Abstract
In addition to their established roles in nucleocytoplasmic transport, the intimate association of nuclear pore complexes (NPCs) with chromatin has long led to speculation that these structures influence peripheral chromatin structure and regulate gene expression. These ideas have their roots in morphological observations, however recent years have seen the identification of physical interactions between NPCs, chromatin, and the transcriptional machinery. Key insights into the molecular functions of specific NPC proteins have uncovered roles for these proteins in transcriptional activation and elongation, mRNA processing, as well as chromatin structure and localization. Here, we review recent studies that provide further molecular detail on the role of specific NPC components as distinct platforms for these chromatin dependent processes.
Introduction
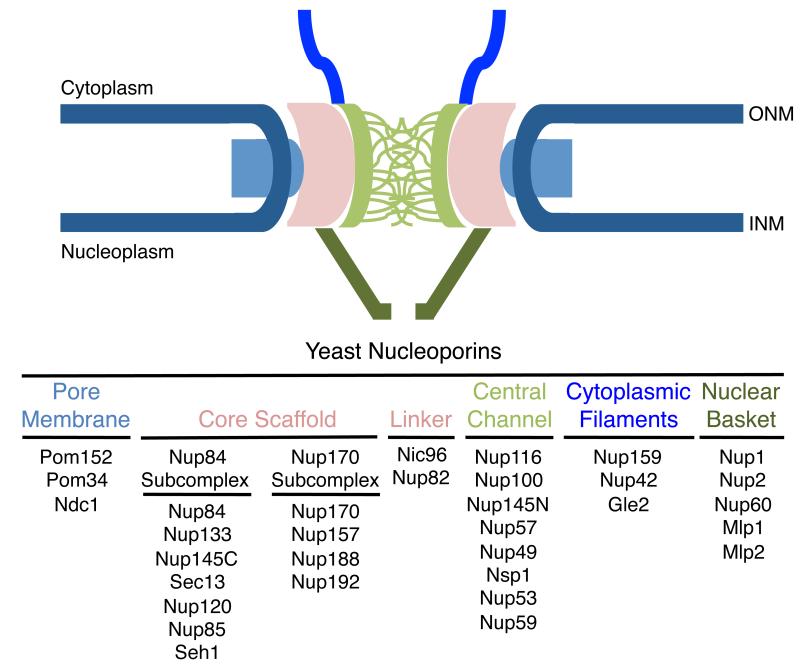
Nuclear pore complexes (NPCs) perforate an otherwise impermeable nuclear envelope (NE) membrane and the primary function long ascribed to these channels is to regulate exchange of water-soluble metabolites and macromolecules between the cytoplasm and the nucleoplasm. NPCs are unlike other transport channels, both in their degree of complexity and the mechanisms by which they move a highly diverse array of cargos across the NE. Cylindrical in geometry, and ~60-100 million Daltons in mass, these evolutionarily conserved structures exhibit a distinguishing octagonal symmetry around a central transport channel. NPCs do not cross the two lipid bilayers of the NE, but rather they extend from the surface of the chromatin and penetrate the NE at pores formed where the inner and outer nuclear membranes are fused. The membrane walls of these pores are attached to the ‘waists’ of cylindrical NPCs (Figure 1) [reviewed in 1,2].
Figure 1.
Structural organization of NPCs. NPCs are embedded within the NE at sites where the outer nuclear membrane (ONM) and inner nuclear membrane (INM) are fused. NPCs are bound to the pore membrane through the integral pore membrane proteins and amphipathic domains of core Nups. The core scaffold Nups can be grouped into two subcomplexes, the Nup84p- and Nup170p-subcomplexes, which bind the linker Nup Nic96. The core scaffold contains multiple binding sites for the FG-containing Nups. The FG portions of these Nups are unstructured and fill the central channel. Filaments also extend from the NPC core into both the cytoplasm (cytoplasmic filaments) and nucleoplasm (nuclear basket).
Despite their large size and elaborate structure, NPCs are composed of relatively few proteins (~30). These nucleoporins (Nups) are present in multiple copies, and specific groups of Nups contribute to distinct repetitive subunits that assemble to form the NPC. On the basis of their structural features and localization within the NPC, Nups can be placed into distinct groups (Figure 1). Integral proteins of the pore membrane interact with complexes of Nups that form the core scaffold of the NPC, which includes the Nup84- and Nup170-subcomplexes. Multiple copies of these subcomplexes are organized into 8-fold symmetrical ring structures that line the circumference of the pore where they interact with the pore membrane proteins and the membrane itself. Interestingly, sequence similarities between some Nups and coat proteins of secretory vesicles suggest these Nups have evolved from similar membrane coating ancestors. The core scaffold also supports Nups containing natively unfolded domains rich in phenylalanine-glycine (FG) residue repeats that occupy the central channel. These FG-Nups play a central role in transport. Among the FG-Nups, several members show a biased or strict localization to the nucleoplasmic or cytoplasmic face of the NPC. This group contributes to filaments that extend from the NPC core into the cytoplasm or nucleoplasm. In addition to FG-Nups, the nuclear fibers (a.k.a. nuclear basket) also consist of the proteins Mlp1 and Mlp2 (termed Tpr in vertebrates) [reviewed in 1,2]. Nuclear filaments likely play a role in transport, however, an accumulating body of data suggests these structures and other Nups exposed to the nucleoplasmic face of the NPC also play important roles in modulating chromatin structure and gene expression [reviewed in 3].
In this review we summarize insights into the functional relationships between NPCs and the regulation of gene expression. It has long been speculated that NPCs are intimately associated with chromatin. Studies have underscored the importance of chromatin in NPC assembly, both in yeast and higher eukaryotes, including an intriguing requirement for chromatin remodeling factors in the assembly of yeast NPCs [4]. Conversely, observations continue to emerge showing the importance of Nups in chromatin structure and the regulation of gene expression. In this regard it is reasonable to view many Nups as chromatin-associated factors that act within the context of the NPC platform to influence genome function.
NPCs associate with transcriptionally active and inactive chromatin
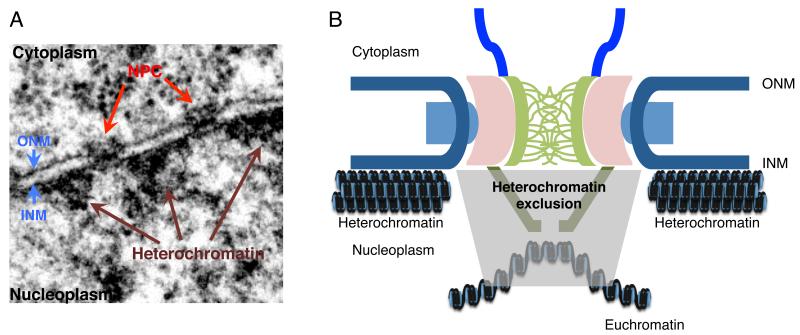
Chromatin is not randomly distributed within the nucleus. Each chromosome occupies a defined nuclear territory, and the chromatin therein localizes to specific spatial domains that are dependent upon distinct structural and functional states, including heterochromatin, which is highly compact and transcriptionally silent, and euchromatin, which is loosely packed and contains transcriptionally active loci [5]. Electron micrographs of nuclei from higher eukaryotes reveal that a portion of their heterochromatin is preferentially localized to the nuclear periphery, in close association with the nucleoplasmic face of the inner nuclear membrane (INM; Figure 2). This peripheral heterochromatic landscape, however, is disrupted at euchromatin channels that extend from NPCs into the nucleoplasm. It appears, at least in part, that these euchromatin channels are maintained by components of the NPC nuclear basket including Tpr in metazoan cells [6]. The association of NPCs with euchromatin has long been interpreted to reflect their association with transcriptionally active genes [7].
Figure 2.
Structural features of the nuclear periphery. A. Shown is an electron micrograph of the nuclear periphery of a HeLa cell. Densely staining heterochromatin is visible adjacent to nucleoplasmic face of the inner nuclear membrane (INM). The continuity of the peripheral heterochromatin is interrupted by lesser straining euchromatin channels at positions along the NE occupied by NPCs. B. A cartoon of the interplay between chromatin and the nuclear periphery is shown. Heterochromatin associates with the INM, but is excluded from regions containing NPCs in a manner dependent upon the nuclear basket. These regions of heterochromatin exclusion contain transcriptionally active euchromatin. ONM - outer nuclear membrane
Chromatin organization in the yeast Saccharomyces cerevisiae shows many similarities to higher eukaryotes, providing a genetically tractable model organism for such studies [reviewed in 8]. Many regions of silenced chromatin reside primarily at the nuclear periphery, including telomeric and subtelomeric chromatin, centromeres, and silenced mating loci, and the basket components Mlp1/2 are implicated in keeping NPCs free from heterochromatin crowding [9••]. By contrast, numerous actively transcribing genes are observed to associate with NPCs. These active genes interact with Nups present in disparate substructures of the NPC, including the nuclear basket, the core scaffold, and the central channel. Interestingly, NPCs are also associated with silenced chromatin and studies suggest that NPCs function in the deposition of these chromatin domains adjacent to the INM. This myriad of interactions suggests NPCs possess binding platforms for diverse chromatin domains and here we discuss emerging details on the interrelationships between chromatin and specific NPC substructures.
Nuclear basket and transcriptional activation
Various studies in yeast have implicated the nuclear basket as a binding platform for numerous highly transcribed housekeeping genes and genes strongly induced by changes in environmental conditions [reviewed in 10-12].
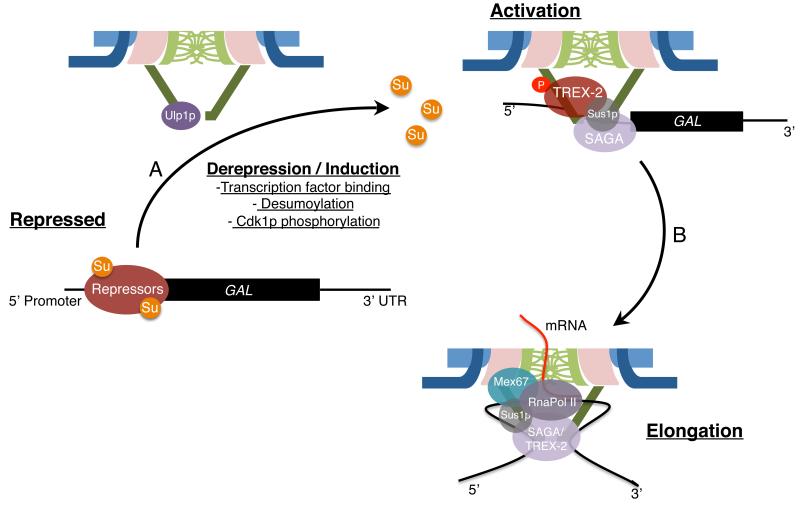
While recruitment of activated genes to NPCs may facilitate mRNA export, recruitment could also serve additional purposes. Among the most studied of the inducible genes are those functioning in galactose metabolism (GAL genes), which are derepressed and actively transcribed when the carbon source for yeast is switched from glucose to galactose. Derepression is accompanied by the translocation of GAL loci to the NPC nuclear basket independent of active transcription, raising the possibility that GAL-NPC associations occur prior to transcription initiation, which suggests that the phenomenon is not simply a consequence of coincident mRNA export and transcription. An early study proposed that the initial interaction with the NPC involves the GAL upstream activating sequences (UAS) with the nuclear basket in a manner dependent on the transcriptional activator Gal4p [13]. However, the specific basket Nups involved were not identified, although Mlp1p has been shown to associate with the UAS of GAL genes [14].
Recently, GAL1 translocation was shown to require Ulp1p, a SUMO-isopeptidase that associates with the nuclear basket in an Mlp-dependent manner [15]. This study showed that GAL recruitment to the NPC is dependent upon desumoylation of the Ssn6p repressor and, possibly, the desumoylation of other transcription associated factors [16••]. These results are consistent with a model in which translocation of promoters to the basket brings promoter-associated proteins proximal to Ulp1p, resulting in their desumoylation. Desumoylation is proposed to uncover binding sites on these proteins and promote the association of transcriptional activators and the assembly of chromatin remodelers, including the SAGA complex, on the promoter [16••].
Assembly of transcriptional complexes at the NPC also appears to employ distinct regions of the basket. For example, Mlp1p binds the SAGA complex [14] and Nup1 interacts with the TREX-2 complex (transcription elongation and RNA export complex) [17]. Furthermore, SAGA and TREX-2 appear to be linked through a common subunit, Sus1p [18]. Disruption of any of these interactions using nup1Δ, mlp1Δ, ada2Δ (SAGA subunit), sac3Δ (TREX-2 subunit) or sus1Δ mutants [14,17,19] or displacement of Ulp1p from the nuclear basket [16••] leads to a loss of GAL tethering. Thus, derepression of GAL genes is accompanied by the formation of a highly interconnected complex, centered on the promoter, which employs distinct regions of the NPC nuclear basket as a binding platform.
Gene tethering and basket Nup phosphorylation
In addition to sumoylation, the regulated phosphorylation of basket Nups has also emerged as a mechanism for modulating NPC-gene associations. For example, a recent study showed that osmostress promotes the nuclear import of the Hog1p kinase, which in turn phosphorylates nuclear targets leading to induced gene expression [20*]. Among the Hog1p targets are the basket Nups, Nup1, Nup2, and Nup60. Intriguingly, phosphorylation of these Nups appears to be required for NPC association of osmostress-induced genes [20*].
Phosphorylation of Nup1 by Cdk1p is also required for the NPC association of actively transcribed GAL1 and INO1 [21]. However, during periods of DNA replication (S-phase), NPC-gene associations are disrupted, presumably to prevent replication fork stalling induced by collisions between NPCs and replication forks [22••]. This loss of gene tethering to NPCs during S-phase appears to be linked to the inhibition of Cdk1p-mediated phosphorylation of Nup1p during this cell-cycle stage. Post-replication, Cdk1p phosphorylation of Nup1p is reestablished and genes reengage with NPCs [21].
The loss of NPC-gene interactions during S-phase also appears to employ the replication checkpoint kinase Rad53p, which phosphorylates multiple Nups including Mlp1p and Nup1p, in response to stalled replication. In contrast to Cdk1p, Rad53p-mediated phosphorylation is thought to inhibit the association of its Nup targets with the SAGA and TREX-2 complexes [22••]. Thus, it appears that context-dependent phosphorylation of basket Nups can positively or negatively affect NPC-gene interactions.
The nuclear basket supports gene clustering
Environmental stimuli often elicit a concerted change in the transcriptional state of functionally linked genes such as those present in an inducible metabolic pathway. A recent study indicates that such co-regulation may be concomitant with clustering of these genes at the nuclear periphery, in association with NPCs [23••]. In particular, cells grown in media lacking inositol exhibit induced gene expression of the INO1 locus that is accompanied by DNA sequence-mediated NPC tethering [24]. These cis-acting DNA elements, termed gene recruitment sequences (GRSs) [25], together with an interacting transcriptional activator Put3p, mediate NPC association. When recruitment of another gene carrying the same GRS was assessed simultaneously with INO1, the two genes were found to occupy the same region along the nuclear periphery. This gene clustering was dependent upon the nuclear basket associated Nup2p, suggesting this Nup contributed to both gene clustering and NPC tethering. Curiously, once activated and clustered, loss of their NPC localization did not disrupt the gene clusters, suggesting that NPCs contribute to the formation of a gene cluster, but they are not required to maintain the organization [23••].
Nups in transcript elongation and processing
Binding of activated genes to NPCs is followed by the initiation of transcription and, transcript elongation. The coupling of these reactions with NPC association appears to reflect the involvement of distinct components of the NPC in various steps of mRNA synthesis, processing, and export [reviewed in 10].
The nuclear basket has been shown to associate with the 5′ and 3′ ends of some activated genes to form NPC tethered gene loops [13,26-28]. Gene loops are also associated with mRNA processing factors that would facilitate mRNA maturation and ultimately export to occur in the vicinity of an NPC. In addition, loops may function to facilitate rapid reuse of RNA pol II post-transcription by placing 5′ and 3′ ends of genes in close proximity with one another such that the polymerase could easily re-engage the promoter [reviewed in 10].
Apart from the nuclear basket, recent studies carried out in C. elegans support a role for the NPC core in mediating transcriptional processes downstream of initiation. Heat shock in C. elegans is accompanied by the induction of an hsp locus and its NPC association. This association is dependent on promoter elements, a transcription factor, (the C.elegans homolog of yeast Sus1p, ENY-2), and RNA pol II, suggesting a mode of NPC-gene association that is homologous to that observed in yeast. Furthermore, ChIP analysis using NPP-13 (Nic96 in yeast, Nup93 in vertebrates), indicated that the NPC interacts with this locus not only at the promoter, but also within the open reading frame and at 3′ ends of the genes, consistent with a role for this core linker Nup in latter stages of transcription [29*].
Further support for a post-initiation role for core NPC components stems from the observed interactions between NPP-13 and RNA pol III transcribed tRNA and snoRNA genes (Ikegami and Lieb, 2013). NPP-13 knockdown experiments revealed that this Nup is not required for the association of these genes at NPCs. Rather, the loss of NPP-13 led to defects in the processing of these RNAs, consistent with ChIP analysis showing NPP-13 interacts with regions of these genes downstream of the transcription start site and, by co-immunoprecipitation, with RNA pol III itself. Together these data point to a role for core associated elements of the NPC in coordinating transcription elongation and RNA processing [30••].
In yeast, the RNA pol II elongation complex may also be linked to NPCs through the mRNA export factor Mex67p [10]. Prior to its incorporation into an mRNP export complex, Mex67p participates in NPC-gene association during transcription elongation [19]. This association appears to be dependent upon Mex67 interactions with both NPCs and the RNA pol II elongation complex. Mex67p is thought to engage the elongation complex, in part, through an interaction with Sus1p, a component of both the TREX-2 and SAGA complexes that also remain associated with the gene during elongation [18].
How Mex67p bridges its interaction with the elongation complex to NPCs remains to be determined. Possibilities include interactions through its C-terminal ubiquitin binding domain, which plays a role in its association with transcribing genes [31]. This domain interacts with ubiquitinated proteins, such as the THO/TREX component Hpr1p and FG-Nups [32,33]. Recent work has also identified numerous mono- and poly-ubiquitinated Nups [34] making these potential Mex67p binding partners. In addition, the Nup84p subcomplex, which also plays a role in transcription elongation [35*], has similarly been shown to interact with Mex67p [36]. Irrespective of how Mex67p may function to bridge NPCs and RNA pol II elongation complexes, these results highlight the close relationship between the transcriptional machinery and NPCs (Figure 3).
Figure 3.
NPC-GAL gene association and transcription. A. Shifting from glucose to galactose-containing media initiates derepression and activation of GAL genes. This process is accompanied by the translocation of the locus from the nucleoplasm to NPCs in a manner dependent upon transcription factor binding to upstream activating elements in the promoter as well as the association of chromatin remodellers (SAGA) and mRNA export factors (TREX-2) with specific Nups. Cdk1p-dependent phosphorylation of Nup1p aids in facilitating its interaction with TREX-2 and stabilizes NPC-gene association. At least for GAL1, formation of this integrated complex is promoted by the removal of SUMO (Su) from Ssn6p, a member of the repressor complex, by the NPC-bound SUMO isopeptidase Ulp1p. B. Once activated, GAL genes remain associated with NPCs. 5′ and 3′ ends of these loci are linked to the nuclear basket, which is thought to induce the formation of gene loops that aide in bringing 5′ mRNA capping and 3′ processing factors to NPCs (not shown). The ORF also associates with NPCs through the formation of a highly integrated complex centered around the RNA pol II elongation complex that includes both SAGA and TREX-2. Mex67p appears to provide a bridge between the elongation complex and NPCs. The TREX-2 component Sus1p links Mex67p to the RNA pol II complex. These interactions contribute to the positioning of the ORF in close proximity to the NPC
FG Nups in transcriptional memory
Various FG-Nups in yeast [37], flies [38,39] and human cells [40] have also been detected in association with transcriptionally active genes. In yeast, the FG-Nups (with the exception of Nup2) are only detected in association with NPCs [41], while in higher eukaryotes many FG-Nups are mobile and also present within the nucleoplasm. Here a subset of FG-Nups, including Nup98, has been shown to regulate gene transcription at intranuclear loci [38-40]. More recently, FG-Nups have also been shown to play a role in transcriptional memory [42,43••].
Transcriptional memory is defined as a process whereby genes that have been recently repressed are poised for a more rapid reactivation than when repressed for extended periods. In yeast, the capability of certain loci to exhibit transcriptional memory has been linked with their continued association with NPCs following repression. Several of the same complexes implicated in gene translocation to NPCs, including basket Nups and members of the SAGA and TREX complexes, appear required for this process [42]. However, there is an additional subset of Nups not required for gene recruitment during activation that instead function specifically in tethering a recently repressed gene to the NPC and facilitating memory. These include members of the Nup84p subcomplex and the FG-containing Nup100p. Loss of Nup100p leads to defects in several events linked to memory, including NPC targeting mediated by a cis element of the INO1 promoter (memory recruitment sequence, MRS), H2A.Z incorporation, and the association of a pre-initiation form of RNA pol II with the promoter [42]. Importantly, transcriptional memory can be passed from mother to daughter cells, and in a recent study Nup100p was shown to be required for maintenance of histone H3 dimethylation (H3K4me2), an epigenetic histone modification that mediates inheritance of the INO1 memory complex [43••].
The phenomenon of transcriptional memory is also observed higher eukaryotes, with a representative example being the transcriptional regulation of many interferon-γ responsive genes. Recently, work by Light and colleagues have established striking parallels between the mechanism of transcriptional memory employed during reactivation of interferon-γ responsive genes and events documented in yeast [43••]. Notably, the mammalian counterpart of yeast Nup100, Nup98, binds to recently expressed HLA-DRA, an interferon-γ induced gene exhibiting transcriptional memory. In cells depleted of Nup98, transcriptional memory is lost and molecular events associated with memory, including RNA polymerase II association with promoters and histone H3 dimethylation are reduced [43••]. An important distinction between Nup98 and Nup100 is that Nup98, like many mammalian FG-Nup, is detectable both at NPCs and in the nucleoplasm. Thus, Nup98 may execute its function in transcriptional memory at loci within the nucleoplasm.
NPC platforms as repressors of transcription
While the majority of studies have investigated the relationships between Nups and active transcription, NPCs also function in transcriptional repression. An observation consistent with this notion showed that the association of the yeast GAL1 gene with NPCs, after a shift from repressive (glucose medium) to activating (galactose medium) conditions, appeared to dampen its transcriptional activation [44••]. Furthermore, mutant cells lacking Nup1p or the SAGA complex component Ada2p, both of which are required for NPC tethering of GAL1, exhibited more rapid induction of GAL1 expression as well as a delayed response to glucose-induced repression. Both observations are consistent with the existence of a repressive environment at the NPC. Similar repressive activities attributed to NPC association have been documented for ribosomal protein (RP) genes. A mutant (arp6Δ) that disrupts the NPC association of RP genes causes their up-regulation [45]. Moreover, RP genes were shown to interact with the NPC core scaffold protein Nup170p, and the loss of Nup170p led to the increased expression of essentially all RP genes [46••]. These results are consistent with specific Nups functioning to suppress the expression of targeted genes.
Nups have also been shown to play an essential role in the nuclear organization and transcriptional repression of silenced heterochromatin in yeast, including silenced mating type loci [47] and subtelomeric chromatin [46••,48-50]. Recently, reports have shown that regions adjacent to the HMR mating type locus interact with Nup60p, which contributes to both silencing and the peripheral nuclear localization of HMR [47]. Furthermore, the Nup84p- [50] and Nup170p- [46••] subcomplexes of the NPC core scaffold have been linked to subtelomeric chromatin structure and gene silencing. In this context, Nup170p was shown to interact with components of the RSC chromatin remodeling complex and subtelomeric chromatin. Furthermore, Nup170p was shown to contribute to the proper assembly of subtelomeric chromatin, including binding of the silencing protein Sir4p, its association with the nuclear periphery, and subtelomeric gene silencing [46••]. As telomeres are not concentrated at NPCs and appear to reside primarily in association with the INM during interphase, NPCs are proposed to transiently associate with subtelomeric chromatin, promoting its assembly and binding to INM proteins. Such a mechanism is supported by the observation that INM-associated proteins that bind subtelomeric chromatin are physically associated with NPCs [9••]. As such, an integrated protein network, engaging NPCs and INM associated proteins, may function together to regulate many key aspects of chromatin structure and function at the nuclear periphery.
Conclusions
It is becoming clear that the NPC is a complex platform where distinct groups of Nups associate themselves with various nuclear functions, including gene transcription and the maintenance of chromatin structure. The field continues to identify distinctive roles for individual Nups in facilitating both gene activation and repression through the characterization of their interactions with components of the transcriptional machinery (e.g. activator and repressor proteins), regulators of chromatin structure (e.g. chromatin remodeling complexes), and protein modifiers (e.g. desumoylases). In organisms such as yeast these Nup activities are likely restricted to the NE, however, in higher eukaryotes the ability of Nups to populate the nucleoplasm allows these proteins to function throughout the nucleoplasm. Their roles as epigenetic regulators of gene expression offers an opportunity to understand and to control the multiple pathologies associated with NPC dysfunction.
Acknowledgements
We thank Jason Brickner (Northwestern University, USA) for critical reading of the manuscript. Funds for this work were provided to R.W.W. by the CIHR (MOP 130404 and MOP 106502), and AIHS, and to J.D.A. by the NIH (P50 GM076547, U54 GM103511, and 1U01GM098256-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aitchison JD, Rout MJ. The yeast nuclear pore complex and transport through it. Genetics. 2012;190:855–883. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 3.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 4.Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Krull S, Dörries J, Boysen B, Reidenbach S, Maginus L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blobel G. Gene gating: A hypothesis. Proc Natl Acad Sci. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taddei A, Gasser SM. Structure and Function in the budding yeast nucleus. Genetics. 2012;192:107–129. doi: 10.1534/genetics.112.140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteosome. Mol Biol Cell. 2013;24:3920–3938. doi: 10.1091/mbc.E13-07-0412. The nuclear basket components Mlp1 and Mlp2 are shown to interact with proteins that link NPCs to an extended peripheral protein network, including spindle organizers, silencing factors, the proteosome, and mRNPs. In addition, Mlps are shown to reduce heterochromatin crowding at NPCs, and contribute to both nuclear envelope morphology and NPC positioning within the nuclear envelope.
- 10.Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci. 2010;123:1989–1999. doi: 10.1242/jcs.053694. [DOI] [PubMed] [Google Scholar]
- 11.Arib G, Akhtar A. Multiple facets of nuclear periphery in gene expression control. Curr Opin Cell Biol. 2011;23:346–353. doi: 10.1016/j.ceb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Steglich B, Sazer S, Ekwall K. Transcriptional regulation at the yeast nuclear envelope. Nucleus. 2013;4:1–11. doi: 10.4161/nucl.26394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Wu C-Y, Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, Stutz F. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell. 2013;51:807–818. doi: 10.1016/j.molcel.2013.08.047. The SUMO isopeptidase Ulp1p, which associates with the NPC nuclear basket, is shown to play a role in GAL1 translocation to NPCs and derepression of this locus through desumoylation of the Ssn6p repressor when cells are transferred form glucose to galactose media.
- 17.Cabal GC, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin J-C, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 18.Pascual-Garcia P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodríguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Regot S, de Nadal E, Rodríguez-Navarro S, González-Novo A, Pérez-Fernandez J, Gadal O, Seisenbacher G, Ammerer G, Posas F. The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J Biol Chem. 2013;288:17384–17398. doi: 10.1074/jbc.M112.444042. Osmostress induces the nuclear import of the Hog1p kinase where it phosphorylates several nuclear basket Nups. The authors also show that promoter association of the osmostress induced gene STL1 with basket Nups is Hog1p dependent, correlating STL1-NPC association with Hog1-dependent phosphorylation.
- 21.Brickner DG, Brickner JH. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol Biol Cell. 2010;21:3421–3432. doi: 10.1091/mbc.E10-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gómez-González B, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. Collisions between replication forks and transcription bubbles at genes associated with NPCs may cause DNA topological strain leading to fork destabilization. The authors present data suggesting that this physical strain on replication forks is counteracted by Rad53p-dependent phosphorylation of nuclear basket Nups, which, in turn, is proposed to disrupt gene-NPC interactions.
- 23••.Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell. 2012;22:1234–1246. doi: 10.1016/j.devcel.2012.03.012. Gene recruitment sequences, when found within the promoter of multiple genes, direct the translocation of these genes to NPCs as well as the clustering of these genes to a region of the nuclear periphery.
- 24.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippe J-P, Singh BN, Krishnamurthy S, Hampsey M. A physiologcal role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcritpional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol. 2013;200:589–604. doi: 10.1083/jcb.201207024. * Heat shock induces NPC association of an hsp locus in C. elegans that is dependent upon a transcription factor, RNA pol II, SAGA and TREX. In addition, NPCs, specifically through the Nic96p homolog NPP-13, are also associated with the coding region of these genes suggesting a role for NPCs in transcript elongation.
- 30••.Ikegami K, Lieb JD. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell. 2013;51:840–849. doi: 10.1016/j.molcel.2013.08.001. The C. elegans nucleoporin NPP-13 is shown to associate with RNA pol III transcribed genes as well as with the RNA pol III transcription machinery itself. This association is required for the processing of tRNAs and sno-RNAs, suggesting that NPP-13 coordinates the transcription and processing of these RNAs.
- 31.Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, Stewart N, Divita G, Dargemont C. Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol Biol Cell. 2007;18:2561–2568. doi: 10.1091/mbc.E07-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gwizdek C, Iglesias I, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. Ubiquitin-Associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl. Acad. Sci. USA. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobeika M, Brockmann C, Gruenssing F, Neuhaus D, Divita G, Stewart M, Dargemont C. Structural requirements for the ubiquitn-associated domain of the mRNA export factor Mex67 to bind its specific targets, the transcription elongation THO complex component Hpr1 and nucleoporin FXFG repeats. J Biol Chem. 2009;284:17575–17583. doi: 10.1074/jbc.M109.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayakawa A, Babour A, Sengmanivong L, Dargemont C. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J Cell Biol. 2012;196:19–27. doi: 10.1083/jcb.201108124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tous C, Rondón AG, García-Rubio M, González-Aguilera C, Luna R, Aguilera A. A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J. 2011;30:1953–1964. doi: 10.1038/emboj.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao W, Lutzmann M, Hurt E. A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J. 2008;27:6–16. doi: 10.1038/sj.emboj.7601947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casolari JM, Brown SK, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;177:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 38.Kalverda B, Pickergill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of Nup98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2 dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–1478. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Light WH, Freany J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickenr JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. Transcriptional memory is shown to be dependent upon the same basic processes in both yeast and human cells highlighting its conservation within eukaryotes. Recently repressed genes associate with an FG-Nup, Nup100 in yeast cells and Nup98 in human cells, that mediates epigenetic histone modification and the binding of inactive RNA pol II at these loci.
- 44••.Green EM, Jiang YJ, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Cell Biol. 2012;23:1367–1375. doi: 10.1091/mbc.E11-06-0547. Deletion mutations that prevent GAL locus association with NPCs increased both the initial expression rate of these genes upon induction and the rate of gene inactivation upon repression. Based on these and other data, the authors propose that the association of these genes with the NPC functions dampen their expression.
- 45.Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, Iwahashi H, Kugou K, Ohta K, Gasser SM, et al. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010;6:e1000910. doi: 10.1371/journal.pgen.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Van de Vosse DW, Wan Y, Lapetina DL, Chen W-M, Chiang J-H, Aitchison JD, Wozniak RW. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 2013;152:969–983. doi: 10.1016/j.cell.2013.01.049. The core NPC component Nup170p is required for nucleosome positioning and transcriptional repression at subtelomeric and ribosomal protein genes. These functions of Nup170p were linked to its association with the RSC chromatin remodeler and Sir4p, a component of the silent information regulator. Nup170p was shown to be required for the assembly of Sir4p on subtelomeric chromatin and its association with the inner nuclear membrane.
- 47.Ruben GJ, Kirkland JG, MacDonough T, Chen M, Dubey RN, Gartenberg MR, Kamakaka RT. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS ONE. 2011;6:e21923. doi: 10.1371/journal.pone.0021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 49.Feuerbach F, Galy V, Trelles-Sticken, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherten H, Nehrbass U. Nuclear architecture and spatial positioning help establish transcritpional states of telomeres in yeast. Nat Cell Biol. 2002;4:214–221. doi: 10.1038/ncb756. [DOI] [PubMed] [Google Scholar]
- 50.Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]