Abstract
The bacterial type IV secretion systems (T4SSs) translocate DNA and protein substrates to bacterial or eukaryotic target cells generally by a mechanism dependent on direct cell-to-cell contact. The T4SSs encompass two large subfamilies, the conjugation systems and the effector translocators. The conjugation systems mediate interbacterial DNA transfer and are responsible for the rapid dissemination of antibiotic resistance genes and virulence determinants in clinical settings. The effector translocators are used by many Gram-negative bacterial pathogens for delivery of potentially hundreds of virulence proteins to eukaryotic cells for modulation of different physiological processes during infection. Recently, there has been considerable progress in defining the structures of T4SS machine subunits and large machine subassemblies. Additionally, the nature of substrate translocation sequences and the contributions of accessory proteins to substrate docking with the translocation channel have been elucidated. A DNA translocation route through the Agrobacterium tumefaciens VirB/VirD4 system was defined, and both intracellular (DNA ligand, ATP energy) and extracellular (phage binding) signals were shown to activate type IV-dependent translocation. Finally, phylogenetic studies have shed light on the evolution and distribution of T4SSs, and complementary structure-function studies of diverse systems have identified adaptations tailored for novel functions in pathogenic settings. This review summarizes the recent progress in our understanding of the architecture and mechanism of action of these fascinating machines, with emphasis on the ‘archetypal’ A. tumefaciens VirB/VirD4 T4SS and related conjugation systems.
Keywords: Conjugation, DNA transfer, pathogenesis, protein translocation, pilus, coupling protein, traffic ATPases, core complex
1. Introduction
The type IV secretion systems (T4SSs) are a diverse group of translocation systems present in nearly all bacterial and some archaeal species [1–6]. These systems translocate DNA and monomeric and multimeric protein substrates to bacterial or eukaryotic target cells. The two main T4SS subfamilies are the conjugation machines and the effector translocator systems, both of which deliver substrates intercellularly by a mechanism requiring establishment of direct cell-to-cell contact [7]. The conjugation machines translocate single-stranded DNA substrates within and across many bacterial species. These systems are responsible for shaping of genomes on an evolutionary time-scale and dissemination of antibiotic resistance genes and virulence determinants in clinical settings [1, 8]. The effector translocators deliver protein substrates directly to eukaryotic cells. These systems are central to infection processes of Gram-negative bacterial pathogens including Agrobacterium tumefaciens, Helicobacter pylori, Brucella spp., Bartonella spp., Rickettsial spp., and Legionella pneumophila [1–6]. A third smaller group of T4SSs is represented by the DNA release system of the Neisseria gonorrhoeae GGI system, the H. pylori ComB competence system and the Bordetella pertussis pertussis toxin export (Ptl) system [9–11]. These systems take up DNA from the milieu or release DNA or protein substrates into the milieu independently of contact with another cell.
The A. tumefaciens VirB/VirD4 system is one of the best-characterized T4SSs in Gram-negative bacteria and here will serve as a reference point for discussion of these fascinating machines. This system is composed of 11 VirB proteins synthesized from the virB operon and the VirD4 subunit from the separate virD operon [12]. It functions to deliver a fragment of the A. tumefaciens genome, oncogenic T-DNA, as well as several effector proteins, e.g., VirE2, VirE3, VirF, to susceptible plant cells, resulting in the tumorous Crown Gall disease [13]. Many T4SSs of Gram-negative bacteria are composed of homologs of most or all of the VirB and VirD4 subunits, reflecting a common ancestry and likely conservation of machine architecture [14, 15]. However, many T4SSs also display striking differences in subunit composition and number compared to the ‘archetypal’ VirB/VirD4 T4SS and related systems. Many Gram-positive conjugation systems, for example, are composed of only a subset of the VirB/VirD4 homologs [15, 16], whereas other systems such as the L. pneumophila Dot/Icm [4] and H. pylori Cag T4SSs [3, 5, 17] are built from nearly 30 subunits of which only a few bear discernible homology to the VirB/VirD4 subunits. These T4SSs almost certainly possess important variations in structure and mechanism of action in comparison to the VirB/VirD4-like systems.
Since publication of a review on the A. tumefaciens VirB/VirD4 system in this journal in 2004 [18], there has been considerable progress on several fronts in structure-function studies of T4SSs. X-ray structures have been solved for homologs of many of the VirB/VirD4 subunits and high-resolution X-ray or cryoelectron microscopy images now exist for several T4SS machine subassemblies. In vivo studies have identified substrate translocation sequences and defined a general translocation route for substrate passage through the VirB/VirD4 T4SS. Intra- and extracellular signals have been shown to activate T4SS channels for secretion of cognate substrates or uptake of bacteriophage that utilize T4SSs as receptors. Finally, phylogenetic studies have shed light on the evolution and distribution of T4SSs in bacteria and archaea, and complementary structure-function studies have unveiled adaptations tailored for assembly of T4SSs across diverse cell envelopes and for a variety of novel applications in pathogenic settings.
This review is intended to update the reader about the structure and function of T4SSs, with an emphasis on systems closely related to the A. tumefaciens VirB/VirD4 system. Due to limitations in space, we will not attempt a comprehensive overview of the literature but instead highlight recent data from studies exploring fundamental mechanisms. We refer the reader to a number of excellent, specialized reviews on topics such as T4SS gene regulation [19–22], the biological and functional diversity of T4SSs [1, 2, 7, 15, 16, 23], and the physiological consequences of T4SS-mediated effector translocation during infection [3, 24–28].
2. General architectural/functional features of paradigmatic VirB/VirD4 T4SSs
There is accumulating evidence that the T4SSs of Gram-negative bacteria evolved through consolidation of four functionally-distinct machine subassemblies: i) the type IV coupling protein (T4CP), a hexameric ATPase related to the SpoIIIE/FtsK DNA translocases that physically couples early DNA and protein substrate processing reactions to the translocation machinery, ii) an inner membrane complex (IMC) responsible for substrate transfer across the inner membrane, iii) an envelope spanning outer membrane complex (OMC) required for substrate passage across the periplasm and outer membrane, and iv) the conjugative pilus, an extracellular organelle that initiates contact with potential recipient cells (Fig. 1). The first three subassemblies interact to form the translocation system. Surprisingly, the physical relationship between the translocation channel and conjugative pilus is not yet specified, nor is it known whether pilus polymerization initiates from a platform at the inner or outer membrane.
Fig. 1.
Schematic depicting subunits and subassemblies of the A. tumefaciens VirB/VirD4 type IV secretion system (T4SS). Bottom: The 11 VirB subunits are synthesized from the virB operon and VirD4 from the separate virD operon. Lower: The VirB/VirD4 subunits are inserted in the inner membrane (IM) or delivered to the periplasm (P) with general topologies/locations as indicated. Upper: The subunits form a network of interactions resulting in four functional subassemblies: i) T4CP substrate receptor, ii) inner membrane translocase (IMC), iii) outer membrane complex (OMC)/core complex, and iv) extracellular pilus. The T4CP, IMC, and OMC interact to form the substrate translocation channel (red arrow). During translocation, the T-DNA forms formaldehyde-crosslinkable contacts with the 6 VirB/VirD4 subunits listed [38]. The IMC and OMC, together with the VirB1 transglycosylase, mediate assembly of the conjugative pilus.
The A. tumefaciens VirB/VirD4 subunits and subassemblies are depicted in Fig. 1. VirD4 is the T4CP, also termed the substrate receptor (where necessary for clarity, we will identify the species or plasmid origin of a protein in subscript, e.g., VirD4At). VirD4-like T4CPs possess canonical Walker A and B nucleoside triphosphate binding motifs [29] and family members have been shown to bind and/or hydrolyze ATP in vitro [30–32]. The IMC is composed of the ATPases VirB4 and VirB11, polytopic membrane proteins VirB3 and VirB6, and bitopic protein VirB8. The OMC consists of a structure termed the core complex and a translocation channel presumptively located within the central chamber of the core complex. The A. tumefaciens core complex is composed of outer membrane-associated lipoprotein VirB7 and VirB9 and the cell-envelope-spanning subunit VirB10, and the channel itself is likely composed of the pilin subunit VirB2 and domains of VirB8 and VirB9 [12, 33]. The core complex of the closely related E. coli pKM101-encoded T4SS is configured as a large barrel composed of 14 copies each of homologs of VirB7, VirB9, and VirB10 that extends across the entire cell envelope [33, 34]. The A. tumefaciens VirB-encoded pilus is composed of the pilin subunit VirB2 [35] and the pilus-tip adhesin VirB5 [36]. The lytic transglycosylase VirB1 is required for pilus biogenesis, but not for assembly of the translocation channel [37].
A formaldehyde (FA)-crosslinking assay termed transfer DNA immunoprecipitation (TrIP) was used to identify close contacts between the A. tumefaciens T-DNA substrate and subunits of the VirB/VirD4 T4SS during translocation [38]. Contacts were detected with the VirD4 T4CP, VirB11 ATPase, inner membrane proteins VirB6 and VirB8, and periplasmic/outer membrane subunits VirB2 and VirB9 (Fig. 1). Further TrIP analyses with virB and virD4 null mutants supplied evidence that DNA substrate - channel subunit contacts are ordered such that the VirD4 T4CP receptor first engages the DNA substrate and then transfers it to VirB11. VirB11 in turn delivers the substrate to a presumptive channel composed of VirB6 and VirB8 for translocation across the inner membrane. The substrate then passes through the periplasm and across the outer membrane via a channel minimally composed of VirB2 and VirB9 [38]. By combining the available structural and biochemical data, it can be postulated that T4SS substrates dock with the VirD4 T4CP and then are transferred to the IMC for further processing and translocation across the inner membrane via a channel composed minimally of VirB6 and VirB8. The IMC sits within or at the base of the OMC/core, forming specific contacts with the N-terminal region of VirB10 and, possibly, the N-terminal region of VirB9. The C-terminal region of VirB8, one or more domains of VirB9, and the VirB2 pilin form the translocation channel within the central chamber of the VirB7/VirB9/VirB10 core complex for conveyance of substrates to the cell surface [14, 39].
2.1. Structural variations/adaptations among T4SSs
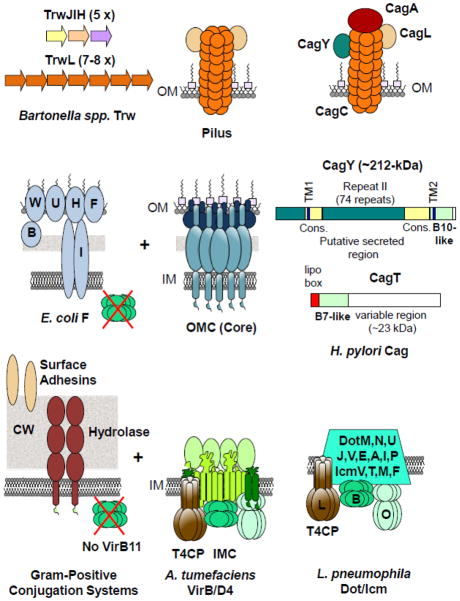
The structural features described to date for the A. tumefaciens VirB/VirD4 T4SS and closely related systems are probably conserved among many T4SSs, yet as a whole this transporter superfamily has evolved considerable mosaicism in terms of subunit composition, and thus overall architecture and function. Variations exist in each of the four T4SS machine subassemblies, as illustrated in Fig. 2 for some of the better-characterized T4SSs. Some T4SSs, including the B. pertussis Ptl and Brucella spp. VirB systems lack a VirD4-like T4CP and therefore utilize alternative mechanisms for substrate recognition. Some systems, including the E. coli F plasmid transfer system and Gram-positive bacterial conjugation systems lack a VirB11 homolog [1, 11, 15, 16, 40, 41]. Indeed, the Gram-positive conjugation machines lack several VirB homologs, including the OMC/core and pilin subunits. These ‘minimized’ systems are composed only of the IMC/T4CP complex for delivery of substrates across their single membranes and one or two extracytoplasmic proteins that presumptively form a channel across the thick cell wall [15, 16]. By contrast, IMCs of the L. pneumophila Dot/Icm (Fig. 2) and H. pylori Cag (not shown) systems are composed of many more subunits than the A. tumefaciens VirB/VirD4 system [4, 5, 17, 42].
Fig. 2.
Examples of T4SS structural adaptations. Bottom: Gram-positive conjugation systems are composed mainly of homologs of the A. tumefaciens T4CP (VirD4) and IMC (VirB3, VirB4, VirB6, VirB8), plus a cell wall hydrolase presumptively for elaboration of a channel across the cell wall. These ‘minimized’ systems lack a VirB11 homolog, the OMC/core, and conjugative pili, and may or may have associated surface adhesins. The T4CP/IMC of the L. pneumophila Dot/Icm system has three channel ATPases, but the membrane channel is compositionally much more complex than that of A. tumefaciens. Middle: The OMC/core can vary in structure/composition. The E. coli F plasmid-encoded OMC consists of a core complex (TraV/K/B), as well as the complex of Tra/Trb proteins shown whose function is implicated in pilus extension and retraction. In the H. pylori Cag system, the presumptive core subunits are much larger than their A. tumefaciens counterparts and possess novel domains that extend beyond the cell surface and associate with extracellular pili. Top: T4SS-encoded surface organelles can vary in composition and structure in comparison to the A. tumefaciens VirB-encoded pilus. The Bartonella spp. Trw system elaborates antigenically-variable pili through use of different pilins, and the H. pylori Cag system elaborates pili that are composed of several different subunits. The sole substrate of the Cag T4SS, CagA, also associates with the Cag pilus. Some T4SSs do not elaborate pili, e.g., Gram-positive conjugation systems, or instead produce novel fibrous or sheathed structures or surface adhesins.
With respect to the OMC/core, although homologs of the A. tumefaciens VirB7, VirB9, and VirB10 subunits are widespread among Gram-negative systems, the core subunits often possess additional novel domains (Fig. 2). In the H. pylori Cag T4SS, for example, only a small C-terminal region of CagY is similar to VirB10. Both the N- and C-terminal regions possess transmembrane domains, and a large central region composed of multiple repeats is surface-displayed and associates with a pilus structure. Similarly, CagT is a predicted VirB7-like lipoprotein, but also possesses a novel, surface-exposed variable region [5, 17]. Structural variation in the OMC composition can also occur through cooption of novel proteins or protein complexes, as exemplified with the E. coli F transfer system (Fig. 2). In addition to the TraV/K/B core complex, the F plasmid-encoded OMC consists of a set of proteins, TraF, -H, -U, -W and TrbB, -I that form an interaction network distinct from the core complex [43–46]. The TraF/H/U/W-TrbB/I complex is implicated in the unique capacity of F-type conjugative pili to dynamically extend and retract and confer efficient mating in liquid media. Finally, many structural variations exist among T4SS surface components and pili. T4SSs including all Gram-positive conjugation systems and the B. pertussis Ptl system lack extracellular pili altogether [15, 16]. Conversely, many effector translocators including the Bartonella spp. Trw and H. pylori Cag systems elaborate antigenically-variable pili or other fibrous or sheathed structures (See [1]). Although such structures are generally considered to mediate target cell attachment and immune evasion, in the H. pylori Cag system, the sole substrate CagA has been shown to associate with the Cag T4SS-encoded pilus (Fig. 2), pointing to a direct contribution of the pilus to substrate delivery to mammalian target cells [17, 47, 48].
3. Substrates/translocation signals/Substrate docking reactions
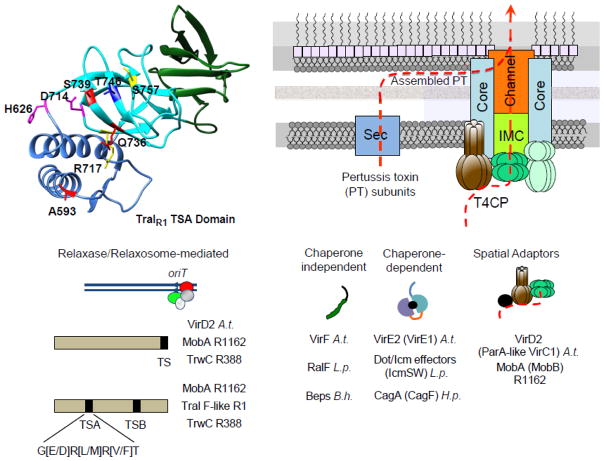
The type IV secretion substrates typically rely on the T4CP receptor for translocation and, accordingly, carry translocation signals required for substrate-T4CP docking. A small number of substrates carry classical N-terminal signal sequences for translocation across the inner membrane via the general secretory (Sec) pathway (Fig. 3).
Fig. 3.
Type IV substrate translocation signals and accessory factors. Most substrates are translocated through the T4CP-dependent pathway. DNA substrates are processed by Dtr processing factors through assembly of the relaxosome at the origin of transfer sequence (oriT). Lower left: Relaxases responsible for nicking at oriT can carry positively-charged C-terminal or internal translocation signals (TS’s) for docking with the T4CP receptor. A conserved sequence found in the TraIR1 TSA motif and present in TSA and TSB motifs of MobAR1162, TraIR1, and TrwCR388 is shown. Upper left: A model of the TSA motif from TraIR1 generated from an X-ray structure [64]. The motif bears structural homologies to SF1B helicase domains. Mutations of residues shown in stick representation and color-coded disrupted TSA translocation function: red, significant impairment; pink, moderate impairment; yellow, no effect; blue, stimulation of TSA translocation function. Molecular graphics and analyses were performed with the UCSF Chimera package [158] using the PDB ID (4L0J) of the TraIR1 TSA domain structure [64]. Lower right: DNA substrates and effector proteins can be recruited directly to the T4CP (Chaperone-independent) or via association with chaperones or spatial adaptors. Protein substrates can carry one or more of a combination of C-terminal, N-terminal or internal translocation signals; chaperones and spatial adaptors are in parantheses and the relevant species or plasmid is in parantheses (A.t., A. tumefaciens VirB/VirD4; L.p., L. pneumophila Dot/Icm; B.h., Bartonella henselae VirB/VirD4; H.p., H. pylori Cag). Upper right: In the T4CP-independent pathway, B. pertussis S1–S5 subunits are secreted across the inner membrane by the Sec system and assemble in the periplasm as the pertussis toxin (PT), which then docks by an unknown mechanism with the Ptl T4SS for extrusion across the outer membrane. Most T4SS substrates dock with the T4CP and are delivered in one step through the IMC/OMC channel.
3.1. T4CP-dependent translocation
Conjugation systems, most effector translocator systems, and the N. gonorrhoeae DNA release system use the T4CP-dependent pathway [1, 23]. Substrates translocated by this pathway bind the T4CP receptor via one or both of the following translocation signals: i) signals located at an unstructured C terminus consisting of clusters of positively-charged or hydrophobic residues or ii) one or more internal signals. Docking of both types of substrates also often requires chaperones or adaptor proteins (Fig. 3).
Studies exploring the nature of type IV secretion signals capitalized on findings that T4SSs can translocate reporter proteins, e.g., Cre recombinase, adenylate cyclase (CyaA), TEM β-lactamase, intercellularly when fused to an effector. In early studies, the A. tumefaciens VirB/VirD4 system was shown to translocate fusion proteins composed of Cre joined to the N termini of VirE2 or VirF effectors to plant and yeast cells [49]. Similarly, the L. pneumophila Dot/Icm T4SS translocated a CyaA-RalF effector to mammalian cells [50]. For both systems, the translocation signals were localized within unstructured C termini of the effectors, and consisted of clusters of positively charged Arg residues (for A. tumefaciens VirB/VirD4 substrates) or hydrophobic residues (for L. pneumophila Dot/Icm substrates) [51, 52].
Many T4SS effectors (or candidates) have now been identified with intercellular translocation assays [53–56]. Effectors typically possess C-terminal translocation signals, but there is also accumulating evidence for the importance of additional motifs for T4CP docking. These signals are located terminally or within the protein (Fig. 2). In Bartonella spp., for example, the Bartonella-translocated effector proteins (Beps) are translocated through a VirB/VirD4-like system. The Beps display a modular architecture with a bipartite secretion signal composed of a positively charged C terminus and at least one internal domain termed a Bep-intracellular delivery domain (BID) [50]. Interestingly, BIDs are also present in relaxases associated with conjugative plasmids carried by various α-proteobacterial species. The TraA relaxase encoded by A. tumefaciens plasmid pATC58 carries both a positively-charged C terminus and BIDs and, strikingly, TraA fragments carrying both types of recognition signals mediate translocation of Cre through a Bartonella henselae VirB/VirD4 system [50]. Thus, an effector translocator system and an ancestrally related conjugation system recognize common substrate signals. In H. pylori, a positively charged C-terminal tail is important for translocation of the CagA substrate through the Cag T4SS, and this tail can even be exchanged with similar motifs from other effectors to yield CagA translocation [57]. However, CagA translocation additionally requires an intact N-terminal motif of unspecified sequence composition [57] as well as internal motifs required for interaction with the specialized secretion chaperone CagF [58–60].
In the conjugation systems, the relaxase component of the DNA transfer intermediate confers DNA substrate specificity (Fig. 3). Several relaxases (MobAR1162, VirD2At, TraApATC58, TrwCR388, TraIR1) were shown to mediate transfer of N-terminally-fused Cre to target cells [50, 51, 61, 62], and domain-mapping studies have identified two or more translocation signals (Fig. 3). MobA R1162 is composed of two domains, an N-terminal relaxase domain and a C-terminal primase domain, and each domain can separately mediate translocation of Cre through the plasmid R751-encoded T4SS. Mutation of the positively charged C terminus of the primase domain or of the catalytic sites of the relaxase or primase domains abolishes translocation, suggesting that MobAR1162 translocation requires a C-terminal signal as well as motifs exposed only in the folded protein [62].
More recently, the F-like relaxase TraIR1 was found to carry two internal translocation signals (TSs), designated TSA and TSB, that mapped within ~300 residue regions located in each half of the protein (Fig. 3) [63]. Further studies identified key conserved residues conforming to a consensus sequence (G[E/D]R[L/M]R[V/F]T) in both TS motifs which, interestingly, were also identified in the MobAR1162 translocation signals. A crystal structure of the TSA region of TraIR1 was solved. It consists of three domains, each with structural homologies to SF1B helicase family domains, and a putative interaction surface was mapped to one of the helicase domains through mutational analysis [64]. These findings lend support to early models whereby helicase-mediated unwinding of the DNA substrate from its template strand is physically coupled with the T4CP and proceeds concomitantly with substrate transfer through the mating channel. These results also establish for the first time that a structural fold can serve as a signal for translocation through the T4CP-dependent pathway.
Substrates also can have distinct translocation signals for docking with different T4SSs (Fig. 3). Translocation of MobAR1162 relaxase through the R751-encoded T4SS channel requires two internal translocation signals, but a positively-charged C-terminal motif suffices for translocation through the A. tumefaciens VirB/VirD4 T4SS [65]. Similarly, translocation of the TrwCR388 relaxase through R388-encoded Trw T4SS requires two internal translocation sequences with consensus motifs resembling those in TraIR1 and MobAR1162, but translocation through the B. henselae VirB/VirD4 T4SS requires only an intact C terminus [66].
3.2 Contributions of other factors to substrate-T4CP docking
In addition to the intrinsic translocation signals discussed above, various accessory factors, adaptor proteins, or chaperones are often required for type IV secretion (Fig. 3).
3.2.1. Dtr accessory factors
In conjugation systems, Dtr (DNA transfer and replication) accessory factors play important roles as components of the relaxosome to stimulate relaxase nicking at oriT. In A. tumefaciens, VirC1 stimulates the nicking activity of the relaxase VirD2At through binding to a DNA sequence termed overdrive located adjacent to the oriT-like T-DNA border sequence [67]. Interestingly, VirC1 was identified as a member of the ParA/Soj/MinD ATPase superfamily [68], and further work established that VirC1 localizes at cell poles and interacts with a second Dtr accessory factor VirC2, the VirD2 relaxase, and the VirD4 T4CP, which is also polarly localized. Through these interactions, VirC1 is postulated to spatially couple the T-DNA substrate and the T-DNA processing reaction at the site of the VirB/VirD4 T4SS (Fig. 3) [69]. ParA homologs are associated with other conjugative plasmids and integrative and conjugative elements (ICEs), and further studies examining their contributions to the substrate-T4CP docking reaction are warranted. Spatial positioning of the MobAR1162 relaxase near a T4SS is also important for R1162 plasmid transfer. Recall that MobAR1162 has two functionally distinct domains, each with a translocation signal. The accessory factor MobBR1162 was found to stimulate translocation efficiencies of both MobA domains by a mechanism dependent on binding of MobBR1162 to the membrane [62]. MobBR1162 thus can be postulated to function analogously to VirC1 by tethering the R1162 substrate at or near a T4CP (Fig. 3). R1162 plasmids are non-selftransmissible plasmids that promiscuously translocate through many different T4SSs, and conceivably MobBR1162 functions to stabilize inherently weak interactions between MobA and different T4CPs [62].
3.2.2. Effector protein chaperones/adaptors
Effector proteins are translocated through their cognate T4SSs by alternative pathways distinguishable by the requirement of an adaptor protein or chaperone (Fig. 3). In A. tumefaciens, translocation of VirF through the VirB/VirD4 T4SS proceeds independently of other known factors, whereas translocation of VirE2 requires cosynthesis of the VirE1 chaperone [70]. VirE1 shares several features of the specialized chaperones associated with type III secretion systems (T3SSs), including a small size, acidic pI, and an amphipathic helix. An X-ray structure solved for a VirE1 chaperone/VirE2 effector complex identified an unusual structural arrangement of N- and C-terminal domains of VirE2 wrapped around the VirE1 chaperone [71] (Fig. 2). VirE1 does not participate directly in mediating the VirE2 interaction with the VirD4 T4CP [72, 73]. Rather, chaperone binding prevents self-aggregation of VirE2 and binding of DNA within the bacterium. Formation of the complex also ensures that VirE2’s disordered C-terminal tail is solvent exposed and accessible for docking with the T4CP [71].
In L. pneumophila, the Dot/Icm T4SS translocates more than ~250 effectors of which a subset are dependent on the IcmSW chaperone complex [74]. A complex of 4 proteins including DotM, DotN, IcmS, and IcmW has been shown to interact with the DotL T4CP. DotM and DotN are membrane associated and stabilize DotL, whereas IcmS and IcmW are small, acidic proteins that appear to function similarly to VirE1 and the T3SS specialized secretion chaperones [42]. IcmS and IcmW heterodimerize and the IcmSW complex appears to promote conformations in cognate secretion substrates required for productive engagement with the DotL T4CP (Fig. 3) [75, 76]. A recent study supplied evidence that a C-terminal domain of DotL directly binds the IcmSW complex and that this interaction is necessary for delivery of IcmSW-dependent effectors through the Dot/Icm channel [77]. The findings prompted the interesting model that IcmSW binds an effector in the cytosol and, like VirE1, maintains it in a translocation-competent state. The effector docks via a C-terminal translocation signal with the DotL T4CP, whereupon IcmSW is released and sequestered through interaction with the DotL C terminus. This permits effector translocation through the Dot/Icm T4SS and, ultimately, recycling of the IcmSW chaperone to the cytosol for another round of substrate binding and translocation [77].
In H. pylori, CagA is the sole known protein substrate of the Cag T4SS [5, 78]. Crystal structures of CagA show that this protein is comprised of 5 domains that contribute in distinct ways to modulation of eukaryotic cell function. In addition to its N- and C-terminal signals, CagA must interact with its chaperone CagF for translocation [58, 59]. CagF has features reminiscent of VirE1 and the IcmS and IcmW chaperones, although it is much larger (~35-kDa) than these chaperones. Recently, it was shown that distinct domains of CagF bind to the five domains of CagA, each with μM affinity. The findings led to a proposal that multiple copies of CagF form CagA domain contacts necessary for maintaining the highly labile CagA effector in a translocation-competent, protease-resistant conformation [60]. CagF is also membrane-associated and has been proposed to mediate delivery of CagA to the Cag T4SS [58].
Why do such a diverse array of intrinsic motifs and extrinsic accessory factors exist for substrate docking with the T4CP receptor? A reasonable hypothesis is that this complexity evolved for coordination of type IV secretion with other cellular activities. Conjugation must be tightly coordinated with the cell cycle to avoid the competing activities of conjugative DNA metabolism and chromosome replication and segregation. Similarly, the effector translocators must precisely time delivery of potentially many different effectors to the eukaryotic host cell during the infection process. While translocation signals carried by relaxases and effector proteins confer substrate specificity by mediating T4CP docking, these or other intrinsic motifs also mediate interactions with accessory factors, adaptors, or chaperones. It can be speculated that complex regulatory circuitries regulate the synthesis or activity of the accessory factors for spatiotemporal control of substrate presentation to the transfer apparatus.
3.3. T4CP-independent translocation
Two effector translocator systems have been shown to translocate secretion substrates via a T4CP-independent pathway. The B. pertussis Ptl system translocates its only substrate, the pertussis toxin (PT), exclusively across the outer membrane (Fig. 3) [11, 79]. The 5 subunits (S1–S5) comprising this A/B toxin each carry canonical N-terminal sec signal sequences and they are exported across the inner membrane via the Sec system. In the periplasm, the S1–S5 subunits fold and oligomerize to form active PT, which then docks with the Ptl machine for translocation across the outer membrane. The Ptl system is unique among the known T4SSs in its capacity to translocate a large, multisubunit toxin across the outer membrane by a cell-contact-independent mechanism.
The Brucella spp. VirB T4SS also lacks a T4CP, but interestingly a number of translocated effector proteins have now been identified and shown to carry translocation signals at either terminus [53, 55, 80]. Effectors with N-terminal sec signals, e.g., BPE123, most probably are exported via the Sec system to the periplasm, where they then engage with the T4SS channel for transit across the outer membrane [55]. However, effectors with C-terminal signals, e.g., VceA and VceC, presumptively are not exported through the Sec system [53]. Interestingly, VceC was shown to translocate via its C-terminal translocation signal through the T4CP-dependent Dot/Icm system of L. pneumophila [53]. This likely represents another example of a substrate showing promiscuity for docking with different T4SSs, but the findings still do not answer the interesting question of how type IV substrates bearing C-terminal signals dock with the Brucella spp. VirB T4SS.
4. The T4CP substrate receptor
The role of T4CPs as substrate receptors is now well supported by genetic and biochemical findings. T4CPs can functionally substitute for one another, in some cases conferring substrate specificity switching [66, 81, 82]. In A. tumefaciens, the TrIP studies showed that the VirD4 T4CP binds the T-DNA substrate independently of the VirB subunits and also by a mechanism dependent on processing of the T-DNA by the VirD2 relaxase [38]. Bacillus fragilis plasmid pLV22a also was shown to form a formaldeyde-crosslinkable contact with the TraGRP4 T4CP [83], and Enterococcus faecalis pCF10 with the PcfCpCF10 T4CP [31]. Complementary in vitro studies have identified interactions between T4CPs and relaxosome components or protein substrates, and also demonstrated stimulatory effects of purified T4CPs on relaxase cleavage reactions [1, 23, 84].
T4CPs characteristically possess an N-terminal transmembrane domain and a large C-terminal cytoplasmic region comprised of a nucleotide-binding domain (NBD), an all-α-domain (AAD), and in some cases a C-terminal extension (CTE) [1, 85, 86]. An X-ray structure of the 50-kDa soluble domain of TrwBR388 revealed a globular hexameric assembly in which each subunit is composed of an NBD and an AAD (Fig. 4). The NBD is composed of a central twisted β-sheet flanked by several helices on both sides, and the AAD contains seven helices with a structure similar to the N-terminal domain of XerD recombinase. The six TrwB protomers assemble to form a globular ring that is ~110 Å in diameter and 90 Å in height, with a ~20 Å-wide channel in the center. This channel narrows to 8 Å at the cytoplasmic pole of the hexamer. Nucleotide binding pockets are located at the interface between the subunits. The N-terminal domain of TrwBR388 is predicted to project across the inner membrane on the basis of electron microscopy imaging [87]. The full-length protein is thus configured as a ball-stem, F1-F0-like structure (Fig. 4) [85, 86]. In view of noted sequence and structural similarities with the FtsK and SpoIIIE DNA translocases and findings that duplex DNA threads through the annulus of the FtsK hexamer [88, 89], it has been suggested that the T4CP might function similarly to drive ssDNA substrate transfer across the inner membrane (see Section 9).
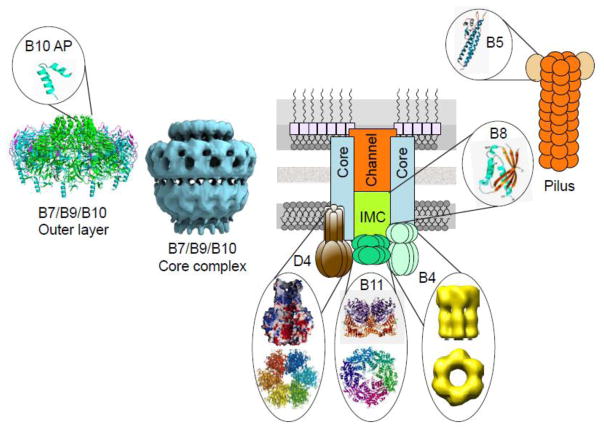
Fig. 4.
X-ray and CryoEM structures of T4SS machine subunits and subassemblies. The schematic shows the T4SS translocation channel and conjugative pilus. Available crystal structures of homologs of individual VirB and VirD4 T4CP subunits are shown. A CryoEM structure of the pKM101-encoded core complex and an X-ray structure of the distal half (outer layer) of the core complex along with a magnified view of the pore-forming antennae projection (AP) are presented. Structures were reproduced with permission as follows: T4CP (TrwBR388), [86]; VirB11 (CagβH.p.), Yeo et al. [111]; VirB4 (TrwKR388), Pena et al. [105]; VirB5 (TraCpKM101), Yeo et al. [159]; VirB8 (VirB8B.s.), [121]; Core complex CryoEM (pKM101 TraN/TraO/TraF), Fronzes et al. [33]; Core complex X-ray (pKM101, O-layer) Chandran et al. [34].
Interesting structural variations exist in T4CP domain architecture [1, 90]. The well-characterized T4CPs from Gram-negative systems, e.g., TraDF, TraGRP4, TrwBR388, VirD4At, typically range in molecular sizes of ~600 – 750 residues and possess a minimum of two predicted N-terminal TM domains with an intervening periplasmic loop of ~30–50 residues. TraG encoded by Salmonella typhi plasmid R27A-encoded TraG and related T4CPs comprise a distinct subfamily of T4CPs with much smaller (~4 residues) periplasmic domains and TraJ interaction partners. TraJ subunits are hydrophobic, multi-pass membrane proteins with sequence relatedness to the N terminus of FtsK translocase. Thus, it was proposed that TraJ and TraG cumulatively represent the domain architecture of the larger FtsK/SpoIIIE DNA translocases [91]. Members of another T4CP subfamily, represented by S. agalactiae pIP501 and B. cenecapacia AU1054, appear to lack N-terminal TM domains entirely, but upstream of the genes for these T4CPs are genes coding for small proteins (~150–200 residues) with 2 – 4 predicted TM domains. These subunit pairs might interact to form novel two-partner T4CPs [1].
Some T4CPs also carry CTEs [1]. The contributions of CTEs to substrate transfer are largely unspecified, with the exception that the CTE of TraDF plays a critical role in the F plasmid docking reaction. The CTE is necessary for efficient F transfer but hinders RSF1010 transfer [92–94]. This region of TraD was shown to bind TraM, which is a component of the relaxosome. An X-ray structure for the TraD CTE bound to the TraM tetramerization domain shows that the TraD peptide forms extensive contacts with each TraM monomer, indicating that as many as four TraD CTEs could simultaneously bind a single TraM tetramer. These findings suggest that a TraD hexamer might establish extensive contacts with TraM in vivo, forming the basis of a highly specific relaxosome - T4CP interaction [95].
The T4CP interacts with the IMC, although as mentioned above substitution of a T4CP with a closely related homolog can yield functional conjugation systems. There is some evidence for T4CP interactions with VirB10-like subunits of the core complex, and the interaction surfaces generally map within the N-terminal regions of both subunits [96]. Results of two-hybrid studies and mutational analyses have pointed to a contribution of the transmembrane helices of both subunits to this interaction [97]. In A. tumefaciens, however, the transmembrane domain (TMD) of VirB10 can be substituted with heterologous TM domains, including a nondimerizing poly-Leu/Ala helix, without loss of channel function, and further mutational analyses implicated the N-terminal periplasmic loop of VirD4 and both cytoplasmic and periplasmic domains of VirB10 in this interaction [98–100]. Given the subunit complexity of the IMC and OMC/core, it is reasonable to predict that the T4CP forms multiple, possibly dynamic, interactions with both subassemblies.
5. The inner membrane complex (IMC)
The A. tumefaciens IMC is composed of the energy ATPases VirB4 and VirB11 in association with two polytopic membrane proteins VirB3 and VirB6 and the bitopic subunit VirB8.
5.1. The ATP energy center
Besides the T4CP, the VirB4 and VirB11 ATPases are required for elaboration of most T4SSs. VirB4 ATPases are ubiquitous components of T4SSs, whereas the VirB11 subunits are missing from some Gram-negative systems, e.g., the F plasmid T4SS, and all Gram-positive systems [1, 90]. VirB4 subunits are integral or tightly-associated membrane proteins [101, 102], and the VirB11 ATPases bind the membrane peripherally and typically co-fractionate with the membrane and cytosol, suggestive of a possible dynamic association with the IMC [103, 104].
The VirB4 ATPases are large (>70-kDa) proteins with consensus Walker A and B NTP-binding domains and a variable number of predicted TMDs. VirB4 ATPases share a common ancestry with the T4CPs [90], and there is now structural evidence by cryoelectron microscopy that VirB4-like TrwKR388 assembles as a hexamer (Fig. 4) [105]. By small-angle X-ray scattering, low-resolution structures were solved for a membrane-extracted dimeric form of pKM101 TraB and a soluble monomeric form of LvhB4 from L. pneumophila [106]. These structural data, and earlier biochemical findings [29], suggest that the VirB4 ATPases might exist in different oligomeric states, possibly transitioning between lower- and higher-order complexes to fulfill distinct activities within the cell. VirB4 homologs form multiple contacts with other IMC subunits as well as the VirB10 component of the core complex [107–109].
Despite the structural advances, it remains unknown how VirB4 subunits catalyze substrate transfer. In A. tumefaciens, VirB4 did not form detectable FA-crosslinks with the T-DNA substrate, but catalytically-active VirB4 was necessary for detection of substrate crosslinks with the VirB6 and VirB8 channel components [38, 107]. These findings led to a proposal that VirB4 mediates substrate transfer indirectly through coordination of its ATP binding/hydrolysis activities with those of VirD4 and VirB11 [107]. However, more recent studies of the E. faecalis pCF10-encoded T4SS showed that VirB4-like PrgJ forms FA-crosslinks with pCF10 in a reaction requiring the pCF10-encoded Dtr factors and PcfC T4CP [31]. Additionally, purified VirB4 homologs encoded by E. coli plasmids pKM101 and R388 recently were shown to bind DNA substrates in vitro [105, 110]. These findings, coupled with recent structural evidence for localization of VirB4 hexamers at the base of the IMC, suggest the possibility that VirB4 participates more directly in substrate transfer than previously envisioned.
VirB11 ATPases are structurally related to a large family of AAA+ ATPases that also include the GspE ATPases associated with type II secretion systems (T2SSs) and type IV pilus assembly systems [111]. Crystal structures were solved for H. pylori HP0525 (now called Cagβ; [5]) and B. suis VirB11, and both present as hexameric, double-stacked rings in which the N-terminal domains (NTDs) interact to form one ring and the C-terminal domains (CTDs) form the second (Fig. 4) [111, 112]. For Cagβ, the CTD adopts a RecA fold, whereas the N-terminal domain is unique. The B. suis VirB11 monomer differs dramatically from that of Cagβ by a large domain swap caused by the insertion of additional sequences into the linker between the NTD and the CTD. The overall assembly of the VirB11 hexamer remains intact compared to Cagβ but the domain organization profoundly modifies the nucleotide-binding site and the interface between subunits. Based on sequence comparisons, it is likely that most VirB11 homologs display a hexameric structure closely resembling B. suis VirB11 [112].
VirB11 subunits are required for substrate transfer and pilus biogenesis. In A. tumefaciens, the T-DNA substrate was shown to crosslink with VirB11, and this contact was dependent on VirD2 nicking at the T-DNA borders and production of the VirD4 T4CP. These findings led to the proposal that the T4CP receptor delivers the T-DNA substrate to VirB11 for further processing [38, 107]. The VirB11 subunits might use the mechanical leverage generated by nucleotide-dependent conformational changes to unfold relaxases and protein substrates and dissociate bound accessory factors, e.g., relaxosome subunits or secretion chaperones [113]. Such an activity would be reminiscent of that described for InvC, a VirB11-like subunit associated with a type III secretion system in Salmonella enterica [114]. VirD4, VirB11, and VirB4 physically and functionally interact to energize substrate transfer [107, 115], but further biochemical studies are needed to decipher the molecular details of ATP-dependent reactions mediating substrate processing and translocation through the IMC.
5.2. The inner membrane channel
In addition to the ATPases, the postulated subunits of the IMC and, specifically, the inner membrane translocon include VirB3, VirB6, and VirB8. A. tumefaciens VirB3 is a small, two-pass membrane protein that binds to VirB4. VirB3 subunits invariably are cosynthesized with VirB4-like ATPases and in some systems the two are fused as a single protein, suggestive of a 1:1 stoichiometry and coordination of function [1]. VirB6 subunits have evolved as two distinct subtypes. Those resembling A. tumefaciens VirB6 are ~300 residues with 5 or more membrane-spanning domains and a large central, periplasmic domain. The ‘extended-VirB6’ subunits are larger (>500 residues) with an N-terminal polytopic membrane domain and a large C-terminal hydrophilic domain [1]. The role of this C-terminal domain is unspecified, although there is evidence in the E. coli F plasmid [116] and Vibrio cholerae SXT integrative and conjugative element (ICE) [117, 118] transfer systems for protrusion or release of this domain to the cell surface and into target cells. In A. tumefaciens, FA-crosslinking of VirB6 with the T-DNA substrate depended on synthesis of catalytically active forms of VirD4 and the VirB4 and VirB11 ATPases, supporting the notion that ATP energy is necessary for substrate engagement with the VirB6 component of the translocon [107, 119].
VirB8 subunits are bitopic proteins with a short cytoplasmic N-terminal domain, TM domain, and large C-terminal periplasmic domain [120]. X-ray structures have been solved for the periplasmic fragments of B. suis [121, 122] and A. tumefaciens [121, 122] VirB8 subunits (Fig. 4). Recent studies have identified VirB8-like subunits as common components of Gram-positive systems, and crystal structures now exist for the periplasmic domains of two VirB8-like subunits, Clostridium perfringens pCW3-encoded TcpC [123] and Streptococcus agalactiae pIP501-encoded TraM [124]. VirB8 domains present as large extended β-sheets with five α-helices, giving rise to an overall globular fold. These subunits packed as dimers or trimers in the crystal structures, and results of mutational analyses suggest that oligomerization is physiologically relevant [125, 126]. This dimer interface has even served as a target for a high-throughput screen aimed at identifying small molecule inhibitors of T4SSs [127]. In A. tumefaciens, VirB8 formed an FA-crosslink with the T-DNA, pointing to a direct role for this subunit in substrate translocation [38]. In the TRIP assays, some VirB6 mutations were shown not to affect substrate crosslinking with VirB6, but did block substrate crosslinking with VirB8 [119]. VirB6 and VirB8 thus appear to participate sequentially in substrate transfer, reflecting a possible architectural arrangement for this portion of the translocation channel.
6. The outer membrane complex (OMC) and core
Early studies of the A. tumefaciens VirB/VirD4 system showed that cosynthesis of the VirB7 lipoprotein, VirB9, and VirB10 were required for stabilization of the other two subunits, as well as most of the other VirB subunits (see [12]). Results of these studies led to the concept of the ‘core complex’, an intrinsically stable VirB7/B9/B10 subassembly that serves as a structural scaffold during machine biogenesis. Existence of this core complex has now been validated by detection of ring-shaped core complexes from A. tumefaciens by transmission electron microscopy [128] and high resolution structural analysis of the related E. coli pKM101-encoded TraN/O/F core complex by cryoelectron microscopy and X-ray crystallography (Fig. 4) [33, 34].
The pKM101 core complex, composed of 14 copies of each subunit, is a 1.05 MDa structure of 185 Å in width and height [33]. It is composed of two layers (I- and O-layers) that form a double-walled ring-like structure. The I-layer, composed of the N-terminal domains of TraO and TraF, is anchored in the inner membrane with a 55 Å diameter hole formed by the TM helices of TraF. The O-layer, composed of TraN and the C-terminal domains of TraO and TraF, has a main body and a narrower cap on the outermost side of the complex that is thought to insert in the outer membrane. A narrow hole (10 Å) exists in the cap, resulting in a channel extending through the entire cylindrical structure. A crystal structure of the O-layer revealed that the 14 copies of a TraF/VirB10 domain comprising two α-helices separated by a loop, termed the antennae projection (AP), form the cap structure (Fig. 4) [34, 98]. In view of biochemical evidence that the N-terminal region of VirB10 stably inserts into the inner membrane [34, 98], it is now evident that the VirB10/TraF subunits are entirely unique bacterial proteins in their capacity to span the entire cell envelope.
The transenvelope topology of the core complex lends itself well to a role as a structural scaffold for the T4SS [14, 98]. The inner membrane ring formed by the TM helices of VirB10 is predicted to encircle or sit on top of the IMC. The central chamber of the core complex is approximately 100 Å in diameter, sufficiently large to house VirB subunits postulated on the basis of the TrIP studies and other findings to include the C-terminal domain of VirB8, the pilin subunit VirB2, and a portion of the N-terminal domain of VirB9 [14, 39]. The entire OMC, consisting of the core complex encircling the central translocation channel, is predicted to interact with the IMC and mediate passage of substrates through the periplasm and outer membrane.
7. Activation of substrate transfer through the distal portion of the translocation channel
Most T4SSs translocate their cargo substrates only in response to establishment of productive contact with target cells. These T4SSs therefore must assemble in a closed state and require specific signals for channel activation. Studies have now shown that a combination of intracellular and extracellular signals activates substrate transfer. These studies also have provided evidence that VirB10 subunits function not only as components of the core structural scaffold but also play a central role in signal perception.
A. tumefaciens VirB10 undergoes a conformational switch, detectable as a change in protease susceptibility, in response to two intracellular signals: i) DNA substrate binding to the VirD4 and VirB11 ATPases and ii) ATP energy use by the VirD4 and VirB11 ATPases [99, 129]. Further work established that this conformational switch is required for passage of the DNA substrate through the distal portion of the translocation channel [38]. The nature of this switch is not yet known, but coupling of these intracellular signals to gating of the outer membrane pore is suggested by isolation of a mutation near the VirB10 AP pore that ‘locks’ VirB10 in the energized conformation and allows for release of secretion substrates to the cell surface independently of target cell contact [130]. Interestingly, it has also been reported that a DNA substrate and a Dtr accessory factor stimulate the ATPase activity of TrwBR388 T4CP in vitro [131]. In a generalized model, therefore, one can postulate a sequence of reactions whereby docking of the DNA substrate with the T4CP and subsequent transfer to VirB11 stimulates the catalytic activities of one or both ATPases. ATP-dependent conformational changes in the ATPases induces a VirB10 conformational switch that is transduced to the C-terminal region of the core complex near or at the cap structure, resulting in channel opening and substrate passage. This overall energy-coupled transfer reaction is reminiscent of that described for the TonB energy coupling system, which also induces channel gating at the outer membrane in response to energy sensing through contacts with an inner membrane energy harvesting complex [129]. Notably, VirB10 senses ligand and ATP energy signals and is a component of the supramolecular core complex, whereas TonB senses the proton motive force and functions as a monomer or dimer [132]. Deciphering the molecular details of energy transduction along the core complex and the mechanism of OM gating remain intriguing areas for future investigation.
In early studies of the E. coli F plasmid transfer system, genetic evidence was presented for the existence of a ‘mating signal’ that was propagated by recipient cells upon contact with donor cells [133]. The nature of this mating signal remains unknown, but recent work showed that an external signal is transduced across the T4SS to the T4CP [134]. These studies explored the requirements for transduction by bacteriophage R17, whose receptor is the conjugative pilus encoded by the IncF plasmid R1. The findings established that R17 infects only piliated cells in which the plasmid R1 relaxosome is docked with the TraDR1 T4CP. Nicking activity of the R1 relaxase is also required, establishing that the relaxosome must be catalytically active. In this system, the TraD T4CP does not display detectable ATPase activity even when bound by relaxosome. However, binding of bacteriophage R17 to the conjugative pilus stimulates TraD activity and, correspondingly, phage uptake. Activation of the R1 T4SS therefore requires relaxosome docking at the T4CP as well as phage binding to the R1-encoded pilus. Although the findings highlight the role of phage binding to the pilus receptor for T4SS activation, the authors suggest that phage binding mimics the natural mating signal propagated upon pilus-mediated contact with the recipient cell [134]. In light of these findings and the transenvelope topology of VirB10 and its demonstrated role in sensing of intracellular ligand and energy signals, it is enticing to suggest that extracellular signals are also transduced along VirB10 and the core complex to the cell interior.
8. The translocation route: one or two steps?
In the T4SS translocation pathway presented here, the T-DNA substrate is delivered in one step through a channel that spans the entire cell envelope. This one-step pathway is supported by considerable biochemical and structural data for conjugation machines. However, as described above (Section 3.3, Fig. 3), there is evidence that the B. pertussis Ptl T4SS and, possibly, the Brucella spp. VirB sysstem, recruit substrates from the periplasm, implying a two-step translocation pathway. In this pathway, substrates are translocated first across the inner membrane to the periplasm and then engage with the OMC for translocation across the outer membrane [135].
The B. pertussis Ptl T4SS is composed of close homologs of all of the VirB proteins except for the VirB5 pilus-tip adhesin. This system does not elaborate detectable pili, but it likely assembles a composite IMC/OMC structure that is architecturally similar to structures visualized for the archetypal conjugation systems [11]. This raises a fundamental question of where the multisubunit PT engages with the Ptl translocation channel. The toxin may enter the core’s chamber through a lateral gate, but this seems unlikely given the extensive network of contacts observed between the TraO/VirB9 and TraF/VirB10 subunits [33, 34, 136]. Upon Sec-mediated secretion, the PT subunits associate with the outer membrane where they undergo disulfide crosslinking and oligomerization. This suggests that the outer membrane might be the site of substrate engagement with the T4SS; if so, PT might dock with the distal end of the OMC at or near the core’s cap structure. This is an appealing model because it dispenses with the need for the large multisubunit toxin to pass through a narrow translocation channel within the core chamber and, instead, simply dock at the distal end of the T4SS. For PT extrusion across the outer membrane, a mechanism analogous to that envisioned for type II secretion systems (T2SSs) might be employed [137, 138]. For type II secretion, substrates delivered to the periplasm via the Sec system are thought to engage with the tip of a pseudopilus structure elaborated by the T2SS. Upon substrate docking, the pseudopilus extends and pushes the substrate through an outer membrane secretin. Substrate is released into the milieu, and the pseudopilus retracts for another round of substrate docking and extrusion. It is interesting to note that B. pertussis PT is a member of the A/B toxin superfamily and that all other multisubunit A/B toxins are exported via T2SSs. It seems reasonable that the type II and Ptl systems have evolved similar extrusion mechanisms for secretion of these large, multisubunit toxins.
Do other T4SSs use a two-step mechanism for translocation? In A. tumefaciens, there is some evidence for accumulation of protein substrates in the periplasm [139, 140], although results of the TrIP studies did not support the notion that the T-DNA transfer intermediate accumulates in this compartment [38]. Presently, there is no structural information about the interface of the T4CP/IMC, and OMC/core complex, and it is conceivable that a lateral gate exists at the junction of the inner membrane and periplasm. The notion that T4SSs employ a two-step translocation route has largely been driven by the demonstrated phylogenetic and structural similarities between the hexameric NBDs of T4CPs and the SpoIIIE and FtsK DNA motor proteins [141]. FtsK and SpoIIIE translocate directionally along DNA [142–144], and T4CPs might similarly thread the ssDNA substrate in a 5’ to 3’ direction through NBD hexamer and transmembrane stem to the periplasm [135, 145]. Whether all T4SS substrates transiently enter the periplasm, via either a T4CP or IMC translocon, before entering the OMC/core remains an intriguing question for further study.
9. The conjugative pilus
Many Gram-negative bacterial T4SSs elaborate extracellular pili. For the VirB/VirD4 and related conjugation systems, the genetic requirements are nearly the same for assembly of these two structures, with the exception that the T4CP is not required for pilus production and the VirB1 transglycosylase is not required for assembly of the translocation channel. It is tempting to depict the T4SS channel and T-pilus as a single, supramolecular organelle, but at this time there is no structural evidence for this association. Nevertheless, the genetic findings suggest that the transenvelope IMC/OMC structure functions as a scaffold both for the channel and pilus (Fig. 5).
Fig. 5.

Postulated biogenesis pathway for the A. tumefaciens virB-encoded pilus. VirB2 pilin is inserted into the membrane and processed in several steps to yield a pool of cyclized pilins. VirB4, aided by VirB11, catalyzes dislocation of mature pilins and feeds the pilin monomers into the site of pilus assembly within the core complex. Pilus polymerization initiates either on an inner membrane platform composed of VirB6 and VirB8 or an outer membrane platform composed of the cap structure of the core complex. An alternative pathway proposes that pilin monomers are shunted through the periplasm to the core’s cap.
There are two well-characterized groups of conjugative pili, F-pili elaborated by the E. coli F plasmid and other plasmids of the IncF, IncH1, and IncH2 incompatibility (Inc) groups and P-pili produced by E. coli plasmids of the P, N, or W Inc groups and the A. tumefaciens VirB/VirD4 T4SS [41, 146]. Studies of these pilus systems have identified physical properties of pilins and pili as well as early-stage reactions required for polymerization (Fig. 4). The F-type pili are typically ~90 Å and flexible, and range in length up to 1 micron. These pili dynamically extend and retract, enabling donor cells to bind and draw recipient cells into physical contact for establishment of the mating junction. The P-type pili are thicker (90–110 Å), more rigid, and much shorter than F-pili although length measurements are complicated by the fact that isolated pili are typically broken [147–149]. These pili do not appear to undergo cycles of extension/retraction, but instead accumulate in the milieu, either through breakage or an active sloughing mechanism. They tend to aggregate as a mesh of polymers, which is thought to promote nonspecific clumping of donor and recipient cells and subsequent formation of mating junctions.
The F- and P-pili are synthesized as pro-proteins with unusually long (~30–50 residues) leader peptides that are cleaved upon insertion into the inner membrane (Fig. 5) [45, 150]. Upon membrane insertion, the pro-pilins are processed further by LepB signal peptidase and additional proteases. The N termini of both F- and P-type pilins are modified, the F-type pilin by N-acetylation and the P-type pilin by covalent linkage to the C terminus in a head-to-tail cyclization reaction [41, 151, 152] Cyclization stabilizes the P-type pilins in the membrane and is essential for pilus assembly. Mature F- and P-type pilins accumulate in the inner membrane, the former as a pool estimated at ~100,000 monomers [153]. Upon receipt of an unknown signal, the pilin monomers are extracted from this pool to build the pilus. A recent study explored the mechanism by which pilins are extracted from the hydrophobic membrane environment by assaying for accessibility of Cys-residues engineered along the length of A. tumefaciens VirB2 to a membrane impermeable thiol-reactive reagent [154]. This study defined the inner membrane topology of VirB2 and also presented evidence that the VirB4 ATPase modulates the structural state of membrane-integrated pilin. Catalytically-active VirB4 induced changes in the thiol accessibility of engineered Cys residues in VirB2 that were consistent with a role for the ATPase in extraction of the pilin from the membrane [154]. Independently, evidence was presented that VirB4 stabilizes subunits required for pilus assembly and is essential for interaction of VirB2 with the pilus-tip protein VirB5, a reaction thought to be essential for pilus nucleation [108, 155]. Taken together, these findings support a model in which VirB4 dislocates mature pilins from the inner membrane and mediates formation of the VirB2-VirB5 nucleation complex for subsequent pilus polymerization (Fig. 5).
Where in the cell envelope do pili polymerize? As mentioned above, the recent structural studies place VirB4 monomers or hexamers at the base of the IMC, in physical association with the core complex [106]. This association might facilitate the lateral translocation of pilin monomers through the membrane and into the chamber of the core complex. Once in the chamber, pilins would then undergo polymerization to build the conjugative pilus (Fig. 5) [1]. The central chamber of the pKM101-encoded core complex is approximately 100 Å in diameter, sufficiently large to house the pilus (90–110 Å). However, the outer membrane ring formed by TraF’s AP domain is at most only 32 A and clearly not large enough to accommodate pili [14]. If pili assemble from a platform of subunits located at the inner membrane, pilus extension through the chamber and across the outer membrane must induce gross structural changes in the distal portion of the core complex. A model for pilus assembly from an inner membrane platform is reminiscent of that described for type IV pili [156]. For F-pilus biogenesis, this is an appealing model because F-pili, like type IV pili, successively recycle membrane-integrated pilins during extension and retraction. Alternatively, pili might assemble at the outer membrane, on a platform formed by the AP domain of the core complex [1, 108]. Accordingly, upon extraction from the inner membrane the hydrophobic pilins would be chaperoned within the core’s chamber or directly across the periplasm to the outer membrane for nucleation. Such an assembly pathway is reminiscent of that described for P or Type I fimbriae [157]. Clearly, answering the long-standing question of whether conjugative pili nucleate from an inner or outer membrane platform remains a central challenge to this field.
10. Summary and Future Directions
In the last decade, there has been striking progress in definition of structural and mechanistic features of type IV secretion. Results with the TrIP assay supplied the first direct evidence that these machines indeed assemble as DNA transfer channels, and the data generated the first architectural view of the VirB/VirD4 channel. The CryoEM structure of the pKM101-encoded TraN/TraO/TraF core complex generated further excitement in the field, followed quickly by an X-ray structure of the outer portion of this complex. Higher resolution studies generated a refined structure of the core complex and allowed the fitting of atomic models. In view of this remarkable progress, we can anticipate a near-atomic resolution structure of an entire T4SS in the very near future. The mechanistic studies also have generated a more detailed understanding of substrate translocation signals and contributions of accessory factors to substrate engagement with the T4CP receptor. In vivo biochemical approaches have enabled formulation of topological models for all the conserved T4SS subunits, and mutant analyses and domain swapping experiments continue to provide insights into specific contributions of individual subunits and domains to channel and pilus assembly and function.
There remain, however, many “black boxes” about type IV secretion. Major gaps in our knowledge exist across the entire machine. At the cytoplasmic entrance to the channel, how do the ATPases coordinate their activities and how does ATP binding and hydrolysis contribute to assembly of the translocation channel and substrate processing or transfer, or biogenesis of the conjugative pilus or other surface structures? At the inner membrane, what constitutes the channel, and in what form – folded or unfolded - is the substrate translocated? In the periplasm, what is the nature of the junction between the IMC and the core complex and does a channel extend from one to the next? Or is a two-step translocation model generally applicable for T4SSs; if so, where does the substrate gain access to the translocation channel for delivery across the outer membrane? Where does the pilus initiate polymerization and what is its physical relationship with the IMC/core complex? Finally, at the outer membrane, what constitutes the outer membrane pore and how is the pore gated by intracellular and extracellular signals? And perhaps the blackest of the boxes pertains to the donor-target cell junction. How do donor cells establish productive contacts with target cells, and what is the basis for the extremely broad host range of some systems such as the A. tumefaciens VirB/VirD4 T4SS which can deliver substrates to many different bacterial, plant, fungal, and human cell types?
Answers to these and other outstanding questions will continue to require structural analyses of machine subassemblies in vitro as well as high-resolution imaging of machines in their native membrane environments. The forthcoming images will provide us with an architectural blueprint, but a comprehensive mechanistic understanding of how these machines actually work will require a combination of creative in vivo genetic and state-of-the art cell biological approaches together with in vitro reconstitution assays. Finally, to fully appreciate the biological diversity of T4SSs and to gain a better understanding of the mechanistic themes and variations, it is imperative that we invest comparable effort in studies of systems very distantly related to the A. tumefaciens VirB/VirD4 T4SS, e.g., effector translocators employed by intracellular pathogens and Gram-positive and archaeal conjugation systems [1].
Highlights.
We review structural information about bacterial type IV secretion systems (T4SSs).
We discuss adaptations to type IV machines that evolved for specialized functions.
We summarize functions of subunits and subassemblies of archetypal T4SSs.
We review mechanisms for substrate presentation to the bacterial T4SSs.
We present models depicting the substrate translocation route through T4SSs.
Acknowledgments
We thank members of the Christie laboratory for helpful discussions. This work was supported by NIH grants R01GM48746 and 1R21AI105454. Chimera software is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron C. From bioremediation to biowarfare: on the impact and mechanism of type IV secretion systems. FEMS Microbiol Lett. 2005;253:163–170. doi: 10.1016/j.femsle.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Backert S, Clyne M. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2011;16(Suppl 1):19–25. doi: 10.1111/j.1523-5378.2011.00876.x. [DOI] [PubMed] [Google Scholar]
- 4.Nagai H, Kubori T. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer W. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 2011;278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 6.Llosa M, Roy C, Dehio C. Bacterial type IV secretion systems in human disease. Mol Microbiol. 2009;73:141–151. doi: 10.1111/j.1365-2958.2009.06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol. 2006;188:882–893. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey ME, Woodhams KL, Dillard JP. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria. Front Microbiol. 2011;2:61. doi: 10.3389/fmicb.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278:4668–4682. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- 12.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelvin SB. Traversing the cell: Agrobacterium T-DNA's journey to the host genome. Front Plant Sci. 2012;3:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid. 2013;70:289–302. doi: 10.1016/j.plasmid.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terradot L, Waksman G. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 2011;278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- 18.Christie PJ. Bacterial type IV secretion: The Agrobacterium VirB/D4 and related conjugation systems. Biochem Biophys Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambow-Larsen AA, Petersen EM, Gourley CR, Splitter GA. Brucella regulators: self-control in a hostile environment. Trends Microbiol. 2009;17:371–377. doi: 10.1016/j.tim.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost LS, Koraimann G. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 2010;5:1057–1071. doi: 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- 21.Dunny GM, Johnson CM. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr Opin Microbiol. 2011;14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laverde-Gomez JA, Sarkar MK, Christie PJ. Regulation of bacterial type IV secretion systems. In: Vasil M, Darwin A, editors. Regulation of Bacterial Virulence. ASM Press; Washington, D. C: 2012. pp. 335–362. [Google Scholar]
- 23.Zechner EL, Lang S, Schildbach JF. Assembly and mechanisms of bacterial type IV secretion machines. Philos Trans R Soc Lond B Biol Sci. 2012;367:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Tekaya H, Gorvel JP, Dehio C. Bartonella and Brucella--weapons and strategies for stealth attack. Cold Spring Harb Perspect Med. 2013;3 doi: 10.1101/cshperspect.a010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voth DE, Broederdorf LJ, Graham JG. Bacterial Type IV secretion systems: versatile virulence machines. Future Microbiol. 2012;7:241–257. doi: 10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge J, Shao F. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cell Microbiol. 2011;13:1870–1880. doi: 10.1111/j.1462-5822.2011.01710.x. [DOI] [PubMed] [Google Scholar]
- 27.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 28.Rikihisa Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev. 2011;24:469–489. doi: 10.1128/CMR.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tato I, Zunzunegui S, de la Cruz F, Cabezon E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA dependent ATPase. Proc Natl Acad Sci U S A. 2005;102:8156–8161. doi: 10.1073/pnas.0503402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Zhang X, Manias D, Yeo HJ, Dunny GM, Christie PJ. Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J Bacteriol. 2008;190:3632–3645. doi: 10.1128/JB.01999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangrez AY, Abajy MY, Keller W, Shouche Y, Grohmann E. Biochemical characterization of three putative ATPases from a new type IV secretion system of Aeromonas veronii plasmid pAC3249A. BMC Biochem. 2010;11:10. doi: 10.1186/1471-2091-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai EM, Kado CI. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Backert S, Fronzes R, Waksman G. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 2008;16:409–413. doi: 10.1016/j.tim.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christie PJ. Structural biology: Translocation chamber's secrets. Nature. 2009;462:992–994. doi: 10.1038/462992b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 41.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 42.Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2012;85:378–391. doi: 10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris RL, Hombs V, Silverman PM. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol Microbiol. 2001;42:757–766. doi: 10.1046/j.1365-2958.2001.02667.x. [DOI] [PubMed] [Google Scholar]
- 44.Harris RL, Silverman PM. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol. 2004;186:5480–5485. doi: 10.1128/JB.186.16.5480-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman PM, Clarke MB. New insights into F-pilus structure, dynamics, and function. Integr Biol (Camb) 2010;2:25–31. doi: 10.1039/b917761b. [DOI] [PubMed] [Google Scholar]
- 46.Arutyunov D, Arenson B, Manchak J, Frost LS. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J Bacteriol. 2010;192:1730–1734. doi: 10.1128/JB.00726-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Path. 2011;7:e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190–1202. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 50.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, Carena I, Dehio C. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vergunst AC, van Lier Aden Dulk-Ras MC, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carey KL, Newton HJ, Luhrmann A, Roy CR. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Path. 2011;7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marchesini MI, Herrmann CK, Salcedo SP, Gorvel JP, Comerci DJ. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell Microbiol. 2011;13:1261–1274. doi: 10.1111/j.1462-5822.2011.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hohlfeld S, Pattis I, Puls J, Plano GV, Haas R, Fischer W. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol. 2006;59:1624–1637. doi: 10.1111/j.1365-2958.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- 58.Couturier MR, Tasca E, Montecucco C, Stein M. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun. 2006;74:273–281. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pattis I, Weiss E, Laugks R, Haas R, Fischer W. The Helicobacter pylori CagF protein is a type IV secretion chaperone-like molecule that binds close to the C-terminal secretion signal of the CagA effector protein. Microbiology. 2007;153:2896–2909. doi: 10.1099/mic.0.2007/007385-0. [DOI] [PubMed] [Google Scholar]
- 60.Bonsor DA, Weiss E, Iosub-Amir A, Reingewertz TH, Chen TW, Haas R, Friedler A, Fischer W, Sundberg EJ. Characterization of the translocation-competent complex between the Helicobacter pylori oncogenic protein CagA and the accessory protein CagF. J Biol Chem. 2013;288:32897–32909. doi: 10.1074/jbc.M113.507657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc Natl Acad Sci U S A. 2005;102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parker C, Meyer RJ. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol Microbiol. 2007;66:252–261. doi: 10.1111/j.1365-2958.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- 63.Lang S, Gruber K, Mihajlovic S, Arnold R, Gruber CJ, Steinlechner S, Jehl MA, Rattei T, Frohlich KU, Zechner EL. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol Microbiol. 2010;78:1539–1555. doi: 10.1111/j.1365-2958.2010.07423.x. [DOI] [PubMed] [Google Scholar]
- 64.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, Zangger K, Zechner EL, Waksman G. Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol Microbiol. 2013;89:324–333. doi: 10.1111/mmi.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Kregten M, Lindhout BI, Hooykaas PJ, van der Zaal BJ. Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal. Mol Plant Microbe Interact. 2009;22:1356–1365. doi: 10.1094/MPMI-22-11-1356. [DOI] [PubMed] [Google Scholar]
- 66.Alperi A, Larrea D, Fernandez-Gonzalez E, Dehio C, Zechner EL, Llosa M. A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system. J Bacteriol. 2013;195:4999–5006. doi: 10.1128/JB.00367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toro N, Datta A, Carmi OA, Young C, Prusti RK, Nester EW. The Agrobacterium tumefaciens virC1 gene product binds to overdrive, a T-DNA transfer enhancer. J Bacteriol. 1989;171:6845–6849. doi: 10.1128/jb.171.12.6845-6849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 2007;26:2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Z, Sagulenko E, Ding Z, Christie PJ. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dym O, Albeck S, Unger T, Jacobovitch J, Branzburg A, Michael Y, Frenkiel-Krispin D, Wolf SG, Elbaum M. Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners. Proc Natl Acad Sci U S A. 2008;105:11170–11175. doi: 10.1073/pnas.0801525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vergunst AC, Van Lier MC, Den Dulk-Ras A, Hooykaas PJ. Recognition of the Agrobacterium tumefaciens VirE2 translocation tignal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 2003;133:978–988. doi: 10.1104/pp.103.029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 75.Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol Microbiol. 2005;55:912–926. doi: 10.1111/j.1365-2958.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- 76.Cambronne ED, Roy CR. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 2007;3:e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Path. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 79.Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 80.de Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual JF, Muller A, Twizere JC, Nkengfac B, Vandenhaute J, Hill DE, Salcedo SP, Gorvel JP, Letesson JJ, De Bolle X. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- 81.Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 82.Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, von Bodman SB, Lanka E, Ream W, Farrand SK. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas J, Hecht DW. Interaction of Bacteroides fragilis pLV22a relaxase and transfer DNA with Escherichia coli RP4-TraG coupling protein. Mol Microbiol. 2007;66:948–960. doi: 10.1111/j.1365-2958.2007.05967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 85.Gomis-Ruth FX, Coll M. Structure of TrwB, a gatekeeper in bacterial conjugation. Int J Biochem Cell Biol. 2001;33:839–843. doi: 10.1016/s1357-2725(01)00060-7. [DOI] [PubMed] [Google Scholar]
- 86.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1- ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 87.Hormaeche I, Alkorta I, Moro F, Valpuesta JM, Goni FM, De La Cruz F. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J Biol Chem. 2002;277:46456–46462. doi: 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- 88.Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 89.Strick TR, Quessada-Vial A. FtsK: a groovy helicase. Nat Struct Mol Biol. 2006;13:948–950. doi: 10.1038/nsmb1106-948. [DOI] [PubMed] [Google Scholar]
- 90.Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2012;30:315–31. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gunton JE, Gilmour MW, Baptista KP, Lawley TD, Taylor DE. Interaction between the co-inherited TraG coupling protein and the TraJ membrane-associated protein of the H-plasmid conjugative DNA transfer system resembles chromosomal DNA translocases. Microbiology. 2007;153:428–441. doi: 10.1099/mic.0.2006/001297-0. [DOI] [PubMed] [Google Scholar]
- 92.Disque-Kochem C, Dreiseikelmann B. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol. 1997;179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol. 2004;186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu J, Frost LS. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J Bacteriol. 2005;187:4767–4773. doi: 10.1128/JB.187.14.4767-4773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol. 2008;70:89–99. doi: 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- 96.de Paz HD, Sangari FJ, Bolland S, Garcia-Lobo JM, Dehio C, de la Cruz F, Llosa M. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiology. 2005;151:3505–3516. doi: 10.1099/mic.0.28410-0. [DOI] [PubMed] [Google Scholar]
- 97.Segura RL, Aguila-Arcos S, Ugarte-Uribe B, Vecino AJ, de la Cruz F, Goni FM, Alkorta I. The transmembrane domain of the T4SS coupling protein TrwB and its role in protein-protein interactions. Biochim Biophys Acta. 2013;1828:2015–2025. doi: 10.1016/j.bbamem.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 98.Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol Microbiol. 2009;71:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cascales E, Atmakuri K, Sarkar MK, Christie PJ. DNA substrate-induced activation of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol. 2013;195:2691–2704. doi: 10.1128/JB.00114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garza I, Christie PJ. A putative transmembrane leucine zipper of Agrobacterium VirB10 is essential for T-pilus biogenesis but not type IV secretion. J Bacteriol. 2013;195:3022–34. doi: 10.1128/JB.00287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dang TA, Christie PJ. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–462. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rabel C, Grahn AM, Lurz R, Lanka E. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J Bacteriol. 2003;185:1045–1058. doi: 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rashkova S, Spudich GM, Christie PJ. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J Bacteriol. 1997;179:583–591. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pena A, Matilla I, Martin-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezon E, Arechaga I. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem. 2012;287:39925–39932. doi: 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wallden K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci USA. 2012;109:11348–11353. doi: 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yuan Q, Carle A, Gao C, Sivanesan D, Aly KA, Hoppner C, Krall L, Domke N, Baron C. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J Biol Chem. 2005;280:26349–26359. doi: 10.1074/jbc.M502347200. [DOI] [PubMed] [Google Scholar]
- 109.Mossey P, Hudacek A, Das A. Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J Bacteriaol. 2010;192:2830–2838. doi: 10.1128/JB.01331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Durand E, Oomen C, Waksman G. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J Bacteriol. 2010;192:2315–2323. doi: 10.1128/JB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 112.Hare S, Bayliss R, Baron C, Waksman G. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol. 2006;360:56–66. doi: 10.1016/j.jmb.2006.04.060. [DOI] [PubMed] [Google Scholar]
- 113.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 115.Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezon E. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol. 2013;195:4195–4201. doi: 10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007;153:442–451. doi: 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- 117.Marrero J, Waldor MK. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell. 2005;8:963–970. doi: 10.1016/j.devcel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 118.Marrero J, Waldor MK. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol. 2007;189:6469–6473. doi: 10.1128/JB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol. 2004;341:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baron C. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem Cell Biol. 2006;84:890–899. doi: 10.1139/o06-148. [DOI] [PubMed] [Google Scholar]
- 121.Terradot L, Bayliss R, Oomen C, Leonard, Baron C, Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:4956–4961. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc Natl Acad Sci U S A. 2006;103:2582–2587. doi: 10.1073/pnas.0511216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, Lyras D, Whisstock JC, Rood JI. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol Microbiol. 2012;83:275–288. doi: 10.1111/j.1365-2958.2011.07930.x. [DOI] [PubMed] [Google Scholar]
- 124.Goessweiner-Mohr N, Grumet L, Arends K, Pavkov-Keller T, Gruber CC, Gruber K, Birner-Gruenberger R, Kropec-Huebner A, Huebner J, Grohmann E, Keller W. The 2.5 A structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem. 2013;288:2018–2028. doi: 10.1074/jbc.M112.428847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paschos A, Patey G, Sivanesan D, Gao C, Bayliss R, Waksman G, O'Callaghan D, Baron C. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc Natl Acad Sci U S A. 2006;103:7252–7257. doi: 10.1073/pnas.0600862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sivanesan D, Baron C. The dimer interface of Agrobacterium tumefaciens VirB8 is important for type IV secretion system function, stability, and association of VirB2 with the core complex. J Biol Chem. 2011;193:2097–2106. doi: 10.1128/JB.00907-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith MA, Coincon M, Paschos A, Jolicoeur B, Lavallee P, Sygusch J, Baron C. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem Biol. 2012;19:1041–1048. doi: 10.1016/j.chembiol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 128.Sarkar MK, Husnain SI, Jakubowski SJ, Christie PJ. Isolation of bacterial type IV machine subassemblies. Methods Mol Biol. 2013;966:187–204. doi: 10.1007/978-1-62703-245-2_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci U S A. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Banta LM, Kerr JE, Cascales E, Giuliano ME, Bailey ME, McKay C, Chandran V, Waksman G, Christie PJ. An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect. J Bacteriol. 2011;193:2566–2574. doi: 10.1128/JB.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tato I, Matilla I, Arechaga I, Zunzunegui S, de la Cruz F, Cabezon E. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J Biol Chem. 2007;282:25569–25576. doi: 10.1074/jbc.M703464200. [DOI] [PubMed] [Google Scholar]
- 132.Freed DM, Lukasik SM, Sikora A, Mokdad A, Cafiso DS. Monomeric TonB and the Ton box are required for the formation of a high-affinity transporter-TonB complex. Biochemistry. 2013;52:2638–2648. doi: 10.1021/bi3016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kingsman A, Willetts N. The requirements for conjugal DNA synthesis in the donor strain during flac transfer. J Mol Biol. 1978;122:287–300. doi: 10.1016/0022-2836(78)90191-2. [DOI] [PubMed] [Google Scholar]
- 134.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol Microbiol. 2011;82:1071–1085. doi: 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 136.Rivera-Calzada A, Fronzes R, Savva CG, Chandran V, Lian PW, Laeremans T, Pardon E, Steyaert J, Remaut H, Waksman G, Orlova EV. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 2013;32:1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ayers M, Howell PL, Burrows LL. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 2010;5:1203–1218. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]
- 138.Campos M, Cisneros DA, Nivaskumar M, Francetic O. The type II secretion system - a dynamic fiber assembly nanomachine. Res Microbiol. 2013;164:545–555. doi: 10.1016/j.resmic.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 139.Chen L, Li CM, Nester EW. Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc Natl Acad Sci U S A. 2000;97:7545–7550. doi: 10.1073/pnas.120156997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 141.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 142.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 143.Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 144.Ptacin JL, Nollmann M, Becker EC, Cozzarelli NR, Pogliano K, Bustamante C. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat Struct Mol Biol. 2008;15:485–493. doi: 10.1038/nsmb.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gomis-Ruth FX, de la Cruz F, Coll M. Structure and role of coupling proteins in conjugal DNA transfer. Res Microbiol. 2002;153:199–204. doi: 10.1016/s0923-2508(02)01313-x. [DOI] [PubMed] [Google Scholar]
- 146.Schroder G, Lanka E. The mating pair formation system of conjugative plasmids - A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 147.Paranchych W, Frost LS. The physiology and biochemistry of pili. Adv Microb Physiol. 1988;29:53–114. doi: 10.1016/s0065-2911(08)60346-x. [DOI] [PubMed] [Google Scholar]
- 148.Bradley DE, Taylor DE, Cohen DR. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980;143:1466–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bradley DE. Morphological and serological relationships of conjugative pili. Plasmid. 1980;4:155–169. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- 150.Kalkum M, Eisenbrandt R, Lurz R, Lanka E. Tying rings for sex. Trends Microbiol. 2002;10:382–387. doi: 10.1016/s0966-842x(02)02399-5. [DOI] [PubMed] [Google Scholar]
- 151.Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 152.Lai EM, Eisenbrandt R, Kalkum M, Lanka E, Kado CI. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J Bacteriol. 2002;184:327–330. doi: 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Silverman PM. Towards a structural biology of bacterial conjugation. Mol Microbiol. 1997;23:423–429. doi: 10.1046/j.1365-2958.1997.2411604.x. [DOI] [PubMed] [Google Scholar]
- 154.Kerr JE, Christie PJ. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol. 2010;192:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aly KA, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology. 2007;153:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- 156.Douzi B, Filloux A, Voulhoux R. On the path to uncover the bacterial type II secretion system. Philos Trans R Soc Lond B Biol Sci. 2012;367:1059–1072. doi: 10.1098/rstb.2011.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Busch A, Waksman G. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci. 2012;367:1112–1122. doi: 10.1098/rstb.2011.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 159.Yeo HJ, Yuan Q, Beck MR, Baron C, Waksman G. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci U S A. 2003;100:15947–15952. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]