Abstract
Recent observations link myeloperoxidase (MPO) activation to neurodegeneration. In multiple sclerosis MPO is present in areas of active demyelination where the potent oxidant hypochlorous acid (HOCl), formed by MPO from H2O2 and chloride ions, could oxidatively damage myelin-associated lipids. The purpose of this study was (i) to characterize reaction products of sphingomyelin (SM) formed in response to modification by HOCl, (ii) to define the impact of exogenously added SM and HOCl-modified SM (HOCl-SM) on viability parameters of a neuronal cell line (PC12), and (iii) to study alterations in the PC12 cell proteome in response to SM and HOCl-SM. MALDI-TOF-MS analyses revealed that HOCl, added as reagent or generated enzymatically, transforms SM into chlorinated species. On the cellular level HOCl-SM but not SM induced the formation of reactive oxygen species. HOCl-SM induced severely impaired cell viability, dissipation of the mitochondrial membrane potential, and activation of caspase-3 and DNA damage. Proteome analyses identified differential expression of specific subsets of proteins in response to SM and HOCl-SM. Our results demonstrate that HOCl modification of SM results in the generation of chlorinated lipid species with potent neurotoxic properties. Given the emerging connections between the MPO–H2O2–chloride axis and neurodegeneration, this chlorinating pathway might be implicated in neuropathogenesis.
Keywords: Chlorinative stress, Chlorohydrins, 2D-DIGE, Glycolysis, Lipotoxicity, Proteomics, MPO–H2O2–chloride system, Free radicals
Oxidative stress has long been implicated in neurodegenerative disorders. Because of its high oxygen demand the mammalian brain is particularly sensitive to oxidative damage [1]. Generation of reactive oxygen species during chronic inflammation in the central nervous system (CNS) is a marker for progression of neurodegenerative diseases [2]. Potential sources of reactive species in the brain are mitochondria, amyloid-β peptides, redox-active iron, the NADPH-oxidase complex, and myeloperoxidase (MPO) [1,3].
Under physiological conditions MPO contributes to host defense [4], whereas chronic activation contributes to tissue injury [5–7]. MPO generates HOCl (the physiological mixture of hypochlorous acid and its anion) and other reactive species that are able to induce protein and lipid modification [8]. Increasing evidence points toward MPO as a disease-amplifying enzyme in neurodegeneration [3]. Recent studies revealed increased expression of MPO and subsequent formation of HOCl-modified proteins in a mouse model of Parkinson disease (PD) and human PD postmortem brain tissue [9]. Findings that knockout of the MPO gene in this PD model mitigates features of the disease emphasize the central role of MPO during disease progression [9]. In postmortem tissue of Alzheimer disease (AD) patients it was demonstrated that microglia and neurons contain MPO [10,11]. Overexpression of human MPO in a mouse model of AD resulted in enhanced cerebral lipid peroxidation, enhanced levels of oxidized phospholipids, and greater spatial memory deficits [12]. In multiple sclerosis, MPO is present in microglia/macrophages at lesion sites [13–15] and cortical demyelination is associated with increased MPO activity [16]. In pattern III lesions MPO expression by microglia in predemyelinating and actively demyelinating lesions was demonstrated [17]. These authors [17] suggested that focal activation of microglia leads to the formation of reactive species causing mitochondrial dysfunction and hypoxia-like degeneration of surrounding neurons. This setting amplifies tissue injury and oligodendrocyte apoptosis and demyelination and augments the inflammatory response. As activation of MPO occurs in the vicinity of axons, one could expect oxidative damage of myelin-associated proteins and/or lipids.
Indeed, HOCl has the capacity to modify lipids that are vital for CNS function. Compared with plasma membranes of other somatic cells the myelin membrane contains particularly high concentrations of cholesterol, phospholipids, and sphingolipids. Chlorination of cholesterol by MPO-derived HOCl leads to the generation of cholesterol chlorohydrins as well as dichlorinated products [18,19]. Phospholipids are subject to HOCl modification involving attack of double bonds of unsaturated fatty acyl residues and/or head-group modification, which can lead to chlorohydrin and/or chloramine generation [20]. In addition, nitrogen-centered radicals, phosphatidylglycoaldehydes, and phosphonitriles originating from HOCl-derived modification of either ethanolamine or serine phospholipids are formed [21,22]. Another phospholipid class in which the sn-1 aliphatic chain is attached to the glycerol backbone by a vinyl ether bond is the plasmalogen family [23]. These ether phospholipids account for approx 80% of total myelin glycerophosphoethanolamines and are particularly sensitive to HOCl attack. This reaction generates 2-chlorohexadecanal [24], a chlorinated fatty aldehyde with potent biological properties [7,25,26].
Within the sphingolipid subfraction, sphingomyelin (SM) takes a central role in axonal and neuronal maturation [27]. Based on the presence of MPO at areas of demyelination and evidence that even subtle perturbations in the sphingolipid content of neurons can disrupt their function in multiple sclerosis [28], this study focused on the accessibility of SM to HOCl modification. We have first analyzed product formation and then investigated the effects of HOCl-modified SM(d18:1/16:0) (HOCl-SM) on intracellular reactive oxygen species formation, mitochondrial function, and apoptosis. Finally we established the impact of SM and HOCl-SM on the PC12 cell (an in vitro model for dopaminergic neurons [29]) proteome.
Materials and methods
Materials
Cell culture supplies were from Gibco (Invitrogen, Vienna, Austria), PAA Laboratories (Linz, Austria), and Costar (Vienna, Austria). Fluorescent two-dimensional–difference gel electrophoresis (2D-DIGE) cyanine dyes (Cy2, Cy3, and Cy5), immobilized pH gradient strips (IPG strips, pH 3–10), and Pharmalyte (pH 3–10) were from GE Healthcare (Amersham Biosciences, Vienna, Austria). SM from chicken egg yolk, N-palmitoyl-d-erythro-sphingosylphosphorylcholine (SM(d18:1/16:0)), dipalmitoylphosphatidylcholine (PC(16:0/16:0)), NaOCl (reagent hypochlorite), colchicine, 2,5-dihydroxybenzoic acid (DHB), methionine (Met), propidium iodide (PI), and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) were from Sigma (Vienna, Austria). Cellulose acetate was from Sartorius AG (Goettingen, Germany). 2′,7′-Dichlorofluorescin diacetate (H2DCFDA) and 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzamidazolocarbocyanin iodide (JC-1) was from Molecular Probes (Invitrogen). Polyclonal rabbit anti-caspase-3 antibody (raised against human full-length caspase-3) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal rabbit anti-poly(ADP-ribose) polymerase (PARP) antibody (raised against a synthetic peptide corresponding to the caspase cleavage site in PARP) was from Cell Signaling Technology (Danvers, MA, USA). Horseradish peroxidase (HRP)-labeled secondary goat anti-rabbit IgG was from Sigma. SuperSignal Western blot detection reagent and BCA protein assay kit were from Pierce (Thermo Scientific, Waltham, MA, USA). MPO was from Planta Naturstoffe (Vienna, Austria). All other chemicals were from Sigma, Roth (Vienna, Austria), and Merck (Darmstadt, Germany).
Cell culture and cell treatment
Rat pheochromacytoma (PC12) cells were cultured in collagen-coated 75-cm2 flasks containing 15 ml RPMI 1640 medium supplemented with 10% (v/v) horse serum, 5% (v/v) fetal calf serum, 0.4% (v/v) Hepes buffer, 0.4% (w/v) sodium pyruvate, 0.4% (w/v) l-glutamine, and 50 μg/ml gentamycin at 37 °C (5% CO2). Passages below 30 were used for experiments.
Modification of brain lipids and SM by reagent HOCl
Female C57BL/6 mice were killed by cervical dislocation. The brains were removed, weighed, snap-frozen in liquid nitrogen, and homogenized in a mortar. Lipid extraction was performed as described [30] and lipids were stored at −70 °C until used. HOCl-mediated brain lipid modification was performed with NaOCl (1, 10, and 50 μg/100 μg lipid) overnight at room temperature (RT). Subsequently, lipids were extracted, dissolved in 1 ml CHCl3:MeOH (1:1, v/v), and analyzed by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS; see below).
HOCl modification of SM(d18:1/16:0) was performed at a molar oxidant:SM ratio of 2.5:1. Liposomes were prepared by dispersing 2 mg SM in 1 ml H2O using sonication (4×20 s on ice). After lipid extraction reaction products were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS) as described below. Residual HOCl in the reaction mixture was quenched by the addition of Met at an oxidant:Met ratio of 1:5 (35.6 mM final concentration).
Modification of SM by the MPO–H2O2–chloride system
SM liposomes (30 μg/ml) in phosphate-buffered saline (PBS; 50 mM, pH 5) supplemented with 140 mM NaCl were incubated in the presence of MPO (70 nM final concentration) at 37 °C for 30 min with shaking. Reaction was started by the addition of H2O2 (200 μM final concentration; eight additions of 25 μM H2O2 at 4-min intervals [31]) and stopped by removing cationic MPO using cellulose acetate. Subsequently, SM modification was allowed to proceed at RT in the dark for a further 24 h. Lipid extracts were dissolved in 20 μl CHCl3 and 1.5-μl aliquots were used for MALDI-TOF-MS analysis.
MALDI-TOF-MS
For lipid analysis, a 0.5 M DHB/methanol solution containing 0.1% trifluoroacetic acid was used as matrix. Lipids dissolved in CHCl3 were directly applied to a gold target plate (Applied Biosystems, Foster City, CA, USA) as 1.5 μl droplets, followed by addition of 1 μl matrix solution. Subsequently, samples were allowed to crystallize at RT or in a warm gas stream. All MALDI-TOF mass spectra were acquired on a Voyager-DE STR BioSpectrometry workstation (Applied Biosystems). The system utilizes a pulsed nitrogen laser emitting at 337 nm, which was operated in positive ion mode. The extraction voltage was 20 kV and the “low mass gate” was turned on to prevent saturation of the detector by ions resulting from the matrix. For each mass spectrum, three single spectra recorded with more than 100 single laser shots were averaged. To enhance the spectral resolution, all spectra were measured in the reflector mode.
Fourier-transform ion cyclotron resonance mass spectrometry
Analysis was performed on an Accela U-HPLC coupled to an LTQ-FT Ultra hybrid mass spectrometer (Thermo Scientific). Lipid extracts were diluted (1:100) in acetonitrile:2-propanol (5:2, v/v) containing 1% ammonium acetate (w/v), 0.1% formic acid (v/v), and 1 pmol/μl d-31 34:1 phosphatidylcholine as internal standard. Lipid samples were separated on a Thermo Hypersil GOLD C18 column (100×1 mm, 1.9 μm particle size). Solvent A was water with 1% ammonium acetate and 0.1% formic acid, and solvent B was acetonitrile:2-propanol (5:2, v/v) with 1% ammonium acetate and 0.1% formic acid. The gradient ran from 35 to 70% B in 4 min and then to 100% B in another 16 min with a hold for 10 min. The flow rate was 250 μl/min. Data acquisition was done by FT-MS full scan at a resolution of 100 k and <2 ppm mass accuracy with external calibration. The spray voltage was set to 5000 V, capillary voltage to 35 V, and the tube lens was at 120 V. Capillary temperature was 250 °C. Peak areas were calculated by QuanBrowser for all lipid species, identified previously by exact mass (<2 ppm) and retention time. Lipids were quantified by correlation with known amounts of internal standard.
Kinetic studies of SM modification with HOCl
Liposomes prepared from chicken egg yolk palmitoyl SM (2 mg/ml stock solution) in PBS (50 mM, 140 mM NaCl, pH 6.5) diluted to the indicated concentrations were incubated with the indicated concentrations of reagent HOCl at 37 °C for 10 s (final volume of 250 μl). The NaOCl solution was diluted 1:1 with PBS (50 mM, 140 mM NaCl, pH 5) before addition to prevent a shift in the pH value. Reactions were stopped by addition of CHCl3:MeOH (2:1, v/v) to extract lipids. The samples were vortexed rigorously for 1 min and then dipalmitoylphosphatidylcholine (3 μg/ml, final concentration) was added as an internal standard before further vortexing for 1 min. The organic phase was removed and dried under a stream of nitrogen. Samples were redissolved in MeOH and analyzed by HPLC electro-spray ionization (ESI)-MS (Aquity UPLC, Synapt HDMS Q-TOF; Waters, Manchester, UK), using a Waters C18 column (2.1 × 50 mm, 1.7 μm) and a gradient from H2O:MeOH (1:1, v/v) to isopropanol. Extracted ion chromatograms of m/z 703.58 (SM) and m/z 734.57 (PC) were integrated using the MassLynx Software (Waters). Reaction rates and second-order rate constants (k2) were calculated by following the time-dependent decrease of the m/z 703.58 peak.
Cell experiments
PC12 cells were incubated in serum-free RPMI 1640 medium supplemented with 0.4% (v/v) Hepes buffer, 0.4% (w/v) sodium pyruvate, 0.4% (w/v) l-glutamine, and 50 μg/ml gentamycin in the absence or presence of SM, HOCl-SM, or reagent HOCl. SM(d18:1/16:0) (stock solution 2.85 mM), HOCl-SM (used within 2 days of modification), and reagent HOCl (aqueous stock solution, 832 mM, stored at 4 °C) were applied to PC12 cells at the indicated concentrations for the indicated time periods. HOCl-SM concentrations were calculated using the molecular weight of the SM(d18:1/16:0)-derived chlorohydrin, which was identified as the dominant HOCl-modified lipid species.
Measurement of ROS
ROS formation was examined using fluorescent DCF (940 nmol N2-dried H2DCFDA was dissolved in 250 μl dimethyl sulfoxide before application). Rather than being a radical-species-selective probe, H2DCFDA (after hydrolysis to dichlorofluorescin and internalization) is converted to fluorescent dichlorofluorescein by several reactive radical species and allows assessment of general oxidative stress [32]. PC12 cells were plated in 12-well plates and grown to 80% confluence. Cells were washed two times with PBS and then incubated in the absence or presence of SM or HOCl-SM at 37 °C at the indicated concentrations for the indicated time periods. Incubations were stopped by washing the cells two times with PBS and, subsequently, H2DCFDA dissolved in HBSS (12.5 μM) was added and the cells were incubated for a further 1 h at 37 °C in the dark. Then, the plates were kept on ice for 10 min and then washed two times with ice-cold PBS, and cell lysis was performed with 100 μl lysis solution (3% Triton X-100 in PBS) on a rotary shaker (1350 rpm) at 4 °C in the dark for 60 min. Afterward, 30 μl absolute ethanol was added to each well and shaking was continued for another 15 min to ensure complete solubilization of deacetylated and oxidized DCF. The cell lysates were transferred to Eppendorf tubes and centrifuged to remove cellular debris (13,000 rpm, 4 °C, 10 min). One hundred microliters of the supernatant was transferred to black 96-well microtiter plates and fluorescence intensity was measured at 484/540 nm (excitation/emission) on a Victor 1420 multilabel counter (Wallac). An aliquot of the supernatant was used for estimation of protein concentration using the BCA assay.
Cell viability (MTT assay)
PC12 cells were plated in 96-well plates (20,000 cells per well; the cells were counted manually using a hemacytometer after trypsinization) and allowed to grow for 24 h before the cells were treated with SM, HOCl-SM, or reagent HOCl in serum-deprived medium at the indicated concentrations and for the indicated time periods. MTT (1.2 mM, dissolved in serum-free medium, 100 μl per well) was added to cells and incubated for 3 h at 37 °C under standard conditions. Cells were washed with PBS, and cell lysis was performed with 100 μl lysis solution (isopropanol:1 M HCl, 25:1 (v/v)) on a rotary shaker (1200 rpm, 15 min [33]). Absorbance was measured at 570 nm on a Victor 1420 multilabel counter and corrected for background absorption (650 nm).
JC-1 assay
To examine the mitochondrial membrane potential (Δψm) of PC12 cells, the JC-1 assay was performed. JC-1 is a cationic, lipophilic, fluorescent dye that can easily enter the electronegative matrix of intact mitochondria. As a monomer, JC-1 exhibits green fluorescence. Concentration-dependent accumulation in mitochondria leads to formation of J-aggregates resulting in a shift from green to red fluorescence. Hence, mitochondrial depolymerization that occurs in the early apoptotic cascade can be observed via a decrease in red and increase in green fluorescence. PC12 cells were plated in black 96-well plates (20,000 cells per well) and allowed to grow for 24 h before addition of SM, HOCl-SM, or reagent HOCl in serum-deprived medium at the indicated concentrations and for the indicated time periods. Cells were then incubated with JC-1 (2.5 μM, dissolved in serum-free medium, 100 μl per well) at 37 °C in the dark for 30 min followed by two washing steps with PBS. Fifty microliters of PBS were then added to each well and the fluorescence intensities (FIs) read at 484/540 nm (ex/em) for detection of the green substrate and 544/590 nm (ex/em) for detection of the red substrate on a Victor 1420 multilabel counter. The results were analyzed in terms of the ratio of red to green FIs. Therefore, results are not linked to the number of viable cells.
Western blot analysis of caspase-3 activation and PARP cleavage
PC12 cells (80% confluency, on 9-cm2 culture plates) were incubated in the absence or presence of SM, HOCl-SM, reagent HOCl, or colchicine at the indicated concentrations and for the indicated time periods. For protein isolation, the cells were washed with ice-cold PBS and then scraped in 70 μl lysis buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM Na3VO4, 1 mM NaF, 1 mM EDTA, 10 μM phenylmethylsulfonyl fluoride, and 1 μg/ml each aprotinin, leupeptin, and pepstatin). After sonication (2 × 2 min on ice) the cell debris was removed by centrifugation (13,000 rpm, 4 °C, 10 min) and the protein content was determined using the BCA assay. Detergent protein extracts were diluted in sample buffer and boiled for 5 min at 95 °C before SDS–PAGE. Proteins were separated by linear PAGE (150 V, reducing conditions) and electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (150 mA). To block nonspecific adsorption, membranes were incubated with 5% (w/v) nonfat milk powder in Tris-buffered saline Tween 20 (TBS-T). Polyclonal rabbit anti-caspase-3 antibody and polyclonal rabbit anti-PARP antibody (diluted 1:200 and 1:500, respectively, both in 5% (w/v) nonfat milk powder in TBS-T, incubation overnight at 4 °C) were used as primary antibodies. Immunoreactive bands were visualized using HRP-conjugated goat anti-rabbit IgG (dilution 1:5000 in 5% (w/v) nonfat milk powder in TBS-T, 2 h incubation) and subsequent SuperSignal development.
Sub-G1 population analysis
Cells on 9-cm2 dishes (80% confluency) were incubated in the absence or presence of SM, HOCl-SM, reagent HOCl, or colchicine at the indicated concentrations and for the indicated times. At the indicated time points, both control and treated cells were harvested (0.25% trypsin–EDTA), resuspended in 500 μl PBS (pH 7.4), fixed with 5 ml ice-cold MeOH, and stored at 4 °C. PI staining was done using DNA-Prep stain (PI at 50 μg/ml) and RNase (4 KU/ml; type III-A, bovine pancrease) [34]). Finally, the cells were analyzed on a Coulter EPICS XL-MCL using System II/3.0 data-analyzing software (Beckman Coulter).
Sample preparation for 2D-DIGE
Confluent PC12 cells on 75-cm2 flasks were incubated for 24 h in the absence or presence of SM or HOCl-SM (47.5 μM). Cells were washed with sucrose (250 mM in 10 mM Tris–HCl, pH 7.4), then scraped in 100 μl lysis buffer (7 M urea, 2 M thiourea, 4% Chaps, 30 mM Tris), and processed for 2D-DIGE. Cell lysates were sonicated (4 × 10 s on ice) and centrifuged (13,000 rpm, 15 °C, 10 min). Lysates from control, SM-treated, and HOCl-SM-treated cells were labeled using Cy dyes according to the manufacturer’s recommendations. Protein lysates of control cells were labeled with Cy2, whereas protein lysates from SM- and HOCl-SM-treated cells were labeled with Cy3 and Cy5, respectively. The protein content was determined using the Bradford assay. Protein (50 μg) from cell lysates was labeled with 400 pmol amine-reactive Cy2, Cy3, or Cy5 N-hydroxysuccinamide ester DIGE dye (Amersham Biosciences), freshly dissolved in anhydrous dimethyl formamide. The labeling mixture was incubated on ice in the dark for 30 min. The reaction was quenched by addition of lysine (10 nmol) followed by incubation on ice for another 10 min. Each respective labeled protein sample was mixed with an equal volume of 2 × sample buffer containing 7 M urea, 2 M thiourea, 4% (w/v) Chaps, 2% (w/v) dithiothreitol (DTT), and 2% (v/v) Pharmalyte (pH 3–10) and subsequently left on ice for 10 min. Afterward, 600 μg of the corresponding unlabeled protein lysate was added to respective labeled protein sample, and the three samples were mixed before isoelectric focusing (IEF) on Immobiline IPG strips.
2D-DIGE and image analysis
The IPG strips (24 cm, linear pH range of 3–10) were rehydrated overnight in ceramic strip holders in 600 μl of reswelling solution containing 1.95 mg of protein lysate from control, SM-treated, and HOCl-SM-treated cells (see above); 7 M urea; 2 M thiourea; 1% (w/v) Chaps; 0.4% (w/v) DTT; 0.5% (v/v) Pharmalyte (pH 3–10); and 0.002% (v/v) bromophenol blue. To prevent adverse IEF of proteins, at least 300 μl of reswelling solution was mixed with the merged protein lysates. IEF was carried out at 20 °C with a maximum current set at 50 μA/strip using an Ettan IPGphor unit (Amersham Biosciences). After IEF (3 h at 150 V, 3 h at 300 V, 3 h at 600 V, 3 h gradient to 8000 V, remaining time at 8000 V to reach a total of 50,000 Vh) the strips were equilibrated in SDS equilibration buffer (6 M urea, 2% (w/v) SDS, 30% (v/v) glycerol, 50 mM (v/v) Tris–HCl, pH 8.8) supplemented either with 1% (w/v) DTT (first equilibration step, 15 min) or with 4.5% (w/v) iodoacetamide (second equilibration step, 15 min), each with gentle shaking. IPG strips were directly applied to self-cast SDS gels (12%). SDS–PAGE was carried out at 18 °C (Ettan Dalt, Amersham Biosciences; 15 mA/gel). Fluorescence imaging was performed on a Typhoon 9400 scanner (Amersham Biosciences; excitation 488/532/633 nm, emission 520/580/670 nm for Cy2, Cy3, and Cy5, respectively). Statistical analysis and quantification of protein spots were carried out using the DeCyder-DIA software (Amersham Biosciences). Differentially expressed protein spots were manually picked and ingel tryptically digested. Fold regulation is expressed as the mean of three gels run from three independent experiments.
Liquid chromatography (LC)-MS/MS analysis of tryptic digests
Peptide extracts were dissolved in 0.1% formic acid and separated on a nano-HPLC system. Seventy microliters was injected and concentrated on the loading column (LC Packings C18 PepMap, 5 μm, 0.3 × 5 mm, 100 Å; LC Packings, Amsterdam, The Netherlands) for 5 min using 0.1% formic acid as the isocratic solvent at a flow rate of 20 μl/min. The column was then switched into the nanoflow circuit, and the sample was loaded on the nanocolumn (LC-Packings C18 PepMap, 0.075 × 150 mm) at a flow rate of 300 nl/min and separated using a gradient from 0.3% formic acid/5% acetonitrile to 0.3% formic acid/50% acetonitrile over 60 min. The sample was ionized in a Finnigan Nano-ESI ion source (Finnigan MAT, San Jose, CA, USA) equipped with NanoSpray tips (PicoTip emitter; New Objective, Woburn, MA, USA) and analyzed in a Thermo Finnigan LCQ Deca XPplus ion trap mass spectrometer. The MS/MS data were analyzed by searching the National Center for Biotechnology Information (NCBI) public database with SpectrumMill version 2.7 (Agilent, Waldbronn, Germany). Acceptance parameters were at least three or more identified distinct peptides.
Clustering of identified proteins
Relative expression patterns (log2(control/SM) and log2(control/HOCl-SM)) for spots within one gel representing the same protein (RefSeq protein identifier) were filtered for outliers (>upper quartile + 1.5 × IQR (interquartile range) or <lower quartile – 1.5 × IQR). The median from the remaining values was averaged over two or three gels resulting in one expression value for each identified protein. Data were combined, grouped, and visualized as heat maps using Genesis [35].
Ingenuity Pathway Analysis (IPA)
Differentially expressed proteins were analyzed using a free trial version of the IPA software to monitor possible global proteomic regulation in response to SM and HOCl-SM. IPA (www.ingenuity.com; https://analysis.ingenuity.com) uses a knowledge base derived from the literature to relate gene products based on their interaction and function. This software is designed to identify biological networks, global canonical pathways, and global functions. The identified proteins and their corresponding fold change and accession numbers were uploaded as .txt files into the Ingenuity software package. Ingenuity uses these data to navigate the Ingenuity Pathways knowledge base and extract overlapping networks between the candidate proteins. A score of better than 2 is usually attributed to a valid network (the score represents the log probability that this network was found by random chance).
Statistical analysis
Data are presented as means ± SD. ANOVA with Bonferroni correction (using the GraphPad Prism package) was used for analysis of statistical significance. All values of p<0.05 were considered significant. For proteome analyses only spots that were regulated by greater than or equal to twofold (up or down; DeCyder software; GE Healthcare) were picked and further processed for identification.
Results
Modification of SM by reagent and MPO-generated HOCl
To establish the accessibility of SM to HOCl modification in a physiologically relevant lipid matrix, brains of C57BL/6 mice were homogenized in liquid nitrogen, and the lipids were extracted and incubated in the presence of increasing HOCl:lipid ratios and then analyzed by FT-ICR-MS. These analyses revealed SM(d18:1/18:0) (N-(octadecanoyl)-sphing-4-enine-1-phosphocholine) and SM(d18:1/24:1) (N-(tetracosenoyl)-sphing-4-enine-1-phosphocholine) as the predominant molecular species (means 950 and 420 nmol/g wet tissue) in the brain sphingomyelin fraction (Fig. 1A). The corresponding N-hexadecanoyl, N-octadecenoyl, and N-eicosanoyl species were present in minor amounts contributing to approx 20 nmol/g wet brain. Modification of mouse total brain lipids with reagent HOCl resulted in depletion of these SM species in a HOCl-dependent manner. Of note, 1 μg NaOCl per 100 μg lipid resulted in the depletion of approx 40% of the original SM content, indicating that this lipid subspecies is highly sensitive to HOCl modification. A 50-fold excess of HOCl over brain lipids resulted in nearly complete disappearance of SM(d18:1/18:0) and (d18:1/24:1) (Fig. 1B).
Fig. 1. FT-ICR-MS analysis of native and HOCl-modified mouse brain lipids.
C57BL/6 mice were killed by cervical dislocation, brains were removed and homogenized in liquid nitrogen, and lipids were extracted using a modified Folch extraction. Extracts were dried under a stream of N2 and defined amounts were modified at the indicated NaOCl concentrations. After another extraction step the lipids were dissolved in CHCl3: MeOH (1:1, v/v) and analyzed by a hybrid linear ion trap FT-ICR-MS in positive ESI mode as described under Materials and methods. (A) Composition of native mouse brain sphingomyelin. (B) Total, SM(d18:1/18:0), and SM(d18:1/24:1) content in mouse brain lipid samples before and after HOCl modification at the indicated ratios. Results are mean values of duplicate determinations.
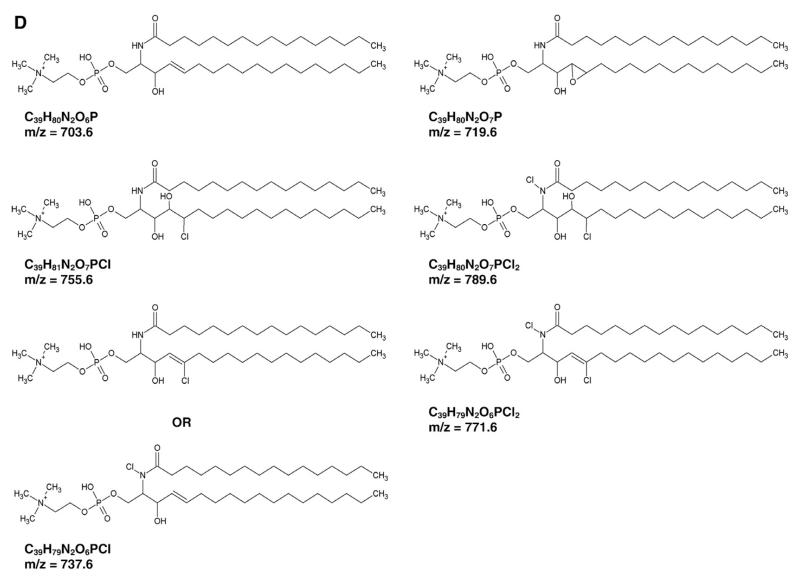
In the next set of experiments product formation in response to HOCl modification of SM(d18:1/16:0) was characterized by MALDITOF-MS. Untreated SM(d18:1/16:0) exhibits two peaks at 703.6 and 725.6 Da, corresponding to the H+ and Na+ adducts (Fig. 2A). In response to HOCl added as reagent (Fig. 2B) or generated by the MPO–H2O2–chloride system (Fig. 2C) two new peaks, shifted by 52 Da, appeared at 755.6 and 777.6 Da. These indicate the formation of the H+ and Na+ adducts of the corresponding monochlorohydrin of SM (d18:1/16:0). Two other products detected at 737.6 and 759.6 Da suggest formation of the H+ and Na+ adducts of a monochlorinated derivative of SM(d18:1/16:0) similar to that described for phosphatidylcholine modified by reagent HOCl [36]. As is evident from Fig. 2C a proportion of unmodified SM(d18:1/16:0) was still detectable when HOCl was generated enzymatically. Peaks detected at 719.6, 771.6, and 789.6 Da are indicative of the formation of an epoxide and mixed chlorohydrin/chloramide SM species (mass assignment is summarized in Supplementary Table I). The proposed structures, chemical formulas, and corresponding m/z values are displayed in Fig. 2D.
Fig. 2. MALDI-TOF analysis of SM and HOCl-modified SM.
SM(d18:1/16:0) was modified with HOCl (either added as reagent or generated enzymatically by the MPO–H2O2–chloride system) as described under Materials and methods. After modification, lipids were extracted and analyzed by MALDI-TOF-MS using DHB as matrix as described under Materials and methods. Representative spectra of (A) SM(d18:1/16:0), (B) HOCl-modified SM(d18:1/16:0), and (C) MPO-modified SM(d18:1/16:0) are shown. (D) Proposed structures and resulting masses of the chlorinated products.
Determination of rate constants for HOCl-mediated SM modification
To get an indication of the susceptibility of SM to HOCl modification, rate constants for the reaction were determined. Experimental conditions, reaction rates, and the resulting rate constants are given in Table 1. These experiments (performed at pH 6.5) revealed a second-order rate constant (k2) of 18.7 ± 3.05 L/mol/s (mean ± SD). This is in a range comparable to that reported for the reaction of plasmalogens with HOCl (55 ± 7 L/mol/s/; Ref. [37]), a lipid species readily modified by MPO in vivo [25].
Table 1. Second-order rate constants at pH 6.5 (37 °C) for the reaction of SM with NaOCl.
| NaOCl (mol/L) | SM0a (mol/L) | SMtb (mol/L) | SMmodc (mol/L) | Rated (mol/L/s) | k2e (L/mol/s) |
|---|---|---|---|---|---|
| 4.00 × 10−3 | 1.20 × 10−3 | 4.04 × 10−4 | 7.96 × 10−4 | 7.96 × 10−5 | 16.58 |
| 4.00 × 10−3 | 2.00 × 10−3 | 1.14 × 10−4 | 1.86 × 10−3 | 1.86 × 10−4 | 23.20 |
| 4.00 × 10−3 | 2.84 × 10−3 | 1.05 × 10−3 | 1.79 × 10−3 | 1.79 × 10−4 | 15.78 |
| 2.84 × 10−3 | 2.84 × 10−3 | 1.21 × 10−3 | 1.64 × 10−3 | 1.64 × 10−4 | 20.23 |
| 5.69 × 10−3 | 2.84 × 10−3 | 2.99 × 10−5 | 2.82 × 10−3 | 2.82 × 10−4 | 17.46 |
| 18.7±3.05 (mean±SD) |
Samples were incubated for 10 s, extracted in CHCl3:MeOH (2:1, v/v), dried under nitrogen, redissolved in methanol, and analyzed by LC-ESI-MS as described under Material and methods. Dipalmitoylphosphatidylcholine was used as internal standard. All experiments were performed in triplicate.
SM0, initial [SM].
SMt, [SM] after 10 s incubation in the presence of NaOCl.
[SMmod]=[SM0]–[SMt].
Rate=[SMmod]/10.
k2=rate/([SM0] [NaOCl]).
HOCl-SM induces the formation of ROS
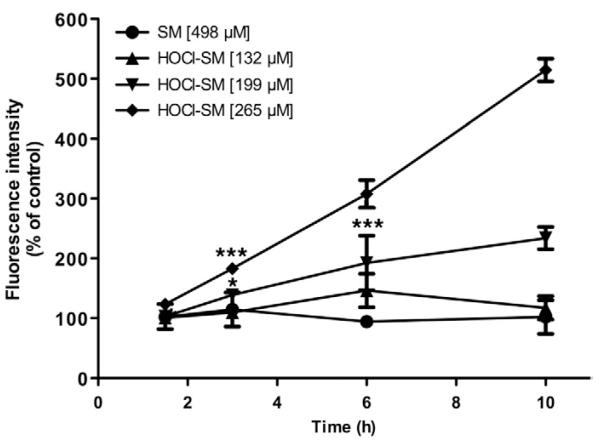
Intracellular radical formation was followed using DCFDA as probe. Cells were incubated in the presence of increasing concentrations of SM and of HOCl-SM in a time-dependent manner (Fig. 3). These experiments revealed that HOCl-SM at concentrations >200 μM resulted in strongly enhanced fluorescence (two- and fivefold over baseline at 10 h) indicative of intracellular formation of reactive oxygen species. Native SM was without effect on fluorescence intensities.
Fig. 3. ROS formation in SM- and HOCl-SM-treated PC12 cells.
Generation of ROS in PC12 cells was examined in assays using the fluorescent tracer DCFDA. Cells were incubated in the absence or presence of SM or HOCl-SM at the indicated concentrations for the indicated times. Results are expressed as fluorescence intensity as % of control and represent the means ± SD (n = 3). *p<0.05; ***p<0.001. For clarity p values are shown only for the first data point reaching significance.
HOCl-SM impairs mitochondrial function
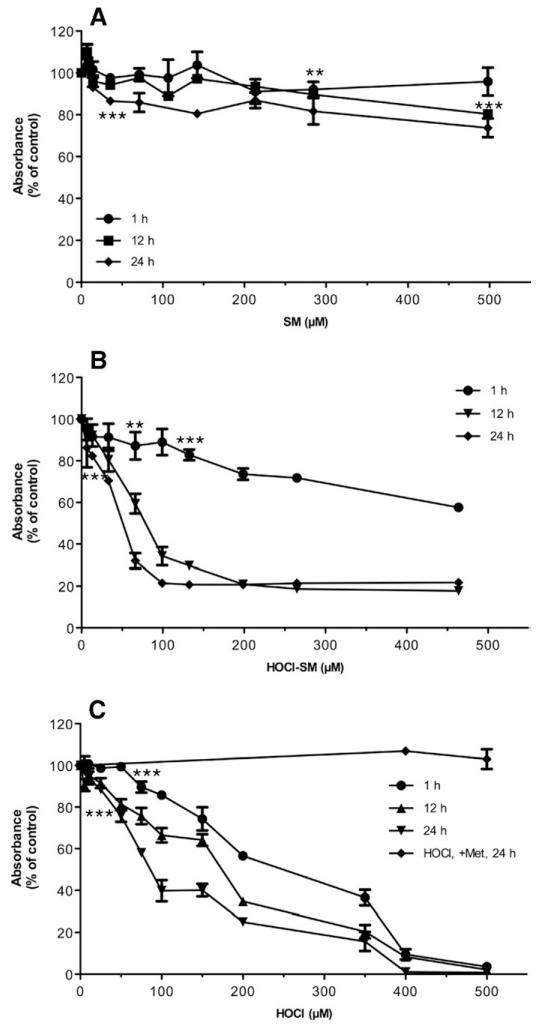
Next, the effects of SM, HOCl-SM, and reagent HOCl on mitochondrial function of PC12 cells were established. These results demonstrate that SM induced a slight reduction in mitochondrial activity (approx 20–30% at the highest concentrations used, 498 μM; Fig. 4A). In contrast, incubation of PC12 cells with HOCl-SM resulted in rapid and extensive loss of mitochondrial function/cell viability, with IC50 values of approx 80 (12 h) and 50 (24 h) μM (Fig. 4B). Also reagent HOCl significantly impaired mitochondrial function with IC50 values of 170 and 90 μM (at 12 and 24 h, respectively; Fig. 4C). The addition of Met (a HOCl scavenger) completely blocked HOCl-mediated cytotoxicity. These data indicate the high lipotoxic potential of HOCl-SM, with IC50 concentrations being almost twofold lower as calculated for the strong oxidant HOCl.
Fig. 4. Viability of SM-, HOCl-SM-, and HOCl-treated PC12 cells.
Viability of PC12 cells was analyzed using the MTT assay. Cells were incubated in the absence or presence of (A) SM, (B) HOCl-SM, or (C) HOCl at the indicated concentrations for the indicated times. Results are expressed as % of control and represent the means ± SD (n = 3). **p<0.005; ***p<0.001. For clarity p values are shown only for the first data point reaching significance.
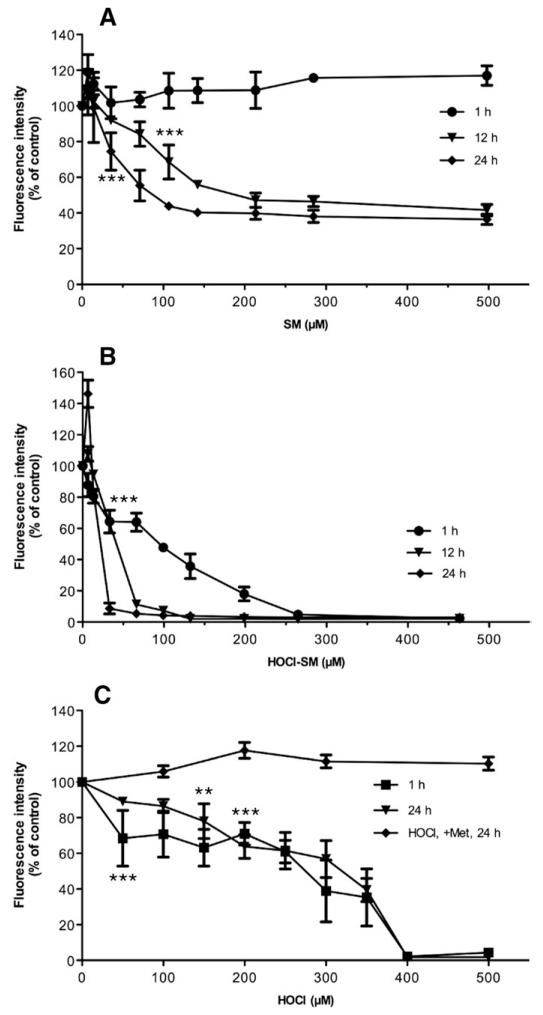
To get an indication of the alterations in Δψm, cells were incubated time dependently in the presence of increasing concentrations of SM, HOCl-SM, and HOCl (Fig. 5). All three of the compounds led to significantly reduced Δψm, but to different degrees: SM had no effect on Δψm after 1 h. At 12 and 24 h the IC50 values were 140 and 70 μM (Fig. 5A), whereas the corresponding values for HOCl-SM were substantially lower (50 and 25 μM at 12 and 24 h; Fig. 5B). Also reagent HOCl induced a strong decrease in Δψm though this was a more linear effect with IC50 values of approx 300 μM. The presence of Met during HOCl treatment completely abolished mitochondrial dysfunction up to concentrations of 500 μM HOCl (Fig. 5C).
Fig. 5. Mitochondrial membrane potential (Δψm) of SM-, HOCl-SM-, and HOCl-treated PC12 cells.
Δψm was analyzed using the JC-1 assay. Cells were incubated in the presence of (A) SM, (B) HOCl-SM, or (C) HOCl at the indicated concentrations for the indicated times. Results were calculated by division of red fluorescence by green fluorescence and are expressed as fluorescence intensity as a % of control and represent the means ± SD (n = 3). **p<0.005; ***p<0.001. For clarity p values are shown only for the first data point reaching significance.
HOCl-SM induces apoptosis
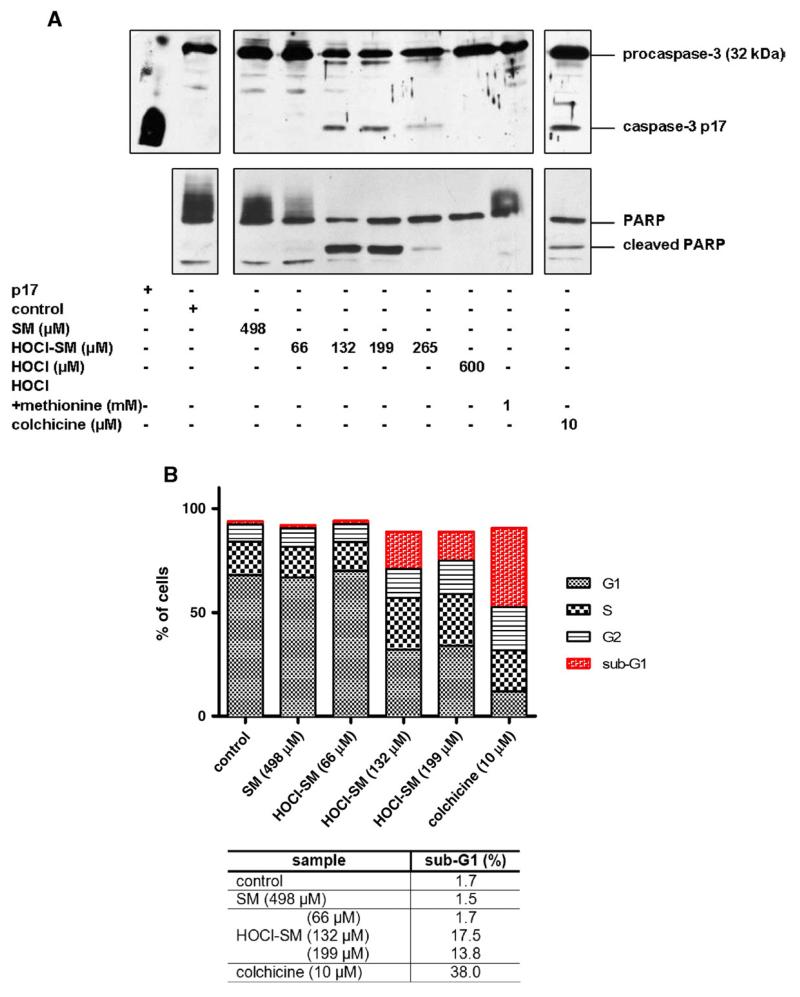
To confirm that exogenous HOCl-SM induces apoptosis, cleavage of procaspase-3 and PARP was analyzed by immunoblotting. Exposure of PC12 cells to HOCl-SM induced procaspase-3 processing and PARP cleavage in a concentration-dependent manner. Caspase-3 and subsequent PARP cleavage was observed at HOCl-SM concentrations >66 μM, whereas neither SM nor HOCl alone resulted in detectable caspase and PARP processing (Fig. 6A).
Fig. 6. Induction of the apoptotic machinery of PC12 cells in response to SM, HOCl-SM, and HOCl.
Cells were incubated in the absence or presence of SM, HOCl-SM, or HOCl at the indicated concentrations for 13 h. Cell treatment with colchicine was performed for 24 h. (A) Protein lysates (40 μg/lane) were run on either 12% (caspase-3 activation) or 8% (PARP cleavage) SDS gels and transferred onto PVDF membranes for subsequent detection using anti-caspase-3 and anti-PARP primary antibody. (B) Cells after treatment with SM, HOCl-SM, or HOCl were processed for PI stain-coupled FACS analysis. Apoptotic cells appear below G1 phase (sub-G1 peak). Results in (B) are expressed as % of cells and represent the mean values from duplicate experiments.
In addition to Western blotting, PI staining coupled to FACS analysis of SM- and HOCl-SM-incubated cells was performed (Fig. 6B). SM at 498 μM was without effect on sub-G1 populations. In contrast, HOCl-SM concentrations of 132 and 199 μM induced formation of sub-G1 peaks (17.5 and 13.8% apoptotic cells, respectively), whereas at higher concentrations the numbers of viable and apoptotic cells were below the detection limit, similar to cells treated with reagent HOCl alone. The effect of reagent HOCl was completely abolished when cells were preincubated with Met (molar ratio Met:HOCl of 5:1; not shown). Colchicine was used as a pharmacological inducer of apoptosis (38% of cells in sub-G1; Fig. 6B).
Proteome analyses of SM- and HOCl-SM-treated cells
Next, the effects of exogenously added SM and HOCl-SM on protein expression in PC12 cells were established by a 2D-DIGE/ESILC-MS approach. Representative gel images are shown in Supplementary Fig. I. Differentially expressed spots (greater than twofold up or down) were picked, digested, and subsequently analyzed by nanoLC-ESI-MS. Supplementary Tables II (proteins regulated in response to SM) and III (proteins regulated in response to HOCl-SM) provide information on spot number, number of acquired spectra, distinct identified peptides, MS/MS search score, percentage amino acid coverage, theoretical and experimental MW, theoretical and experimental pI, and fold regulation. Protein identifications are based on multiple peptide sequences with a minimum number of three peptides. Changes in protein expression patterns in response to SM or HOCl-SM are summarized in Fig. 7 and presented as heat maps (values are expressed as log2 of fold regulation). Exogenously added SM induced upregulation of 21 and downregulation of 42 proteins. In response to HOCl-SM, 9 proteins were up- and 22 were down-regulated. From these, a total of 12 proteins were coregulated by SM and HOCl-SM (2 up and 10 down).
Fig. 7. Changes in protein expression in response to SM and HOCl-SM.
Lysates of PC12 cells incubated in the absence or presence of SM or HOCl-SM were labeled with Cy2 (control), Cy3 (SM), or Cy5 (HOCl-SM). After addition of the corresponding unlabeled protein extracts the three samples were pooled and applied to isoelectric focusing on IPG strips. Subsequently, second-dimension SDS–PAGE was carried out on 12% SDS gels at 18 °C. After laser scanning of the gels, twofold differentially expressed spots were identified using DeCyder software, manually picked, digested, and analyzed by LC-MS/MS. MS data were analyzed by searching the NCBI database using SpectrumMill version 2.7, and log2 expression patterns of proteins (control vs SM and control vs HOCl-SM) identified in two or three gels were calculated and averaged. Data were combined and are visualized as heat maps.
Pathway analysis
To examine potential relationships between proteins with altered expression levels, IPA was applied. IPA builds hypothetical networks from the identified proteins and proteins from the database that are needed to complete a meaningful biological network. Input proteins that map to the networks are called focus proteins. Network generation ranked by score is optimized for inclusion of as many proteins from the input expression profile as possible and aims for highly interrelated networks, which are likely to represent significant biological functions. Supplementary Fig. II shows the pathways generated during IPA of proteins regulated in response to SM or HOCl-SM (identified proteins from Supplementary Tables II and III and their fold regulation were used as input data).
Pathway identification in response to SM
The Energy Production, Molecular Transport, Nucleic Acid Metabolism pathway was identified by IPA with a score of 44 (indicating that there is a chance of 1:1044 of another randomly occurring network containing the same number of focus genes; Supplementary Fig. IIA), followed by Small Molecule Biochemistry, Molecular Transport, Free Radical Scavenging with a score of 26 (Supplementary Fig. IIB). In the first network SLC2A4 is a major hub and we have identified eight focus genes (ECSH1, MDH2, CNN3, EIF4A1, PRDX2, ENO1, and ALDOC) in the immediate neighborhood of SLC2A4, which is identical to insulin-sensitive GLUT4. Within the latter network, Small Molecule Biochemistry, Molecular Transport, Free Radical Scavenging, several upregulated proteins are products of genes that are under the control of Nrf2. Among these are PRDX1 and PRDX2, SOD, and GSTO1 (Supplementary Table II and Supplementary Fig. IIB) conferring cellular protection against oxidative damage. This might be a reasonable explanation for the absence of intracellular ROS formation in SM-treated PC12 cells.
Pathway identification in response to HOCl-SM
For HOCl-SM the highest score was obtained for Carbohydrate Metabolism, Energy Production, Molecular Transport (score 48; Supplementary Fig. IIC), followed by Cell-Mediated Immune Response, Cellular Growth and Proliferation, Hematological System Development and Function (score 15; not shown). Also in this network SLC2A4 (insulin-sensitive GLUT4) is a central hub, though not identified during our proteomic analyses. However, of the total of 34 interaction partners in this network, 20 focus genes were identified in our proteome approach (Supplementary Table III and Fig. IIC). Some of them are direct interactors of GLUT4 (ECHS1, HADH, MDH, VDAC1 and VDAC2, OAT, AAT, and ALDOA). Of the regulated proteins within this network five (HADH, CACYBP, HNRNPC, AK2, and NME1) were upregulated, whereas all the other enzymes/transporters related to glycolysis, energy production, and molecular transport were downregulated, clearly indicating severe interference of HOCl-SM with cellular energy homeostasis.
Discussion
The results of this study revealed that modification of SM(d18:1/16:0) by HOCl (added as reagent or produced by the MPO–H2O2–chloride system) generated chlorinated compounds with high neurotoxic potential. Exogenously added HOCl-SM, in contrast to SM, severely impaired cell viability, resulted in dissipation of Δψm, and induced the apoptotic machinery. On the proteome level addition of SM and HOCl-SM exerted comparable downregulation of glycolytic enzymes acting downstream of fructose 1,6-biphosphate. In contrast to HOCl-SM, SM induced several proteins involved in the Nrf2-mediated oxidative stress response.
Using MALDI-TOF-MS the major modification products were identified as the corresponding chlorohydrin and unsaturated, monochlorinated SM species. This is reminiscent of what was reported for HOCl modification of unsaturated phospholipids: During a series of isotopic labeling experiments Spalteholz and colleagues [36] demonstrated that an increase of 34 Da corresponds to a (formal) exchange of a hydrogen for a chlorine atom (and not the formation of a glycol moiety [38]) leading to an unsaturated, monochlorinated phospholipid species by a hitherto unknown reaction mechanism. Although not experimentally verified during the present study, we suggest that a comparable pathway could lead to the formation of unsaturated, monochlorinated SM species. Analysis of the SM species present in mouse brain lipid extracts revealed the presence of C18:1 with C16:0, C18:0, C18:1, C20:0, and C24:1 acyl chains; of these, C18:0 and C24:1 are the most abundant molecular species, similar to what was reported for SM composition of cortical neurons [39]. Of note, SM species in total mouse brain lipid extracts were also susceptible to HOCl-mediated oxidation (Fig. 1).
Both (reagent) HOCl and HOCl-modified lipids are potent inducers of apoptosis/cell death (reviewed in [3]). Here we provide evidence for time- and concentration-dependent apoptosis elicited by HOCl-SM. Collapse of Δψm is indicative of mitochondrial permeability transition pore formation (MTP), which is considered a major contributor to disease progression in many neurological disorders (for reviews see [40,41]). MTP leads to release of cytochrome c, formation of the apoptosome, and induction of the caspase cascade. MTP was shown to occur in response to HOCl in hepatocytes and isolated mitochondria, resulting in consumption of intracellular GSH and subsequent production of ROS, chromatin condensation, and procaspase-3 processing [42]. This series of events is reminiscent of what was observed during the present study in response to HOCl-SM, namely formation of ROS, dissipation of Δψm, activation of caspase-3 and PARP cleavage, nuclear fragmentation, and nuclear pore formation. Some of these proapoptotic events were also reported for HOCl, fatty acid- and phospholipid-derived chlorohydrins [43,44], and oxidized phospholipids [45], which are abundantly produced in brains of mice with astrocyte-specific overexpression of human MPO [12]. Interestingly, unmodified SM also induced a decrease in Δψm (however, to a lower extent than HOCl-SM) but was without effect on ROS generation, cell viability, activation of caspase-3, and PARP cleavage. Whether this is due to intracellular ceramide generation (via the activation of sphingomyelinases [45,46]) and subsequent induction of mitochondrial dysfunction (as described, e.g., in TNFα-treated breast cancer cells [47]) is currently not clear.
Neuronal activity requires energy that is provided almost exclusively by oxidation of glucose. It is becoming increasingly clear that astrocytes play a critical role in the brain metabolic response to neural activity via metabolic coupling by a pathway termed the neuron–astrocyte lactate shuttle [48]. After neuronal activation astrocytes increase their glycolytic rates and generate lactate, which is then transported to neurons where it is converted back to pyruvate (by means of LDH) and enters oxidative phosphorylation [49]. Downregulation of glycolytic enzymes by SM and HOCl-SM as observed during this in vitro study would impair neuronal oxidative glycolysis during the initial phase and probably also nonoxidative glycolysis in astrocytes. Downregulation of LDH as observed here would also decrease lactate oxidation and ATP production, a prerequisite for glutamate recycling. These would be classical signs of energy deprivation as observed in many neurological diseases. Along this line it is important to note that application of a small molecule, chlorohydrin (3-Cl-propanediol, also termed S-α-chlorohydrin), in rats induces dysfunction of the blood–brain barrier and causes astrocyte death and morphological transition of neurons, further emphasizing the neurotoxic properties of chlorohydrins [50].
The mechanism(s) by which SM and HOCl-SM alter protein expression levels is a major question arising from our study. One potential explanation is ceramide generation via sphingomyelinase-mediated hydrolysis of SM and HOCl-SM. In LPS-treated rats it was demonstrated that the inflammatory response results in impaired glycolysis and a net reduction of glycogen biosynthesis, effects abolished by pharmacological inhibition of ceramidase [51]. Based on these findings the authors [51] proposed that the SM–ceramide pathway might be responsible for inhibited cardiac glycolysis in LPS-treated rats. In vitro studies have demonstrated that ceramide inhibits activation of Akt, antagonizes insulin-dependent glucose transport [52], and suppresses phosphorylation of insulin receptor substrate-1 [53]. In neurons inhibition of hexokinase activity (via inhibition of Akt) was reported in response to C2-ceramide [54]. Finally, analysis of alterations in gene expression accompanying C2-ceramide-induced apoptosis in PC12 cells revealed significantly decreased (3-fold) expression of aldolase C expression at the mRNA level [55], which is consistent with our proteome findings (downregulation of aldolase C by SM and HOCl-SM between 2.2- and 3.5-fold).
However, also with regard to protein expression, some distinct differences between SM and HOCl-SM became apparent. Whereas SM induced the expression of peroxiredoxins 1 and 2 (PRDX1 and 2), mitochondrial superoxide dismutase 2, proteasome subunit α type-1, and glutathione S-transferase ω-1 (which are all involved in Nrf2-mediated oxidative stress response), this type of response was not found with HOCl-SM. This might be a plausible explanation for the absence of intracellular ROS generation in SM-treated PC12 cells. A recent report identified erythrocyte peroxiredoxin 2 as a target for HOCl-dependent oxidation [56].
In summary we propose that SM and HOCl-SM act via two distinct pathways in neuronal cells: first, HOCl-SM is clearly proapoptotic and neurotoxic under the experimental conditions applied during this study, and SM is not. This suggests that increased MPO activity might be a driving factor in the progression of neurological disorders; thus MPO could represent a novel therapeutic target in neurodegeneration [57]. Second, both SM and HOCl-SM are potent lipid modulators of protein expression in neuronal cells. Downregulation of glycolytic enzymes and lactate dehydrogenase would result in energy deprivation as observed in many neurodegenerative diseases. The possibility that the different cellular responses to SM and HOCl-SM could be ascribed to intracellular, sphingomyelinase-mediated formation of chlorinated ceramide species (that could display different signaling properties compared to natural ceramides) is currently being investigated.
Supplementary Material
Acknowledgments
The work was supported by the Molecular Medicine Ph.D. program of the Medical University of Graz, the Austrian Science Fund (F3007, F3009, P19074-B05), and the Austrian Research Promotion Agency (Bridge P820107). C.N. was funded by the PhD program Molecular Medicine of the Medical University of Graz, A.Ü. and S.W. are funded by the Austrian Science foundation within the PhD program Molecular Medicine of the Medical University of Graz. We are grateful for the opportunity to use a free trial version of the IPA program.
References
- [1].Halliwell B. Oxidative stress and neurodegeneration: where are we now? J. Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- [2].Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- [3].Yap YW, Whiteman M, Cheung NS. Chlorinative stress: an under appreciated mediator of neurodegeneration? Cell Signal. 2007;19:219–228. doi: 10.1016/j.cellsig.2006.06.013. [DOI] [PubMed] [Google Scholar]
- [4].Klebanoff SJ. Myeloperoxidase: friend and foe. J. Leukoc. Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- [5].Hazell LJ, Arnold L, Flowers D, Waeg G, Malle E, Stocker R. Presence of hypochlorite-modified proteins in human atherosclerotic lesions. J. Clin. Invest. 1996;97:1535–1544. doi: 10.1172/JCI118576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Malle E, Marsche G, Arnhold J, Davies MJ. Modification of low-density lipoprotein by myeloperoxidase-derived oxidants and reagent hypochlorous acid. Biochim. Biophys. Acta. 2006;1761:392–415. doi: 10.1016/j.bbalip.2006.03.024. [DOI] [PubMed] [Google Scholar]
- [7].Thukkani AK, Martinson BD, Albert CJ, Vogler GA, Ford DA. Neutrophil-mediated accumulation of 2-ClHDA during myocardial infarction: 2-ClHDA-mediated myocardial injury. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2955–2964. doi: 10.1152/ajpheart.00834.2004. [DOI] [PubMed] [Google Scholar]
- [8].Pattison DI, Davies MJ. Reactions of myeloperoxidase-derived oxidants with biological substrates: gaining chemical insight into human inflammatory diseases. Curr. Med. Chem. 2006;13:3271–3290. doi: 10.2174/092986706778773095. [DOI] [PubMed] [Google Scholar]
- [9].Choi DK, Pennathur S, Perier C, Tieu K, Teismann P, Wu DC, Jackson-Lewis V, Vila M, Vonsattel JP, Heinecke JW, Przedborski S. Ablation of the inflammatory enzyme myeloperoxidase mitigates features of Parkinson’s disease in mice. J. Neurosci. 2005;25:6594–6600. doi: 10.1523/JNEUROSCI.0970-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reynolds WF, Rhees J, Maciejewski D, Paladino T, Sieburg H, Maki RA, Masliah E. Myeloperoxidase polymorphism is associated with gender specific risk for Alzheimer’s disease. Exp. Neurol. 1999;155:31–41. doi: 10.1006/exnr.1998.6977. [DOI] [PubMed] [Google Scholar]
- [11].Green PS, Mendez AJ, Jacob JS, Crowley JR, Growdon W, Hyman BT, Heinecke JW. Neuronal expression of myeloperoxidase is increased in Alzheimer’s disease. J. Neurochem. 2004;90:724–733. doi: 10.1111/j.1471-4159.2004.02527.x. [DOI] [PubMed] [Google Scholar]
- [12].Maki RA, Tyurin VA, Lyon RC, Hamilton RL, Dekosky ST, Kagan VE, Reynolds WF. Aberrant expression of myeloperoxidase in astrocytes promotes phospholipid oxidation and memory deficits in a mouse model of Alzheimer disease. J. Biol. Chem. 2009;284:3158–3169. doi: 10.1074/jbc.M807731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nagra RM, Becher B, Tourtellotte WW, Antel JP, Gold D, Paladino T, Smith RA, Nelson JR, Reynolds WF. Immunohistochemical and genetic evidence of myeloperoxidase involvement in multiple sclerosis. J. Neuroimmunol. 1997;78:97–107. doi: 10.1016/s0165-5728(97)00089-1. [DOI] [PubMed] [Google Scholar]
- [14].Gray E, Thomas TL, Betmouni S, Scolding N, Love S. Elevated myeloperoxidase activity in white matter in multiple sclerosis. Neurosci. Lett. 2008;444:195–198. doi: 10.1016/j.neulet.2008.08.035. [DOI] [PubMed] [Google Scholar]
- [15].Chen JW, Breckwoldt MO, Aikawa E, Chiang G, Weissleder R. Myeloperoxidase-targeted imaging of active inflammatory lesions in murine experimental autoimmune encephalomyelitis. Brain. 2008;131:1123–1133. doi: 10.1093/brain/awn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gray E, Thomas TL, Betmouni S, Scolding N, Love S. Elevated activity and microglial expression of myeloperoxidase in demyelinated cerebral cortex in multiple sclerosis. Brain Pathol. 2008;18:86–95. doi: 10.1111/j.1750-3639.2007.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Marik C, Felts PA, Bauer J, Lassmann H, Smith KJ. Lesion genesis in a subset of patients with multiple sclerosis: a role for innate immunity? Brain. 2007;130:2800–2815. doi: 10.1093/brain/awm236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Heinecke JW, Li W, Mueller DM, Bohrer A, Turk J. Cholesterol chlorohydrin synthesis by the myeloperoxidase–hydrogen peroxide–chloride system: potential markers for lipoproteins oxidatively damaged by phagocytes. Biochemistry. 1994;33:10127–10136. doi: 10.1021/bi00199a041. [DOI] [PubMed] [Google Scholar]
- [19].Hazen SL, Hsu FF, Duffin K, Heinecke JW. Molecular chlorine generated by the myeloperoxidase–hydrogen peroxide–chloride system of phagocytes converts low density lipoprotein cholesterol into a family of chlorinated sterols. J. Biol. Chem. 1996;271:23080–23088. doi: 10.1074/jbc.271.38.23080. [DOI] [PubMed] [Google Scholar]
- [20].Spickett CM. Chlorinated lipids and fatty acids: an emerging role in pathology. Pharmacol. Ther. 2007;115:400–409. doi: 10.1016/j.pharmthera.2007.06.002. [DOI] [PubMed] [Google Scholar]
- [21].Kawai Y, Kiyokawa H, Kimura Y, Kato Y, Tsuchiya K, Terao J. Hypochlorous acid-derived modification of phospholipids: characterization of aminophospholipids as regulatory molecules for lipid peroxidation. Biochemistry. 2006;45:14201–14211. doi: 10.1021/bi0610909. [DOI] [PubMed] [Google Scholar]
- [22].Flemmig J, Spalteholz H, Schubert K, Meier S, Arnhold J. Modification of phosphatidylserine by hypochlorous acid. Chem. Phys. Lipids. 2009;161:44–50. doi: 10.1016/j.chemphyslip.2009.06.144. [DOI] [PubMed] [Google Scholar]
- [23].Gorgas K, Teigler A, Komljenovic D, Just WW. The ether lipid-deficient mouse: tracking down plasmalogen functions. Biochim. Biophys. Acta. 2006;1763:1511–1526. doi: 10.1016/j.bbamcr.2006.08.038. [DOI] [PubMed] [Google Scholar]
- [24].Albert CJ, Crowley JR, Hsu FF, Thukkani AK, Ford DA. Reactive chlorinating species produced by myeloperoxidase target the vinyl ether bond of plasmalogens: identification of 2-chlorohexadecanal. J. Biol. Chem. 2001;276:23733–23741. doi: 10.1074/jbc.M101447200. [DOI] [PubMed] [Google Scholar]
- [25].Thukkani AK, McHowat J, Hsu FF, Brennan ML, Hazen SL, Ford DA. Identification of alpha-chloro fatty aldehydes and unsaturated lysophosphatidylcholine molecular species in human atherosclerotic lesions. Circulation. 2003;108:3128–3133. doi: 10.1161/01.CIR.0000104564.01539.6A. [DOI] [PubMed] [Google Scholar]
- [26].Marsche G, Heller R, Fauler G, Kovacevic A, Nuszkowski A, Graier W, Sattler W, Malle E. 2-Chlorohexadecanal derived from hypochlorite-modified high-density lipoprotein-associated plasmalogen is a natural inhibitor of endothelial nitric oxide biosynthesis. Arterioscler. Thromb. Vasc. Biol. 2004;24:2302–2306. doi: 10.1161/01.ATV.0000148703.43429.25. [DOI] [PubMed] [Google Scholar]
- [27].Ledesma MD, Brugger B, Bunning C, Wieland FT, Dotti CG. Maturation of the axonal plasma membrane requires upregulation of sphingomyelin synthesis and formation of protein–lipid complexes. EMBO J. 1999;18:1761–1771. doi: 10.1093/emboj/18.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wheeler D, Bandaru VV, Calabresi PA, Nath A, Haughey NJ. A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis. Brain. 2008;131:3092–3102. doi: 10.1093/brain/awn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Blanchard-Fillion B, Prou D, Polydoro M, Spielberg D, Tsika E, Wang Z, Hazen SL, Koval M, Przedborski S, Ischiropoulos H. Metabolism of 3-nitrotyrosine induces apoptotic death in dopaminergic cells. J. Neurosci. 2006;26:6124–6130. doi: 10.1523/JNEUROSCI.1038-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Goti D, Balazs Z, Panzenboeck U, Hrzenjak A, Reicher H, Wagner E, Zechner R, Malle E, Sattler W. Effects of lipoprotein lipase on uptake and transcytosis of low density lipoprotein (LDL) and LDL-associated alpha-tocopherol in a porcine in vitro blood–brain barrier model. J. Biol. Chem. 2002;277:28537–28544. doi: 10.1074/jbc.M203989200. [DOI] [PubMed] [Google Scholar]
- [31].Bergt C, Oettl K, Keller W, Andreae F, Leis HJ, Malle E, Sattler W. Reagent or myeloperoxidase-generated hypochlorite affects discrete regions in lipid-free and lipid-associated human apolipoprotein A-I. Biochem. J. 2000;346:345–354. [PMC free article] [PubMed] [Google Scholar]
- [32].Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br. J. Pharmacol. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kratzer I, Wernig K, Panzenboeck U, Bernhart E, Reicher H, Wronski R, Windisch M, Hammer A, Malle E, Zimmer A, Sattler W. Apolipoprotein A-I coating of protamine–oligonucleotide nanoparticles increases particle uptake and transcytosis in an in vitro model of the blood–brain barrier. J. Control. Release. 2007;117:301–311. doi: 10.1016/j.jconrel.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hrzenjak A, Moinfar F, Kremser ML, Strohmeier B, Staber PB, Zatloukal K, Denk H. Valproate inhibition of histone deacetylase 2 affects differentiation and decreases proliferation of endometrial stromal sarcoma cells. Mol. Cancer Ther. 2006;5:2203–2210. doi: 10.1158/1535-7163.MCT-05-0480. [DOI] [PubMed] [Google Scholar]
- [35].Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- [36].Spalteholz H, Wenske K, Panasenko OM, Schiller J, Arnhold J. Evaluation of products upon the reaction of hypohalous acid with unsaturated phosphatidylcholines. Chem. Phys. Lipids. 2004;129:85–96. doi: 10.1016/j.chemphyslip.2003.12.005. [DOI] [PubMed] [Google Scholar]
- [37].Skaff O, Pattison DI, Davies MJ. The vinyl ether linkages of plasmalogens are favored targets for myeloperoxidase-derived oxidants: a kinetic study. Biochemistry. 2008;47:8237–8245. doi: 10.1021/bi800786q. [DOI] [PubMed] [Google Scholar]
- [38].Arnhold J, Osipov AN, Spalteholz H, Panasenko OM, Schiller J. Effects of hypochlorous acid on unsaturated phosphatidylcholines. Free Radic. Biol. Med. 2001;31:1111–1119. doi: 10.1016/s0891-5849(01)00695-5. [DOI] [PubMed] [Google Scholar]
- [39].Tyurin VA, Tyurina YY, Feng W, Mnuskin A, Jiang J, Tang M, Zhang X, Zhao Q, Kochanek PM, Clark RS, Bayir H, Kagan VE. Mass-spectrometric characterization of phospholipids and their primary peroxidation products in rat cortical neurons during staurosporine-induced apoptosis. J. Neurochem. 2008;107:1614–1633. doi: 10.1111/j.1471-4159.2008.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Norenberg MD, Rao KV. The mitochondrial permeability transition in neurologic disease. Neurochem. Int. 2007;50:983–997. doi: 10.1016/j.neuint.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Whiteman M, Rose P, Siau JL, Cheung NS, Tan GS, Halliwell B, Armstrong JS. Hypochlorous acid-mediated mitochondrial dysfunction and apoptosis in human hepatoma HepG2 and human fetal liver cells: role of mitochondrial permeability transition. Free Radic. Biol. Med. 2005;38:1571–1584. doi: 10.1016/j.freeradbiomed.2005.02.030. [DOI] [PubMed] [Google Scholar]
- [43].Vissers MC, Carr AC, Winterbour CC. Fatty acid chlorohydrins and bromohydrins are cytotoxic to human endothelial cells. Redox Rep. 2001;6:49–55. doi: 10.1179/135100001101536030. [DOI] [PubMed] [Google Scholar]
- [44].Dever G, Wainwright CL, Kennedy S, Spickett CM. Fatty acid and phospholipid chlorohydrins cause cell stress and endothelial adhesion. Acta Biochim. Pol. 2006;53:761–768. [PubMed] [Google Scholar]
- [45].Loidl A, Sevcsik E, Riesenhuber G, Deigner HP, Hermetter A. Oxidized phospholipids in minimally modified low density lipoprotein induce apoptotic signaling via activation of acid sphingomyelinase in arterial smooth muscle cells. J. Biol. Chem. 2003;278:32921–32928. doi: 10.1074/jbc.M306088200. [DOI] [PubMed] [Google Scholar]
- [46].Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell. Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- [47].Birbes H, Luberto C, Hsu YT, El Bawab S, Hannun YA, Obeid LM. A mitochondrial pool of sphingomyelin is involved in TNFalpha-induced Bax translocation to mitochondria. Biochem. J. 2005;386:445–451. doi: 10.1042/BJ20041627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pellerin L, Bouzier-Sore AK, Aubert A, Serres S, Merle M, Costalat R, Magistretti PJ. Activity-dependent regulation of energy metabolism by astrocytes: an update. Glia. 2007;55:1251–1262. doi: 10.1002/glia.20528. [DOI] [PubMed] [Google Scholar]
- [49].Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- [50].Willis CL, Nolan CC, Reith SN, Lister T, Prior MJ, Guerin CJ, Mavroudis G, Ray DE. Focal astrocyte loss is followed by microvascular damage, with subsequent repair of the blood–brain barrier in the apparent absence of direct astrocytic contact. Glia. 2004;45:325–337. doi: 10.1002/glia.10333. [DOI] [PubMed] [Google Scholar]
- [51].Tessier JP, Thurner B, Jungling E, Luckhoff A, Fischer Y. Impairment of glucose metabolism in hearts from rats treated with endotoxin. Cardiovasc. Res. 2003;60:119–130. doi: 10.1016/s0008-6363(03)00320-1. [DOI] [PubMed] [Google Scholar]
- [52].Summers SA, Garza LA, Zhou H, Birnbaum MJ. Regulation of insulin-stimulated glucose transporter GLUT4 translocation and Akt kinase activity by ceramide. Mol. Cell. Biol. 1998;18:5457–5464. doi: 10.1128/mcb.18.9.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kanety H, Hemi R, Papa MZ, Karasik A. Sphingomyelinase and ceramide suppress insulin-induced tyrosine phosphorylation of the insulin receptor substrate-1. J. Biol. Chem. 1996;271:9895–9897. doi: 10.1074/jbc.271.17.9895. [DOI] [PubMed] [Google Scholar]
- [54].Arboleda G, Huang TJ, Waters C, Verkhratsky A, Fernyhough P, Gibson RM. Insulin-like growth factor-1-dependent maintenance of neuronal metabolism through the phosphatidylinositol 3-kinase-Akt pathway is inhibited by C2-ceramide in CAD cells. Eur. J. Neurosci. 2007;25:3030–3038. doi: 10.1111/j.1460-9568.2007.05557.x. [DOI] [PubMed] [Google Scholar]
- [55].Decraene C, Brugg B, Ruberg M, Eveno E, Matingou C, Tahi F, Mariani J, Auffray C, Pietu G. Identification of genes involved in ceramide-dependent neuronal apoptosis using cDNA arrays. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0042. RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Stacey MM, Peskin AV, Vissers MC, Winterbourn CC. Chloramines and hypochlorous acid oxidize erythrocyte peroxiredoxin 2. Free Radic. Biol. Med. 2009;47:1468–1476. doi: 10.1016/j.freeradbiomed.2009.08.022. [DOI] [PubMed] [Google Scholar]
- [57].Malle E, Furtmuller PG, Sattler W, Obinger C. Myeloperoxidase: a target for new drug development? Br. J. Pharmacol. 2007;152:838–854. doi: 10.1038/sj.bjp.0707358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.