Abstract
The monoclonal antibody S9.6 binds DNA-RNA hybrids with high affinity, making it useful in research and diagnostic applications, such as in microarrays and in the detection of R-loops. A single-chain variable fragment (scFv) of S9.6 was produced, and its affinities for various synthetic nucleic acid hybrids were measured by surface plasmon resonance (SPR). S9.6 exhibits dissociation constants of approximately 0.6 nM for DNA-RNA and, surprisingly, 2.7 nM for RNA-RNA hybrids that are AU-rich. The affinity of the S9.6 scFv did not appear to be strongly influenced by various buffer conditions or by ionic strength below 500 mM NaCl. The smallest epitope that was strongly bound by the S9.6 scFv contained six base pairs of DNA-RNA hybrid.
Keywords: Monoclonal antibody, scFv, antibody sequence, DNA-RNA hybrids
Introduction
Monoclonal antibody S9.6 was originally generated in mice by immunization with a ΦX174 bacteriophage-derived synthetic DNA-RNA antigen. It was characterized as having high specificity and affinity for DNA-RNA hybrids [Boguslawski et al. 1986]. Initially, S9.6 was used as an enzyme-labeled reagent in solution-based nucleic acid hybridization assays [Yehle et al. 1987; Miller et al. 1988; Casebolt and Stephensen 1992]. A study of small Escherichia coli RNAs by DNA microarray techniques showed the utility of S9.6 for detecting DNA-RNA hybridization without needing to amplify the RNA samples [Hu et al. 2006]. An extensive transcriptome analysis in Schizosaccharomyces pombe by microarrays depended on S9.6 for hybrid detection, which eliminated the need for, and potential bias introduced by, reverse transcription and PCR amplification [Hu et al. 2006; Dutrow et al. 2008]. More recently, S9.6 has been found to be useful in identifying and localizing DNA-RNA hybrids in normal and stressed eukaryotic cells. For example, S9.6 was used to show the presence of extensive DNA-RNA hybrid intermediates during replication of mitochondrial DNA [Pohjoismaki et al. 2010]. Several investigators have used S9.6 to detect and image R-loops, which are regions of genomic DNA that are annealed to RNA and that have been linked to genomic instability and the prevention of silencing of CpG island promoters by CpG methylation [Szekvolgyi et al. 2007; El Hage et al. 2010; Gan et al. 2011; Skourti-Stathaki et al. 2011; Ginno et al. 2012]. Additionally, several groups have used S9.6 detection of DNA-RNA hybrids as a system for the development of biosensor systems [Sipova et al. 2010; Qavi et al. 2011]. Polyclonal DNA-RNA hybrid specific antibodies are a key component of human papillomavirus (HPV) molecular diagnostics developed by Digene and now marketed by Qiagen. Because of the renewed use of mAb S9.6 as a research tool and its potential use in diagnostic reagents, we sought to fully characterize S9.6. In this work, we cloned and sequenced the S9.6 cDNA, and then produced a monovalent scFv S9.6 construct through expression in bacteria. The interactions of the S9.6 scFv with nucleic acid hybrids of various lengths and compositions were measured in a variety of conditions.
Materials and Methods
Synthesis of nucleic acid hybrids
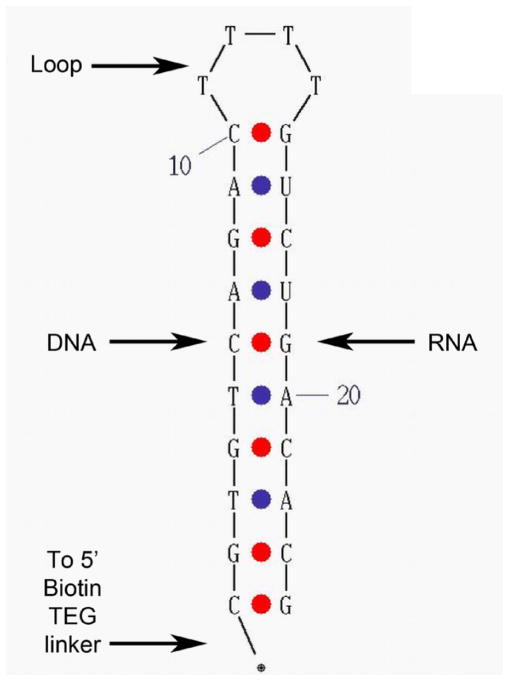
DNA-DNA hybrids were synthesized by the FDA Facility for Biotechnology Resources (Bethesda, MD). RNA-RNA and DNA-RNA hybrids were synthesized by Integrated DNA Technologies (Coralville, IA). All hybrids were synthesized with a 5′ biotin-tetra-ethylene glycol (TEG) linker. Oligonucleotide sequences and descriptions are provided in Table 1. The image in Figure 1 was generated using the UNAFold software on the Integrated DNA Technologies website (http://www.idtdna.com/UNAFold).
Table 1.
Descriptions and sequences of nucleic acid hybrids used in SPR
| Hairpin Length (bp) | Content | %GCa | Hybrid nameb | Oligonucleotide Sequencec,d |
|---|---|---|---|---|
| 0 | Equal Mix | 50 | 30ssD | CGGTGTGGTCGCTGTAATCAGAAAACGTGT |
| 23 | AT-rich | 17.4 | 23ATDD | AATTACATTGATAGAATTATTAGTTTTCTAATAATTCTATCAATGTAATT |
| 23 | GC-rich | 82.6 | 23GCDD | GGCGGTCCCAGCGAGGCCGCCGATTTTTCGGCGGCCTCGCTGGGACCGCC |
| 23 | AU-rich | 17.4 | 23AURR | rArArUrUrArCrArUrUrGrArUrArGrArArUrUrArUrUrArGrUrUrUrUrCrUrArArUrArArUrUrCrUrArUrCrArArUrGrUrArArUrU |
| 23 | GC-rich | 82.6 | 23GCRR | rGrGrCrGrGrUrCrCrCrArGrCrGrArGrGrCrCrGrCrCrGrArUrUrUrUrUrCrGrGrCrGrGrCrCrUrCrGrCrUrGrGrGrArCrCrGrCrC |
| 23 | AT-rich | 17.4 | 23ATDR | AATTACATTGATAGAATTATTAGTTTTrCrUrArArUrArArUrUrCrUrArUrCrArArUrGrUrArArUrU |
| 23 | GC-rich | 82.6 | 23GCDR | GGCGGTCCCAGCGAGGCCGCCGATTTTrUrCrGrGrCrGrGrCrCrUrCrGrCrUrGrGrGrArCrCrGrCrC |
| 23 | Equal Mix | 52.2 | 23MDR | CGGTGTGGTCGCTGTAATCAGAATTTTrUrUrCrUrGrArUrUrArCrArGrCrGrArCrCrArCrArCrCrG |
| 15 | Equal Mix | 53.3 | 15MDR | CGGTGTGAATCAGACTTTTrGrUrCrUrGrArUrUrCrArCrArCrCrG |
| 10 | Equal Mix | 60 | 10MDR | CGTGTCAGACTTTTrGrUrCrUrGrArCrArCrG |
| 8 | Equal Mix | 50 | 8MDR | GAGAACTGTTTTrCrArGrUrUrCrUrC |
| 6 | Equal Mix | 50 | 6MDR | GAACTGTTTTrCrArGrUrUrC |
| 4 | Equal Mix | 50 | 4MDR | GACTTTTTrArGrUrC |
GC content is calculated for the hybrid portion of the oligonucleotides, which does not include the four thymidine bases encompassing the turn. “Equal mix” denotes 40-60% GC content.
For example, “30ssD” is an abbreviation of “30-nucleotide single stranded DNA”.
AT = AT-rich, GC = GC-rich, DD = DNA-DNA hairpin, RR = RNA-RNA hairpin, DR = DNA-RNA hairpin, M=equal mix of AT and GC.
Thymidines and uracils (T, rU) in bold and underline do not participate in base pairing.
5′(Biotin-TEG) linker is at the 5′ end of all sequences
Figure 1.
Schematic of 10MDR, one of the 10-base nucleic acid hybrids used in this study. Ten bases of DNA-RNA hybrid extend from the base of the loop to the 5′ and 3′ ends of the oligonucleotide. Four thymidines, that remain single-stranded, form the hairpin loop. Other hybrids are similar in design, but differ in hybrid length, in percent GC content, and whether they are DNA-DNA, RNA-RNA, or DNA-RNA.
Isolation and expression of the S9.6 IgG and scFv
The hybridoma that produces the S9.6 IgG, HB-8730, was purchased from the ATCC (Manassas, VA); this cell line is not being distributed by the ATCC at this time. S9.6 hybridoma cells were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM Glutamax, 1 mM sodium pyruvate, and 50 μg/mL gentamycin (Life Technologies, Carlsbad, CA). S9.6 IgG was purified from culture supernatants using HiTrap Protein G columns (GE Healthcare Life Sciences, Piscataway, NJ) as previously described [Hu et al. 2006]. For cloning of the antibody genes and construction of an scFv expression vector, hybridoma cells were grown to 80–90% confluence, then were centrifuged at 1000 rpm for 10 min, supernatant was decanted, and the cell pellet was resuspended in RNAlater (Life Technologies) and placed at 4°C overnight. Total RNA was isolated using TRIzol (Life Technologies), and reverse transcription was performed using avian myeloblastosis virus reverse transcriptase (AMV-RT) with an anchored oligo(dT)20 primer from Integrated DNA Technologies. The VL and VH genes were amplified and fused into the scFv format in the vector pAK400 as previously described [Krebber et al. 1997]. The two initial PCR reactions to amplify the variable genes used a pool of 23 degenerate primers for amplification of the VL gene, and another 23 degenerate primer pool was used to amplify the VH gene in a separate reaction. The third, subsequent PCR reaction fused the two variable genes together in the order of VL-linker-VH into an scFv construct. The primers and the linker sequence were used as previously described [Krebber et al. 1997]. The ligation mixture was electroporated into E. coli strain XL1-Blue, and 48 individual colonies were picked and inoculated into 200 μL/well Luria Broth (LB) with 2% glucose (w/v) and 30 μg/mL chloramphenicol in a 96-well plate, placed at 30°C, and shaken overnight. The next day, a fresh plate was prepared with a 1:10 inoculation of the overnight cultures into the same medium and it was shaken at 30°C for 3 h. The plate was then centrifuged, and the bacterial pellets were resuspended in 200 μL of LB with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 30 μg/mL chloramphenicol and shaken at 25°C overnight. After centrifugation the following morning, bacterial pellets were resuspended in 20% BugBuster HT (EMD Millipore, Billerica, MA) in PBS and shaken for 1 h at 25°C. Cell debris was pelleted by centrifugation, and the scFv-containing supernatant was transferred to a new plate and stored at 4°C until screening, as described below. A bacterial clone selected as described below was used for large-scale S9.6 scFv protein purification by immobilized metal affinity chromatography and size exclusion chromatography, as described previously [Radjainia et al. 2010]. The S9.6 heavy and light chains of the original S9.6 hybridoma cells were also independently determined by sequencing of the hybridoma RNA (LakePharma, Belmont, CA). Heavy and light chains of the S9.6 Fab, which were produced by papain cleavage of the IgG, were subjected to N-terminal sequencing by Edman degradation (Research Technologies Branch, NIAID/NIH).
Surface plasmon resonance analysis
Surface plasmon resonance (SPR) experiments were performed using a ProteOn XPR36 instrument (Bio-Rad, Hercules, CA). The standard running buffer used in experiments and analysis was 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, pH 7.4 (1x HBS-T). Other buffers used to test the effect of pH are described in Table 3. Screening of E. coli cell lysates from potential S9.6 scFv-producing clones was accomplished by a 60 sec injection of the supernatant at 50 μL/min across surfaces of a neutravidin SPR chip (NLC chip, Bio-Rad, Hercules, CA) coated with oligonucleotide 23MDR (Table 1) immobilized at four different densities, and dissociation was measured over a 300-second period. Single stranded DNA (30ssD, Table 1) and a neutravidin surface lacking bound oligonucleotides were negative controls. Regeneration was accomplished with a 30-sec injection of 1 M NaCl at 50 μL/min.
Table 3.
Running buffers used to test the effects of differing ionic strength, divalent cations, and pH on binding in SPR.
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, pH 7.4 -- standard |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, 1 mM EDTA, pH 7.4 |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, 4 mM MgCl2, pH 7.4 |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, 4 mM CaCl2, pH 7.4 |
| 10 mM Hepes, 300 mM NaCl, 0.005% Tween 20, pH 7.4 |
| 10 mM Hepes, 500 mM NaCl, 0.005% Tween 20, pH 7.4 |
| 10 mM Tris HCl, 15 mM MgCl2, 0.005% Tween 20, pH 7.4 (1x Tris-Mg) |
| 1.06 mM KH2PO4, 5.60 mM Na2HPO4, 154 mM NaCl, 0.005% Tween 20, pH 7.4 (1 × PBS-T) |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, pH 6.5 |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, pH 7.0 |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, pH 8.0 |
| 10 mM Hepes, 150 mM NaCl, 0.005% Tween 20, pH 8.5 |
For kinetic analysis of purified S9.6 scFv, all flow channels were first rinsed and equilibrated with running buffer. Each nucleic acid hybrid was separately resuspended in doubly-deionized water and then diluted to a concentration of 6.25 nM in running buffer. These biotinylated hybrids were immobilized at a flow rate of 50 μL/min for 30 sec onto separate vertical lanes of a neutravidin-coated chip (NLC chip, Bio-Rad, Hercules, CA). Purified S9.6 scFv was diluted in running buffer to concentrations of 6.25, 12.5, 25, 50, and 100 nM, and the concentration series and a buffer blank was applied across horizontal lanes at 100 μL/min for 60 sec with a dissociation time of 600 sec, allowing for the simultaneous detection of S9.6 scFv interactions with multiple types of hybrids. Regeneration was accomplished with a 30-sec injection of 1 M NaCl at 50 μL/min, followed by equilibration in running buffer until the baseline stabilized. The neutravidin surface lacking oligonucleotides between each immobilized lane, termed the “interspot,” was used as a control for analysis. ProteOn Manager software was used to measure kinetic parameters using the Langmuir model of interaction.
Results
Production and purification of the S9.6 scFv
After the S9.6 variable regions were placed in the scFv format, it was found that supernatants from 28 of the 48 clones tested showed specific binding to the 23MDR DNA-RNA hybrid. After sequencing the 28 plasmids from the clones, one of them was picked that represented the consensus protein sequence of the samples. The scFv protein sequence encoded by this plasmid is presented in Figure 2 and this plasmid was used for expression, purification, and all further analyses. Large scale expression and purification of the S9.6 scFv from the E. coli strain XL1-Blue resulted in 555 μg of purified protein from 1 L of bacterial culture.
Figure 2.

Sequence of the S9.6 scFv protein displayed with the light and heavy variable domains, numbered by the Kabat scheme [Kabat et al. 1991] above the amino acid sequence (light chain residues 1 to 107 and heavy chain residues 1 to 113). The framework regions (FR), complementarity determining regions (CDR), and linker region (LINKER) are drawn above the numbered sequence.
To confirm the fidelity of this sequence, the S9.6 cDNA from the S9.6 hybridoma was independently sequenced. The light chain cDNA sequence predicted a V domain that was almost identical to the one selected as a consensus, with the N-terminal sequence of DVLMTQTP (bolded and underlined residues indicate differences from the consensus of Figure 2). The two altered amino acids observed in the scFv clone of DIVMTQTP were likely introduced by the degenerate primers. However, two different heavy chain V domains were detected. These heavy chain variants showed differences only in framework region 1 (FR1) and were identical from amino acid position 15 onward. The first heavy chain variant sequence began with EVQLQQSGPELVKP, and thus differs from the scFv consensus sequence only at the E at the N-terminus. The corresponding N-terminal Q in the scFv sequence was likely introduced by the degenerate primers used during the initial PCR amplification step while constructing the scFv. The second heavy chain variant sequence began with QVHLQQSGPEMVRL. The variant amino acid residues at positions 3, 11, 13, and 14 in this sequence occur at low frequencies in these positions in all antibodies, as determined by reference to the Abysis database (http://www.bioinf.org.uk/abysis/). In contrast, the residues present at the same positions in the first heavy chain variant sequence are the most common at each location. Subsequent N-terminal sequencing of the S9.6 variable heavy and light chains revealed that only the first heavy chain sequence, EVQLQQSGPELVKP, was produced by the hybridoma. Therefore, the scFv and IgG VH sequences are identical except for the N-terminal amino acid. The N-terminal sequence of the VL domain was confirmed as DVLMTQTP, which differs from the scFv sequence at positions 2 and 3, as mentioned earlier.
S9.6 scFv specificity and affinity
Nucleic acid hybrids of varying sizes and sequence were synthesized for use in characterizing the binding of the S9.6 scFv (Table 1 and Figure 1). Each hybrid consisted of a single strand of deoxyribo- and ribo- nucleotides with a biotin residue at the 5′ end. Four thymidines (uracils in RNA-RNA) were located between the complementary sequences so that they formed a loop of non-pairing nucleotides. The name of each hybrid molecule includes the number of base pairs formed in the hybrid.
SPR was performed between the purified S9.6 scFv and 23-base pair DNA-DNA, RNA-RNA, and DNA-RNA hybrids in standard running buffer (1x HBS-T). All three types of hybrids were synthesized and tested in A-T (A-U) and G-C-rich forms to assess whether nucleotide composition affects affinity. For these two 23-base pair DNA-RNA hybrids, addition of S9.6 scFv in 1x HBS-T generated sensorgrams characteristic of high affinity binding. As expected, neither of the DNA-DNA hybrids nor the GC-rich RNA-RNA hybrid showed binding to the scFv. However, S9.6 scFv bound AU-rich RNA-RNA, although with a weaker affinity than to the DNA-RNA hybrids. When S9.6 IgG, instead of the scFv, was used under the same conditions, it also displayed binding to AU-rich RNA-RNA (data not shown).
The 23-base pair AT-rich DNA-RNA, GC-rich DNA-RNA, and AU-rich RNA-RNA hybrids were further analyzed by SPR to measure S9.6 scFv affinity and to assess the influence of their nucleotide composition. For the interaction of S9.6 scFv with each hybrid, data from three trials were combined to calculate the average association rates (ka), dissociation rates (kd), and the equilibrium dissociation constants (KD). KD values for the AT-rich, GC-rich, and mixed 23-base pair DNA-RNA hybrids were 0.6 ± 0.1 nM, 0.8 ± 0.2 nM, and 0.47 ± 0.08 nM, respectively (Table 2). The KD for AU-rich RNA-RNA was 2.7 ± 0.4 nM.
Table 2.
Kinetic-Langmuir approximations of the affinity of S9.6 scFv binding to hybrids obtained via SPR.
| Nucleic Acid Hybrid | ka (× 106 1/M·s)a | kd (× 10−3 1/s) | KD (× 10−9 M, nM) |
|---|---|---|---|
| 23AURR | 0.44 ± 0.03 | 1.19 ± 0.07 | 2.7 ± 0.4 |
| 23ATDR | 2.2 ± 0.3 | 1.2 ± 0.4 | 0.6 ± 0.1 |
| 23GCDR | 1.3 ± 0.3 | 1.0 ± 0.1 | 0.8 ± 0.2 |
| 23MDR | 3.22 ± 0.06 | 1.5 ± 0.3 | 0.47 ± 0.08 |
| 15MDR | 2.6 ± 0.3 | 1.4 ± 0.2 | 0.54 ± 0.02 |
| 10MDR | 3.2 ± 0.1 | 1.6 ± 0.3 | 0.50 ± 0.06 |
| 8MDR | 3.1 ± 0.4 | 1.6 ± 0.2 | 0.51 ± 0.01 |
| 6MDR | 1.00 ± 0.06 | 1.81 ± 0.04 | 1.82 ± 0.09 |
Averages and standard deviations for all parameters are derived from 3 trials.
S9.6 scFv affinity and epitope length
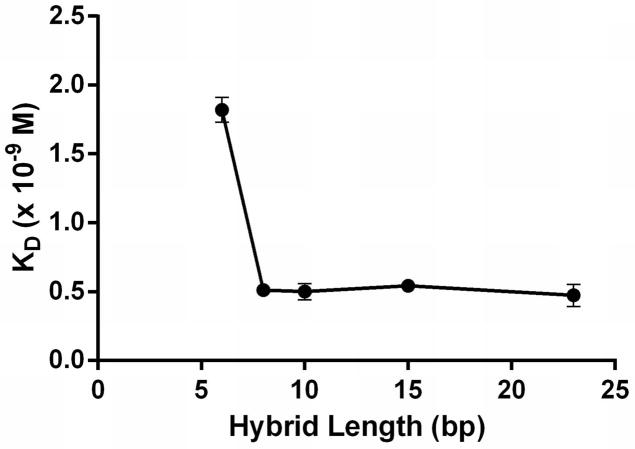
DNA-RNA hybrids of six different lengths were tested via SPR to determine the minimum size of the epitope that S9.6 would bind. The S9.6 scFv displayed similar affinities to hybrids having 8, 10, 15 and 23 base pairs, with affinities averaging 0.5 nM (Table 2 and Figure 3), consistent with those noted above for the hybrids having 23 base pairs. The affinity decreases to 1.9 nM for a hybrid of six base pairs, while no binding could be detected for the four base pair hybrid (Table 2, Figure 3).
Figure 3.
Effect of DNA-RNA hybrid length on S9.6 scFv affinity as determined by SPR in 1x HBS-T buffer. The smallest detectable hybrid had six base pairs (6MDR in Table 1). A four-base pair hybrid was not detectable (4MDR in Table 1). Each point represents the average of three measurements. Error bars denote standard deviation.
Affinity in various ionic, pH, and buffer conditions
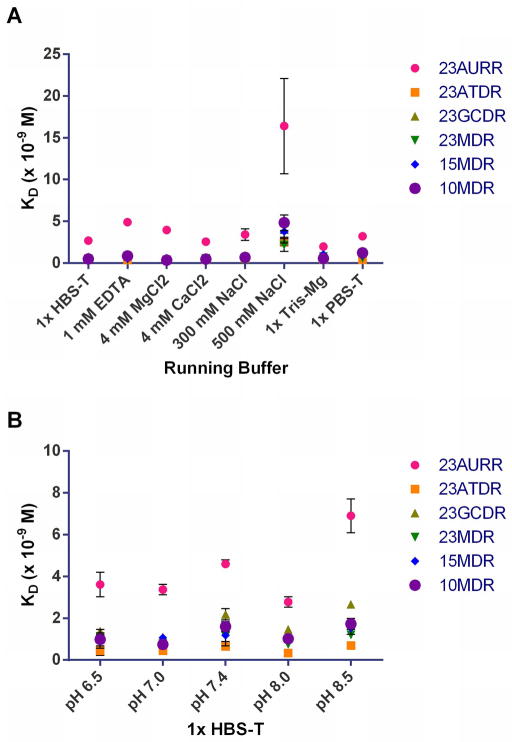
Several other running buffers were tested to characterize solvent effects on the affinity of S9.6 scFv for nucleic acid hybrids. The buffers tested included ionic or pH variations of the standard 1x HBS-T and two other buffer systems at pH 7.4 (Table 3). Nearly all the hybrids that previously had shown binding to S9.6 scFv were included, with the exceptions of the two shortest hybrids. In general, there was minimal variation in KD for the DNA-RNA hybrids between buffers prepared at pH 7.4 and slightly more variation across different levels of pH. The affinities were significantly weaker for the AU-rich RNA-RNA hybrid across all conditions relative to DNA-RNA hybrids (Figure 4A and B). All samples resulted in higher KD values (weaker binding) in 1x HBS-T containing 500 mM NaCl, which was expected in a higher salt environment.
Figure 4.
Comparison of S9.6 scFv affinities under several conditions obtained via SPR. (A) Binding between S9.6 scFv and nucleic acid hybrids was measured in ionic strength variations of 1x HBS-T buffer, in the presence of Mg/Ca, in Tris-Mg, and 1x PBS-T. The leftmost lane shows the binding in the standard 1x HBS-T buffer. Note that the RNA-RNA hybrid (23AURR) displayed lower affinity binding under every condition when compared to the DNA-RNA hybrids. (B) Binding interactions were tested in individual experiments with standard 1x HBS-T prepared at different levels of pH. Each point represents the average of three measurements. Error bars denote standard deviation.
Discussion
Elucidation of the protein sequence of the S9.6 scFv allows for analysis of this particular antibody in relation to previously characterized antibodies. CDR H3, which is known to significantly affect the specificity of antibodies, is ten amino acids in length in S9.6. This corresponds well with a previous analysis of mouse antibodies, which revealed that the peak length of CDR H3 was eleven residues for DNA binding antibodies [Johnson and Wu 1998]. When comparing the amino acid sequences of the S9.6 scFv against the Kabat database, only two residues are distinctive at their given positions. The lysines located at light chain position 106A and heavy chain position 85 are unusually low in their frequency at these positions (0.5% and 0.4%, respectively) [Martin 1996]. Previous work has shown that although there is considerable variability in the protein sequences of CDR loops, the main chains of the hypervariable loops are found to inhabit a relatively few structural conformations [Chothia and Lesk 1987; Chothia et al. 1989]. The set of conformations for each CDR are referred to as Chothia canonical classes, and the analysis of the CDR loops of S9.6 by online resources provided by Dr. A.C.R. Martin (http://www.bioinf.org.uk/abs/chothia.html) reveal that CDR L2, L3, and H1 each conform to the Chothia canonical class one. CDR L1 is most similar to Chothia canonical class four, except that light chain residue 27B is isoleucine rather than leucine. CDR H2 is most similar to Chothia canonical class two, with heavy chain residues Leu52A, Asp55, and Ser71 differing from the expected residues.
The binding of the S9.6 scFv and IgG to AU-rich RNA-RNA hybrids was an unanticipated result that confirmed a previous report of the direct detection of RNA-RNA hybrids with the S9.6 IgG [Kinney et al. 1989]. This type of cross-reactivity has been reported for the siRNA binding protein, P19, isolated from tombusviruses [Silhavy et al. 2002]. P19 has been determined to bind to RNA-RNA hybrids with low nanomolar affinity [Jin et al. 2010], but it has also been used to detect DNA-RNA hybrids [Gullerova and Proudfoot 2012].
The kinetic measurements presented here confirm that S9.6 displays a strong preference for binding DNA-RNA hybrids, while also showing a weaker affinity for certain RNA-RNA hybrids. The measured KD of about 0.6 nM (or 600 pM) for DNA-RNA hybrids appears quite different from the KD of 12 pM originally reported for the bivalent IgG in a competition ELISA done in solution with the high molecular weight ΦX174 DNA-RNA hybrid [Boguslawski et al. 1986]. However, that assay was subject to complications due to avidity effects and to its different format, making a comparison of the affinities problematic.
The S9.6 scFv appears to interact with a nucleic acid duplex of at least six base pairs, with full occupancy of the antibody binding site occurring only for the duplex having eight base pairs. This can be rationalized structurally, as an A-form DNA-RNA hybrid of six or eight base pairs is 16.9 and 22.5 Å, respectively, in length and distances on a typical scFv molecule that span the entire binding face or only the CDR3 loops, both light and heavy, are on the order of 30 and 18 Å, respectively. Thus, when the DNA-RNA hybrid length drops below the prototypical core size of an scFv, the affinity is reduced. It is likely that S9.6 recognizes the A-form of nucleic acid hybrids, as DNA-RNA and RNA-RNA hybrids are believed to exist predominantly in this conformation. The viral protein P19 similarly cross-reacts with both of these hybrids, and the crystal structure of the P19-double stranded RNA complex reveals that P19 binds to the A-form of double stranded RNA in a sequence-independent manner [Vargason et al. 2003].
Conclusions
We examined the specificity and affinity of the monoclonal antibody S9.6 using its monovalent scFv fragment. Isolation of the S9.6 scFv and SPR binding analyses provided a value for the monovalent affinity of S9.6 in the range of 600 pM, as compared to the originally determined KD of 12 pM. Because RNA-RNA hybrids were a secondary target, with five-fold lower affinity, it may be worthwhile to engineer the S9.6 scFv to exhibit more stringent specificity. The sequence of the S9.6 scFv, as presented here, should aid in that process.
Acknowledgments
The authors thank Dr. Larry Lantz for purification of S9.6 IgG from hybridoma cells. This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
References
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. Journal of immunological methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- Casebolt DB, Stephensen CB. Monoclonal antibody solution hybridization assay for detection of mouse hepatitis virus infection. Journal of clinical microbiology. 1992;30:608–612. doi: 10.1128/jcm.30.3.608-612.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. Journal of molecular biology. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nature genetics. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & development. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & development. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Molecular cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nature structural & molecular biology. 2012;19:1193–1201. doi: 10.1038/nsmb.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic acids research. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cid M, Poole CB, McReynolds LA. Protein mediated miRNA detection and siRNA enrichment using p19. BioTechniques. 2010;48:xvii–xxiii. doi: 10.2144/000113364. [DOI] [PubMed] [Google Scholar]
- Johnson G, Wu TT. Preferred CDRH3 lengths for antibodies with defined specificities. International immunology. 1998;10:1801–1805. doi: 10.1093/intimm/10.12.1801. [DOI] [PubMed] [Google Scholar]
- Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. U.S. Dept. of Health and Human Services, Public Health Service, National Institutes of Health; Bethesda, Md. (Bethesda, 20892): 1991. [Google Scholar]
- Kinney JS, Viscidi RP, Vonderfecht SL, Eiden JJ, Yolken RH. Monoclonal antibody assay for detection of double-stranded RNA and application for detection of group A and non-group A rotaviruses. Journal of clinical microbiology. 1989;27:6–12. doi: 10.1128/jcm.27.1.6-12.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber A, Bornhauser S, Burmester J, Honegger A, Willuda J, Bosshard HR, Pluckthun A. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. Journal of immunological methods. 1997;201:35–55. doi: 10.1016/s0022-1759(96)00208-6. [DOI] [PubMed] [Google Scholar]
- Martin AC. Accessing the Kabat antibody sequence database by computer. Proteins. 1996;25:130–133. doi: 10.1002/(SICI)1097-0134(199605)25:1<130::AID-PROT11>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Miller CA, Patterson WL, Johnson PK, Swartzell CT, Wogoman F, Albarella JP, Carrico RJ. Detection of bacteria by hybridization of rRNA with DNA-latex and immunodetection of hybrids. Journal of clinical microbiology. 1988;26:1271–1276. doi: 10.1128/jcm.26.7.1271-1276.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Holmes JB, Wood SR, Yang MY, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. Journal of molecular biology. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qavi AJ, Kindt JT, Gleeson MA, Bailey RC. Anti-DNA:RNA antibodies and silicon photonic microring resonators: increased sensitivity for multiplexed microRNA detection. Analytical chemistry. 2011;83:5949–5956. doi: 10.1021/ac201340s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radjainia M, Hyun JK, Leysath CE, Leppla SH, Mitra AK. Anthrax toxin-neutralizing antibody reconfigures the protective antigen heptamer into a supercomplex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14070–14074. doi: 10.1073/pnas.1006473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. The EMBO journal. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipova H, Zhang S, Dudley AM, Galas D, Wang K, Homola J. Surface plasmon resonance biosensor for rapid label-free detection of microribonucleic acid at subfemtomole level. Analytical chemistry. 2010;82:10110–10115. doi: 10.1021/ac102131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Molecular cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekvolgyi L, Rakosy Z, Balint BL, Kokai E, Imre L, Vereb G, Bacso Z, Goda K, Varga S, Balazs M, et al. Ribonucleoprotein-masked nicks at 50-kbp intervals in the eukaryotic genomic DNA. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14964–14969. doi: 10.1073/pnas.0702269104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Yehle CO, Patterson WL, Boguslawski SJ, Albarella JP, Yip KF, Carrico RJ. A solution hybridization assay for ribosomal RNA from bacteria using biotinylated DNA probes and enzyme-labeled antibody to DNA:RNA. Molecular and cellular probes. 1987;1:177–193. doi: 10.1016/0890-8508(87)90026-0. [DOI] [PubMed] [Google Scholar]