Abstract
The use of epigenetic modifiers, such as histone deacetylase inhibitors and DNA methyltransferase inhibitors, has been explored increasingly as a technique to induce the production of additional microbial secondary metabolites. The application of such molecules to microbial cultures has been shown to upregulate otherwise suppressed genes, and in several cases has led to the production of new molecular structures. In this study, the proteasome inhibitor bortezomib was used to induce the production of an additional metabolite from a filamentous fungus (Pleosporales). The induced metabolite was previously isolated from a plant, but the configuration was not assigned until now; in addition, an analogue was isolated from a degraded sample, yielding a new compound. Proteasome inhibitors have not previously been used in this application and offer an additional tool for microbial genome mining.
Introduction
Fungal secondary metabolites are produced via several key pathways, including those involving polyketide synthases (PKSs), non-ribosomal peptide synthases (NRPSs), hybrid PKS-NRPS (HPN), as well as pathways coding for terpenes and indole alkaloids.1 In fungi, these genes are often found in clusters, a fact that has implications on the transcription and regulation of secondary metabolite pathways. With the recent completion of fungal genome sequences,2–5 it has become clear that the number of gene clusters encoding these pathways greatly outnumbers the known secondary metabolites for these organisms.6 A study on the sequence of Aspergillus niger demonstrated that less than 30% of its PKS-NRPS- and HPN-encoding gene clusters were transcriptionally active.7 This transcriptional suppression has led to a variety of studies delving into the mechanisms of transcriptional regulation, as well as developing techniques to induce these suppressed pathways.
Efforts have been made to manipulate the epigenetic regulation of gene transcription by growing fungi in the presence of various small-molecule modifiers. Such research has focused mainly on HDAC (histone deacetylase) inhibitors8–14 and DNA methyltransferase (DNMT) inhibitors,11–17 in order to make more transcriptionally available the genes that these proteins normally help to suppress. Treatment of A. alternata and Penicillium expansum with the HDAC inhibitor trichostatin A resulted in increased production of numerous secondary metabolites.9 A similar study using the HDAC inhibitor suberoylanilidehydroxamic acid (SAHA) to treat A.niger resulted in the isolation of a new metabolite, nygerone A, containing a unique 1-phenylpyridin-4(1H)-one core that previously had not been reported from any natural source.10 Treatment of P. citreonigrum with the DNMT inhibitor 5-azacytidine (5-AZA) produced ten additional secondary metabolites, including two new compounds.15 New compounds have also been isolated from a SAHA-treated culture of Cladosporium cladosporioides and a 5-AZA-treated culture of a Diatrype species.12 The effects of SAHA and 5-AZA on A. niger gene expression were further characterized by using real-time quantitative reverse-transcription PCR to analyze the change in expression of PKS, NRPS, and HPN pathways when treated with the epigenetic modifiers;11 all but seven of these 55 gene clusters showed increased transcriptional rates.
This study examined the capability of bortezomib, a proteasome inhibitor, to induce the production of secondary metabolites in a fungus (MSX 63935, order Pleosporales) that had been shown previously to biosynthesize a series of resorcylic acid lactones of polyketide origin.18 Proteasomes are protein complexes responsible for the degradation of proteins by proteolysis. Among the many proteins degraded by the proteasome pathway, several transcriptional regulators have been identified,19, 20 implicating proteasomes as a crucial player in gene transcription. Growing MSX 63935 in the presence of bortezomib induced the production of an additional secondary metabolite. An analogue was also isolated after degradation of the original metabolite in solution, yielding a new compound.
Results and Discussion
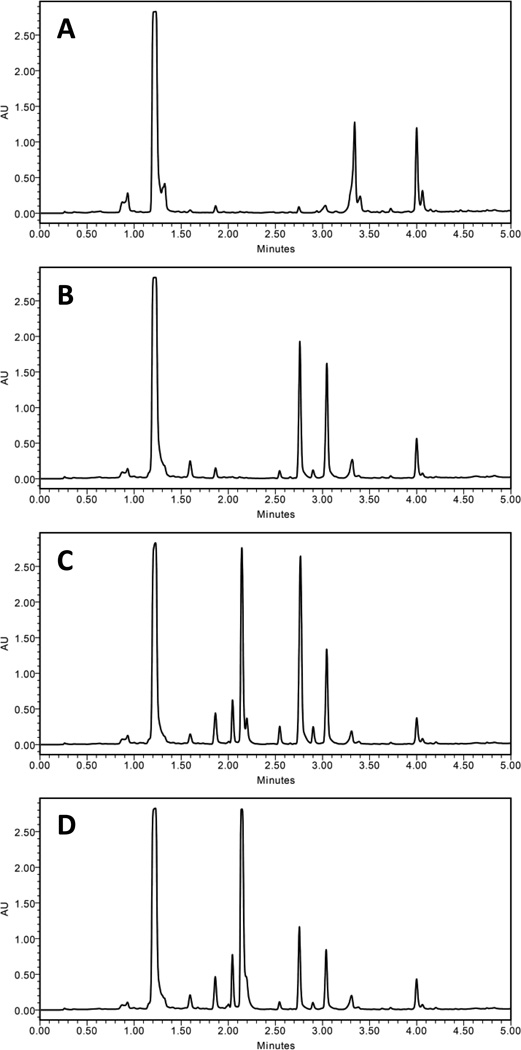
Preliminary experiments tested the effects of an HDAC inhibitor (SAHA), a DNMT inhibitor (5-AZA), and a proteasome inhibitor (bortezomib), on the secondary metabolite production of MSX 63935 in several growth media. Initial experiments with solid media (rice) showed poor results, regardless of very high doses of the epigenetic modifiers (as much as 100 mg per flask). Tests with liquid media, including Czapek Dox broth and potato dextrose broth (PDB), were more promising. Increased production to the expected metabolites as well as a number of additional chromatographic peaks was observed in cultures dosed with epigenetic modifiers (Fig. 1). Secondary metabolite production was greater in PDB than Czapek Dox (Fig. S1, Supplementary Information), so subsequent experiments used PDB as the culture medium. Extractions of media without any fungal growth were used to confirm that the chromatographic peaks were due to secondary metabolites and not the media (Fig. S2, Supplementary Information).
Figure 1.
Comparison of control (A) and dosed (B–D) growths of MSX 63935 grown in potato dextrose broth. The culture shown in (B) was grown with 50 µg/mL SAHA, (C) with 100 µg/mL 5-AZA, and (D) with 50 µg/mL bortezomib. The separation was performed via UPLC-PDA (235 nm), using a C18 column and a gradient increasing linearly from 10% CH3CN (H2O) at 0.0 min to 100% at 4.5 min, held at 100% for an additional 0.5 min. All extracts were solubilized at 0.2 mg/mL and injected at a volume of 6 µL.
Of the three inhibitors, bortezomib was chosen for further experiments for two reasons. First, the number of additional metabolites in preliminary experiments was greater when the fungus was grown with bortezomib than with SAHA. Secondly, while secondary metabolite production by bortezomib-dosed cultures was similar to that observed with 5-AZA-dosed cultures, bortezomib had not been previously studied for this application.
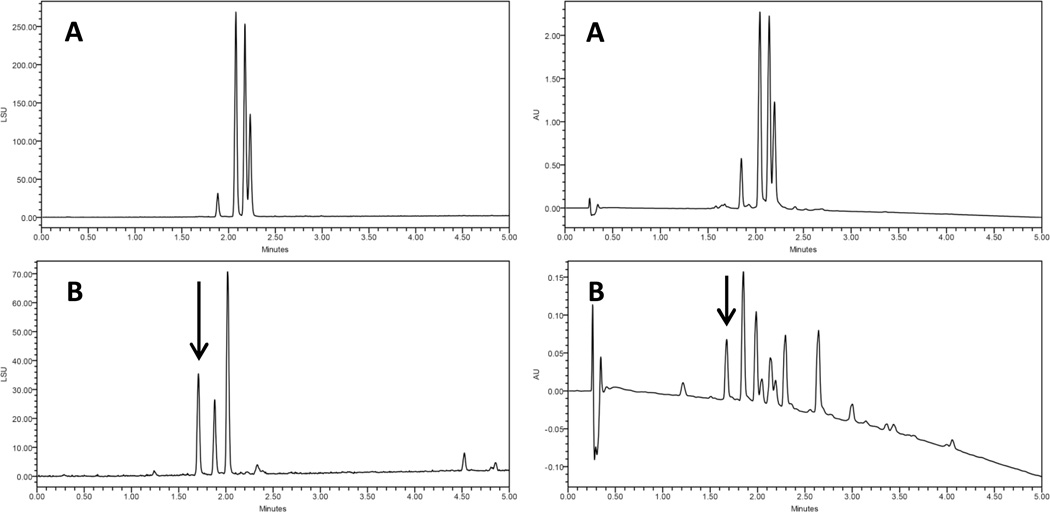
Triplicate growths of the fungus in PDB containing 0, 25, 50, 75, 100, and 125 µg/mL bortezomib were extracted and their organic extracts compared via gradient UPLC-PDA-ELSD (Fig. S3, Supplementary Information). Relative levels of the resorcylic acid lactones18 and several fatty acids varied between replicate growths. In addition to those expected compounds, a new peak was evident in the chromatograms of the bortezomib-dosed growths (arrow, Fig. 2). Growths containing this peak were combined; the compound was isolated via preparative high pressure liquid chromatography (HPLC), and the structure analyzed.
Figure 2.
Comparison of control (A) and dosed (B) growths of MSX 63935, grown in potato dextrose broth with 75 µg/mL bortezomib. The arrow indicates a peak that does not appear in extracts of control growths. The peaks in (B) after 4.5 minutes were attributed to fatty acids. The separation was performed via UPLC-PDA-ELSD, using a C18 column and a gradient increasing linearly from 10% CH3CN (H2O) at 0.0 min to 100% at 4.5 min, held at 100% for an additional 0.5 min. ELSD data are shown on the left and PDA (235 nm) data on the right.
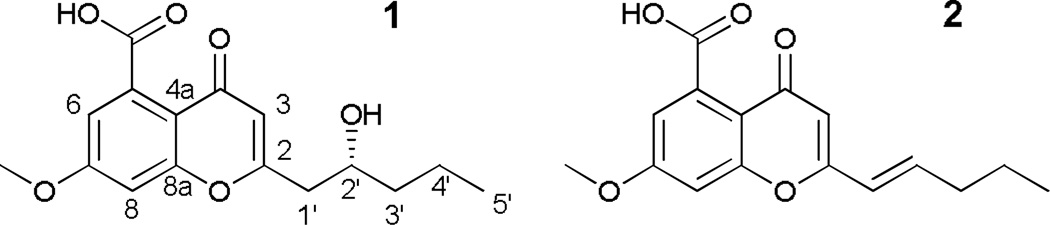
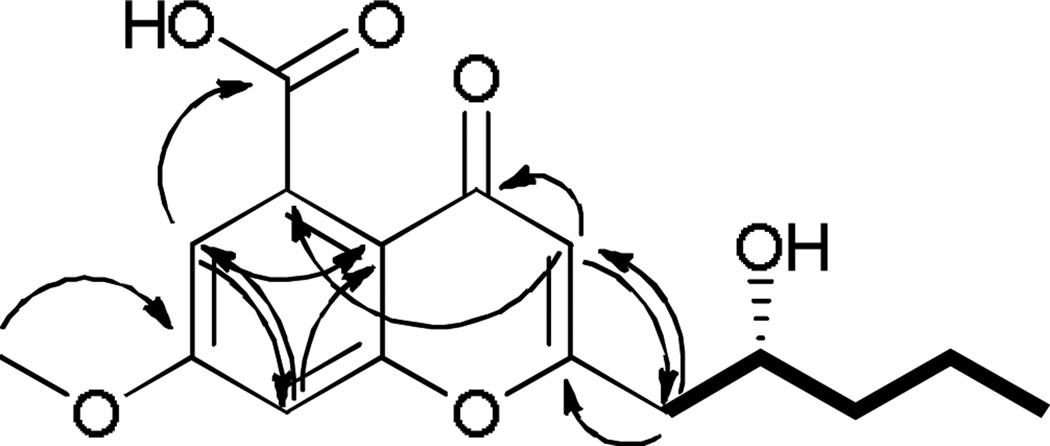
Compound 1 (Fig. 3, Table 1) was obtained as a white powder. Using HRESIMS a molecular ion was measured at 307.1165 [M+H]+ (calcd for C16H19O6, 307.1176), indicating a molecular formula of C16H18O6 with eight degrees of unsaturation. The 1H NMR spectrum displayed two aromatic m-coupled protons at δH 7.20 (H-6) and δH 7.22 (H-8), a singlet at δH 6.28 (H-3), a methoxy signal at δH 3.96, and signals for six protons corresponding to the hydroxylated aliphatic side chain at δH 4.06 (H-2′), 2.88 and 2.70 (H-1′a/b), 1.56 (H-3′), 1.56 and 1.45 (H-4′), and 0.98 (H-5′). The structure of this side chain was assigned based on examination of COSY data (Fig. 4). HMBC correlations (Fig. 4) between H-3 and carbons C-2 (δ 167.0) and C-1′ (δC 43.3), as well as between the diastereotopic H-1′ protons and the C-2 and C-3 (δC 112.0) carbons, determined the C-2/C-1′ bond. Further analysis of the HMBC and HSQC data and a search of the literature suggested the chromone skeleton, which accounted for seven degrees of unsaturation.21–23 An HMBC correlation between the methoxy singlet and the C-7 carbon (δC 165.4) established its connectivity to the aromatic ring. The mass indicated a carboxylic acid moiety as the remaining substituent, accounting for the last degree of unsaturation; this was corroborated by a correlation between the H-6 proton and the carboxylic acid carbon (δC 171.9). A four-bond correlation between the H-3 proton and C-5 (δC 136.8), established the position of the carboxylic acid moiety. The absolute configuration was assigned using a modified Mosher’s ester analysis (Fig. 5).24
Figure 3.
Compound 1 was isolated from bortezomib-dosed growths of MSX 63935, and 2 was isolated from a degraded solution of 1.
Table 1.
NMR Data (400 MHz, MeOH-d4) of Compounds 1 and 2.
| 1 | 2 | |||||
|---|---|---|---|---|---|---|
| position | δH, mult. (J in Hz) | δC, type | HMBCa | δH, mult. (J in Hz) | δC, type | HMBCa |
| 2 | 167.0, C | 161.8, C | ||||
| 3 | 6.28, s | 112.0, CH | 2, 4, 4a, 5, 1′ | 6.21, s | 107.3, CH | 2, 4, 4a, 5, 1′ |
| 4 | 179.4, C | 177.6, Cc | ||||
| 4a | 114.9, C | 112.9, C | ||||
| 5 | 136.8, C | 138.5, Cc | ||||
| 6 | 7.20, d (2.3) | 103.3, CH | 4a, 8, 5-COOH | 7.13, d (2.3) | 100.8, CH | 4a, 8, 5-COOH |
| 7 | 165.4, C | 163.4, C | ||||
| 8 | 7.22, d (2.3) | 116.8, CH | 4a, 6, 7, 8a | 7.20, d (2.3) | 114.0, CH | 4a, 7, 8a |
| 8a | 160.6, C | 157.9, C | ||||
| 1′a | 2.88, dd (4.1, 14.7) | 043.3, CH2 | 2, 3, 2′, 3′ | 6.33, dd (1.4, 15.6) | 121.5, CH | 2, 3′ |
| 1′b | 2.70, dd (9.2, 14.7) | 2, 3, 2′, 3′ | ||||
| 2′ | 4.06, m | 070.0, CH | 6.98, dt (7.3, 15.6) | 141.8, CH | 2, 3′, 4′ | |
| 3′ | 1.56, mb | 040.8, CH2 | 1′, 2′, 4′, 5′ | 2.31, ddt (1.4, 7.3, 7.3) | 034.0, CH2 | 1′, 2′, 4′, 5′ |
| 4′ | 1.56, mb / 1.45, m | 020.1, CH2 | 2′, 3′, 5′ | 1.58, sextet (7.3) | 020.9, CH2 | 2′, 3′, 5′ |
| 5′ | 0.98, t (6.9) | 014.5, CH3 | 3′, 4′ | 1.00, t (7.3) | 012.1, CH3 | 3′, 4′ |
| 7-OCH3 | 3.96, s | 057.1, CH3 | 7 | 3.95, s | 054.9, CH3 | 7 |
| 5-COOH | 171.9, C | 171.5, Cc | ||||
HMBC correlations are from the proton(s) listed to the indicated carbons.
Overlapping signals
The quaternary 13C signals for C-4, C-5, and 5-COOH were very weak, and their δC values were corroborated by HMBC data.
Figure 4.
Key HMBC (→) and COSY (▬) correlations of compound1.
Figure 5.
ΔδH values [Δδ(in ppm) = δS – δR] obtained for (R)- and (S)-MTPA esters (1a and 1b, respectively) in pyridine-d5.
A compound with the same planar structure as 1 was isolated from the plant Stratiotes aloides in 2009.22 Those authors did not assign the absolute configuration, possibly due to paucity of material (less than 200 µg).22Due to signal overlap of the DMSO-d6 solvent, the data reported here were obtained in MeOH-d4, though 1H and 13C NMR spectra were also acquired in DMSO-d6. Key differences between NMR signals in DMSO-d6 were noted at positions H-3, H-6, H-8, and C-5 (Table S1, Supplementary Information). However, both the NMR data and the UV maxima25 at 233 and 291 supported the chromone skeleton, and the structure of the alkyl side chain was firmly assigned by 2D NMR. Coupling of the H-6 and H-8 protons (J = 2.3) established the meta positions of the aromatic protons, and the strong four-bond H-3 to C-5 correlation confirmed the position of the carboxylic acid.
During an unsuccessful attempt to crystalize 1, the molecule degraded in solution, yielding several analogues, one in quantities sufficient to assign a structure. Compound 2 (Fig. 3), which was new, represented the dehydro analogue of 1, which was supported by an 18 amu difference in the HRMS data and an increase by one in the index of hydrogen deficiency. COSY data, especially a correlation between H-2′ (δH 6.98) and H-3′ (δH 2.31), helped assign the alkene side chain. The large vicinal coupling constant between H-2′ and H-3′ (J = 15.1) established the trans configuration.
Both 1 and 2 were tested for cytotoxicity against human melanoma and colon cancer cells and were found to be inactive (i.e., IC50 values > 20 µM).
To our knowledge, this is the first use of a proteasome inhibitor in epigenetic modification studies to induce additional secondary metabolites from microorganisms, and may inspire new methods for the up-regulation of silent metabolite genes.
Experimental Section
Culture Methods
Mycosynthetix fungal strain 63935 was isolated in 1992 by Dr. Barry Katz of MYCOsearch from leaf litter. The culture was stored on a malt extract slant and was transferred periodically. A fresh culture was grown on malt extract agar in a 10 cm diameter Petri plate at 22 °C for 20 d, until the cultures were approximately 2.5 cm in diameter. The cultures were cut into small squares, which were used to inoculate 250-mL Erlenmeyer flasks containing 50 mL of potato dextrose broth (PDB). At the time of inoculation, 0.5 mL aliquots of DMSO-dissolved bortezomib was added in triplicate to 15 flasks, resulting in final concentrations in the liquid media of 25, 50, 75, 100, and 125 µg/mL bortezomib. Three flasks were treated with 0.5 mL DMSO (vehicle control), and three flasks were untreated (negative control). The cultures were grown with shaking (100 rpm) at 22 °C for 14 d.
Extraction and Isolation
The cultures were shaken overnight with 60 mL of 1:1 CHCl3:MeOH and filtered the next day. To the filtrate was added 90 mL of CHCl3 and 100 mL of deionized H2O to produce a 4:1:5 mixture of CHCl3:MeOH:H2O (including the 50 mL of H2O from the liquid media). This mixture was stirred for 1 h, then transferred to a separatory funnel. The organic portions were dried in vacuo, then shaken with a 1:1:2 mixture of CH3CN:MeOH:hexanes. The CH3CN:MeOH layers were separated, and all extracts were dried in vacuo, transferred to scintillation vials, then dried completely under air. The extracts of the cultures exhibiting the presence of the new metabolite (Fig. 2) were combined (90.85 mg) and subjected to preparative C18 HPLC using an isocratic 40:60 CH3CN:H2O method over 25 min, followed by a 100% CH3CN column wash (10 min). Compound 1 eluted at ~14 min (6.32 mg).
Instrumentation
Optical rotation was measured with a Rudolph Autopol III polarimeter; UV spectra were obtained using a Varian Cary 3 UV-vis spectrophotometer. NMR experiments were performed on a JEOL ECS 400 MHz NMR equipped with a high sensitivity JEOL Royal probe and an autosampler. Selected samples were measured using a Bruker Avance-III 600 MHz NMR. HRESIMS data were acquired using a Thermo LTQ Orbitrap XL mass spectrometer equipped with an electrospray ionization source. UPLC was performed using a Waters Acquity system, equipped with a photodiode array detector (PDA) and evaporative light scattering detector (ELSD) with data collected and analyzed using Empower software. A Waters BEH C18 column (1.7 µm; 50 × 2.1 mm) was used with a 0.6 mL/min flow rate. Preparative HPLC was performed using a Varian Prostar HPLC system equipped with ProStar 210 pumps and a Prostar 335 PDA, with data collected and analyzed using Galaxie Chromatography Workstation software (version 1.9.3.2). A YMC ODS-A C18 column (5 µm; 250 × 20 mm) was used with a 9.45 mL/min flow rate.
(R)-2-(2-hydroxypentyl)-5-carboxy-7-methoxychromone (1). white powder; [α]20D −26 (c 0.179, MeOH); UV(MeOH) λmax (log ε) 233 (3.01), 291 (2.85); 1H NMR (400 MHz, MeOH-d4) see Table 1 and Fig. S4; 13C NMR (400 MHz, MeOH-d4) see Table 1 and Fig. S5; HRESIMS m/z 307.1165 (calcd for C16H19O6, 307.1176) [M+H]+.
(E)-2-(pent-1-en-1-yl)-5-carboxy-7-methoxychromone (2). white powder; UV(MeOH) λmax (log ε) 256 (3.28), 317 (3.31); 1H NMR (400 MHz, MeOH-d4) see Table 1 and Fig. S6; 13C NMR (400 MHz, MeOH-d4) see Table 1 and Fig. S7; HRESIMS m/z 289.1069 (calcd for C16H17O5, 289.1071) [M+H]+.
Preparation of (R)- and (S)-MTPA ester derivatives of (1)
To 1.00 mg of compound 1, 1000 µL of pyridine-d5 were added; from this, 500 µL were transferred to two separate NMR tubes. To initiate the reaction, 10 µL of S-(+)-α-methoxy-α-(trifluoromethyl)phenylacetyl (MTPA) chloride were added to one NMR tube and shaken. The reaction was monitored immediately by 1H NMR at the following time points: 0, 15, and 30 min. At 30 min, an additional 10 µL of S-MTPA chloride were added and the tube shaken. The reaction was monitored further at 40, 45, and 55 min, at which point it was determined to be complete, yielding the mono (R)-MTPA ester derivative (1a) of 1. To the second NMR tube was added 20 µL of the (R)-MTPA chloride; the reaction was shaken, then monitored immediately by 1H NMR at the following time points: 0, 15, 30, and 40 min. The reaction was found to be complete within 40 min, yielding the mono (S)-MTPA ester derivative (1b) of 1. 1H NMR (500 MHz, pyridine-d5) of 1a: δH0.80 (3H, t, H-5′), 1.28 (2H, m, H-4′), 1.71 (2H, m, H-3′), 3.14 (2H, m, H-1′), 5.78 (1H, m, H-2′), 6.62 (1H, s, H-3). The signals from H-6, H-8, and the methoxy group were obscured by solvent and reagent peaks. 1H NMR (500 MHz, pyridine-d5) of 1b: δH0.87 (3H, t, H-5′), 1.43 (2H, m, H-4′), 1.77 (2H, m, H-3′), 5.77 (1H, m, H-2′), 6.45 (1H, s, H-3). The signals from H-1′, H-6, H-8, and the methoxy group were obscured by solvent and reagent peaks.
Cytotoxicity Assays
Human melanoma cancer cells designated MDA-MB-435 and human colon cancers designated HT-29 were purchased from the American Type Culture Collection (Manassas, VA). The cell lines were propagated at 37 °C in 5% CO2 in RPMI 1640 medium supplemented with fetal bovine serum (10%), penicillin (100 units/mL), and streptomycin (100 µg/mL). Cells in log phase growth were harvested by trypsinization followed by two washings to remove all traces of enzyme. A total of 5,000 cells were seeded per well of a 96-well clear, flat-bottom plate and incubated overnight (37 °C in 5% CO2). Samples dissolved in DMSO were then diluted and added to the appropriate wells (concentrations: 20, 4, 0.8, 0.16, and 0.032 µM; total volume: 100µL; DMSO: 0.5%). The cells were incubated in the presence of test substance for 72 h at 37°C and evaluated for viability with a commercial absorbance assay (CellTiter 96®AQueousOne Solution Cell Proliferation Assay, Promega Corp, Madison, WI) that measured viable cells. IC50 values were expressed by µM relative to the solvent (DMSO) control. The positive control was vinblastine tested at 1 nM in MDA-MB-435 cells, yielding 16% viable cells or 10nM in HT-29 cells, which yielded 39% viable cells. The purity of the compounds was established via UPLC prior to cytotoxicity testing (Fig. S8, Supplementary Information).
Supplementary Material
Acknowledgement
This research was supported by program project Grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. Selected NMR data were obtained by Dr. Kevin Knagge at the David H. Murdock Research Institute Kannapolis, NC, USA. The authors thank T.N. Graf at the University of North Carolina at Greensboro for technical assistance.
References
- 1.Keller NP, Turner G, Bennett JW. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 2.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 3.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latge JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Cordoba S, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 4.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 5.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 6.Scherlach K, Hertweck C. Org. Biomol. Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 7.Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Waegele H, Cichewicz RH. J. Ind. Microbiol. Biot. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- 8.Vervoort HC, Draskovic M, Crews P. Org. Lett. 2011;13:410–413. doi: 10.1021/ol1027199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Eukaryot. Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. Org. Biomol. Chem. 2009;7:435–438. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]
- 11.Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Waegele H, Cichewicz RH. J. Ind. Microbiol. Biotechnol. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- 12.Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Org. Biomol. Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 13.Yakasai AA, Davison J, Wasil Z, Halo LM, Butts CP, Lazarus CM, Bailey AM, Simpson TJ, Cox RJ. J. Am. Chem. Soc. 2011;133:10990–10998. doi: 10.1021/ja204200x. [DOI] [PubMed] [Google Scholar]
- 14.Beau J, Mahid N, Burda WN, Harrington L, Shaw LN, Mutka T, Kyle DE, Barisic B, van OA, Baker BJ. Mar. Drugs. 2012;10:762–774. doi: 10.3390/md10040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XR, Sena JG, Hoover AR, King JB, Ellis TK, Powell DR, Cichewicz RH. J. Nat. Prod. 2010;73:942–948. doi: 10.1021/np100142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kritsky MS, Filippovich SY, Afanasieva TP, Bachurina GP, Russo VEA. Appl. Biochem. Microbiol. 2001;37:243–247. [Google Scholar]
- 17.Chung Y-M, Wei C-K, Chuang D-W, El-Shazly M, Hsieh C-T, Asai T, Oshima Y, Hsieh T-J, Hwang T-L, Wu Y-C, Chang F-R. Bioorg. Med. Chem. 2013;21:3866–3872. doi: 10.1016/j.bmc.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carcache dBEJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmermann J, Erdmann D, Lalande I, Grossenbacher R, Noorani M, Furst P. Oncogene. 2000;19:2913–2920. doi: 10.1038/sj.onc.1203606. [DOI] [PubMed] [Google Scholar]
- 20.Szutorisz H, Georgiou A, Tora L, Dillon N. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 21.Cagide F, Borges F, Gomes LR, Low JN. J. Mol. Struct. 2013;1049:125–131. [Google Scholar]
- 22.Conrad J, Foerster-Fromme B, Constantin M-A, Ondrus V, Mika S, Mert-Balci F, Klaiber I, Pfannstiel J, Moeller W, Rosner H, Foerster-Fromme K, Beifuss U. J. Nat. Prod. 2009;72:835–840. doi: 10.1021/np800769g. [DOI] [PubMed] [Google Scholar]
- 23.Sun R-R, Miao F-P, Zhang J, Wang G, Yin X-L, Ji N-Y. Magn. Reson. Chem. 2013;51:65–68. doi: 10.1002/mrc.3903. [DOI] [PubMed] [Google Scholar]
- 24.Hoye TR, Jeffrey CS, Shao F. Nat. Protoc. 2007;2:2451–2458. doi: 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- 25.Wiley PF. J. Am. Chem. Soc. 1952;74:4326–4328. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.