhere in the interval between ZFA and HSR1 in the Prdm9 gene occurred in the past.
Figure 5.
Phylogeny of the Prdm9 intronic sequence (HSR1) and Tcp1. (A) Upper diagram, map of the chromosomal region containing Tcp1 and Prdm9. Lower diagram, view of the exon–intron organization of Prdm9. HSR1 is located in an intron between two exons that encode the PR/SET domain. (B) Phylogenetic tree of Prdm9 HSR1. (C) Phylogenetic tree of Tcp1 sequence in introns 8–11. M. caroli was used as the outgroup for both trees. An asterisk (*) to the right of a strain name (ZFA variant) indicates that the samples are heterozygous for Prdm9 HSR1 (B) and Tcp1 intronic (C) sequences.
4. Discussion
The results of this study support that the mouse Prdm9 polymorphism is highly polymorphic. Indeed, the degree of the polymorphism of Prdm9 is comparable with that of MHC, which may be maintained by selective advantage.32 Prdm9 polymorphisms converged in the ZF repeats and in amino acid positions at −1, 3, and 6 of the α-helix domain, which are extremely variable and are thought to recognize DNA sequences. A lower level of variation was found for amino acid positions −2 and 1 in both mouse and human ZF repeats. We infer that all these positions have been subjected to positive selection to increase the diversity of Prdm9 polymorphism,17 as non-synonymous substitutions preferentially occur at positions −2 and 1, both in mice and in humans. In humans, 19 unique ZF repeats have been identified by extensive analysis of genome sequences from various human populations.5,7,10,17,33 This number is lower than that in mouse (Fig. 2B). It is likely explained by the shorter time of divergence between different human populations, in contrast to a longer period of divergence between different mouse subspecies (i.e. 0.5–1.0 million years).20
The first and last ZF repeats showed a lower level of variation when compared with internal repeats. In the first repeat, the first cysteine, which is involved in tertiary structure of the C2H2-class ZF, has been lost. As a consequence, it may not function as authentic ZF (Fig. 2A and B). For the last repeat, the C2H2-class ZF is conserved and amino acids at positions 3 and 6 are variable in mice (Fig. 2A). These positions show a higher ratio of Ka/Ks value (4 : 1), suggesting that they participate in DNA recognition.
The most prominent feature of ZFA polymorphism is repeat number variation. In mouse populations, the minimum number of repeats is 9, including the first and last repeats (Fig. 3). This number was found in three mouse subspecies. Longer repeats (15 and 16) are enriched in the MUS territory, although some MUS mice have shorter repeats. In the MUS territory, although mice inhabiting the border with CAS territory carry such shorter repeats, they share ZFA characteristic of the MUS type.
The results of our phylogeograpical study clearly show that M. musculus as a species holds extensive Prdm9 polymorphism, owing to large degrees of variation in the ZFA. Within a subspecies lineage, the ZFA variant types tend to be similar, with the exception of the CAS lineage. Even though MUS territory extends long distances, reaching from the Northern Eurasian continent to Eastern Europe and the Far East, most mice collected in this range are exclusively included in Group 1 (Fig. 4A and B). The Japanese population, M. m. molossinus, is a hybrid of two subspecies, MUS and CAS, but its genome is overwhelmingly derived from MUS.34 We found that the majority of mice collected in different localities in Japanese islands have a single ZFA variant type (Ma5) of Group 1. Another variant type (Ca2) in Group 2 (CAS) is carried by one mouse sample with the wm7 MHC haplotype, which was first used for identification of Prdm9 as the hotspot determinant.7 Thus, the present result supports a hybrid origin of M. m. molossinus.34–36
Recent studies suggested that Southwest Asia and North India are the likely places of origin of M. musculus.37–39 In this region, three major subspecies lineages, DOM, MUS, and CAS, likely separated from one another in subdivided regions and diverged over a relatively long evolutionary time period of 0.5–1.0 million years. Subsequently, the three lineages dispersed to their present ranges, probably associated with agricultural dispersal by humans.22,37–39 The latter event is estimated to have occurred relatively recently, 10–20 thousand years ago.22,40,41 Eastward MUS lineages from the origin of M. musculus may have reached to the Far East, then migrated to Japanese islands 2–3 thousand years ago through the Korean peninsula as stowaways during the transportation of rice, following preceding CAS migration from Southeast Asia, which might have occurred 5–10 thousand years ago.22 If these evolutionary episodes of M. musculus are correct, then it appears that a large degree of Prdm9 diversity is not always required for survival in natural populations. A single version of the hotspot repertoire has been sufficient to maintain the Japanese population in a time span of at least 2–3 thousand years. Likewise, local populations in MUS and DOM territories have survived for 10–20 thousand years with a limited polymorphism at Prdm9. Regarding the CAS lineage, the high heterogeneity of ZFA in these mice is consistent with data suggesting that CAS consists of multiple sublineages, which show a relatively large degree of genome divergence from one another.22 Given that the CAS territory includes the likely place of origin of M. musculus,41 its heterogeneity is reminiscent of the African human population, which shows greater diversity of Prdm9 ZFA.9 Thus, overall, the phylogeographical features of Prdm9 polymorphism in M. musculus are similar to what has been inferred from information about many other genes.21,22,42 Furthermore, we infer that the requirement for hotspot diversity depends on geographical range and time span in evolution, and that Prdm9 polymorphism has not been maintained by a simple balanced selection in the population of each subspecies.
The results of this study show that subspecies-specific Prdm9 variant groups prevail in the demarcated territories of M. musculus subspecies; however, the data also revealed intermingled variant groups in areas where the territory of one subspecies borders another (Fig. 4B). Moreover, except for Group 1, each of the major ZFA variant groups contains small numbers of mice collected in the territory of other subspecies. A more prominent example of intermingled Prdm9 alleles was observed in the case of t-haplotype.
Mouse t-haplotype harbours three inversions in chromosome 17.43–45 The nucleotide sequences of t-haplotype associated genes are largely diverged from those of the wild-type chromosome of M. musculus.44,46 At present, in mouse natural populations, t-haplotype is observed in all subspecies of M. musculus at 10–40% of frequency.47,48 It is inferred that t-haplotype has been introgressed into all subspecies of M. musculus 10–20 thousand years ago as a single event, and expanded to all subspecies during agricultural dispersal, due to high distortion of the transmission ratio of t-haplotype relative to wild-type chromosome.44,46 This study clearly shows that t-haplotype mice have unique and characteristic Prdm9 ZFA and intronic (HSR1) sequences. These data support the idea that the t-haplotype is of monophyletic origin. In our phylogenetic trees, Tcp1 and Prdm9 HSR1 in t-haplotype appear in the same clade as those from mice collected in the CAS territory. This suggests that t-haplotype originated in a sublineage of the CAS subspecies and rapidly introgressed into all subspecies of M. musculus.
Our phylogenetic analysis also revealed that the topology of the ZFA tree differs from that of the HSR1 tree. This implies that intragenic recombination occurred in the interval between these two regions, despite the fact that they are only 9 kb apart. If ZFA contains a recombination hotspot, an associated frequent and unequal recombination might give rise to repeat number variation of ZFs.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported in part by Grant-in-Aid for Scientific Research on Innovative Areas titled ‘Systematical studies of chromosome adaptation’ and ‘Non-coding DNA’ from MEXT, Japan, to T.S. and K.O., respectively, and a grant from the Japan Society for the Promotion of Science (JSPS) Research Fellowship to H.K.
Supplementary Material
Acknowledgements
We thank P. Vogel, K.P. Aplin, E. Nevo, L.V. Yakimenko, L.V. Frisman, and A. Kryukov for providing us with wild-captured mice, and K. Artzt and J. Nadeau for providing us with t-haplotype mice. We also thank T. Takada for discussion of phylogenetic analysis of Prdm9 and J. Galipon for editing the manuscript.
Footnotes
Edited by Dr Minoru Ko
References
- 1.Petes T.D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2001;2:360–9. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 2.Kauppi L., Jeffreys A.J., Keeney S. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 2004;5:413–24. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 3.Shiroishi T., Sagai T., Moriwaki K. A new wild-derived H-2 haplotype enhancing K-IA recombination. Nature. 1982;300:370–2. doi: 10.1038/300370a0. [DOI] [PubMed] [Google Scholar]
- 4.Shiroishi T., Sagai T., Hanzawa N., Gotoh H., Moriwaki K. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 1991;10:681–6. doi: 10.1002/j.1460-2075.1991.tb07997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parvanov E.D., Petkov P.M., Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers S., Bowden R., Tumian A., et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–9. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudat F., Buard J., Grey C., et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groeneveld L.F., Atencia R., Garriga R.M., Vigilant L. High diversity at PRDM9 in chimpanzees and bonobos. PLoS ONE. 2012;7:e39064. doi: 10.1371/journal.pone.0039064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg I.L., Neumann R., Sarbajna S., Odenthal-Hesse L., Butler N.J., Jeffreys A.J. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc. Natl. Acad. Sci. USA. 2011;108:12378–83. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg I.L., Neumann R., Lam K.W., Sarbajna S., Odenthal-Hesse L., May C.A., Jeffreys A.J. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 2010;42:859–63. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brick K., Smagulova F., Khil P., Camerini-Otero R.D., Petukhova G.V. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–5. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandor C., Li W., Coppieters W., Druet T., Charlier C., Georges M. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet. 2012;8:e1002854. doi: 10.1371/journal.pgen.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi K., Yoshida K., Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 14.Mihola O., Trachtulec Z., Vlcek C., Schimenti J.C., Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–5. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 15.Flachs P., Mihola O., Simecek P., et al. Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS Genet. 2012;8:e1003044. doi: 10.1371/journal.pgen.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas J.H., Emerson R.O., Shendure J. Extraordinary molecular evolution in the PRDM9 fertility gene. PloS ONE. 2009;4:e8505. doi: 10.1371/journal.pone.0008505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver P.L., Goodstadt L., Bayes J.J., et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choo Y., Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl. Acad. Sci. USA. 1994;91:11168–72. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billings T., Parvanov E.D., Baker C.L., Walker M., Paigen K., Petkov P.M. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol. 2013;14:R35. doi: 10.1186/gb-2013-14-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H., Shimada T., Terashima M., Tsuchiya K., Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 2004;33:626–46. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Boursot P., Auffray J.C., Britton-Davidian J., Bonhomme F. The evolution of house mice. Annu. Rev. Ecol. Evol. Syst. 1993;24:119–52. [Google Scholar]
- 22.Suzuki H., Nunome M., Kinoshita G., et al. Evolutionary and dispersal history of Eurasian house mice Mus musculus clarified by more extensive geographic sampling of mitochondrial DNA. Heredity. 2013;111:375–90. doi: 10.1038/hdy.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keane T.M., Goodstadt L., Danecek P., et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yonekawa H., Moriwaki K., Gotoh O., et al. Origins of laboratory mice deduced from restriction patterns of mitochondrial DNA. Differentiation. 1982;22:222–6. doi: 10.1111/j.1432-0436.1982.tb01255.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang H., Bell T.A., Churchill G.A., Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007;39:1100–7. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 26.Morita T., Kubota H., Murata K., et al. Evolution of the mouse t haplotype: recent and worldwide introgression to Mus musculus. Proc. Natl. Acad. Sci. USA. 1992;89:6851–5. doi: 10.1073/pnas.89.15.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 29.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Wang J.R., Didion J.P., et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 2011;43:648–55. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes A.L., Ota T., Nei M. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol. Biol. Evol. 1990;7:515–24. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 33.Hussin J., Sinnett D., Casals F., et al. Rare allelic forms of PRDM9 associated with childhood leukemogenesis. Genome Res. 2013;23:419–30. doi: 10.1101/gr.144188.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yonekawa H., Moriwaki K., Gotoh O., et al. Hybrid origin of Japanese mice “Mus musculus molossinus”: evidence from restriction analysis of mitochondrial DNA. Mol. Biol. Evol. 1988;5:63–78. doi: 10.1093/oxfordjournals.molbev.a040476. [DOI] [PubMed] [Google Scholar]
- 35.Yonekawa H., Sato J.J., Suzuki H., Moriwaki K. Evolution of the House Mouse. Cambridge: Cambridge University Press; 2012. Origin and genetic status of Mus musculus molossinus: a typical example of reticulate evolution in the genus Mus. [Google Scholar]
- 36.Takada T., Ebata T., Noguchi H., et al. The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Res. 2013;23:1329–38. doi: 10.1101/gr.156497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Din W., Anand R., Boursot P., et al. Origin and radiation of the house mouse: clues from nuclear genes. J. Evol. Biol. 1996;9:519–39. [Google Scholar]
- 38.Prager E.M., Tichy H., Sage R.D. Mitochondrial DNA sequence variation in the eastern house mouse Mus musculus: comparison with other house mice and report of a 75-bp tandem repeat. Genetics. 1996;143:427–46. doi: 10.1093/genetics/143.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boursot P., Din W., Anand R., et al. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J. Evol. Biol. 1996;9:391–415. [Google Scholar]
- 40.Gündüz I, Rambau R.V., Tez C., Searle J.B. Mitochondrial DNA variation in the western house mouse (Mus musculus domesticus) close to its site of origin: studies in Turkey. Biol. J. Linn. Soc. 2005;84:473–85. [Google Scholar]
- 41.Bonhomme F., Orth A., Cucchi T., et al. Genetic differentiation of the house mouse around the Mediterranean basin: matrilineal footprints of early and late colonization. Proc. R. Soc. Lond. B Biol. Sci. 2011;278:1034–43. doi: 10.1098/rspb.2010.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonhomme F., Searle J.B. Evolution of the House Mouse. Cambridge: Cambridge University Press; 2012. House mouse phylogeography. [Google Scholar]
- 43.Lyon M.F. Transmission ratio distortion in mice. Annu. Rev. Genet. 2003;37:393–408. doi: 10.1146/annurev.genet.37.110801.143030. [DOI] [PubMed] [Google Scholar]
- 44.Pilder S.H., Hammer M.F., Silver L.M. A novel mouse chromosome 17 hybrid sterility locus: implications for the origin of t haplotypes. Genetics. 1991;129:237–46. doi: 10.1093/genetics/129.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silver L.M. Peculiar journey of a selfish chromosome: mouse t-haplotype and meiotic drive. Trends Genet. 1994;9:250–4. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- 46.Hammer M.F., Silver L.M. Phylogenetic analysis of the alpha-globin pseudogene-4 (Hba-ps4) locus in the house mouse species complex reveals a stepwise evolution of t haplotypes. Mol. Biol. Evol. 1993;10:971–1001. doi: 10.1093/oxfordjournals.molbev.a040051. [DOI] [PubMed] [Google Scholar]
- 47.Lenington S., Franks P., Williams J. Distribution of t-haplotypes in natural populations of wild house mice. J. Mammal. 1988;69:489–99. [Google Scholar]
- 48.Ruvinsky A., Polyakov A., Agulnik A., Tichy H., Figueroa F., Klein J. Low diversity of t haplotypes in the eastern form of the house mouse, Mus. musculus L. , Genetics. 1991;127:161–8. doi: 10.1093/genetics/127.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.