Abstract
Breast cancer is one of the most commonly diagnosed cancers in women. While there are several effective therapies for breast cancer and important single gene prognostic/predictive markers, more than 40,000 women die from this disease every year. The increasing availability of large-scale genomic datasets provides opportunities for identifying factors that influence breast cancer survival in smaller, well-defined subsets. The purpose of this study was to investigate the genomic landscape of various breast cancer subtypes and its potential associations with clinical outcomes. We used statistical analysis of sequence data generated by the Cancer Genome Atlas initiative including somatic mutation load (SML) analysis, Kaplan–Meier survival curves, gene mutational frequency, and mutational enrichment evaluation to study the genomic landscape of breast cancer. We show that ER+, but not ER−, tumors with high SML associate with poor overall survival (HR = 2.02). Further, these high mutation load tumors are enriched for coincident mutations in both DNA damage repair and ER signature genes. While it is known that somatic mutations in specific genes affect breast cancer survival, this study is the first to identify that SML may constitute an important global signature for a subset of ER+ tumors prone to high mortality. Moreover, although somatic mutations in individual DNA damage genes affect clinical outcome, our results indicate that coincident mutations in DNA damage response and signature ER genes may prove more informative for ER+ breast cancer survival. Next generation sequencing may prove an essential tool for identifying pathways underlying poor outcomes and for tailoring therapeutic strategies.
Keywords: Mutation load, Breast cancer, DNA damage repair, Estrogen receptor
Introduction
Breast cancer is the second leading cause of cancer-related death in women [1]. Comprehensive gene expression analyses of breast cancer confirmed the presence of the following three histopathologically identified subsets: (1) estrogen receptor (ER)-positive, progesterone receptor (PR)-positive; (2) human epidermal growth factor receptor 2 (HER2)-enriched; and (3) triple-negative (lacking ER, PR, and HER2) [2]. ER+ breast cancers account for approximately 70 % of breast tumors diagnosed, and while effective targeted endocrine therapies have been identified, ~25 % of these tumors develop resistance over time and consequently undergo relapse [3]. Analysis of aromatase inhibitor-treated ER+ breast tumors using whole exome sequencing identified associations between endocrine resistance and mutations in ER-related genes, including GATA3, CBFB, TBX3, RUNX1, and PIK3CA [4]. Similarly, loss of PR in ER+ breast tumors associates with loss of estrogen-dependence, increased endocrine resistance, and diminished overall survival [5]. The discovery of underlying targetable pathways of resistance in this subgroup is required for the identification of markers and the development of tailored therapeutic strategies.
Several prognostic markers have been identified for breast cancer including lymph node involvement, tumor stage, TP53 status, PAM-50 subtype, ER status, PR status, and HER2-enrichment. Mutations in DNA damage response (DDR) pathways are also implicated in clinical outcome of breast cancer. Mutations in BRCA1 (a double-strand break (DSB) repair gene) and TP53 (a DDR checkpoint gene), for instance, are associated with triple-negative breast cancer and poor clinical outcome [6, 7]. Mutations in other DDR genes including ATM, ATR, and BRCA2 (all DSB repair genes) have been associated with increased susceptibility to breast cancer [8]. While some of these markers have contributed significantly to the tailoring of therapeutic strategies, they do not comprehensively predict resistance or increased mortality. Moreover, despite much effort no further globally significant single genes have been identified as predictors of breast cancer clinical outcome, and it is unlikely that many such genes remain to be discovered. In non-breast cancers, DNA damage affects tumor somatic mutation load (SML), and mutations in DDR genes can be predictive of clinical outcomes, such as overall and relapse-free survival [9, 10]. In this context, we postulate that genome-wide phenotypic signatures might have a wide impact on breast cancer prognosis and prediction.
In support of this idea, increased genomic instability in tumors has been associated with the basal-like tumor subtype [11] and with metastasis-free survival in lymph node-negative luminal breast tumors although this analysis was limited by its sample size [12]. This genomic instability score was found to be highly associated with TP53 mutations and proliferative indices. However, genomic instability in this group was restricted to a very small number of tumors, indicating a potential limitation of its scope for use as a prognostic/predictive marker. Recent whole exome sequencing of colorectal cancer by the TCGA initiative identified a high SML subset associated with microsatellite instability, mutations in mismatch repair (MMR) pathway genes, and favorable outcome [13]. However, the effects of SML on breast cancer have not yet been elucidated and we postulated that, as in colorectal cancer, SML of a breast tumor would influence patient survival. To test this hypothesis, we analyzed whole exome sequencing data recently generated by the The Cancer Genome Atlas (TCGA) initiative from breast tumors [14].
Materials and methods
Informatics
Whole exome somatic variants, gene expression, clinical, and epidemiological data were downloaded from The Cancer Genome Atlas Breast Invasive Carcinoma data portal (https://tcga-data.nci.nih.gov/tcga/tcgaCancerDetails.jsp?diseaseType=BRCA&diseaseName=Breast%20invasive%20carcinoma). Details of sample acquisition, DNA sequencing, and RNA expression analyses have been described in the original TCGA publication [14]. Data processing and statistical analysis were carried out using the R statistical software suite [15] and custom scripts written in Perl.
Statistics
t tests were used to determine p-values for continuous data, with Holm’s adjustment for multiple comparisons as required. For data with non-normal distribution, the Wilcoxon Rank Sum test was used. Fisher’s exact tests were used to determine significance of categorical data. Survival analyses used log-rank tests, and Kaplan–Meier curves were plotted using R. Due to the low median follow-up time of the TCGA cohort (575 days), all survival analyses extend only 10 years. Proportional hazards were calculated using the Cox regression model, and the coxph function in R was used to confirm that the dataset met the assumptions for the Cox regression analysis.
Clinical information
ER status provided in the publically available TCGA dataset was used to sort the tumors into ER+ and ER− groups. Age at diagnosis of >50 years was used as a surrogate indicator of postmenopausal status.
Gene lists
Lists of genes within the specific DNA damage response, MAPK, NFkB, and T-cell marker pathways were generated using the KEGG database (keywords: DNA damage repair; base excision repair (BER); nucleotide excision repair (NER); MMR; homologous recombination (HR); non-homologous end joining (NHEJ); DDR checkpoint; MAPK signaling; NFkB signaling; T-cell marker) (see Tables 1, 2). Genes with known prognoses [4, 16–18] were generated from the previous literature (see Table 5). A consensus ER+ breast cancer signature gene list (ABCA3, ACADSB, ALDH3B2, AR, ANXA9, BCL2, CA12, CCND1, CGA, DNAJC12, ESR1, ERBB4, FOXA1, GATA3, GJA1, GREB1, HPN, IGFBP4, IL6ST, KRT18, LRBA, MYB, NAT1, NRIP1, PGR, PTPRT, RABEP1, RARRES1, RERG, RET, SEMA3B1, SLC27A2, SLC39A6, SULT2B1, TFF1, TFF3, XBP1) was generated from five independent studies profiling ER+ (luminal) breast tumors and cell lines [19–23].
Table 1.
KEGG-generated list of genes from three cancer-related pathways
| Gene name | Pathway |
|---|---|
| FOS, JUN, MAP2K3, MAP2K4, MAPK8, MAPK8I-3, MAP3K1, MAP3K7, TNF, MAPK3, MAPK6, MAPK12, MAPK13, MAPK14, MAPK9, MST1, MAP4K1, MAP2K7, MAP2K2, MAP2K6, MAP3K4 | MAPK |
| CASP8, CHUK, IKBKB, IL1B, MAP4K4, NFKB1, NFKB2, NFRKB, REL, IRAK1 | NFkB |
| CD4, CREBBP, CTLA4, FASLG, IL15, JAK2, LAG3, MAPK8, TGFB3, TNFRSF8, TNFRSF9, TYK2, CD27, CD40LG, CD80, PTPRC, CCR5, CXCR3, IL12B, IL12RB2, IL18R1, IL1RL1, IL27, IL7R, STAT1, STAT4, TBX21, TLR4, CCL11, CCL5, CCL7, CCR4, GATA3, GF1I, ICOS, IL13RA1, IL1R1, IL25, IRF4, JAK1, MAF, NFATC1, NFATC2, PCGF2 | T-cell regulation |
Table 2.
KEGG-generated list of DNA damage repair genes
| Base excision repair (BER) | DDR pathway |
|---|---|
| APEX1, APEX2, CCNO, FEN1, LIG3, MBD4, MPG, MUTYH, NEIL1, NEIL2, NEIL3, PARP1, PARP2, PARP3, PCNA, POLB, SMUG1, TDG, UNG, XRCC1 | Base excision repair (BER) |
| ATXN3, CCNH, DDB1, DDB2, ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, ERCC6, ERCC8, MMS19, PNKP, POLL, RAD23A, RAD23B, RPA1, RPA3, SLK, XAB2, XPA, XPC | Nucleotide excision repair (NER) |
| MLH1, MLH3, MSH2, MSH3, MSH4, MSH5, MSH6, PMS1, PMS2, POLD3, TREX1 | Mismatch repair (MMR) |
| ATM, BLM, BRCA2, DMC1, H2AFX, MUS81, POLD1, RAD51, RAD51C, RAD52, RAD54B, RAD54L, RPA2, TP53BP1 | Homologous recombination (HR) |
| DLCRE1C, DNTT, LIG4, MRE11A, NBN, POLM, PRKDC, RAD50, XRCC2, XRCC5, XRCC6 | Non-homologous end joining (NHEJ) |
| ABL1, ATR, BRIP1, CEP63, CHEK1, CHEK2, CHKA, CLSPN, DBF4, E2F1, FOXN3, GRP, HUS1B, MAD2L2, MAPK14, MYH1, PDP1, PIN4, PNPN11, RAD1, RFC4, TIPIN, TP53, WEE1, ZAK | DDR checkpoint |
| ATRIP, ATRX, BARD1, BAX, BBC3, BRCA1, CDC25A, CDC25C, CDK7, CDKN1A, CIB1, CRY1, CSNK2A2, DDIT3, EXO1, FANCA, FANCD2, FANCG, GADD45A, GADD45G, LIG1, MAPK12, MCPH1, MDC1, MGMT, NTHL1, OGG1, PPM1D, PP1R15A, RAD17, RAD18, RAD21, RAD51B, RAD9A, RBBP8, REV1, RFC1, RNF168, RNF8, SIRT1, SMC1A, SUMO1, TOP3A, TOPSBP1, TP73, XRCC3, XRCC6BP1 | Multiple (Other) |
Table 5.
List of ER signature genes with prognostic mutational status in breast cancer
| Gene name | Mutational prognosis | Reference |
|---|---|---|
| GATA3 | Good | Ellis et al. [4] Nature |
| MAP3K1 | Good | Ellis et al. [4] Nature |
| MAP2K4 | Good | Ellis et al. [4] Nature |
| PIK3CA | Good | Cizkova et al. [16] Br Canc Res |
| CDKN1B | Poor | Depowski et al. [17] Mod Pathol |
| RB1 | Poor | Ellis et al. [4] Nature |
| PTEN | Poor | Alkarain et al. [18] J Mamm Gl Neopl |
| TP53 | Poor | Ellis et al. [4] Nature |
Results
Mutation load distribution is different between ER+ and ER− breast cancer
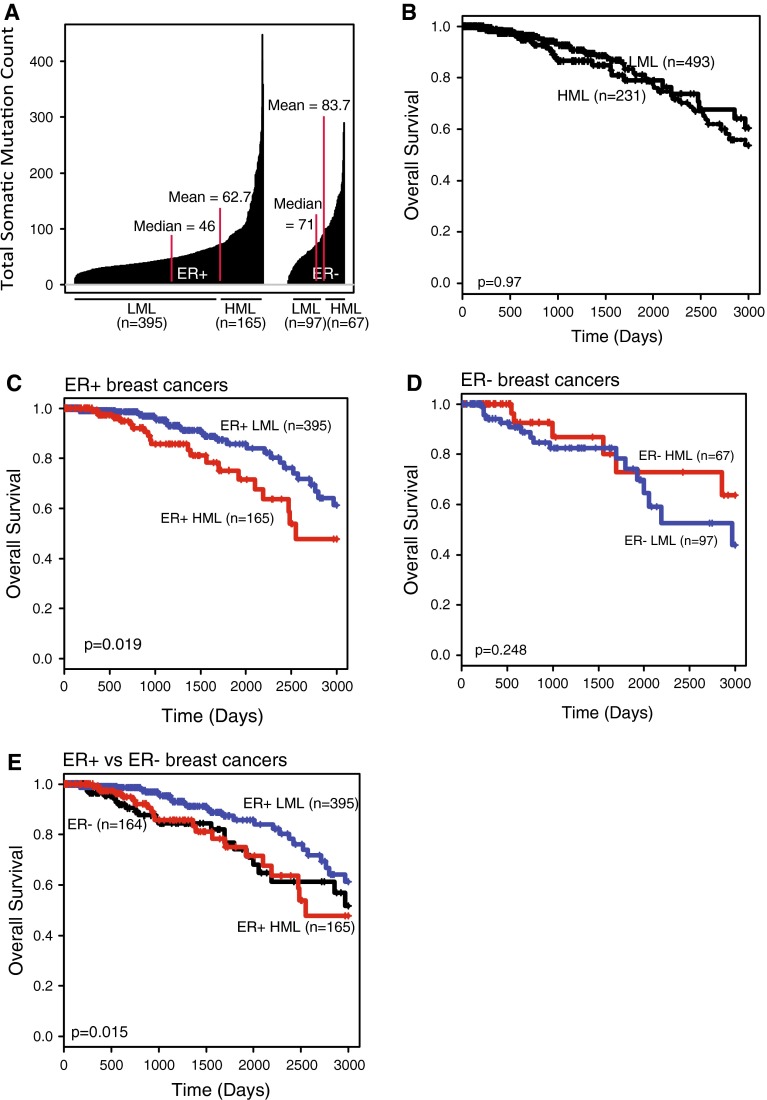
Our sample set comprises 762 invasive breast tumors from the TCGA dataset. Immunohistochemical analysis shows that the majority of these tumors (73.4 %) are ER+ (Table 3). The mean SML is 67.23 mutations per tumor (Table 3); however, ER− tumors have a significantly higher SML than ER+ tumors (p < 0.0001; Fig. 1a). Furthermore, ER+ and ER− tumors are characterized by marked differences in SML distribution. ER+ tumors have a median SML of 46 (Fig. 1a) and a mean SML of 62.7, with a small subset of these tumors carrying significantly high mutation loads (HMLs) (Fig. 1a). Conversely, ER− tumors lack a distinct high mutation subset, instead almost half (42 %) of the tumors carry mutation loads higher than the mean SML (Fig. 1a).
Table 3.
Descriptive characteristics of TCGA dataset used in the study
| Characteristic | Mean | Standard deviation |
|---|---|---|
| Age at diagnosis (years) | 57.97 | 13.15 |
| Mean mutation count (n) | 67.23 | 52.79 |
| Mean overall survival (days) | 901.53 | 1069.441 |
| Total number (n) | 762 | |
| Tumor size at diagnosis (n) | ||
| T1 | 201 | |
| T2 | 441 | |
| T3+ | 109 | |
| Unknown | 11 | |
| Nodal involvement at diagnosis (n) | ||
| N0 | 364 | |
| N1 | 249 | |
| N2+ | 138 | |
| Unknown | 11 | |
| ER status of tumor (n) | ||
| ER-positive | 559 | |
| ER-negative | 165 | |
| Triple-negative | 116 | |
| Unknown | 38 | |
| HER2 status of tumor (n) | ||
| HER2-positive | 105 | |
| HER2-negative | 621 | |
| Unknown | 36 | |
| Vital status of patient (n) | ||
| Alive | 671 | |
| Deceased | 91 | |
Fig. 1.
HML subset of ER+ breast tumors is associated with poor clinical outcome. a Index plot. Median and mean SMLs of each population are indicated with red lines. b–e Kaplan–Meier survival curves of all breast tumors (b) and the HML (red) and LML (blue) subsets of: c ER+ breast cancer; d ER− breast cancer; and e a comparison between ER+ HML, ER+ LML, and ER− (black) breast cancer. The log-rank test was used to determine p-values
SML associates with ER+ breast cancer survival
Associations have been found between genomic instability, mutations in specific DNA damage genes, and clinical outcome in various cancers, including the breast [8], but there have been few previous reports on the effect of mutation load on any type of cancer. One report identified an association between high SML and good clinical outcome in colorectal cancer [13]; however, there are no such associative findings reported for breast cancer. To test whether mutation load affected survival in breast cancer, we divided all breast cancers into HML and low mutation load (LML) groups based on the mean SML across all breast cancers. We found that SML had no effect on breast cancer survival when all tumors were considered (Fig. 1b) in accord with the previous TCGA report [14]. However, we postulated that SML might differentially affect breast cancer outcomes based on ER status, as suggested by the distinct distribution of SML between ER+ and ER− breast tumors (Fig. 1a). Therefore, we next analyzed the effect of SML on overall survival independently in the ER+ and ER− subsets of breast cancer by defining tumors as LML or HML based on mean SML for each ER subtype. We found that patients with ER+ HML tumors exhibit significantly shorter overall survival than do patients with ER+ LML tumors (p = 0.02, Fig. 1c), and conversely, overall survival is not affected by SML in patients with ER− tumors (p = 0.25, Fig. 1d). In addition, the overall survival curve of ER+ HML tumors is virtually identical to the survival curve of ER− tumors (Fig. 1e), emphasizing the significantly poor overall survival observed in the HML subset of ER+ breast tumors. For most of the remaining analyses, we focused on the effects of mutation load on ER+ breast cancer.
We next used a Cox regression model that assessed effect of SML on survival in the presence of known prognostic/predictive factors including PR status, HER2 enrichment, tumor stage, and lymph node involvement. Our results showed that mutation load was an independent prognostic factor in ER+ tumors (p = 0.04, Table 4) with a hazard ratio (HR = 2.02) higher than that of all other factors considered except nodal status. In fact PR status no longer contributed significantly to survival (p = 0.15) although lymph node status remained significant in the multivariate analysis (p = 0.02). The fact that tumor stage did not affect clinical outcome significantly in the Cox analysis is likely due to the small number of patients and the short follow-up time in this study (see “Materials and methods” section). The HML subset overall was enriched for HER2+ tumors (36/395 LML tumors were HER2+ vs 36/105 HML tumors; p < 0.001), and the average SML of HER2-enriched tumors (79.5 ± 55.9) was higher than HER2-negative tumors (65.6 ± 53.5; p = 0.02). However, HER2 enrichment did not contribute significantly to overall survival (Table 4) indicating that HML may be a more compelling contributor to survival in ER+ breast cancer than HER2 status.
Table 4.
Proportional hazards table identifying mutation load as an independent prognostic factor for ER+ breast cancer
| Factor | Hazard Ratio | CI | p-Value |
|---|---|---|---|
| Mutation load | |||
| LML | Ref. | ||
| HML | 2.02 | 1.02–4.00 | 0.04 |
| HER2 status | |||
| Negative | Ref. | ||
| Positive | 1.65 | 0.66–4.12 | 0.29 |
| PR status | |||
| Negative | Ref. | ||
| Positive | 0.55 | 0.25–1.24 | 0.15 |
| Tumor stage | |||
| Stage I | Ref. | ||
| Stage II | 1.11 | 0.34–3.65 | 0.86 |
| Stage III+ | 0.38 | 0.05–2.57 | 0.32 |
| Nodal involvement | |||
| N0 | Ref. | ||
| N1 | 1.81 | 0.75–4.35 | 0.32 |
| N2+ | 8.35 | 1.43–48.68 | 0.02 |
The bolding just highlights the factors that significantly affect breast cancer survival
* p-Value generated by Cox Regression Analysis for Proportional Hazards
DNA damage repair pathways are mutated in tumors with HML
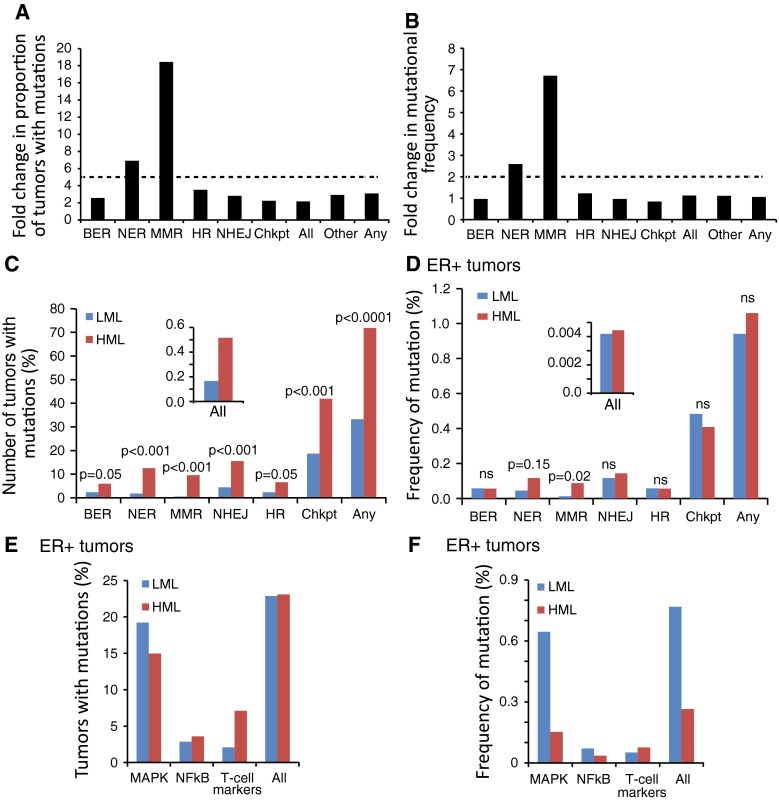
To investigate the pathways underlying the HML phenotype, we next investigated whether HML associated with inactivation of DDR pathways by assessing the mutational status of genes from the DDR checkpoint, as well as from each of the five major DDR pathways: BER; NER; MMR; HR; and NHEJ (see “Materials and methods” section and Table 2). We analyzed the proportion of tumors with mutations in at least one gene from each pathway in HMLs vs LMLs, and the mutational frequency (i.e., the number of non-silent mutations in genes of a specific pathway over total number of mutations in all genes) (Fig. 2a, b).
Fig. 2.
HML in ER+ cancers associates with mutations in DDR, but not checkpoint, genes. a, b Bar graphs representing the fold change in HMLs over LMLs of mutations in specified DDR pathway genes, DDR checkpoint genes (Chkpt), genes that are common to multiple DDR pathways (Other), all DDR-related genes included in the analysis (All), and any non-DDR gene in the genome (Any) in terms of: a proportion of tumors with at least one mutation in each pathway; and b frequency with which every gene of each pathway is mutated. The dotted line represents the threshold fold change calculated from baseline levels graphed in c–d, inset and e–f. c, d Bar graphs. Fisher’s exact test was used to generate p-values. Inset depicts bar graphs representing tumors with mutations in all genes other than DDR-related genes. e, f Percentage of tumors with mutations in genes from three randomly selected cancer-related pathways (e), and the frequency of mutations in genes from these pathways in both HML (red) and LML (blue) tumors (f). Fisher’s exact test was used to determine p-values. Gene lists were generated from KEGG database and from the previous literature and are reproduced in Tables 1 and 2
Mutational analysis in this study is confounded by the fact that HML tumors are theoretically more likely to mutate any given gene than LML tumors. To account for this bias, we calculated baseline statistics of (1) the proportion of tumors with mutations in any given gene and (2) the frequency of mutations in any given gene for both HML and LML subset tumors. We found that the baseline proportion of tumors that have a mutation in any gene is 2.5-fold higher in the HMLs relative to the LMLs as would be expected of tumors with significantly higher mutation load (Fig. 2c, inset). However, we found that the baseline mutational frequency of any gene was similar between the HMLs and LMLs suggesting that the likelihood of any random gene being mutated was comparable between the HML and LML subsets (Fig. 2d, inset). An independent calculation of these same baseline parameters on genes from three randomly selected KEGG-generated pathways revealed no significant increase in mutational proportion or frequency in HMLs (Fig. 2e, f), indicating that high mutation load does not necessarily enrich for mutations in every pathway. Based on these analyses, we set the threshold to find mutational enrichment in the HML subset as twice the baseline difference between HMLs and LMLs. This means that in order to find mutational enrichment in DDR genes in HMLs, 5-fold more tumors would need to have these genes mutated at 2-fold higher frequencies than LMLs.
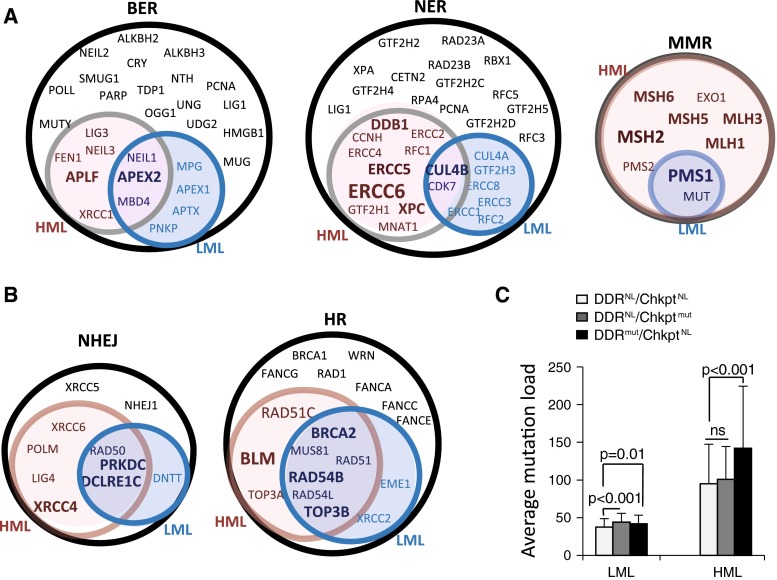
Using these conservative thresholds, we found no significant enrichment for DDR mutations overall in HMLs over LMLs (Fig. 2a, b). However, mutations in MMR pathway genes occurred in 16-fold more tumors and occurred at 7-fold higher frequency in HML than in LML ER+ tumors indicating significant enrichment over and above our set thresholds (Fig. 2a, b). Uniquely, every gene specific to the MMR pathway was mutated at least once in the HML subset of ER+ tumors (Fig. 3a). Genes from the single-strand break repair pathway, NER, were also mutated in 7-fold more HML tumors and at 2.5-fold higher frequency relative to the LML ER+ tumors (Fig. 3a). Notably, there was no significant enrichment in the HMLs in DNA damage checkpoint genes (Fig. 2a, b). Some genes from the double-strand break repair pathways, e.g., BLM and XRCC4, are mutated at higher frequencies and in more tumors in the HML subset than in the LML subset, but this enrichment is not significant (Figs. 3b; 2a, b).
Fig. 3.
ER+ HML tumors are enriched for mutations in MMR and NER pathway genes. a, b Venn diagrams indicating genes from the specified DDR pathway that are mutated in either the HML (red) or LML (blue) subset of ER+ tumors, in both (purple) and in neither (white). Increasing font size indicates an increasing proportion of tumors with mutations in the specific gene. c Bar graph depicting the average SML in tumors with specified mutational status. Student’s t test with Holm’s adjustment for multiple comparisons was used to define p-values. Chkpt, genes from the DNA damage checkpoint; NL, tumors with no identified mutations in genes from the specified pathway; mut, tumors with identified non-silent mutations in genes from the specified pathway; ns not significant
In addition, we found a 50 % increase in mean SML in ER+ HML tumors with mutations in DDR pathway genes, while mutations in DDR checkpoint genes did not affect SML (Fig. 3c). Especially striking is the observation that mutations in TP53 occur in a significant fraction of breast tumors and were previously reported to affect genomic instability [11] but are not enriched over the set threshold in the HML group (0.96-fold for mutational frequency and 2.97-fold for tumor proportion) relative to the LML group. While mutations in DDR genes resulted in increased mutation load within LML subset tumors (Fig. 3c), the extremely small effect size limits the biological relevance of this finding. Together, these results indicate that the HML subset of ER+ tumors is associated with mutations in DDR pathway genes, specifically in MMR and NER genes, but not with mutations in DDR checkpoint and double-strand break repair genes.
Mutations in known prognostic genes do not affect survival
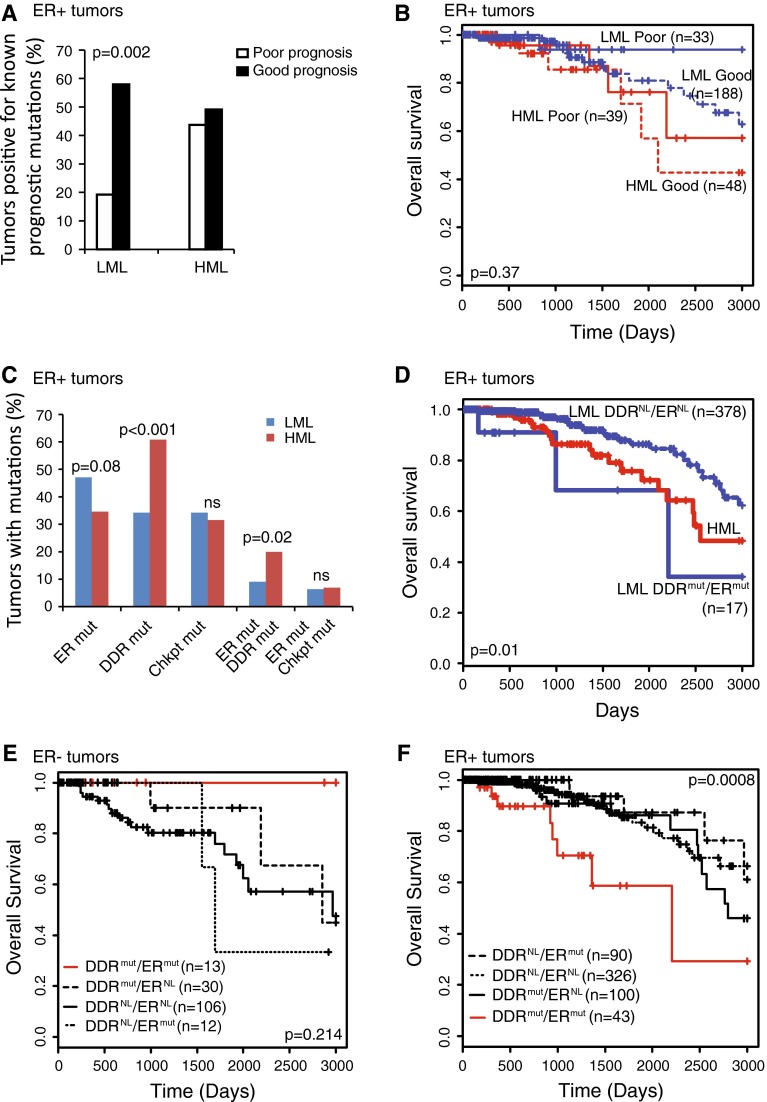
Next, we investigated potential pathways underlying the poor survival phenotype associated with HML tumors using a candidate approach. To determine whether the HML subset of ER+ tumors is enriched for mutations associated with poor prognosis, we generated a list of known prognostic genes mutated at >10 % frequency in human breast cancer based on the existing literature [4, 16–18] (see “Materials and methods” section and Table 5). We assessed the proportion of tumors with mutations in these genes in both the HML and LML ER+ subsets. Our results demonstrate that the LML subset has a significantly higher proportion of good prognostic mutations than poor prognostic mutations (p = 0.002), (Fig. 4a). However, there were no significant associations found between these known prognostic mutations and overall survival in either HML or LML subsets (Fig. 4b). These data indicate that mechanisms other than those associated with known prognostic genetic mutations mediate the association between SML and breast cancer survival.
Fig. 4.
Coincident mutations in DDR and ER signature genes associate with poor survival irrespective of mutation load. a Percentage of tumors with mutations in genes associated with either good or poor prognosis in specified subsets. Fisher’s exact test was used to determine the p-value. b Kaplan–Meier survival curves of indicated groups. Log-rank test was used to generate p-values. c Bar graph depicting the percentage of tumors with mutations in the specified pathways. Fisher’s exact test was used to identify p-values. The list of ER signature genes is presented in “Materials and methods” section. d–f Kaplan–Meier survival curves of indicated groups. Log-rank test was used to determine p-values. ER, ER signature genes; DDR, genes from the five major DNA damage response pathways; Chkpt, genes from the DNA damage checkpoint; mut, tumors with non-silent mutations in genes from the specified pathway; NL, tumors with no identified mutations in genes from the specified pathway; ns, not significant
Coincident mutations in ER and DDR genes are enriched in HML breast tumors and associate with poor patient survival
We next hypothesized that inactivation of DDR increases the frequency of genetic mutations in ER pathways thereby decreasing dependence on ER signaling and potentially increasing resistance to therapy. To test this hypothesis, we assessed the mutational frequency of ER signature genes in HML and LML tumors (see “Materials and methods” section), and the correlation between mutations in ER signature, DDR checkpoint, and DDR pathway genes. Mutations in ER signature and DDR checkpoint genes occurred at comparable rates between LML and HML tumors, both singly and in combination (p > 0.9; Fig 4c). However, when we compared tumors with coincident mutations in DDR pathway and ER signature genes, we observed significant enrichment in the HML subset tumors (~20 %) compared to LML subset tumors (<10 %; p = 0.03; Fig. 4c).
We next evaluated the clinical outcome of women with tumors having mutations in both DDR and ER genes. As predicted by our hypothesis, HML tumors with mutations in both DDR pathway and ER signature genes associate with worse overall survival than all other HML tumors (p = 0.007, data not shown). Notably, even LML tumors with mutations in genes of both the DDR and ER pathways associate with significantly worse overall survival than all other LML tumors (p = 0.01; Fig 4d). Further, ER+ tumors with coincident mutations in DDR pathway and ER signature genes (~10 % of all ER+ tumors) associate with significantly worse overall survival than all other ER+ tumors independent of mutation load (p = 0.0008; Fig. 4f), unlike ER− tumors (Fig. 4e). These data indicate that coincident mutations in DDR and ER signature genes could constitute an indicator of poor prognosis in ER+ breast tumors.
Discussion
Mutation load and cancer outcome association in breast cancer is unique
The results presented here indicate that in ER+ breast cancer high SML may contribute to poor breast cancer survival, contrary to previous reports in colorectal cancer. Our results suggest the hypothesis that ER+ tumors with mutations in both DDR and ER signature genes are inherently less dependent on ER signaling than ER-driven tumors. This hypothesis may also explain the dichotomous behavior between ER+ and ER− breast tumors with respect to mutation load. Therefore, tumors characterized by coincident mutations in DDR and ER genes may be resistant to current therapies, especially anti-estrogen-based therapies. To advance this field it will be necessary to reinvestigate the effects of mutation load on ER− breast cancer as both the number of sequenced tumors as well as the length of patient follow-up in the TCGA sample set increases.
MMR gene mutations affect breast cancer survival
Large-scale studies like the TCGA have reported few new genes that have global impact on breast cancer prognosis or prediction. New discoveries will, therefore, most likely arise through pathway level, rather than gene level, analyses. In alignment with this idea, the HML subset of ER+ tumors described here is enriched for somatic mutations in MMR pathways, rather than individual genes.
While deleterious mutations in MMR genes have been identified in primary breast tumors as well as in adjacent neoplastic tissue [24, 25], we describe here a correlation between MMR genetic mutations and poor clinical outcome of patients with ER+ breast tumors. In contrast to our results, a recent publication analyzing mutational signatures of various cancers was unable to identify any correlation between MMR deficiency and mutational signature in breast cancer [26]. This discrepancy likely arose because this prior analysis examined all breast cancers as a single group instead of considering ER+ and ER− breast cancer individually. This highlights the importance of incorporating knowledge of tumor biology into analyses rather than relying on pure analytics alone.
Clinical significance of mutation load and sequencing strategies in breast cancer
Our results identify mutation load as a quantitative genomic phenotype, rather than a genotype, associated with clinical outcome. Using mutation load for prediction/prognosis enables easy, quantitative estimation, and may have a greater global impact on breast cancer clinical outcomes than many single genes which are currently considered important. Moreover, mutation load may be indicative of the increased potential of an ER+ breast tumor to quickly become resistant to endocrine therapy by mutating individual pathways that can be discovered through mutational analysis. Therefore, our discovery that high SML may serve as a marker for poor survival in a subset of breast tumors indicates that genome wide sequencing can offer important clinically relevant information for ER+ breast cancer.
Conclusions
Our data indicate a novel association between SML and clinical outcome in breast cancer. Our data also implicate somatic mutations in DDR pathway genes and in ER-related genes as predictive of poor clinical outcome for ER+ breast cancer. It is important to acknowledge the small number of samples and the short follow-up time in this dataset which warrant a larger study to ascertain the contribution of mutation load to clinical outcome. However, approximately one-third of the ER+ tumors used in this study were characterized as HML (>65 mutations). This indicates that a significant proportion of ER+ breast cancer patients could benefit from SML characterization of their tumors. As the cost of DNA sequencing steadily decreases [27], analysis of SML could become a reasonable and useful prognostic marker to help select patients with aggressive and/or endocrine-resistant ER+ tumors, who may benefit from aggressive therapy targeting non-hormonal pathways.
Acknowledgments
Financial Support: This work was supported by Cancer Center Support Grant (5 P30 CA016672-38, P.H.B.) Norman Brinker Award for Research Excellence (P.H.B.), John Charles Cain Distinguished Chair Award (P.H.B.), Mariam Rogers Fund for Breast/Women’s Cancer Research (P.H.B.), and The University of Texas MD Anderson Cancer Center Halliburton Fellowship in Cancer Prevention Fund (S.H.). The authors would like to acknowledge Michelle Savage for editing the manuscript, Jennifer Drummond for technical assistance with data analysis, and Shawn Zhang and Sarah Hein for assistance with downloading data.
Conflict of interest
PHB is a consultant/advisor board member of Susan G. Komen for the Cure. MNB is the CEO and founder of Codified Genomics, LLC, Houston, TX. No potential conflicts of interest were disclosed by other authors.
Abbreviations
- BER
Base excision repair
- DDR
DNA damage response
- ER
Estrogen receptor
- HML
High mutation load
- HR
Homologous recombination
- LML
Low mutation load
- MMR
Mismatch repair
- NER
Nucleotide excision repair
- NHEJ
Non-homologous end joining
- PR
Progesterone receptor
- SML
Somatic mutation load
- TCGA
The Cancer Genome Atlas
Contributor Information
Svasti Haricharan, Email: sharicharan@mdanderson.org.
Matthew N. Bainbridge, Email: matthew.bainbridge@codifiedgenomics.com
Paul Scheet, Email: pascheet@mdanderson.org.
Powel H. Brown, Phone: 713-792-4509, Email: phbrown@mdanderson.org
References
- 1.Abell K, et al. Stat3-induced apoptosis requires a molecular switch in PI(3)K subunit composition. Nat Cell Biol. 2005;7(4):392–398. doi: 10.1038/ncb1242. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 4.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arpino G, et al. Estrogen receptor-positive, progesterone receptor-negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. J Natl Cancer Inst. 2005;97(17):1254–1261. doi: 10.1093/jnci/dji249. [DOI] [PubMed] [Google Scholar]
- 6.Lakhani SR, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20(9):2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi H, et al. Hereditary breast and ovarian cancer susceptibility genes (Review) Oncol Rep. 2013;30(3):1019–1029. doi: 10.3892/or.2013.2541. [DOI] [PubMed] [Google Scholar]
- 9.Mar VJ et al. (2013) BRAF/NRAS wild-type melanomas have a high mutation load correlating with histological and molecular signatures of UV damage. Clin Cancer Res 19(17):4589–4598 [DOI] [PubMed]
- 10.Yu JL, et al. Effect of p53 status on tumor response to antiangiogenic therapy. Science. 2002;295(5559):1526–1528. doi: 10.1126/science.1068327. [DOI] [PubMed] [Google Scholar]
- 11.Kwei KA, et al. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol. 2010;4(3):255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnet F, et al. An array CGH based genomic instability index (G2I) is predictive of clinical outcome in breast cancer and reveals a subset of tumors without lymph node involvement but with poor prognosis. BMC Med Genomics. 2012;5:54. doi: 10.1186/1755-8794-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donehower LA, et al. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J Pathol. 2013;229(1):99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R-Development-Core-Team (2008) R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria
- 16.Cizkova M, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14(1):R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Depowski PL, Rosenthal SI, Ross JS. Loss of expression of the PTEN gene protein product is associated with poor outcome in breast cancer. Mod Pathol. 2001;14(7):672–676. doi: 10.1038/modpathol.3880371. [DOI] [PubMed] [Google Scholar]
- 18.Alkarain A, Jordan R, Slingerland J. p27 deregulation in breast cancer: prognostic significance and implications for therapy. J Mammary Gland Biol Neoplasia. 2004;9(1):67–80. doi: 10.1023/B:JOMG.0000023589.00994.5e. [DOI] [PubMed] [Google Scholar]
- 19.Loi S, et al. Gene expression profiling identifies activated growth factor signaling in poor prognosis (Luminal-B) estrogen receptor positive breast cancer. BMC Med Genomics. 2009;2:37. doi: 10.1186/1755-8794-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider J, et al. Identification and meta-analysis of a small gene expression signature for the diagnosis of estrogen receptor status in invasive ductal breast cancer. Int J Cancer. 2006;119(12):2974–2979. doi: 10.1002/ijc.22234. [DOI] [PubMed] [Google Scholar]
- 21.Abba MC, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frasor J, et al. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144(10):4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 23.Jagannathan V, Robinson-Rechavi M. Meta-analysis of estrogen response in MCF-7 distinguishes early target genes involved in signaling and cell proliferation from later target genes involved in cell cycle and DNA repair. BMC Syst Biol. 2011;5:138. doi: 10.1186/1752-0509-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balogh GA, Heulings RC, Russo J. The mismatch repair gene hPMS2 is mutated in primary breast cancer. Int J Mol Med. 2006;18(5):853–857. [PubMed] [Google Scholar]
- 25.Poplawski T, et al. Polymorphisms of the DNA mismatch repair gene HMSH2 in breast cancer occurence and progression. Breast Cancer Res Treat. 2005;94(3):199–204. doi: 10.1007/s10549-005-4793-7. [DOI] [PubMed] [Google Scholar]
- 26.Alexandrov LB et al (2013) Signatures of mutational processes in human cancer. Nature 500(7463):415–421 [DOI] [PMC free article] [PubMed]
- 27.Bainbridge MN, et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11(6):R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]