Abstract
Members of plant WRKY gene family are ancient transcription factors that function in plant growth and development and respond to biotic and abiotic stresses. In our present study, we have investigated WRKY family genes in Brachypodium distachyon, a new model plant of family Poaceae. We identified a total of 86 WRKY genes from B. distachyon and explored their chromosomal distribution and evolution, domain alignment, promoter cis-elements, and expression profiles. Combining the analysis of phylogenetic tree of BdWRKY genes and the result of expression profiling, results showed that most of clustered gene pairs had higher similarities in the WRKY domain, suggesting that they might be functionally redundant. Neighbour-joining analysis of 301 WRKY domains from Oryza sativa, Arabidopsis thaliana, and B. distachyon suggested that BdWRKY domains are evolutionarily more closely related to O. sativa WRKY domains than those of A. thaliana. Moreover, tissue-specific expression profile of BdWRKY genes and their responses to phytohormones and several biotic or abiotic stresses were analysed by quantitative real-time PCR. The results showed that the expression of BdWRKY genes was rapidly regulated by stresses and phytohormones, and there was a strong correlation between promoter cis-elements and the phytohormones-induced BdWRKY gene expression.
Keywords: WRKY, Brachypodium distachyon, evolution, abiotic stresses, biotic stresses
1. Introduction
Grasses (Poaceae), including rice, wheat, and sorghum, are the most important plant species on the earth, and are a major source of nutrition and sustainable energy.1 To study Poaceae genome will help scientists better understand the mechanisms of how genes control physiological events in Poaceae, and help discover and make use of functional genes from the large amount of Poaceae plants, especially from those able to survive under extreme conditions. Recently, Brachypodium distachyon has been used as a new model organism for Poaceae grass, as it is much more closely related to several economically important Poaceae species such as rice, sorghum, wheat, and turf grasses.
The WRKY family genes are plant transcription activators in various physiological processes; they were regarded as the first isolated regulatory genes from plants.2,3 WRKY transcription factors (TFs) are conserved in evolutional history throughout the plant kingdom. Members of this family contain at least one conserved DNA-binding domain with a highly conserved WRKYGQK heptapeptide sequence, followed by a C2H2- or C2HC-type of zinc finger motifs. These conserved sequences have been designated as the WRKY domains, and function in W-box DNA motif (C/T)TGAC(C/T)-binding activation.4 In Arabidopsis thaliana, a total of 72–74 members of the WRKY TFs can be divided into three major groups with several subgroups, based on their sequences in the WRKY domain and their relationships in the phylogenetic clades.4,5 The Group I WRKY TFs contain two WRKY domains, one at the C- and the other at the N-terminal of the protein. These two WRKY domains seem to be functionally redundant.6 Peptide sequences outside the C-terminal WRKY domain contribute significantly to the overall strength of DNA binding; the N-terminal WRKY domain might participate in the binding process by increasing the affinity or specificity to their targets.7–9 In contrast, most Group II and Group III WRKY TFs only contain a single WRKY domain; this domain is more similar in sequence to the C-terminal than to the N-terminal WRKY domain of Group I proteins, suggesting that the C-terminal WRKY domain in Group I WRKY TFs and single WRKY domains in Group II and Group III WRKY TFs are functionally equivalent and constitute the major DNA-binding domain.4 The differences between Groups II and III are in their C-terminal zinc domain.
Previous studies have demonstrated that WRKY TFs play essential roles in various physiological processes, including senescence, root development, sugar signalling, and germination.3,10 Furthermore, WRKY TFs have been shown to be involved in responses to various biotic stresses caused by viruses,11 bacterial pathogens,12,13 fungi,14 abiotic stresses,3,15,16 and some signalling substances such as salicylic acid (SA)/benzothiadiazole,17–19 jasmonic acid (JA),18–20 gibberellin,21 and abscisic acid (ABA).22,23 In Arabidopsis, the majority of the 74 WRKY genes are transcriptionally inducible upon pathogen infection and other defence-related stimuli.24 For example, it has been proven that AtWRKY25 functioned as a negative regulator of SA-mediated defence responses to Pseudomonas syringae.13 In Boea hygrometrica leaves, BhWRKY1 is proven to be a regulator in an ABA-dependent signal pathway to regulate BhGolS1 expression.23 Using northern blotting analysis, Qiu et al.15 revealed in rice that 10 of 13 OsWRKY genes were differentially regulated in response to abiotic stress factors NaCl, polyethylene glycol (PEG), cold, and heat. Under a salinity stress, a microarray analysis using 70-mer oligonucleotide probes representing 23 686 genes revealed that 18 AtWRKY genes were induced in A. thaliana root treated with 150 mM NaCl.16 Furthermore, numerous studies have shown that many WRKY genes were responsive to drought, heat, cold, and so on. On the other hand, a single WRKY gene often showed transcription activity in response to several stress factors, indicating that it has different regulatory function in diverse stress responses. For example, the expression of AtWRKY25 and AtWRKY33 were responsive to both heat and salt stress.25,26 Thus, a genome-wide analysis of B. distachyon WRKY genes should help to reveal the underlying complex molecular mechanisms of WRKY proteins in response to various stresses.
In our study, 86 WRKY genes were identified from the B. distachyon Bd21 genome and classified according to their homology with known WRKY genes in Oryza sativa. We investigated the evolutionary relationship of B. distachyon WRKY TFs with their counterparts from monocot O. sativa and dicot A. thaliana. Subsequently, we used quantitative real-time PCR (qRT-PCR) to examine their transcript profiles in different tissues and in response to several biotic or abiotic stresses and phytohormone treatments. Since BdWRKYs showed various expression patterns and expression levels under a series of abiotic stresses and phytohormone treatments, we checked if there are correlations between the differences in the WRKY domain and their spatial and temporal expression patterns in response to stress treatments. We have also done detailed correlation analyses between promoter cis-elements and the genes expression pattern. Our study provided genome-wide evolutionary characterization and expression analysis of WRKY genes in B. distachyon, an important step for further investigation into the functions of these genes.
2. Materials and methods
2.1. Sequence retrieval
We performed a BLAST search among sequenced genomes of land plants in plantTFDB,27 GramineaeTFDB,28 Superfamily, and Phytozome (http://www.Phytozome.net) using well-known plant WRKY proteins as queries. The database of UniProt (http://www.uniprot.org/blast/) and GeneBank (http://www.ncbi.nlm.nih.gov/) were used for searching the WRKY proteins in red and green algae. To verify the reliability of our results, all putative non-redundant sequences were assessed with UniProt and SMART (http://smart.embl-heidelberg.de/) analyses, respectively.
2.2. Identification of WRKY protein in B. distachyon
To identify B. distachyon genes encoding WRKY proteins with at least one possible WRKY domain, we performed a GeneBank BLASTP search, UniProt, and B. distachyon genome Database (http://www.brachypodium.org/), using the amino acid sequences of the WRKY domain. The Brachy WRKY Database (http://www.igece.org/WRKY/BrachyWRKY/BrachyWRKYIndex.html) was used as a referral for verifying the reliability of our results.29 We also obtained information of the chromosome locations of each gene from the results of BLASTP at the B. distachyon genome Database. A total of 86 BdWRKY genes were found in B. distachyon (Supplementary Table S1). Furthermore, to avoid confusion, we used the same numbering system as Tripathi et al.29
2.3. Sequence analysis
To analyse the sequence of the 86 typical identified B. distachyon WRKY proteins, we performed multiple alignment analyses of the WRKY domains sequence by ClustalW (www.ebi.ac.uk/clustalw/).30
2.4. Phylogenetic analysis
A neighbour-joining (NJ) tree was constructed using the MEGA version 5 software,31 based on the alignment of WRKY domains in O. sativa, A. thaliana, and B. distachyon. To determine the statistical reliability, we conducted bootstrap analysis with the following parameters: p-distance and pairwise deletion. Bootstrap analysis was performed with 1000 replicates.
2.5. Protein motifs and structure analysis
Analysis for conserved motifs in the WRKY proteins was carried out using MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi).32 The settings were: any number of repetitions of a single motif, the minimum width of a motif with six amino acids, the maximum width of a motif with 80 amino acids, and the maximum number of motifs up to 15 amino acids. Subsequently, the MAST program was used to search detected motifs in protein databases.33 The details of sequence logo of motifs were shown in Supplementary Fig. S1.
2.6. Cluster analysis of expression data
The 2-week-old seedlings (Bd21) were used for harvesting leaf, stem, and root samples. For phytohormone analysis, 2-week-old seedlings were treated in MS liquid medium containing 100 µM methyl jasmonate (MeJA), 100 µM ABA, 1 mM SA, and 20 µM 6-Benzylaminopurine (6-BA) for 3 h, respectively. For abiotic stress treatment, 2-week-old seedlings were treated in MS liquid medium containing 20% PEG, 200 mM NaCl, and 10 mM H2O2 for 3 h, respectively. Cold and heat treatments were achieved by placing 2-week-old seedlings in MS liquid medium at 4 or 45°C for 3 h, respectively. For phytopathogen treatment, 2-week-old seedlings were sprayed with Fusarium graminearum (F0968) and two strains of Magnaporthe grisea (Guy11, avirulent ACE1 genotype; PH14, virulent ACE1 genotype) for 4 or 12 h. The BdWRKY array constituted of 86 primer sets representing all members of the B. distachyon WRKY gene family. The primer sets are listed in Supplementary Table S2. The expression of the 86 BdWRKY genes was assessed upon the qPCR result analysis. Each experiment was repeated three separate times. The expression profile was calculated from the –ΔΔCT value [−ΔΔCT = (CTcontrol.gene − CTcontrol.actin) - (CTtreat.gene − CTtreat.actin)], and obtained by the PermutMatrixEN vesion 1.9.3 software, and shown by a green-red gradient. The data were statistically analysed using an OriginPro 7.5 software. The up-regulated genes were defined as a fold change greater than 1.5 with a P-value of <0.05, and with a fold change of ≤0.66 was used to define down-regulated genes when the P-value of <0.05.
2.7. Promoter analysis
The 1500 bp promoter sequences of BdWRKY genes were obtained from the B. distachyon genome Database. PLANT CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to determine the cis-acting regulatory elements and to analyse the BdWRKY gene promoter sequences.34
3. Results and discussion
3.1. Distribution of WRKY domain-containing proteins in plant kingdom
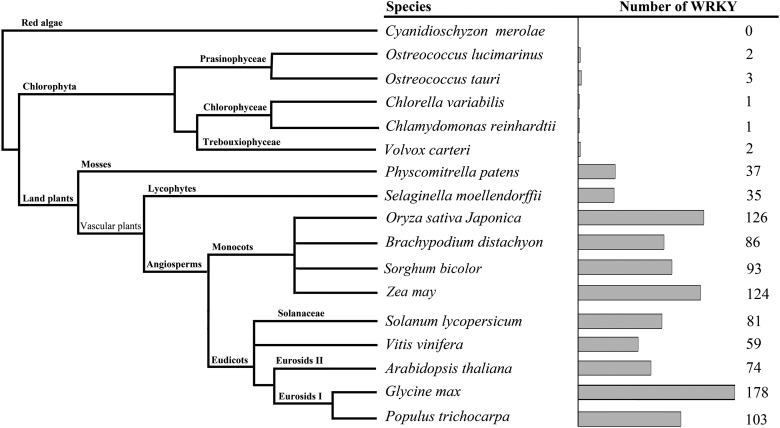
WRKY domain-containing proteins are extensively found in plants, some fungi, bacteria, and slime moulds. Here, we searched for WRKY genes in six comprehensive datasets, GenBank, UniProt, plantTFDB, GramineaeTFDB, Superfamily, and Phytozome of plant species. In this study, we focused our search and analyses on six major types of model organisms whose genomes have been already sequenced, including red alga, the chlorophytes, the moss, the lycophyte, the eudicots, and the monocots.35,36 The result showed that, as a gene super family that plays important roles in regulation of defence response pathways, WRKY TFs conservatively existed in plant kingdom (Fig. 1). In general, only a few of WRKY homologous genes could be found in algae genome, while plants possess a large number of WRKY genes (Fig. 1). The results indicated that the earliest evolutionary origin of the gene containing the WRKY was from unicellular green algae of chlorophyta, suggesting that WRKY proteins arose before plants transitioned from water to land. With the evolution of species, the land plants have developed a series of highly sophisticated mechanisms that help them to adapt to changing environmental conditions,37 and hence, the number of WRKY TFs increased and they were extensively found in land plants in response to the environmental stimuli and regulation of physiological reactions.
Figure 1.
Distribution of the WRKY domain-containing proteins in Plantae. The total number of WRKY homologous genes found in each genome is indicated on the right.
3.2. Chromosomal distribution and duplication events of BdWRKY genes
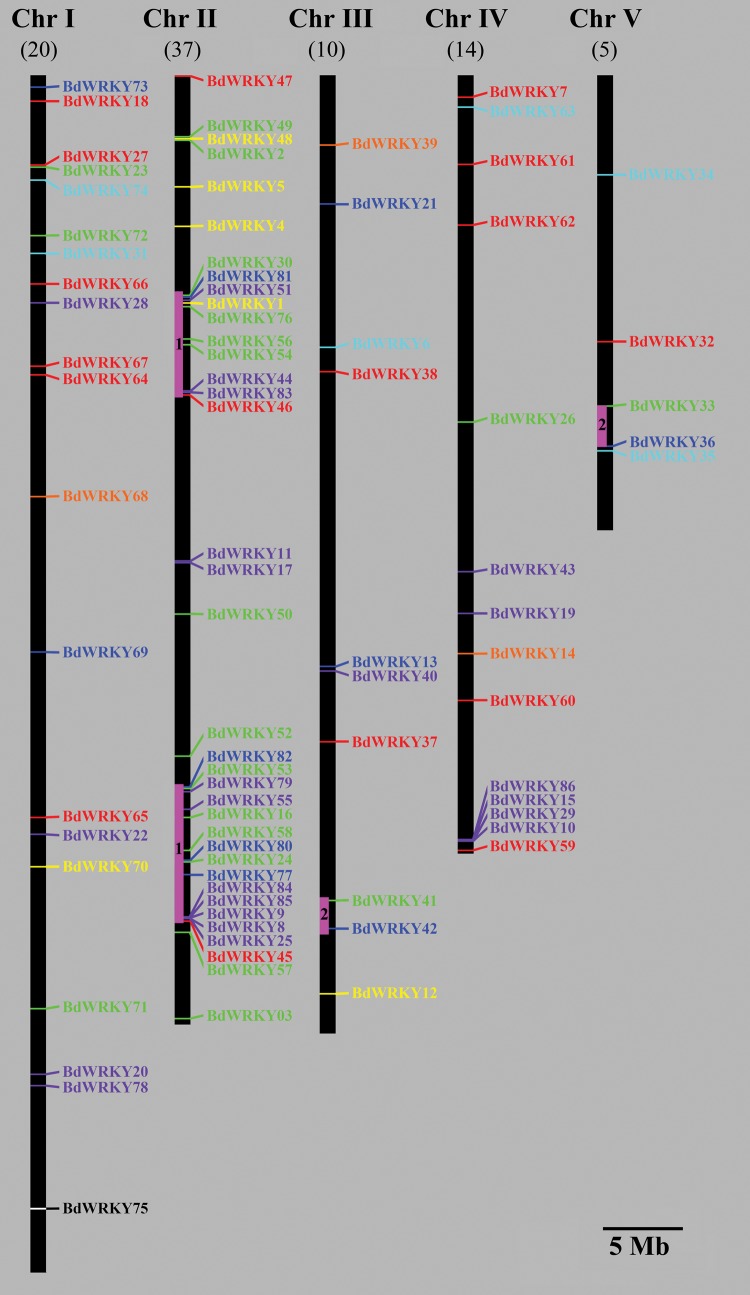
To date, the information regarding expansion events of the BdWRKY gene family in B. distachyon remains unclear. To investigate the relationship between genetic divergence and gene duplication within the BdWRKY gene family in B. distachyon, we determined the chromosomal locations of BdWRKYs based on the information from the B. distachyon genomic database (http://www.brachypodium.org/). The result showed that the BdWRKYs were distributed throughout all the five B. distachyon chromosomes, most BdWRKYs were distributed on Chromosomes 1 and 2 (Fig. 2). Then, the distribution appeared to be uneven. Relatively high densities of BdWRKYs were observed on the top and at the bottom arms of Chromosome 2. In contrast, low densities were detected in Chromosomes 3, 4, and 5. Subsequently, we analysed the gene cluster expansion events of BdWRKYs in the B. distachyon genome. Based on the phylogenetic relationship and sequence similarity, we identified 15 pairs of BdWRKY genes with high levels of protein sequence similarity. For instance, the entire protein sequences of BdWRKY33 and BdWRKY41 shared 71% similarity, whereas those of BdWRKY81 and BdWRKY82 shared 64% similarity. Among BdWRKY genes with a high degree of homology, 8 (53%) pairs of BdWRKYs reside within chromosomal segments that have clear relatives in the B. distachyon genome, suggesting that they may have evolved from duplication events. As shown in Fig. 2, two of those multiple pairs of duplicated regions were located at Chromosome 2, and the others distributed on Chromosomes 3 and 5 (Fig. 2, bars with numbers).
Figure 2.
Chromosomal locations and regional duplication for B. distachyon WRKY genes. The chromosomal position of each BdWRKY was mapped according to the B. distachyon genome. The chromosome number is indicated at the top of each chromosome. The number below indicates the number of BdWRKYs in each chromosome. The scale is 5 Mb. The bars with numbers on the chromosomes indicate the four predicted duplication regions.
In general, plants can integrate alternative developmental pathways during evolution, and then choose suitable pathways in their growth and development in response to different environmental cues.38 It is believed that multiple members of a specific gene family that form a large regulative network to control complicated physiological processes were a result of the long evolutionary history of a particular species.39,40 The individual members of a gene family represent a succession of genomic rearrangements and expansions during the process of evolution.41 In this study, we found at least four putative segmental duplication events in the B. distachyon genome; and those duplications influenced the distribution of BdWRKY genes in B. distachyon. Particularly, the putative duplications between BdWRKY33/36 and BdWRKY41/42 were highly similar. Moreover, on the putative segmental duplications of the Chromosome 2, the order of the BdWRKY genes (including BdWRKY30/81/51/56/46) arrangement on the top arm of Chromosome 2 was similar to those of the BdWRKY genes (including BdWRKY82/53/16/58/45) at the bottom arm of this chromosome. BdWRKY33 and BdWRKY36 are on a segment of Chromosome 5, and this segment is likely a duplicate of a segment on Chromosome 3 where BdWRKY41 and BdWRKY42 are located (Fig. 2). The motif structure of BdWRKY33 is identical with BdWRKY41, while there were only a few differences between BdWRKY36 and BdWRKY42, suggesting that the C-terminal of the segmental duplications on Chromosomes 3 and 5 might diverge to perform new functions during the process of evolution (Fig. 2). Thus, it is inferred that the new gene initially resulted from the duplication, and thereafter diverge from a series of synonymous and/or non-synonymous mutations.
3.3. Characteristics of BdWRKY domains
The gene family of TFs usually contain highly conserved domain or domains involved in DNA binding.42 Assigning structural domains to protein sequences is important in performing a comprehensive analysis of highly divergent sequences in large gene families.43 Based on both the number of WRKY domains and the features of their zinc finger-like motif, the BdWRKY can be classified into three main groups, consistent with the previous report.29 The WRKY TFs with two WRKY domains belong to the Group I, while most proteins with one WRKY domain belong to the Group II (Supplementary Fig. S2). Generally, Group I and Group II WRKY TFs share the same type of zinc finger-like motif with a C2H2 zinc ligand (C–X4–5–C–X22–23–H–X1–H; Supplementary Fig. S2). There is a small subset of BdWRKY TFs containing a C2HC motif (C–X7–C–X 23–H–X1–C; Supplementary Fig. S2), and this subset is assigned to Group III. Although the WRKYGQK heptapeptide sequence was highly conserved in BdWRKY TFs, sequence similarity beyond the domains is quite low among most genes. As we know, a protein domain is considered as an evolutionary unit of protein function and the domain coding sequence can be duplicated and/or recombined.44 From recent research on genomes analysis, new protein functionalities appear to arise from the addition or exchange of protein domains by duplicating one or more domains, recombining fragments of DNA from different organisms, and diverging duplicated sequences by base substitutions, deletions, and insertions.41,45 Therefore, the whole family of BdWRKY TFs, which might result from long-time evolutionary history, represented divergent WRKY domains, even in much closely related gene pairs, such as BdWRKY33/41, BdWRKY24/54, BdWRKY81/82, and so on.
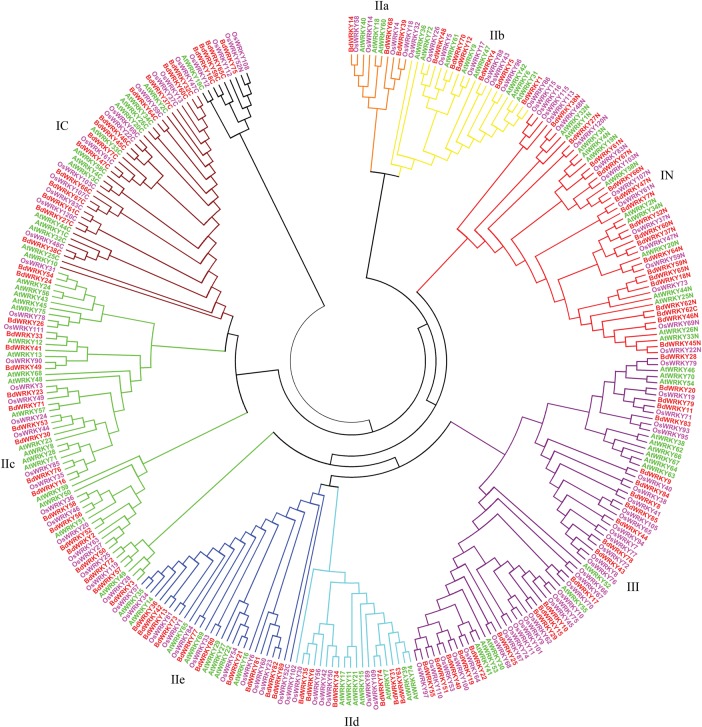
To further investigate the evolutionary relationships among the WRKY domains from different species, we estimated the phylogeny by using the NJ program from MEGA 5 for the WRKY domains from O. sativa, A. thaliana, and B. distachyon. All subgroups were present in monocots and eudicots (Fig. 3), indicating that the appearance of most WRKY TFs in plants predates the divergence of monocot/eudicots. Meanwhile, no species-specific subgroups and/or clades were observed in O. sativa, A. thaliana, or B. distachyon, implying that WRKY family genes were more conserved during evolution. In addition, WRKY domains from the same lineage tend to cluster together in the phylogenetic tree, suggesting that they experienced duplications after the lineages diverged (Fig. 3). Furthermore, WRKY phylogenetic tree showed almost the same clustering patterns in O. sativa and B. distachyon (Fig. 3 and Supplementary Table S3). In total, about 62 pairs of WRKY domains from O. sativa and B. distachyon were clustered as pairs, indicating that they might be the orthologous WRKY domains (Fig. 3). For example, the WRKY domains of BdWRKY54 and OsWRKY31 are highly similar, indicating that some consensus in domain may have existed before the divergence of B. distachyon and O. sativa. Meanwhile, only two pairs of WRKY domains from B. distachyon and A. thaliana could be clustered as pairs, suggesting that the BdWRKY domains are evolutionarily more closely related to OsWRKY domains, which is consistent with the notion that both B. distachyon and O. sativa belong to monocots. The phylogenetic similarity found in O. sativa and B. distachyon WRKY domain suggests that they may have evolved conservatively.
Figure 3.
NJ analyses of 301 WRKY domains from O. sativa, A. thaliana, and B. distachyon, containing 262 plant WRKY proteins. The domains clustered into eight major subgroups, IN, IC, IIa, IIb, IIc, IId, IIe, and III.
3.4. Protein structure and tissue-specific expression pattern of BdWRKY
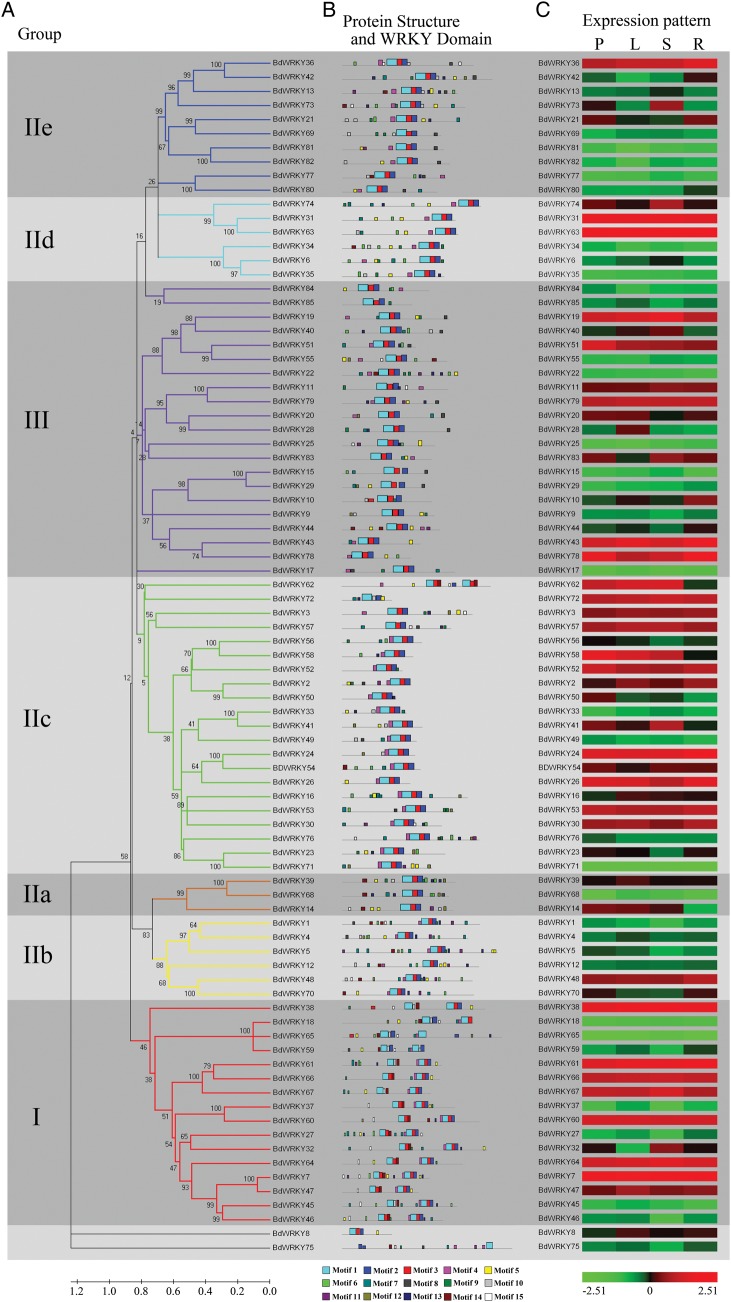
Based on sequence similarity and protein structure, we also divided the 86 members of the BdWRKY TFs into seven subgroups (I, IIa–e, and III) (Fig. 4A and B). Remarkably, the WRKY domains were almost identical, even though the lengths of the coding region of the WRKY genes were different, and the cluster result based on whole BdWRKY sequences was different from the clustering based on BdWRKY domains. A schematic representing the structure of all members of BdWRKY TFs was constructed from the MEME motif analysis results (Fig. 4B).32 Most members of the BdWRKYs shared three motifs, Motif 3, Motif 2, and Motif 1 linked in order. A few members, such as BdWRKY8, BdWRKY65, BdWRKY72, and BdWRKY75, showed quite different protein structures compared with other members (Fig. 4B). Interestingly, many of the motifs were selectively distributed among the specific clades in the phylogenetic tree, for example, Motif 8 in Group IIe, and Motif 15 in Group I. The clustered BdWRKY pairs, i.e. BdWRKY81/82, BdWRKY31/63, showed highly similar motif distribution (Fig. 4B). The motifs and their arrangement in the BdWRKYs are similar among proteins within subfamilies, demonstrating that the protein architecture is remarkably conserved within a specific subfamily. The biological functions of many WRKYs remain to be elucidated. The above findings may facilitate the identification of functional units in BdWRKYs and lead to the discovery of their roles in plant growth and development.
Figure 4.
Phylogenetic relationships and subgroup designations in WRKY proteins with tissue-specific expression profile from B. distachyon. (A) The phylogenetic tree was constructed from the amino acid sequences using the NJ program from MEGA 5, representing relationships among 86 WRKY proteins from B. distachyon. The proteins are clustered into seven subgroups, which are designated with a subgroup number (e.g. IIe) and marked with a different background to facilitate subfamily identification with a high predictive value. The numbers beside the branches represent bootstrap support values (>50%) from 1000 replications. (B) Structure of WRKY proteins and the WRKY domains in B. distachyon. The details of sequence logo of motifs were shown in Supplementary Fig. S1. (C) Expression patterns of WRKY genes in B. distachyon in different tissues. P for seedling, L for leaf, S for stem, R for root. In this expression pattern analysis, the 2-week-old seedlings were used for harvesting different tissues including leaf, stem, and root. The BdWRKY array was constituted of 86 primer sets representing all members of the B. distachyon WRKY gene family. The expression values of the 86 BdWRKY genes were assessed upon the qPCR result analysis.
TFs usually harbour many types of DNA-binding domains and they can be grouped into a handful of different, often large, gene families.46 By forming intricate networks, TFs control the expression of genes in a genome at the transcriptional level.47 It has been noted previously that many TF gene families exhibit great disparities in abundance among different organisms and different tissues to exert different physiological functions. Thus, gene expression patterns can provide important clues for gene function. To further analyse the tissue specificity of the WRKY gene family members, we confirmed their transcription levels in three different tissues, leaves, stems, and roots. The expression of all of the WRKY gene family members was detected in all three tissues (Fig. 4C). The results revealed that several BdWRKY genes, including BdWRKY7, −24, −31, −38, −61, and −64, showed higher expression levels than other members of the WRKY family in all the tissues tested. The expression of BdWRKY78 was highly induced in the root while its expression level was relatively low in the leaf and stem. BdWRKY32, −41, −73, and −74 showed higher expression levels in the stem than that in the leaf and root (Fig. 4C). The expression pattern of these genes suggested that BdWRKYs were involved in the growth and development of organs or tissues under specific conditions. Interestingly, most of clustered gene pairs showed the same expression pattern, such as BdWRKY31/63, BdWRKY81/82, BdWRKY77/80, and so on. On the other hand, gene pairs BdWRKY37/60, BdWRKY36/42, BdWRKY56/58, and other clustered pairs exhibited different expression patterns (Fig. 4C). These results indicated that most of clustered gene pairs had more similarities in the WRKY domain and shared similar expression patterns; they might be functionally redundant. The BdWRKY pairs that showed different expression levels may be involved in different signalling pathways. Since the expression of genes was regulated by a series of TFs, the disparities in abundance of BdWRKY gene among different tissues suggested that the BdWRKY genes, although are TFs themselves, were also regulated by other TFs in different tissues.
3.5. Expression profiles of BdWRKY upon multiple phytohormone treatments and abiotic or biotic stresses
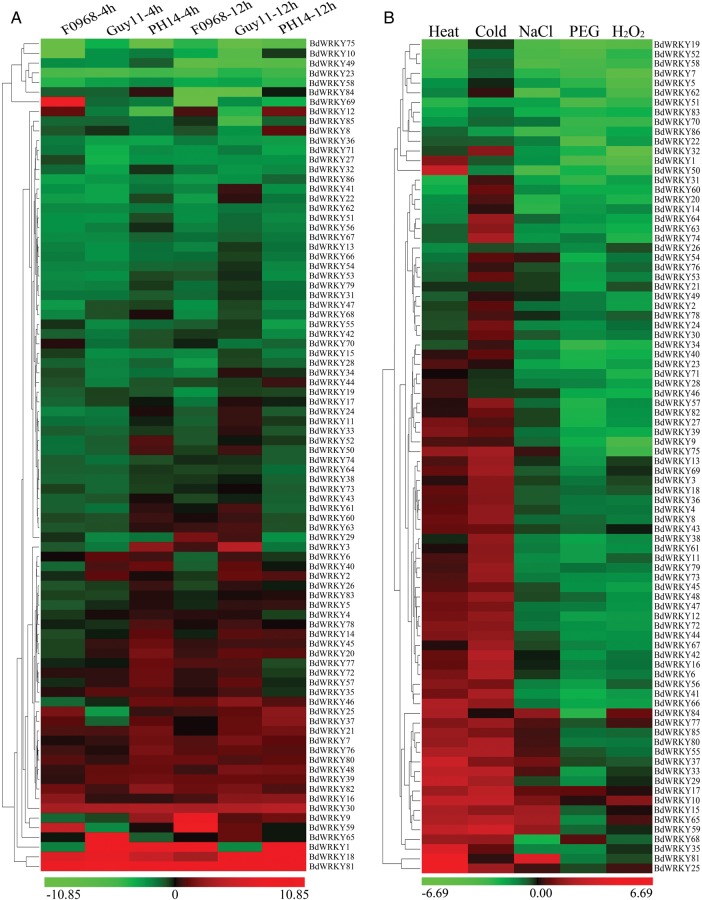
It has been demonstrated that WRKY genes were not only involved in the activation of plant defence systems,48 but also played key roles in the control of plants' response to environmental stimuli.3 Since it has been thought that BdWRKY genes are responsive to plant defence-related phytohormones, we investigated the expression profiles of the WRKY family genes in B. distachyon after phytopathogen treatments. A total of three phytopathogens, including F. graminearum (F0968) and two strains of M. grisea (Guy11, avirulent ACE1 genotype; PH14, virulent ACE1 genotype), were used to inoculate Bd21 seedling in this study. The expression profiles of the BdWRKY family genes at 4 hpi (hour post-inoculation) and 12 hpi were shown in Fig. 5A. The data demonstrated that a large number of BdWRKY genes were rapidly and significantly up-regulated after inoculation of phytopathogen within 4 h. At least 15 BdWRKY genes were up-regulated by all three phytopathogens treated, while nine BdWRKY genes were induced after single phytopathogen inoculation, such as BdWRKY8, −34, −50, −69, −70, and so on. As shown in Fig. 5A, the expression levels of BdWRKY21, −37, −69, and −70 increased remarkably at 4 hpi, and decreased at 12 hpi after F0968 treatment. However, several BdWRKY genes (BdWRKY1, −9, −29, etc.) were up-regulated 12 h after F0968 inoculation. These data suggested that BdWRKY21, −37, −69, and −70 were the early response TFs upon phytopathogen F0968 attack, while BdWRKY1, −9 and −29 were induced at the second stage of the F0968 infection. Interestingly, numbers of BdWRKY genes (e.g. BdWRKY3, −72 and -77) were induced faster by PH14 than by Guy11. They were up-regulated at 4 hpi after infection by PH14, but at 12 hpi, they were down-regulated by PH14 and up-regulated by Guy11. Since the pathogenic ability of virulent ACE1 genotype (PH14) was stronger than the wild-type Guy11, these results suggested that the expression of BdWRKY genes were very sensitive to biotic stress and the regulation of BdWRKYs were important to plant defence. BdWRKY as TF genes were first induced or repressed by phytopathogen, and then, were involved in the regulation of plant defence gene expression.
Figure 5.
Expression profiles of BdWRKY genes under biotic and abiotic stresses. (A) The 2-week-old seedlings were sprayed with different pathogens. (B) Clustering of BdWRKY genes according to their expression profiles in the seedling of B. distachyon after different stress treatments. The BdWRKY array was constituted of 86 primer sets representing all members of the B. distachyon WRKY gene family. The expression of the 86 BdWRKY genes was assessed upon the qPCR result analysis.
Similarly, the expression profiles of the BdWRKY family genes under different stress conditions were also examined using the qRT-PCR in our study. A total of five stress types, i.e. heat, cold, NaCl, PEG, and H2O2, were tested in this study. Detailed expression profiles of the WRKY family genes under different stress conditions were provided in Supplementary Table S4. Heatmap representation of expression profiles of these WRKY family genes under different stress conditions are shown in Fig. 5B. The data revealed that 60 and 80% of BdWRKY genes were up-regulated under heat and cold stress conditions, respectively. More than 50% of the BdWRKY genes were up-regulated under more than one stress conditions. For examples, BdWRKY10, −33, −59, and −65 were up-regulated in both heat and cold treatments, while BdWRKY81 showed a high up-regulation under heat and salt stresses. It has been reported that the severity of the stress and the metabolic status of the plant affected the capacity of plant to tolerate abiotic stress.49 ABA as a phytohormone plays an important role in integrating various abiotic or biotic stress signals and controlling downstream stress responses.49 Here, our data indicated that almost 50% of the BdWRKY genes were down-regulated under three or more stress conditions, which is consistent with the results of most BdWRKY gene down-regulated by ABA treatment. For example, BdWRKY19, −22, −51, and −52 were down-regulated by PEG (drought stress), and similarly, their expression levels were very low after ABA treatment. These correlations of BdWRKY genes expression levels between abiotic stress and phytohormone treatment suggest that BdWRKY regulation of downstream gene expression may be linked to stress-induced phytohormone alteration.
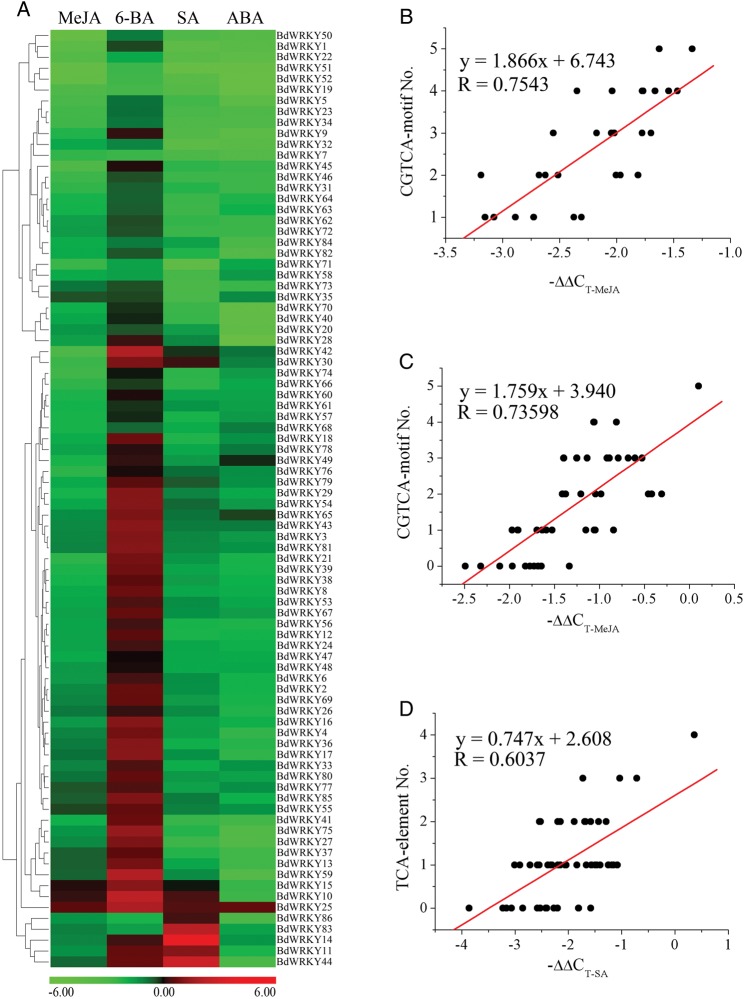
Recent studies of the OsWRKY genes have also shown that many of OsWRKY genes were responsive to JA, SA, and ABA treatments.50,51 It has also been reported that WRKY TFs were key factors for the increased transcript abundance of SA- and JA-responsive genes.52–54 To investigate the hormonal control mechanisms underlying BdWRKY gene expression, we treated Bd21 seedlings with four phytohormones, MeJA, SA, 6-BA, and ABA, respectively and analysed the changes in transcript abundance of these 86 BdWRKY genes using qRT-PCR. Our results demonstrated that most of BdWRKY genes were repressed by ABA after 3 h of treatment (Fig. 6A). In contrast, 52 of the 86 BdWRKY genes were up-regulated within 3 h treatment of 6-BA. Only three and eight BdWRKY genes exhibited increased expression levels in response to MeJA and SA treatments, respectively. Seven of 86 BdWRKY genes exhibited positive modulation of expression in response to two phytohormones, while only BdWRKY25 showed up-regulation in response to all four phytohormones. Interestingly, BdWRKY14 showed very high expression after SA treatment, even though its expression level was decreased after MeJA or ABA treatment. It has been reported that the expression of two genes, OsWRKY45 and −62 (OsWRKY71 and −14 in this paper, respectively), were increased in SA (SA)-treated rice leaves.50 Similarly, BdWRKY11 and −14 (the homologous genes of OsWRKY45 and −62) also showed a rapid increase after SA treatment. The results indicated that some homologous genes between O. sativa and B. distachyon share functional conservation. On the other hand, our data also revealed that a number of B. distachyon homologous of rice WRKY genes (e.g. OsWRKY48/BdWRKY38) did not show similar expression patterns, suggesting that the functions of some genes were altered during the evolution. According to the statistical analysis, there is a good correlation between the number of MeJA-inducible cis-element and the expression levels in most BdWRKY genes after 3 h MeJA treatment (Fig. 6B and C, and Supplementary Fig. S4). Similarly, the number of SA-inducible cis-element showed a good correlation with the expression levels of most BdWRKY genes after 3 h SA treatment (Fig. 6D). These results indicated that BdWRKY TFs were regulated by exogenous phytohormones and then bind to the W-box in promoters of downstream genes and regulate their expressions.
Figure 6.
The expression profiles of BdWRKY genes under hormone treatment. (A) Clustering of BdWRKY genes according to their expression profiles in the seedling of B. distachyon after different phytohormone treatments. (B and C) The relevance analysis between MeJA-related elements and MeJA-induced BdWRKY gene expression. (D) The relevance analysis between SA-related elements and SA-induced BdWRKY gene expression.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grant for Starting Package to the Research Group of Plant Abiotic Stress and Plant Genome Evolution in Shanghai Chenshan Plant Science Research Centre, Chinese Academy of Sciences, and Shanghai Chenshan Botanic Garden from Shanghai Landscaping Administrative Bureau (No. F0112423 and F0122415), the Fund for National Key Laboratory of Plant Molecular Genetics (Y109Z11161), and the Special Fund for Chinese Academy of Sciences (CZBZX-1).
Supplementary Material
Acknowledgements
We thank contributors of the B. distachyon genome Database, which is convenient for searching BdWRKY genes, and thank Tripathi et al. for sharing their work on BdWRKY genes through the Brachy WRKY Database website, which was used as a reference for verifying the reliability of our results. We thank Prof. Weihua Tang, Prof. Zuhua He (Institute of Plant Physiology and Ecology, SIBS, CAS, China), and Prof. Huaigu Chen (Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, China) for providing F. graminearum (F0968) and M. grisea (Guy11 and PH14). We are grateful to Prof. Nanfei Xu from Shanghai Center for Plant Stress Biology, CAS for careful and thorough editing of this manuscript.
Footnotes
Edited by Dr Kazuo Shinozaki
References
- 1.Bevan M.W., Garvin D.F., Vogel J.P. Brachypodium distachyon genomics for sustainable food and fuel production. Curr. Opin. Biotechnol. 2010;21:211–7. doi: 10.1016/j.copbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Berri S., Abbruscato P., Faivre-Rampant O., et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta. 2012;18,19:120–8. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 5.Dong J.X., Chen C.H., Chen Z.X. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003;51:21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 6.Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Pater S., Greco V., Pham K., Memelink J., Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24:4624–31. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulgem T., Rushton P.J., Schmelzer E., Hahlbrock K., Somssich I.E. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–99. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeo K., Hayashi S., Kojima-Suzuki H., Morikami A., Nakamura K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotech. Biochem. 2001;65:2428–36. doi: 10.1271/bbb.65.2428. [DOI] [PubMed] [Google Scholar]
- 10.Rushton P.J., Somssich I.E., Ringler P., Shen Q.X.J. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Park C.J., Shin Y.C., Lee B.J., Kim K.J., Kim J.K., Paek K.H. A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to Tobacco mosaic virus and Xanthomonas campestris. Planta. 2006;223:168–79. doi: 10.1007/s00425-005-0067-1. [DOI] [PubMed] [Google Scholar]
- 12.Xu X.P., Chen C.H., Fan B.F., Chen Z.X. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–26. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Z.Y., Mosher S.L., Fan B.F., Klessig D.F., Chen Z.X. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007;7:2. doi: 10.1186/1471-2229-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Z.Y., Abu Qamar S., Chen Z.X., Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 15.Qiu Y.P., Jing S.J., Fu J., Li L., Yu D.Q. Cloning and analysis of expression profile of 13 WRKY genes in rice. Chin. Sci. Bull. 2004;49:2159–68. [Google Scholar]
- 16.Jiang Y.Q., Deyholos M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006;6:25. doi: 10.1186/1471-2229-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C.H., Chen Z.X. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002;129:706–16. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X.Q., Bai X.Q., Wang X.J., Chu C.C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007;164:969–79. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Qiu D.Y., Xiao J., Ding X.H., et al. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 2007;20:492–9. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- 20.Mao P., Duan M.R., Wei C.H., Li Y. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol. 2007;48:833–42. doi: 10.1093/pcp/pcm058. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z.L., Xie Z., Zou X.L., Casaretto J., Ho T.H.D., Shen Q.X.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004;134:1500–13. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Z., Zhang Z.L., Zou X.L., Yang G.X., Komatsu S., Shen Q.X.J. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006;46:231–42. doi: 10.1111/j.1365-313X.2006.02694.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., Zhu Y., Wang L.L., et al. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta. 2009;230:1155–66. doi: 10.1007/s00425-009-1014-3. [DOI] [PubMed] [Google Scholar]
- 24.Eulgem T. Dissecting the WRKY web of plant defense regulators. Plos Pathog. 2006;2:e126. doi: 10.1371/journal.ppat.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S.J., Fu Q.T., Huang W.D., Yu D.Q. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009;28:683–93. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y.Q., Deyholos M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Jin J., Tang L., et al. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39:D1114–1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. In silico analysis of transcription factor repertoires and prediction of stress-responsive transcription factors from six major gramineae plants. DNA Res. 2011;18:321–32. doi: 10.1093/dnares/dsr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi P., Rabara R.C., Langum T.J., et al. The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics. 2012;13:270. doi: 10.1186/1471-2164-13-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey T.L., Baker M.E., Elkan C.P. An artificial intelligence approach to motif discovery in protein sequences: application to steriod dehydrogenases. J. Steroid Biochem. Mol. Biol. 1997;62:29–44. doi: 10.1016/s0960-0760(97)00013-7. [DOI] [PubMed] [Google Scholar]
- 33.Bailey T.L., Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 34.Lescot M., Dehais P., Thijs G., et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du H., Wang Y.B., Xie Y., et al. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013;20:437–448. doi: 10.1093/dnares/dst021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Maruyama S., Sekimoto H., Sakayama H., Nozaki H. An extended phylogenetic analysis reveals ancient origin of ‘non-green’ phosphoribulokinase genes from two lineages of ‘green’ secondary photosynthetic eukaryotes: Euglenophyta and Chlorarachniophyta. BMC Res. Notes. 2011;4:330. doi: 10.1186/1756-0500-4-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy A.S., Ali G.S., Celesnik H., Day I.S. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23:2010–32. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teale W.D., Ditengou F.A., Dovzhenko A.D., et al. Auxin as a model for the integration of hormonal signal processing and transduction. Mol. Plant. 2008;1:229–37. doi: 10.1093/mp/ssn006. [DOI] [PubMed] [Google Scholar]
- 39.Moore R.C., Purugganan M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005;8:122–8. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Lima M.D., Eloy N.B., Pegoraro C., et al. Genomic evolution and complexity of the Anaphase-promoting Complex (APC) in land plants. BMC Plant Biol. 2010;10:254. doi: 10.1186/1471-2229-10-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang C.Z., Gu X., Peterson T. Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp indica. Genome Biol. 2004;5:R46. doi: 10.1186/gb-2004-5-7-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaestner K.H., Knochel W., Martinez D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Gene Dev. 2000;14:142–6. [PubMed] [Google Scholar]
- 43.Vogel C., Bashton M., Kerrison N.D., Chothia C., Teichmann S.A. Structure, function and evolution of multidomain proteins. Curr. Opin. Struc. Biol. 2004;14:208–16. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Chothia C., Gough J., Vogel C., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–3. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 45.Xing H., Pudake R.N., Guo G., et al. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics. 2011;12:178. doi: 10.1186/1471-2164-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riechmann J.L., Ratcliffe O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000;3:423–34. doi: 10.1016/s1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- 47.Du H., Yang S.S., Liang Z., et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eulgem T., Somssich I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–71. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 49.Tuteja N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007;2:135–8. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryu H.S., Han M., Lee S.K., et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell. Rep. 2006;25:836–47. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- 51.Ramamoorthy R., Jiang S.Y., Kumar N., Venkatesh P.N., Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008;49:865–79. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- 52.Penninckx I.A.M.A., Eggermont K., Terras F.R.G., et al. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–23. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penninckx I.A.M.A., Thomma B.P.H.J., Buchala A., Metraux J.P., Broekaert W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. The Plant Cell. 1998;10:2103–13. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durrant W.E., Dong X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.