Abstract
The study of genetics, genes, and chromosomal inheritance was initiated by Thomas Morgan in when the first visible mutations were identified in fruit flies. The field expanded upon the work initiated by Herman Muller in 1926 when he used X-rays to develop the first balancer chromosomes. Today, balancers are still invaluable to maintain mutations and transgenes but the arsenal of tools has expanded vastly and numerous new methods have been developed, many relying on the availability of the genome sequence and transposable elements. Forward genetic screens based on chemical mutagenesis or transposable elements have resulted in the unbiased identification of many novel players involved in processes probed by specific phenotypic assays. Reverse genetic approaches have relied on the availability of a carefully selected set of transposon insertions spread throughout the genome to allow the manipulation of the region in the vicinity of each insertion. Lastly, the ability to transform Drosophila with single copy transgenes using transposons or site-specific integration using the ΦC31 integrase has allowed numerous manipulations, including the ability to create and integrate genomic rescue constructs, generate duplications, RNAi knock-out technology, binary expression systems like the GAL4/UAS system as well as other methods. Here, we will discuss the most useful methodologies to interrogate the fruit fly genome in vivo focusing on chemical mutagenesis, transposons and transgenes. Genome engineering approaches based on nucleases and RNAi technology are discussed in following chapters.
1. Introduction
Drosophila melanogaster is an important model organisms long used to answer questions related to basic biology and human disease (Pandey and Nichols, 2011). The sequencing of its entire genome (Adams et al., 2000) was the foundation for the development of a plethora of recent genetic technologies previously impossible to achieve (Venken and Bellen, 2005; Venken et al., 2011b; Venken and Bellen, 2012; St Johnston D., 2013; Venken and Bellen, 2007). These technologies have dramatically changed the landscape by which we approach biological questions in fruit flies. It is now possible to mutate, modify, and tag virtually every fly gene. Here we will discuss three technological approaches that have been instrumental towards scientific discovery using genome interrogation in the fruit fly. These are chemical mutagenesis, transposons and transgenes.
First, because we anticipate that there will be a renewed interest in unbiased mutagenic approaches, we will cover forward genetic screens using chemical mutagens, i.e. ethylmethane sulfonate (EMS) mutagenesis (Bokel, 2008). EMS screens have been used in numerous forward genetic screens based on a variety of phenotypic assays (St Johnston, 2002). We provide outlines of typical one (F1), two (F2) and three (F3) generation screens and define recent developments that have made these screens more powerful. The section will end with strategies to map the mutant lesions and assign phenotypes to the appropriate gene.
Next, we will summarize transposon technologies. Transposons or transposable elements (TEs), historically dominated by the P element transposon, have been instrumental to the advancement of experimental fly biology (Hummel and Klambt, 2008). To broaden the number of genes that can be manipulated by transposons, TEs with different insertional specificities, such as piggyBac (Handler and Harrell, 1999), Minos (Loukeris et al., 1995), and Tol2 (Urasaki et al., 2008), have been introduced (Bellen et al., 2011). We then discuss the different TE collections generated by individual labs and the Gene Disruption Project (Bellen et al., 2004; Bellen et al., 2011). We expand on recent additions to the latter collection based on the Minos mediated integration cassette, better known as MiMIC (Venken et al., 2011a). We outline how transposons are used in genetic screens and how they are mapped. In this section, we also explain how TEs can be used to generate large deletions with the help of the Flp recombinase (Parks et al., 2004; Ryder et al., 2004; Ryder et al., 2007; Cook et al., 2010a), or modified by recombinase mediated cassette exchange (RMCE) (Bateman et al., 2006) or site-specific integration (Groth et al., 2004; Bischof et al., 2007), both catalyzed by the ΦC31 integrase.
Finally, we discuss transgenic methods based on random transposition or site-specific integration mediated by the ΦC31 integrase at defined attP sites distributed over the entire genome (Groth et al., 2004; Bischof et al., 2007; Venken et al., 2006; Markstein et al., 2008; Szabad et al., 2012). We focus on the latter system since it is highly efficient and dampens position effects (Groth et al., 2004; Bischof et al., 2007). attP sites have been tested and optimized for specific purposes, including genomic rescue of available mutations (Ejsmont et al., 2009; Venken et al., 2006; Venken et al., 2009; Venken et al., 2010; Szabad et al., 2012), genome-wide efforts to generate reagents for RNAi knockdown (Dietzl et al., 2007; Ni et al., 2008; Ni et al., 2009; Ni et al., 2011), interrogation of genomic regulatory elements (Pfeiffer et al., 2008; Jenett et al., 2012; Jory et al., 2012; Manning et al., 2012), and gain-of function analysis through overexpression using different binary activation systems (Bischof et al., 2013; Schertel et al., 2013).
This review will not cover ends-in (Rong and Golic, 2000) or ends-out (Gong and Golic, 2003) gene targeting methods based on in vivo remobilization, nor will it cover custom nuclease-mediated genome editing (i.e. Zn finger, TALEN or CRISPR) (Bibikova et al., 2002; Liu et al., 2012; Gratz et al., 2013). Moreover, RNAi technologies based on double stranded RNA (Kennerdell and Carthew, 2000) or microRNA backbones (i.e. short hairpin RNAi or shRNAi) (Haley et al., 2008) will only briefly be discussed in the context of chemical and transposon mutagenesis. Gene targeting and RNAi strategies will be covered in depth in the following chapters: Genome Engineering Using Nucleases (Beumer and Carroll, 2014), and RNAi Screens (Mohr et al., 2014).
2. Chemical Interrogation
2.1 Ethylmethane sulfonate
Forward genetic screens based on chemically induced mutations are the least biased way to identify new players in many biological processes (St Johnston, 2002). The most commonly used chemical mutagen is ethylmethane sulfonate (EMS) (Lewis and Bacher, 1968; St Johnston, 2002), which produces random DNA mutations through nucleotide substitution (Bokel, 2008). EMS is an alkylating agent that targets guanine and primarily produces point mutations. EMS is most commonly used in flies because it is effective and less toxic than most other chemical mutagens. The dose-response, germ cell sensitivity and effects of sperm storage in females have been well characterized (Ashburner, 1989). In recent years EMS screens have reduced in popularity and this is largely due to the perception that mapping EMS-induced mutations is arduous (Bokel, 2008). Yet when gauged on cost, efficiency, scope, and lack of bias, EMS screens are still the most efficient of all screening technologies. Other advantages of EMS screens are their potential to be saturating and their ability to be performed in nearly any Drosophila lab (St Johnston, 2002).
On the contrary, genetically encoded RNAi is targeted to candidate regions of the mRNA (i.e. biased), and often results in partial knock-down and off-targeting effects as well (Mohr and Perrimon, 2012). Many transposons have inherent insertional specificity that commonly ignore significant portions of genes and genomes (i.e. they are biased), and 80% of the available TE insertions are weak loss of function alleles, possibly due to internal elements that can act as leaky promoters activating gene transcription downstream of the interruptive transposon insertion (Lafave and Sekelsky, 2011) (see also Section 3).
Hence, EMS is the preferred mutagen for true unbiased genotypic probing of the genome and phenotypic screening of your favorite phenotype. EMS is often used at a concentration of 25 mM (Bokel, 2008), though lower concentrations (e.g., 10 mM) result in fewer second site hits (Yamamoto et al., 2012). Hexamethylphosphoramide (HMPA), an alternative chemical mutagen, introduces small deletions via cross-linking, but it is not commonly used (Nairz et al., 2004). X-rays and gamma rays have previously been used extensively as they induce genomic rearrangements, including inversions, deletions and duplications. However irradiation produces many fewer mutations than EMS and is therefore not so popular anymore. N-ethyl-N-nitrosourea (ENU), though frequently used in other organisms, is not favored in Drosophila, probably because it is too toxic (Ashburner, 1989). Other mutagens are discussed elsewhere (Ashburner, 1989). In the next few sections, we discuss the phenotypic assays used to identify EMS-induced mutations as well as the three commonly used types of EMS screens: the single (F1), two (F2), and three (F3) generation screens.
2.2 Phenotypic Assays
The phenotypic assay used in any forward unbiased EMS screen is paramount to its success, an often underappreciated fact. In most cases the goal of the screen is to isolate as many mutations in as many different genes as possible that affect the phenotype of interest, ideally identifying at least two alleles for each gene. Although we recommend using low concentrations of EMS (e.g., 10 mM) to reduce the number of second site mutations (which can hinder mapping and cloud interpretation at later stages), this will raise the number of flies to be screened although it is already often in the many thousands to tens of thousands range. Hence, primary screening assays should be quick and easy to perform, enabling one to screen hundreds or even thousands of chromosomes per week. In a well-designed screen, the primary phenotypic assay should be followed by secondary screens. These can be more tedious than the primary screen assay as typically no more than 1–2% of flies from the primary screen will be retained.
Simple phenotypic assays are the most powerful because they are easy to perform and can be completed quickly. A classical example is the assay developed to identify flies that are impaired in phototaxis (Benzer, 1967). One hundred mutagenized flies are loaded into a countercurrent phototaxis tool, tapped down toward the dark side of the apparatus, and then allowed to walk towards a light source. Flies that walk to the light source are separated from those that do not, and the process is repeated six times. When combined with an electroretinogram-based secondary screen, this assay previously permitted the isolation of genes that affect synaptic transmission (Verstreken et al., 2003). Another powerful assay is based on flight capabilities. One hundred mutagenized flies are added to a cylinder coated with oil. Flies that were unable to fly fell to the bottom of the container and were collected for analysis of mutations affecting flight muscles (Koana and Hotta, 1978). Given the plethora of genetic tools that are currently available (Venken et al., 2011b), more sophisticated screens can be designed that allow for equally simple phenotypic probing. They can be based on the localization of a GFP in a specific cell or cellular compartment. For example, by expressing a CD8-GFP-Shaker fusion protein at the Drosophila neuromuscular junction (Zito et al., 1999), third instar larva were rapidly screened through the cuticle for morphological defects of the NMJ (Aberle et al., 2002).
2.3 Single (F1) Generation EMS Screens
The simplest and fastest EMS screens are single (F1) generation screens. These screens can be used to isolate, (1) behaviorally defective flies that carry visible X chromosome-linked mutations (Benzer, 1967), (2) dominant suppressors and enhancers of existing mutations (Simon et al., 1991), or (3) new mutations on single chromosome arms via clonal analysis (Stowers and Schwarz, 1999; Newsome et al., 2000).
To isolate viable mutations on the X-chromosome, mutagenized males are crossed to attached X chromosome females (Lindsley and Zimm, 1992), yielding male progeny with a paternally-derived X-chromosome. Flies with visible mutations or defective behaviors can be easily isolated (Benzer, 1967).
In suppressor and enhancer screens, one typically starts with a known mutation that causes a subtle phenotype but which can easily be enhanced or suppressed. A classical example was performed by using a temperature sensitive mutation of sevenless (Simon et al., 1991). Normal development of the R7 photoreceptor occurred at 24°C but not at higher temperatures. Loss of a single copy of another gene could then enhance the phenotype, observed during an F1 screen. The screen was very successful and permitted isolation of seven genes required in photoreceptor development. Similar screens can be used to identify suppressors.
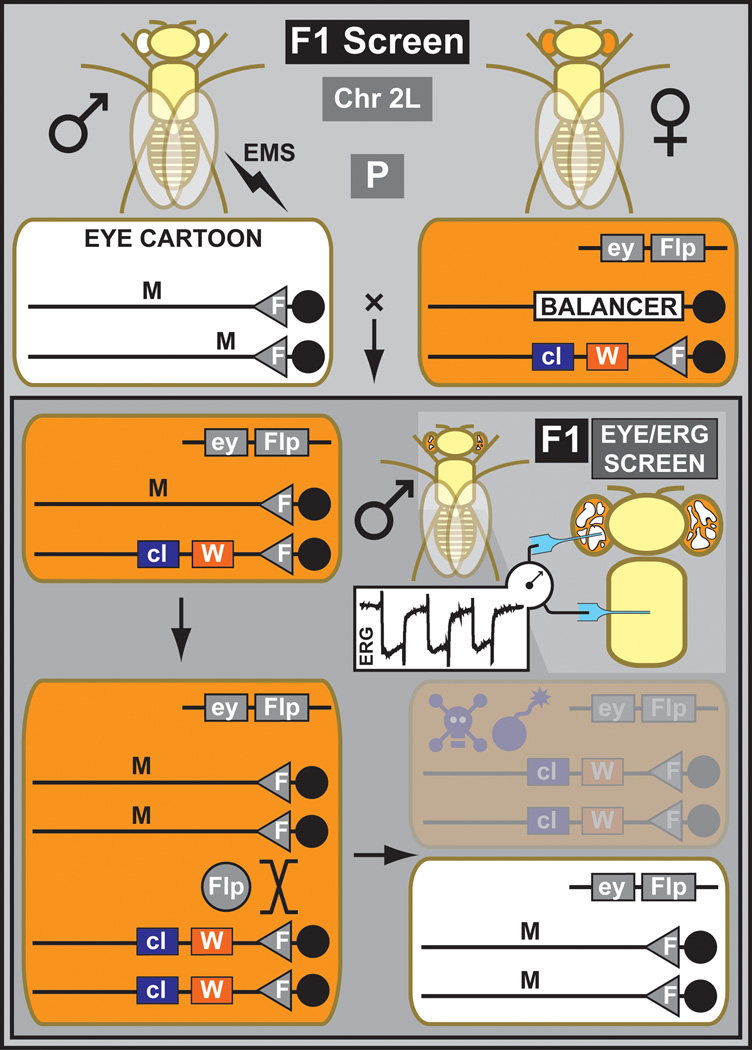
Currently, most F1 screens are performed for the autosomal chromosome arms (2L, 2R, 3L, 3R) that carry an FRT site in close proximity of the respective centromeres (Xu and Rubin, 1993) (Figure 1). Isogenized males carrying the appropriate FRT-containing chromosome arm are mutagenized with EMS and crossed to females carrying the same FRT chromosome and a transgene driving FLP recombinase (Golic and Lindquist, 1989) in a tissue of interest. Subsequent FLP-mediated mitotic recombination results in the exchange of entire chromosome arms during cell division. This permits the generation of homozygous mutant tissue in an otherwise heterozygous animal, therefore limiting the effect of a potentially detrimental mutant phenotype at early developmental stages. The progeny of mutagenized males can be directly screened for a phenotype in the next generation. Desired mutations are isolated and immediately balanced to generate stable stocks. F1 screens are typically based on organ or cell-specific FLP expression driven by tissue-specific regulatory elements (Newsome et al., 2000) or through the binary GAL4/UAS system (Brand and Perrimon, 1993), i.e. an organ- or tissue-specific GAL4 driving a UAS-FLP transgene (Stowers and Schwarz, 1999). The most widely used tissue is the eye since it is large, easily scorable, and excellent drivers like eyeless-FLP are available (Newsome et al., 2000). To obtain clones of sufficient size, it is important to use a driver expressed early in development. The eyeless regulatory elements fulfill this requirement since the gene is already expressed in embryos. The size of the mutant clone can be enlarged by using a cell lethal mutation or a Minute (i.e. a mutation often in ribosomal proteins affecting protein translation and therefore general cell health) on the homologous chromosome so that wild type clones are unable to propagate equally well (Newsome et al., 2000). The large clones in the eye allow to screen for defects during eye development (Newsome et al., 2000), tissue overgrowth (Kango-Singh et al., 2002), bristle development (Tien et al., 2008), synaptic transmission (Verstreken et al., 2003) and neuronal wiring (Mehta et al., 2005). Other tissues can be analyzed using this technology as well, e.g. the Ubx-Flp transgene generates clones in all the imaginal discs among other tissues (Jafar-Nejad et al., 2005).
Figure 1. Outline of a mitotic F1 screen for chromosome 2L.
Mutagenized males containing a chromosome 2 with an FRT site on arm 2L are crossed to females containing the same FRT carrying chromosome but with an additional dominant eye color marker, white+ (W), and recessive cell lethal (cl). In addition, the females carry a transgene that drives the FLP protein under the control of eyeless promoter (ey). This cross is the parental (P) generation. In the next generation (F1), the FLP generates mitotic clones exclusively in the eye. Due to the presence of a recessive cell lethal, homozygous WT clones die, and many ommatidia become homozygous mutant, facilitating phenotypic analyses based on morphology or an electrophysiological assay like the elecroretinogram (ERG). Flies are recovered, and propagated for stock keeping and further analysis.
The FLP/FRT F1 screens are fast but require that heterozygous animals can be used to breed. Hence, the screen cannot be intrusive. Another disadvantage of F1 screens is that not all induced mutations breed true as DNA repair can occur in germ cells and the somatic mutations are therefore not always inherited (Ashburner, 1989). The advantages are that F1 screens are fast and comprehensive if the primary assay is simple. A high throughput and simple assay is extremely important for powerful screens and isogenization of the chromosome of interest prior to mutagenesis is paramount (Bokel, 2008).
2.4 Two (F2) Generation EMS Screens
Two (F2) generation screens are often designed to generate specific mutations and are thus more targeted than screens examining whole chromosomes or chromosome arms. The exception is the X-chromosome. Males only carry a single copy and hence F2 screens have been used to isolate viable behavioral mutations on the X-chromosome (Konopka and Benzer, 1971). For the latter, mutagenized males are crossed to attached X females and single male progeny are backcrossed to attached X females to expand the flies carrying the mutagenized chromosome. The male progeny is then scored for behavioral phenotypes like diurnal rhythmicity (Konopka and Benzer, 1971), learning and memory (Dudai et al., 1976), or defects in vision based on ERGs (Pak et al., 1970; Hotta and Benzer, 1970).
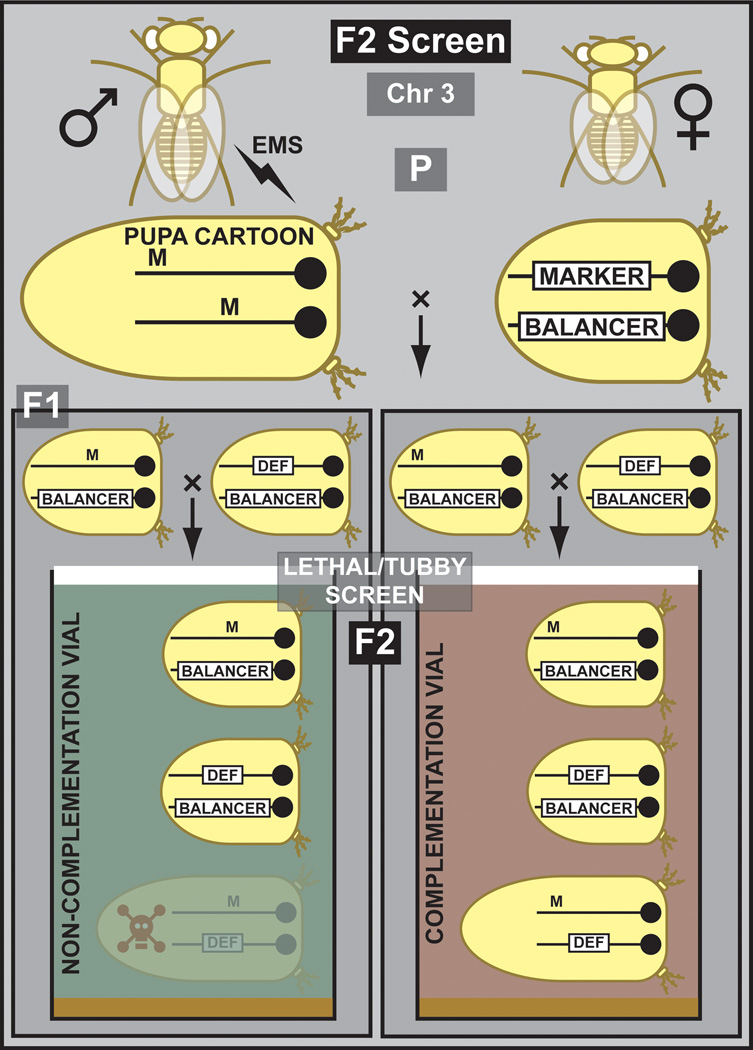
In many F2 screens, the purpose is to isolate new EMS induced mutations that fail to complement genes uncovered by a deletion (Woodruff and Ashburner, 1979b; Woodruff and Ashburner, 1979a) or another mutation, to isolate other alleles of a gene and to initiate structure function analyses (Figure 2). Males are mutagenized and crossed to females that carry a second or third chromosome balancer. Single males are then crossed to a stock carrying a mutation or a deletion (and a balancer) in which the gene of interest is uncovered. Failure to complement the deletion or the mutation permits identification of specific alleles of a gene that are located within the deletion (Woodruff and Ashburner, 1979b; Woodruff and Ashburner, 1979a; Littleton and Bellen, 1994; Diantonio et al., 1993). The inclusion of easily scorable markers (e.g. Tubby) can facilitate the screening process tremendously. Through complementation tests and sequencing of the gene of interest new alleles can be identified. This allows an unbiased recovery of new alleles that permit a structure function analysis (Littleton et al., 1994).
Figure 2. Outline of a lethal phenotype F2 screen for chromosome 3.
Mutagenized males are crossed to balanced third chromosome Tubby females. In the next generation, balanced mutants are crossed to a balanced third chromosome deficiency to isolate mutations that fail to complement the deficiency. In the second generation (F2), progeny are scored for prepupal lethality. The balancer is marked with Tubby, hence the short Tubby females and pupae. The inclusion of the Tubby (Tb) marker facilitates this process since scoring can be performed immediately by looking at the pupae in the vial. The green vial carries the mutation of interest.
2.5 Three (F3) Generation EMS Screens
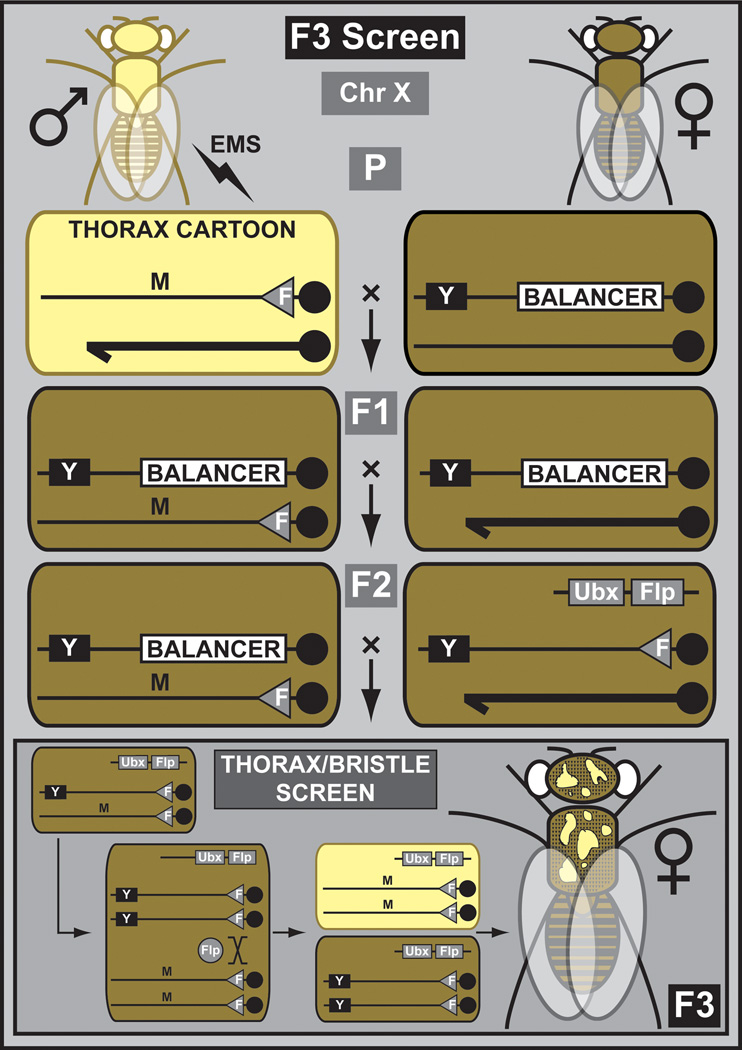
Three (F3) generation screens are the foundation of some of the most probing experiments in developmental biology and neuroscience (Nusslein-Volhard and Wieschaus, 1980; St Johnston, 2002; Bellen et al., 2010). They remain a very powerful, if somewhat underappreciated, tool. With the advent of whole genome sequencing (WGS), they are likely to resurge in popularity since mutations can be identified more easily. Recently, an extensive large-scale F3 mosaic screen was performed for the X chromosome (Yamamoto et al., 2012) (Figure 3). Isogenized males carrying an y w FRT chromosome were mutagenized with lower EMS concentrations than are typically recommended (10 mM versus 25–50 mM) (Bokel, 2008) and crossed to balanced females. Individual mutant balanced female progeny were crossed back to balanced males to establish stocks. Chromosomes that carried homozygous lethal mutations were retained and screened in different assays by creating mutant clones in the eye or thorax. Mutations in numerous genes not previously identified or characterized on the X chromosome were discovered (H.J.B., unpublished data).
Figure 3. Outline of a lethal phenotype F3 screen for the X chromosome.
Mutagenized males containing chromosome X with an FRT site are crossed to balanced X chromosome females. Mutant balanced females are backcrossed to balanced males to generate stocks. Stocks are used to establish X chromosome lethality. Balanced lethal mutant females are crossed to males containing the same FRT chromosome and a dominant body color marker, yellow+ (Y). In addition, males have a transgene that drives the FLP protein under the control of an imaginal disc regulatory element (Ubx). In the next generation (F3), the FLP generates mitotic clones exclusively in the imaginal discs that can be analyzed for a morphological phenotype (e.g. bristle phenotype).
2.6 Mapping of EMS Mutations
Mapping EMS mutations has historically been tedious. This is no longer the case since mapping can now be done on a large scale. Mapping a few dozen genes from an EMS screen should be within reach of almost any fly lab. Depending on the chromosomal location, alleles are mapped by a variant of meiotic mapping (for the 2nd and 3rd chromosome) (Zhai et al., 2003) or a variant of duplication mapping (for the X chromosome) (Cook et al., 2010b; Venken et al., 2010).
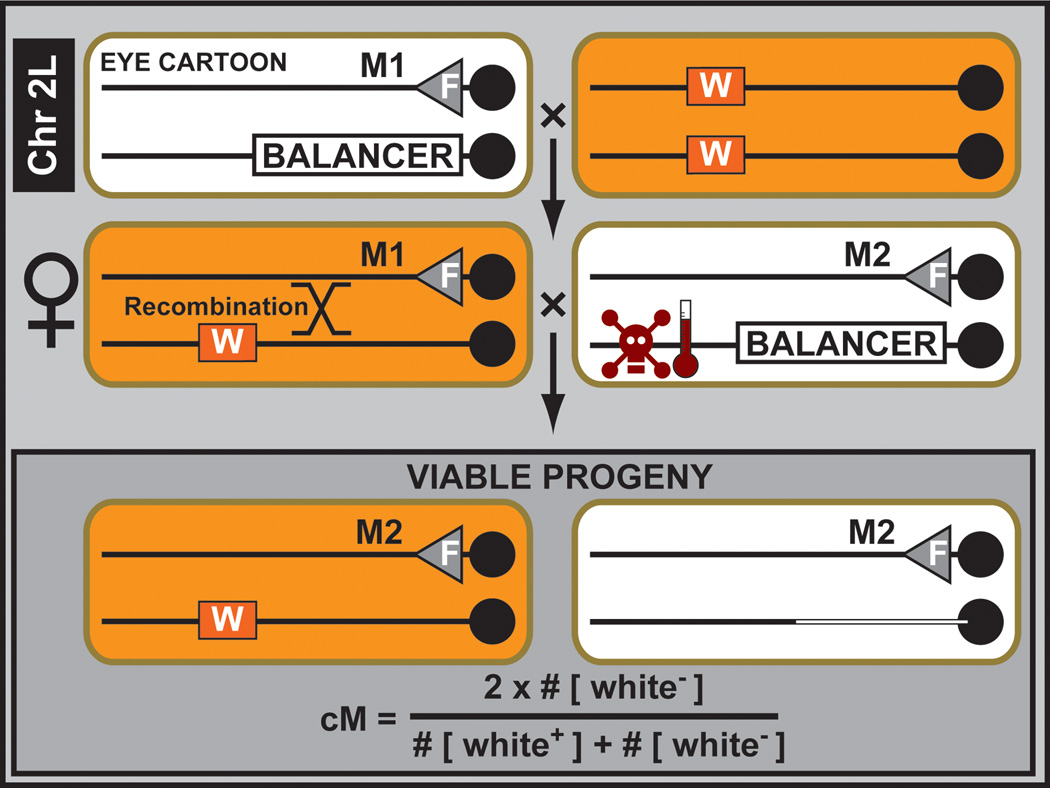
Meiotic mapping is very easy to perform with a set of defined P element insertions (Zhai et al., 2003) (Figure 4). To map a mutation located on a known chromosome arm, two rounds of meiotic mapping are performed. In a first mapping round (i.e. rough mapping), the locus can be mapped to a 400 kb interval using 10–15 defined P elements whose mapping position is known. Stocks for rough mapping of mutations on the second and third chromosomes have been carefully selected so that they show strong expression of the transgenic white+ marker (see also Section 3.2), resulting in a red adult eye which facilitates screening when scoring large populations. Mapping kits for each autosomal chromosome arm (i.e. 2L, 2R, 3L and 3R) are available from the BDSC (http://flystocks.bio.indiana.edu/Browse/misc-browse/Baylor-kits.htm). A second round of mapping permits pinpointing the mutation to a physical interval of less than 100 kb (Zhai et al., 2003), but this mapping resolution is no longer required as one can use next generation sequencing approaches to analyze the data of the 400 kb interval.
Figure 4. Meitoic mutation mapping using P element insertions.
A balanced unmapped lethal mutation (M1), located on chromosome arm 2L, is crossed to a mapped viable P element insertion marked by the dominant eye color marker white+ (W). Next, female balanced mutant progeny are crossed to males containing a second lethal mutation (M2) belonging to the same complementation group maintained over a conditionally lethal balancer chromosome (i.e. temperature induced lethality). During early development of the progeny of this cross, a heat shock removes all unwanted progeny containing the conditionally lethal balancer chromosome. The transheterozygous M1/M2 flies die as well. Since recombination between the unmapped lethal mutation (M1) and the mapped viable P element insertion can occur in the female germ line, the ratio of white− flies compared to the total number of flies in the resulting progeny is a measure of the genetic distance between the unmapped mutation and the mapped P element insertion. Performing this crossing scheme with ~10 mapped P elements located on the same chromosome arm allows mapping to within 100 kb-1Mb depending on the region of the chromosome where the mutation maps. One can then proceed with WGS for mutation identification.
Although meiotic mapping with P elements is feasible for the X chromosome, it becomes more complicated for lethal mutations since they cannot be recovered in males. This problem is easily circumvented by duplication mapping. Two independent duplication kits have been generated for the X chromosome (Venken et al., 2010; Cook et al., 2010b). One duplication kit was generated by sophisticated chromosomal engineering of an attached X-Y chromosome (Cook et al., 2010b) (Figure 5A), while the second duplication kit was generated by site-specific integration of an overlapping set of P[acman] clones into an attP site located on the third chromosome (Venken et al., 2010) (Figure 5B) (see also Section 4.3). The two duplication collections complement each other well. Using the large duplications (attached-XY) (Cook et al., 2010b), mutations can be mapped to a 1 Mb region. Subsequent use of smaller duplications, deficiencies, and the tiled P[acman] array of duplications (Venken et al., 2010), allows one to easily narrow this interval to less than 50 kb. Sanger sequencing of overlapping PCR products ultimately reveals the gene of interest (Zhai et al., 2003). This strategy has proven to be efficient in mapping loci derived from an ongoing, large-scale X chromosome mutagenesis effort. However, the majority of mutations of this screen were mapped using WGS after rough mapping to an area of about 1–1.5 Mb, thereby greatly accelerating the gene identification process (HJB, Unpublished data).
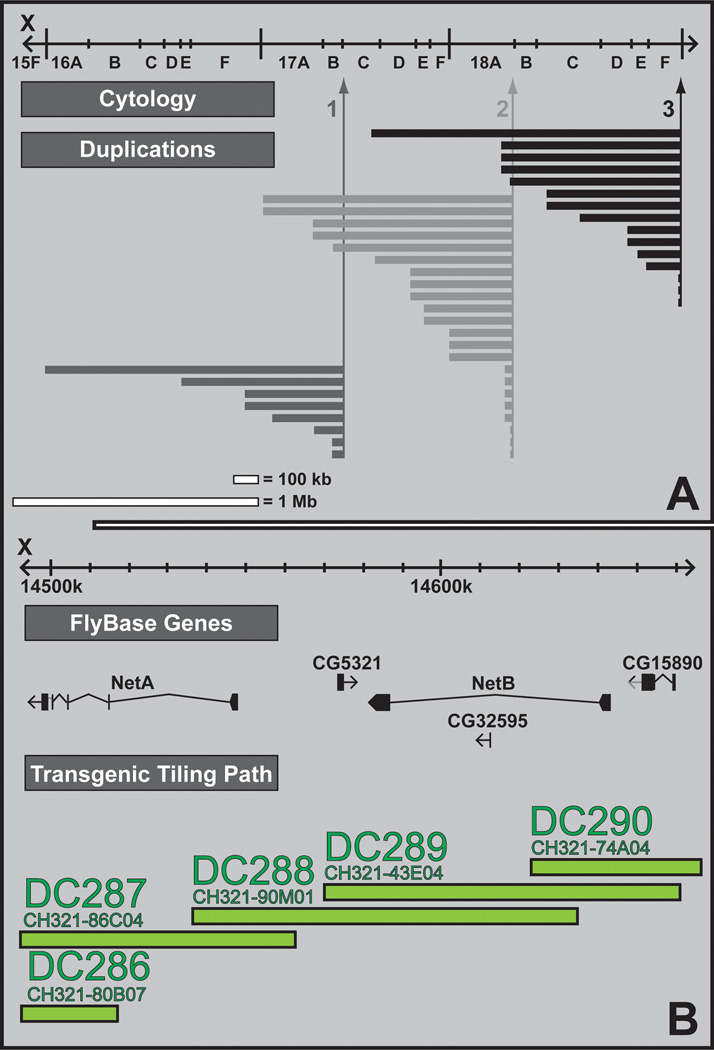
Figure 5. Mutation mapping using duplication complementation.
A. Nested duplication mapping. A nested set of duplicated segments is generated from the same progenitor (e.g. 1). Different progenitors (i.e. 1, 2 and 3) then result in overlapping sets of nest duplications that cover majority of the X chromosome. Mutations can then be mapped by the large segments in a first round of genetic mapping and fine mapped by a nested set of duplications in a second round of genetic mapping. B. Transgenic duplication mapping. The CHORI-321 P[acman] library contains DNA clones (CH321) that have an average insert size of 83 kb. A minimum tiling path (indicated in green) of 408 overlapping P[acman] clones for the entire 22 megabase X chromosome of Drosophila was selected and integrated in the same docking site on chromosome arm 3L, using the ΦC31 integrase. This transgenic duplication kit can be used to map uncharacterized X chromosome mutations. Illustrated is a small portion of the transgenic duplication kit surrounding the Netrin-A and Netrin-B genes.
3. Transposon Interrogation
3.1 Transposons
TEs are insertional mutagens that can be easily mapped using a variety of molecular methods (see also Section 3.2). The most commonly used TE in the fly field is the P element, followed by piggyBac, and Minos (Ryder and Russell, 2003). Wild type P elements are composed of two 31-mer terminal inverted repeats, P transposase (the enzyme required to catalyze transposition), as well as other sequences essential for transposition (Castro and Carareto, 2004) (Figure 6A). Physical separation of the P transposon backbone from the P transposase has permitted the development of a binary transformation system (Rubin and Spradling, 1982) (Figure 6B), allowing controlled transposition of engineered transposons into the host genome using enzyme provided in trans, e.g. as a helper plasmid during microinjection or as a transgene through a genetic crossing scheme (Venken and Bellen, 2007). Although proven to be incredibly useful, P elements also have drawbacks. The integration sites of P elements are strongly biased toward origin of replication sites (Spradling et al., 2011) and the 5’ ends of genes (Bellen et al., 2011). This insertional specificity reduces the effectiveness of mutagenesis screens and limits the number of insertions that can be used to create gene- or protein-traps (Buszczak et al., 2007; Quinones-Coello et al., 2007; Lukacsovich et al., 2001). Additionally, many insertions do not or only partially affect the genes near which they are inserted (Norga et al., 2003; Lafave and Sekelsky, 2011). The latter is also a very important benefit in reverse genetic strategies. The lack of phenotype permits one to assess the loss of function associated with an imprecise excision of the P element that affects or removes the nearby gene without having to worry too much about second site mutations present on many chromosomes.
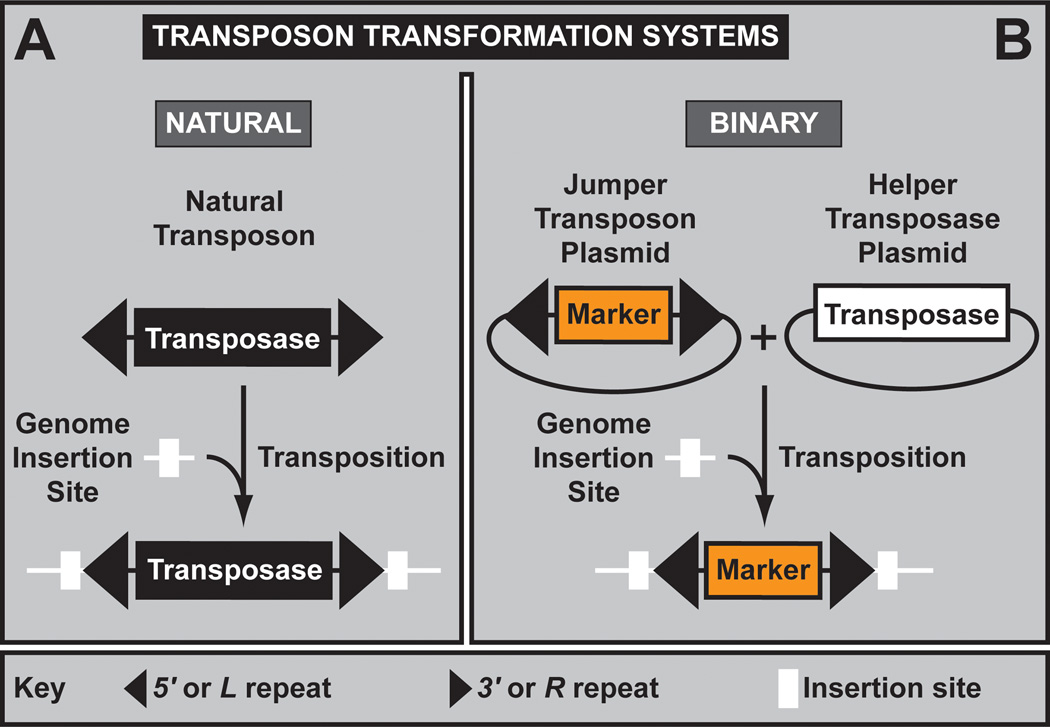
Figure 6. Transposon Interrogation. (A) Transposon transformation systems and transposons.
A. An active natural transposon. Active natural transposons are mobile elements that consist of two inverted terminal repeats (black) that flank an open reading frame encoding a transposase. Both features are required for transposition. Transposition results in a duplication of the insertion site (white box). B. The binary transformation system. The transposon inverted repeats and transposase can be physically separated, resulting in the binary vector/helper transposon transformation system that allows the regulated transposition of transgenes into the genome. Only when both components are provided together can productive transposition occur.
The features of P elements have lead to the exploration of the use of other TE such as piggyBac (Handler and Harrell, 1999) and Minos (Loukeris et al., 1995) (Figure 7). Unlike P elements, piggyBac preferentially targets TTAA sequences whereas Minos, a Tc1/Mariner-like transposon, inserts at random (Spradling et al., 2011). The major drawback of piggyBacs, unlike P elements and Minos, is that they do not excise imprecisely (Witsell et al., 2009), and hence do not permit the creation of other mutations in the gene of interest. Both piggyBac and Minos have been used to generate genome-wide TE collections. The Tol2 TE has also been demonstrated to mobilize in flies (Urasaki et al., 2008) but has not been used in any genome wide application. To be useful for genome wide screens, a transposon has to be integrated into the genome first using its corresponding transposase, before it can be used as ammunition for subsequent remobilization mutagenesis (Figure 8).
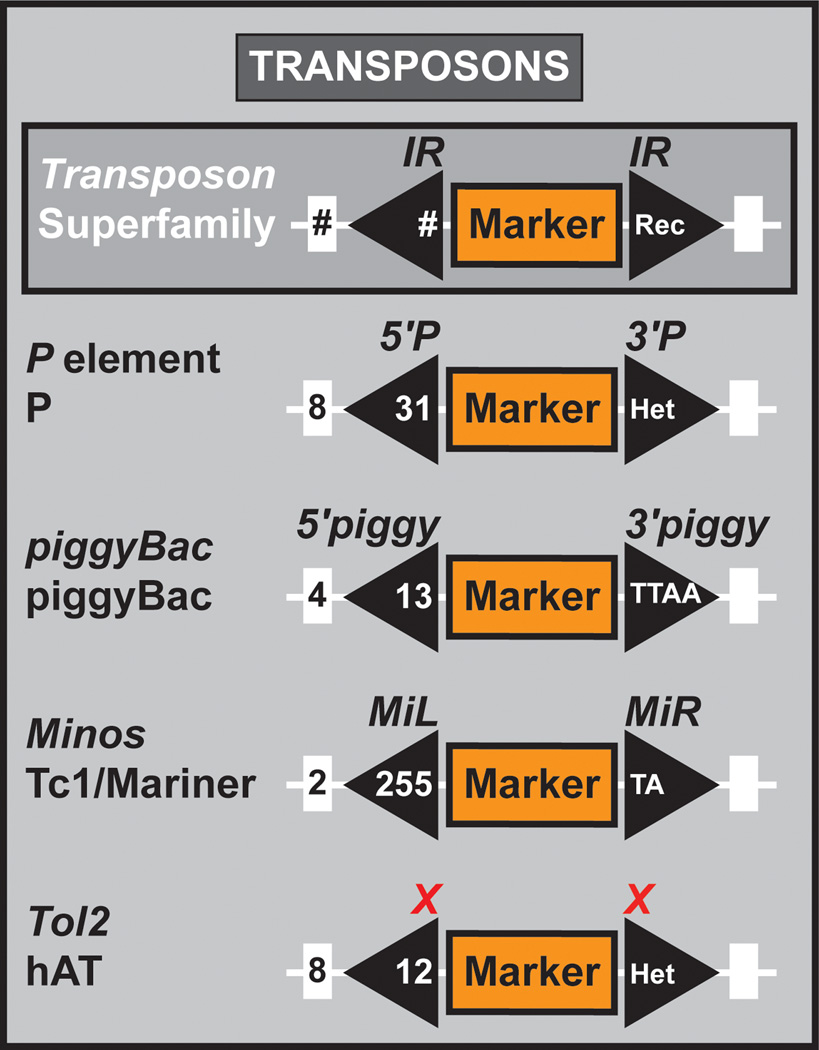
Figure 7. Transposons.
Genetic constitution of a key member of different transposon superfamilies used for genetic analysis in Drosophila. Transposon superfamily, size of the duplicated genome insertion site, size of the inverted repeat, and nucleotide constitution of the inverted repeat are indicated for the P element, piggyBac, Minos, and Tol2.
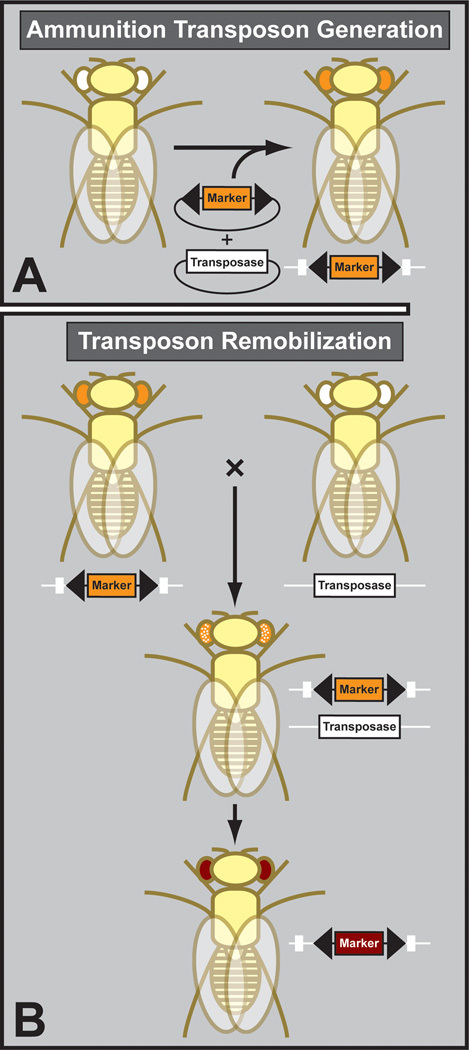
Figure 8. Transposon mutagenesis.
A. The initial ammunition chromosome is often generated by microinjection of a plasmid containing the engineered transposon. B. Subsequent transposon hopping is done by genetic crosses, bringing transposon and transposase together in the same fly. In the next generation, novel insertion sites are identified and the transposase segregated away.
The transposition rates of the various transposable elements vary widely. For P elements, the transposition rate typically ranges from 10 to 65%, depending on the P element construct that is used and the level of transposase (Bellen et al., 2004). For piggyBacs, the transposition rate is also dependent on the nature of the element and the transposase source; it varies widely from 50–100% (Horn et al., 2003; Thibault et al., 2004). However, in our hands piggyBac transposition occurred at much lower frequencies, in the 5–10% range. For Minos, the transposition rate is typically 5–10% (H.J.B., unpublished data), although higher rates have also been recorded previously (Venken et al., 2011a; Metaxakis et al., 2005). High transposition rates are not necessarily beneficial, as they are associated with greater degrees of second site mutations (Liebl et al., 2006). Therefore the Gene Disruption Project has saved a very substantial number of insertions that are homozygous viable.
3.2 Dominant Transposon Markers
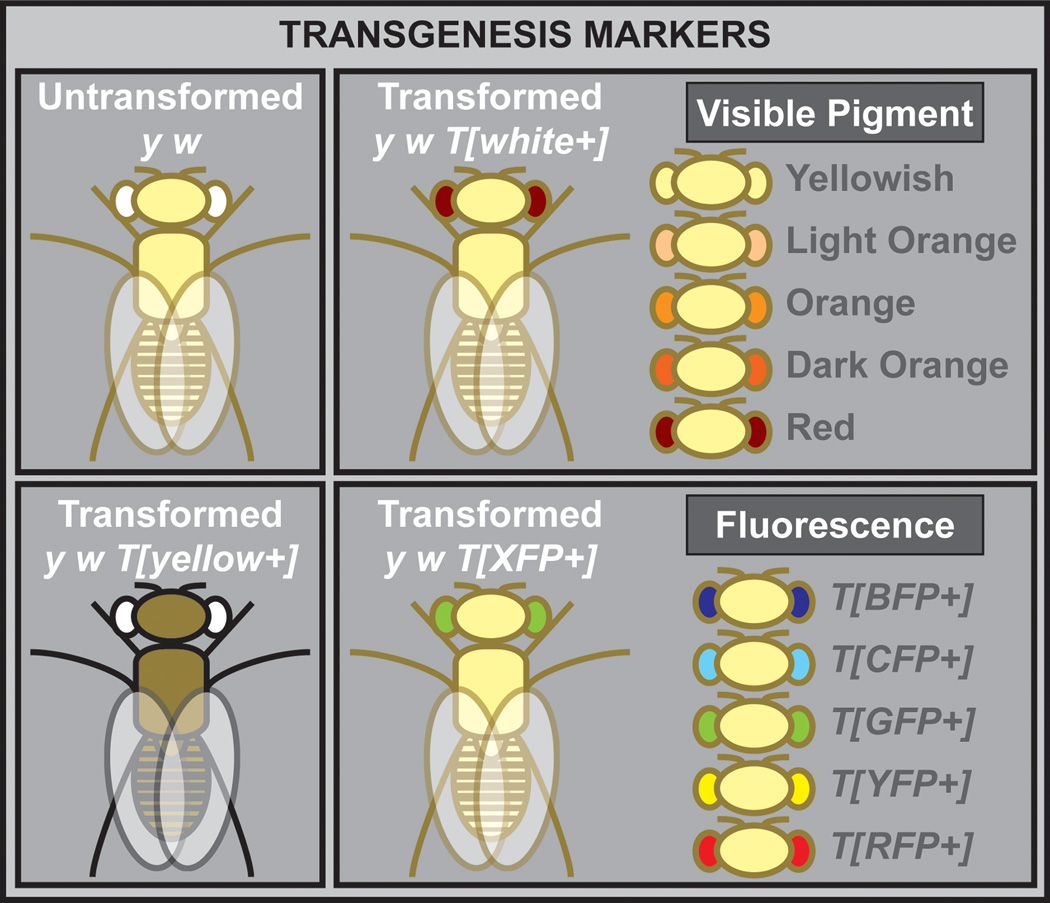
Identification of novel integration events upon the hopping or integration of a TE is dependent on the incorporation of dominant genetic markers (Venken and Bellen, 2007) (Figure 9). The most popular markers are those easily identified under a regular stereoscope. They include white+, a dominant eye color marker that leads to red eye pigmentation (Pirrotta, 1988), and yellow+, a dominant body color marker that turns the yellowish body of a yellow mutant into one more tan in color (Patton et al., 1992). Other commonly used markers are those in which fluorescent proteins have been fused to an artificial eye-specific promoter (e.g., 3xP3) (Berghammer et al., 1999; Horn and Wimmer, 2000). These markers are relatively easy to score but require a dissection microscope equipped with a UV light source and appropriate fluorescent filters.
Figure 9. Commonly used dominant genetic markers.
A commonly used marker is the dominant visible eye color marker white+, which is highly under the influence of nearby genomic position effects, i.e. eye color can vary dependent on the transgene insertion site, resulting in yellowish, light orange, orange, dark orange, or WT red eye color pigmentation. Other markers often used are the dominant visible body color marker yellow+, and the dominant fluorescent markers driven by an artificial eye promoter, i.e. blue (BFP), cyan (CFP), green (GFP), yellow (YFP) or red fluorescent protein (RFP).
3.3 Genome-wide Transposon Insertions
Details about the different TEs and TE markers, as well as the specific features of the more commonly used transposons (P elements, piggyBac, and Minos), are available through the Gene Disruption Project (GDP; http://flypush.imgen.bcm.tmc.edu/pscreen/) (Bellen et al., 2011). This project started in 1992 with the primary goal of generating at least one transposon insertion for every fly gene (Bellen et al., 2011; Bellen et al., 2004; Spradling et al., 1999; Venken et al., 2011a; Spradling et al., 1995). Initially, the project focused on P elements engineered by members of the community and gathered P element and piggyBac collections generated by others (Bellen et al., 2004; Bellen et al., 2011; Spradling et al., 1999). However, the GDP subsequently built a transposon collection using the EY P element which contains a UAS site, thus permitting ectopic expression of the gene near which the transposon is inserted (Bellen et al., 2004). By 2004 the diminishing returns of generating additional P element insertions to tag new genes became cost prohibitive. Indeed, towards the end of the project, for every 100 new insertions generated and sequenced, only 2 P elements could be identified in genes not already tagged. Currently, about 52% of all annotated Drosophila genes contain a P element insertion, and it is unlikely that this figure will change. All TE strains generated by the GDP are available through public stock centers, namely the Bloomington Drosophila Stock Center (BDSC) (http://flystocks.bio.indiana.edu/Browse/in/GDPtop.htm).
As an alternative to P elements, the GDP explored other TEs, including MiET, a Minos derived transposon (Metaxakis et al., 2005). A small pilot study suggested that MiET inserted at random in the genome (Metaxakis et al., 2005), an aspect later confirmed with a larger screen of ~12,000 insertions, indicating that very few loci were refractory to insertion (Bellen et al., 2011). Therefore, a new version of the Minos transposon, the Minos-mediated integration cassette (MiMIC) was designed. MiMIC incorporates two inverted attP sites for recombinase-mediated cassette exchange (RMCE) (see also Section 3.7), allowing a broad array of versatile applications after a transposon insertion is identified (Venken et al., 2011a; Neumuller et al., 2012). Currently more than 12,000 MiMIC insertion strains have been generated and sequenced, half of which are available from the BDSC (http://flypush.imgen.bcm.tmc.edu/pscreen/). This collection consists of insertions that tag about 3, 200 genes so far. Other important collections based on P elements and piggyBac have also been generated—for example, versions carrying FRT recombination sites (Thibault et al., 2004; Parks et al., 2004; Ryder et al., 2004; Ryder et al., 2007). These transposons have been used for genome-wide applications, including the development of molecularly mapped deletion and duplication kits (Cook et al., 2010b) (see also Section 3.6).
3.4 Transposon Mutagenesis
Three basic TE-based approaches can be used to screen for mutations affecting specific biological processes. In the first approach a collection of TE insertions is created and subsequently screened for phenotypes, as shown for P elements (Kania et al., 1995) and piggyBacs (Schuldiner et al., 2008). The TE insertions are then mapped (see also Section 3.5) and molecular lesions assessed. This strategy is labor intensive and relatively inefficient compared to EMS screens. For example, while EMS can induce hundreds of mutations on each chromosome, the goal of TE hopping is often to obtain a single new insertion per chromosome. Thus, TE screens will sample less than 1% of the mutations generated in an EMS screen. Additionally, most TEs do not insert at random. P elements preferentially insert near the transcription start site and have hot spots, genes hit very frequently, thereby diminishing returns (Bellen et al., 2011). piggyBacs also have a slight preference for the transcription start site. Hence, the choice of TE may strongly bias the screen. Other weaknesses of this strategy include the fact that many TE insertions only cause a partial loss of function. This is especially true for P elements that insert in regulatory regions, typically within 200 nucleotides of the transcription start site (Spradling et al., 2011; Bellen et al., 2011). Finally, the main advantage of TEs—the ability to quickly map TEs using inverse PCR—does not always result in the identification of the gene of interest. Indeed, the introduction of transposases, as well as the accumulation of spurious lethals in the genetic background of some stocks, may lead to an elevated level of second site mutations not due to the TE insertion itself (Liebl et al., 2006). Consequently, rescue with a genomic fragment (Venken et al., 2010; Venken et al., 2009; Venken et al., 2010; Szabad et al., 2012) or failure to complement a deletion (Cook et al., 2010a) should be considered in the early phases of mapping project to ascertain whether the phenotype corresponds to the presumable TE insertion.
A second TE-based strategy uses a select number of TE insertion stocks already present in private collections or at the BDSC (http://flystocks.bio.indiana.edu/). Currently, TE insertions in about 11,000 genes have been created by the GDP and are available from the BDSC (Bellen et al., 2004; Bellen et al., 2011; Venken et al., 2011a). These stocks can be screened for specific phenotypes. Numerous screens have used this strategy for bristle number variation (Norga et al., 2003), synaptic transmission (Liebl et al., 2006), hypoxia tolerance (Azad et al., 2012), and olfactory behavior (Tunstall et al., 2012), among others. Once interesting phenotypes have been identified, one needs to ascertain that the TE insertion causes the phenotype, preferably by doing three experiments: precise excision of the TE and reversion of the phenotype (Hummel and Klambt, 2008), failure to complement the phenotype with a deficiency (Cook et al., 2010a), and rescue with a P[acman] transgene encompassing the genomic fragment (Venken et al., 2006; Venken et al., 2009; Venken et al., 2010; Szabad et al., 2012). Importantly, one need not screen all the stocks available. Rather, they can screen just a specific subtype, e.g. lethal insertions. As mentioned above, many P element insertions are hypomorphic and located in regulatory elements (Lafave and Sekelsky, 2011). Consequently, only 18% (~2,000) of all currently available TE insertion strains are homozygous lethal. The mutagenicity of different TEs, however, varies widely: the BG P element, which includes two gene terminating cassettes (i.e. a gene and a polyA trap) (Lukacsovich et al., 2001), only causes 8% lethality; the KG P element, which contains Su(Hw) sites engineered to interfere with gene function by buffering neighboring enhancers (Roseman et al., 1995), causes 22% lethality; and the EY P element (Bellen et al., 2004), which does not include any additional gene interruptive elements apart from the transposon itself, causes 16% lethality. MiMIC (Venken et al., 2011a) has the highest mutagenic index, with 28% of the insertions associated with lethality. Thus, the number of stocks to screen varies depending if one is interested in viable or lethal mutations.
The third screening strategy uses P element collections in which neighboring genes can be overexpressed using UAS sites present in the TE, i.e. notably the EP (Rorth, 1996) and EY P elements (Bellen et al., 2004). By integrating a UAS site in the TE, one can perform screens in which a ubiquitous or tissue-specific GAL4 driver is used to overexpress a gene near the P element insertion (Rorth, 1996). This approach has been used successfully (Staudt et al., 2005; Molnar et al., 2006), but relatively few novel genes have been discovered based on this strategy. Indeed it has proven difficult to associate the gain of function phenotypes with the phenotypes associated with knock-down or loss-of-function alleles needed to interpret the data (DeCelis and Molnar, 2010). Indeed, the main caveats of this type of screen are that overexpression may lead to numerous false positives and that gain-of-function phenotypes in many tissues do not correlate with gene loss.
3.5 Insertion Site Determination
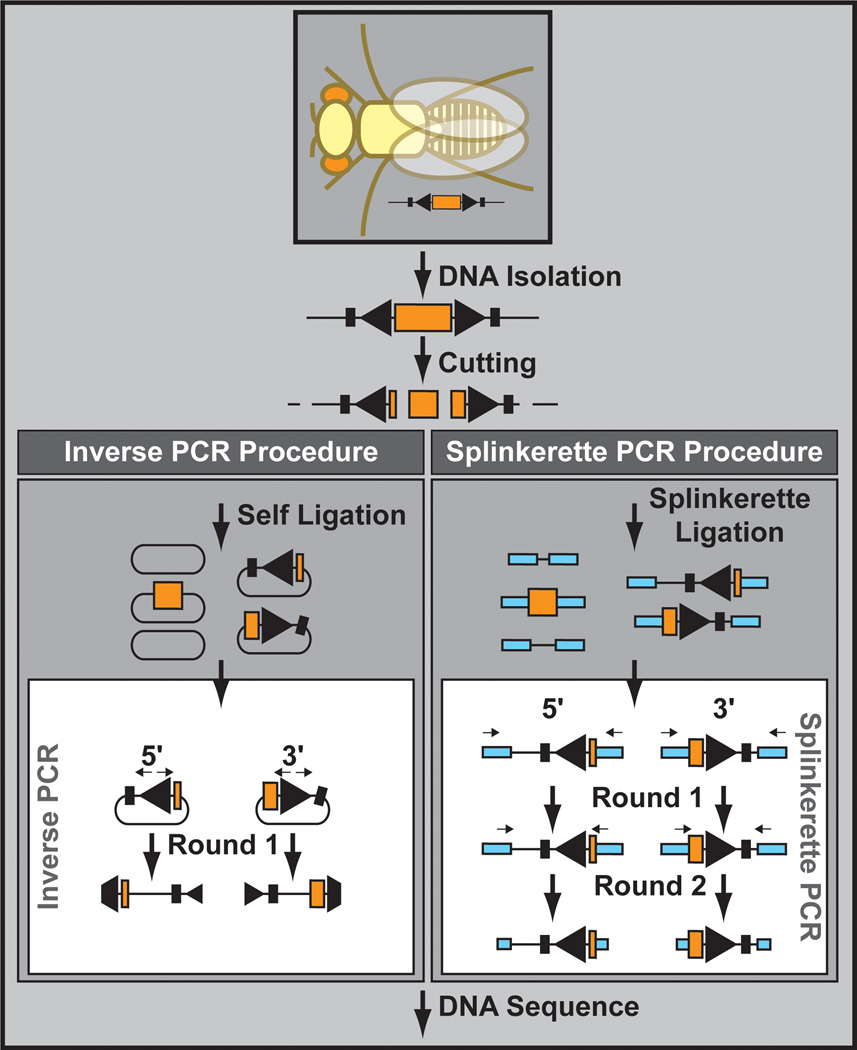
A major advantage of TEs is that they are genetically encoded and can therefore easily be mapped to the genome using a variety of molecular methods. These methods are based on two alternative but partially overlapping procedures: inverse PCR Ochman et al., 1988) and splinkerette PCR (Potter and Luo, 2010) (Figure 10). In inverse PCR genomic DNA is isolated, digested with a restriction enzyme, and self-ligated. A subsequent PCR reaction using primers that go out of the transposon backbone (hence the name “inverse PCR”) and over the unknown genomic fragment is used to determine the sequence, and ultimately the location, of the transposon insertion site. Alternatively, with splinkerette PCR common adapters are ligated to the unknown site after digestion of the genomic DNA. PCR is then conducted using one primer in the known adapter and another primer in the transposon backbone. Often a second, nested, PCR is required to amplify enough material for Sanger sequencing. Although both inverse PCR and splinkerette PCR have been successful for Drosophila, inverse PCR has been used to determine the genomic location of hundreds of thousands of TE insertion sites, illustrating the feasibility and ease of the technology (Bellen et al., 2011; Venken et al., 2011a).
Figure 10. Insertion site determination.
Transposon insertions sites can be molecularly determined by inverse PCR or splinkerette PCR. In both cases, DNA is isolated and cut with a frequently cutting restriction enzyme. Next, using inverse PCR, self-ligated DNA is used as a PCR template. Alternatively, splinkerette adapters are ligated to the cut DNA, and ligated DNA is used as a PCR template in the splinkerette PCR procedure. In both case, DNA sequence of the PCR fragments are determined to retrieve the transposon insertion site.
3.6 Transposon Induced Deletions
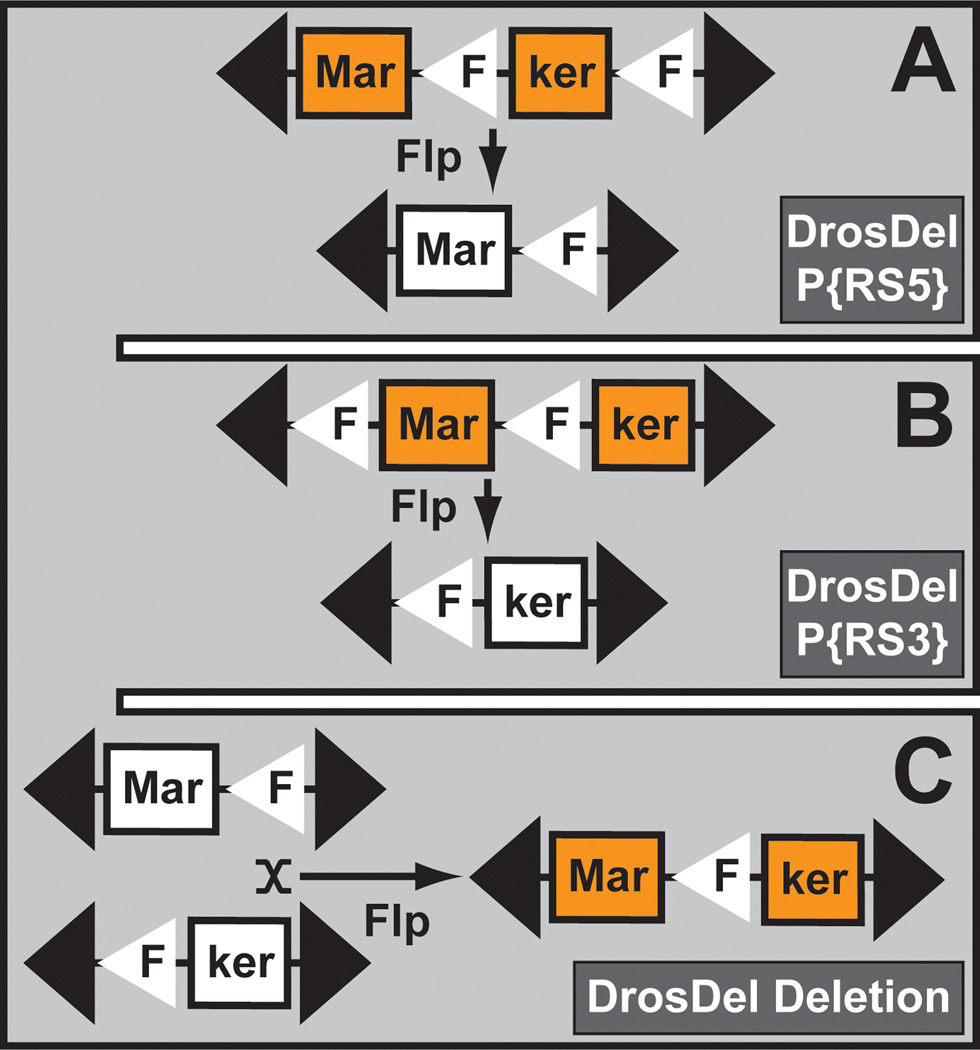
Deletions are some of the most valuable reagents in the fly community (Cook et al., 2010a; Cook et al., 2012). They serve as a reference for null alleles and are frequently used to map genes due to their failure to complement encompassing mutations. Several strategies have been designed to generate deletions in the fly genome (Venken and Bellen, 2005). Currently the methodology employed most often is that based on the FLP/FRT system (Figure 11). When two nearby mapped TEs—each containing an FRT site in identical orientation—are brought in trans, a precise deletion can be generated in the presence of the FLP recombinase. Two independently generated genome-wide transposon collections contain FRT sites (Thibault et al., 2004). One set was obtained by hopping a previously tested FRT-containing P element (Golic and Golic, 1996), generating several thousand new insertions sites. These TEs were then used to create hundreds of defined deficiencies, commonly known as the DrosDel project (http://www.drosdel.org.uk/) (Ryder et al., 2004; Ryder et al., 2007). The other FRT transposon collection was generated with both P element and piggyBac (Thibault et al., 2004) and resulted in hundreds of deficiencies (http://flystocks.bio.indiana.edu/Browse/df/dfextract.php?num=all&symbol=exeldef) (Parks et al., 2004). These two deficiency sets formed the basis of a collection that was compiled at the BDSC (http://flystocks.bio.indiana.edu/Browse/df/dftop.htm) (Cook et al., 2010a; Cook et al., 2012). These deficiencies permit mutation mapping via complementation tests, and their coverage represents true null alleles for 98% of all fly genes.
Figure 11. Transposon induced deletion.
The DrosDel deletions are generated by white+ marker reconstitution. In a first step, part of the white+ markers of two transposon insertions (P{RS5} and P{RS3}) is reduced to invisible but complementary white+ halves. In a next step, both invisible white+ halves are reconstituted into a full length phenotypically visible white+ marker, after FLP-mediated mitotic recombination.
3.7 In Vivo Transposon Upgrading
Since transposons are genetically encoded, they can be engineered to encompass elements that allow future upgrades either through microinjection or through genetic crosses. For example, P elements permit transposon conversion (i.e., in vivo replacement of the DNA internal to the terminal inverted repeats). This is achieved by providing a transposase source in the presence of the original acceptor P element and a donor DNA P element, each containing a different dominant marker. The transposase enzyme promotes the excision of acceptor sequences and its subsequent replacement with donor sequences, precisely swapping the content of the P elements (Gloor et al., 1991; Sepp and Auld, 1999). In doing so, a P element containing one property (e.g. LacZ) can be upgraded with a novel P element containing a second, more sophisticated, property (e.g. the GAL4 binary transactivator). This property is so far unique to P elements and has not yet been reported for other transposons.
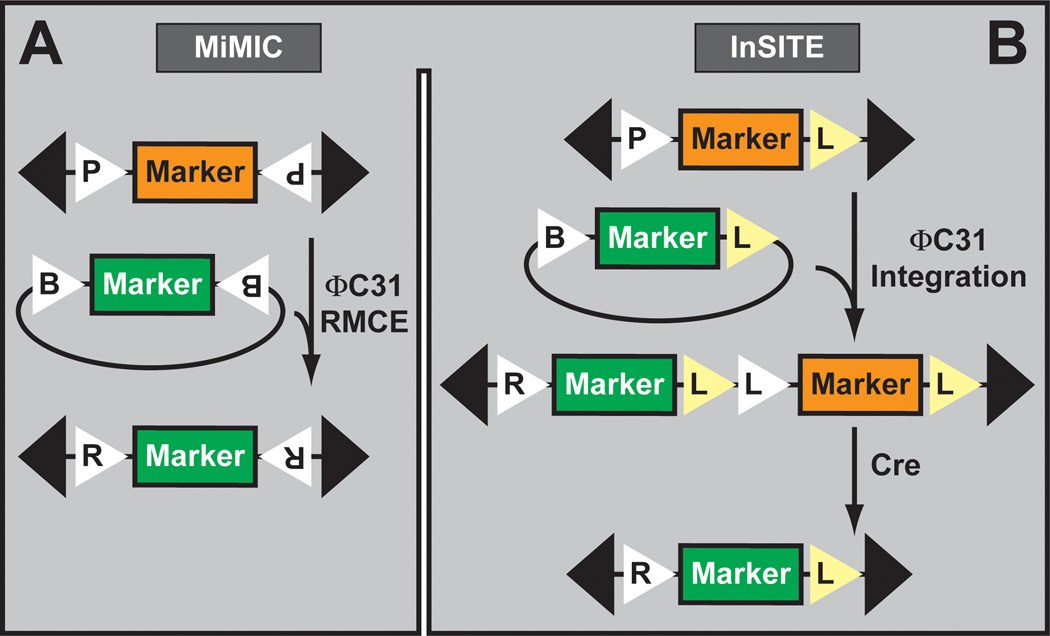
An alternate (and less labor intensive) strategy is to include two recombination (e.g. FRT site) (see also Section 3.6) or integration sites (e.g. attP) in the transposon backbone. Two transposon platforms have been engineered for limitless transposon upgrading. The first uses MiMIC transposon insertions and upgrades the locus through recombinase mediated cassette exchange (Venken et al., 2011a; Neumuller et al., 2012) via the ΦC31 integrase (Groth et al., 2004; Bischof et al., 2007) (Figure 12A). The exchange replaces the existing MiMIC gene trap cassette located in 5’UTR or coding intron with a correction cassette that reverts lethality. Alternatively, MiMIC insertions located in a 5’UTR intron can be upgraded with a novel gene trap that incorporates new properties under the form of a fluorescent marker (e.g. Cherry red fluorescent protein), a binary factor (e.g. GAL4 or QF), or a recombinase (e.g. FLP) for subsequent manipulations. Most interestingly, MiMIC insertions located in coding intronic insertions can be upgraded with artificial exons flanked by an upstream splice acceptor site and a downstream splice donor site. The exons can encode protein tags (i.e. fluorescent markers, peptide affinity tags, or a combination of both) for microscopy visualization or protein purification.
Figure 12. In vivo transposon upgrading.
A. The MiMIC transposon insertions are upgraded by ΦC31 integrase catalyzed Recombinase Mediated Cassette Exchange. B. InSITE (shown) and G-MARET (not shown) transposons are upgraded by ΦC31 integrase catalyzed site-specific integration followed by deletion of unnecessary genetic material.
The second upgradable TE platform uses P element or piggyBac insertions and upgrades the locus via site-specific transgenesis (Figure 12B). These platforms are InSITE (Integrase Swappable In Vivo Targeting Element) (Gohl et al., 2011) and G-MARET (GAL4-based mosaic-inducible and reporter-exchangeable enhancer trap) (Yagi et al., 2010). Both have been used to upgrade GAL4-containing enhancer trap insertions to novel binary factors such as LexA and QF. Interestingly, InSITE was engineered to allow in vivo exchange by simple genetic crosses avoiding microinjection experiments (Gohl et al., 2011). The FLP recombinase releases the modification construct from one location (i.e. the donor transposon), which then integrates into the acceptor transposon using the ΦC31 integrase. Finally, the Cre recombinase is used to remove the undesired portion of the acceptor transposon (e.g. GAL4), leaving the novel property behind (e.g. LexA).
4. Transgene Libraries to Assess Gene Function
4.1 Transgenes and Transgenic libraries
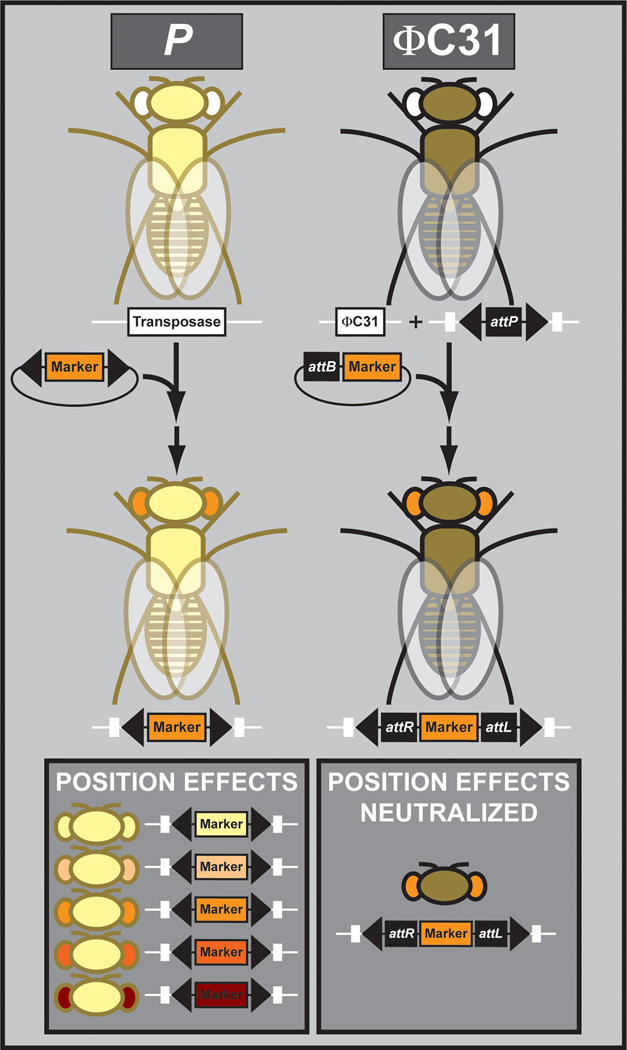
In the past, transgenesis in Drosophila solely relied on the P element transposon (Venken and Bellen, 2007). Unfortunately, the insertion site of transposons can not be controlled (see also Section 3.1), resulting in variable position effects caused by the surrounding genome, making it difficult to compare differently mutagenized transgenes (Figure 13). On the contrary, the site-specific integration of transgenes via the use of a site-specific integrase eliminates this issue. More specifically, the ΦC31 integrase allows for site-specific integration of attB containing transgenes at defined attP docking sites in the genome. Since all constructs can be integrated at the same genomic location, the influence of position effects on the integrated transgene is neutralized. The ΦC31 system additionally allows for the integration of small to very large fragments. Hence, the ΦC31 system is ideally suited to compare transgenes obtained from genome-wide libraries generated for specific purposes. Hence, due to these advantages, this overview will focus on those libraries generated using vectors compatible with the ΦC31 site-specific integrase system. Currently, ΦC31-based libraries are available for several genome-wide interrogation paradigms: genomic rescue (Venken et al., 2009; Ejsmont et al., 2009; Szabad et al., 2012), RNAi (Dietzl et al., 2007; Ni et al., 2009; Ni et al., 2011), regulatory element interrogation (Jenett et al., 2012; Pfeiffer et al., 2008; Manning et al., 2012; Jory et al., 2012), and cDNA overexpression (Bischof et al., 2013; Schertel et al., 2013). These are outlined below.
Figure 13. Transgenesis platforms.
P element transposase catalyzed transgenesis results in uncontrolled transgene integration and variable position effects, while ΦC31 integrase catalyzed transgenesis results in site-specific integration and the neutralization of position effects.
4.2 Genomic Rescue and Recombineering
Genomic rescue refers to the use of genomic DNA fragments to revert or “rescue” phenotypes associated with a given mutation (Figure 14). Upon evidence of phenotype recovery one can manipulate the rescue construct for various downstream applications. For example, rescue constructs can be tagged to help identify cells in which the gene of interest is expressed or determine the subcellular protein distribution. One can also create numerous mutations in the rescue construct to initiate structure function analysis, testing protein variants in a null mutant background.
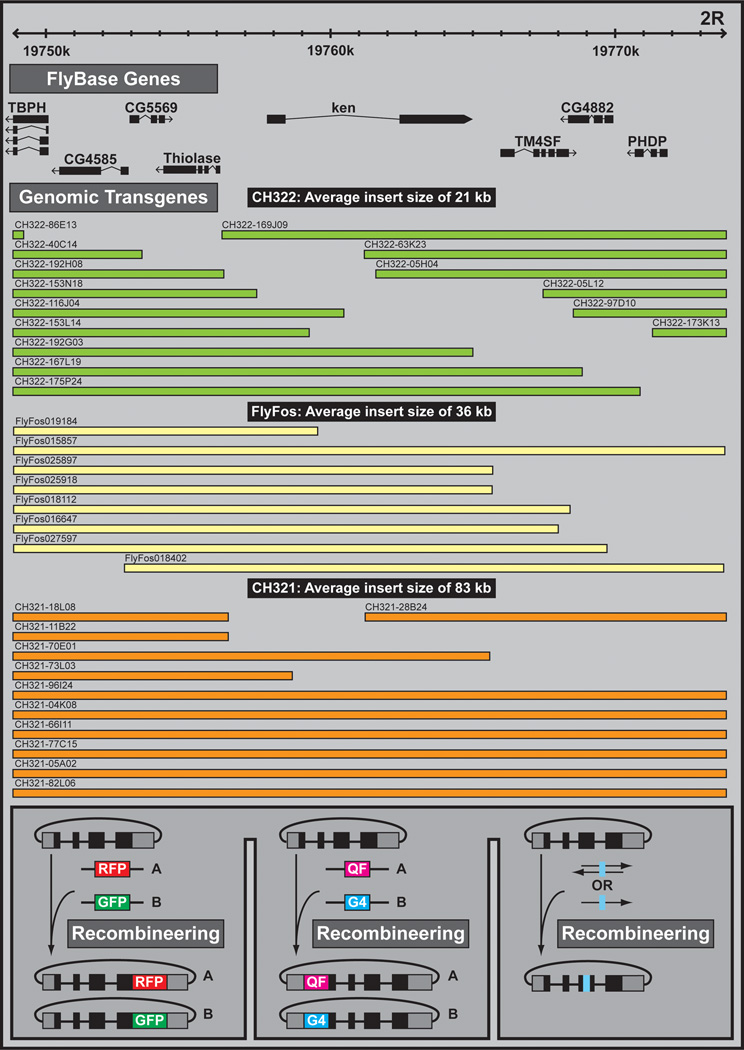
Figure 14. Genomic rescue transgenes and recombineering.
Genomic transgene fragments can be chosen from three genomic DNA libraries: CH322 clones with an average insert size of 21 kb (green), FlyFos clones with an average insert size of 36 kb (yellow), or CH321 clones with an average insert size of 83 kb (orange). Shown is a region surrounding the gene ken and barbie (ken). Subsequently, recombineering can be used to modify the genomic rescue fragment, to include insertions (e.g. fluorescent tags for protein visualization, and binary factors for host gene cellular visualization) or point mutations.
The scope of genomic rescue expanded significantly with the development of the P[acman] transgenesis platform (P/ΦC31 artificial chromosome for manipulation) (Venken et al., 2006). P[acman] combines three powerful technologies: recombineering (Sharan et al., 2009), conditionally amplifiable BACs (Wild et al., 2002), and ΦC31 site-specific integration (Groth et al., 2004; Bischof et al., 2007). First, recombineering, which allows the incorporation of virtually any genetic alteration, was used in the P[acman] system to retrieve DNA fragments up to 102 kb in size from existing BAC libraries via a single gap-repair (Hoskins et al., 2000) (https://bacpac.chori.org/). A double gap-repair procedure, wherein two pieces of overlapping BACs were joined together, allowed for the generation of a 133 kb fragment, reconstituting the Tenascin major gene (Venken et al., 2006). Second, DNA fragments recovered from a BAC are maintained at low-copy number in a conditionally amplifiable plasmid, ensuring the stability of large constructs under normal conditions. Addition of a simple sugar solution induces plasmid replication and results in high copy number, which facilitates cloning and DNA preparation (Wild et al., 2002). Third, the ΦC31 integrase allows the integration of P[acman] vectors into defined attP docking sites located in the fly genome (Venken et al., 2006; Groth et al., 2004; Bischof et al., 2007; Markstein et al., 2008). While the ΦC31 integrase is able to integrate plasmids up to at least 146 kb in vivo (Venken et al., 2006; Zheng et al., 2011), recently we were able to integrate large BACs derived from previously generated libraries (Hoskins et al., 2000) with an upper size limit of 212 kb. These integrations followed appropriate BAC retrofitting (Kondo et al., 2009) and included the necessary elements for P[acman] transgenesis (OriV conditional origin of replication for conditional plasmid copy number amplification, attB site for site-specific integration using the ΦC31 integrase, and the white+ dominant eye color transgenesis marker) (Venken et al., 2006) (Venken, unpublished data).
P[acman] methodology has been used to create three genome-wide Drosophila P[acman] BAC DNA libraries: the CHORI-322 P[acman] library (average insert size of 21 kb), the CHORI-321 P[acman] library (average insert size of 83 kb in P[acman] (http://www.pacmanfly.org/) (Venken et al., 2009)), and the FlyFos library (Ejsmont et al., 2009) (average insert size of 36 kb in a fosmid instead of the P[acman] backbone) (http://transgeneome.mpi-cbg.de/). The three libraries have been end sequenced and annotated onto the reference fly genome. While the FlyFos library provides a 3.3-fold genome coverage, the CHORI-321 and CHORI-322 libraries represent a 12-fold coverage and allow rescue of more than 95% of annotated fly genes. Thus, the libraries largely eliminate the need for gap-repair for genomic rescue. Compared to gap-repair, which requires additional cloning steps and allows defined genomic fragments to be analyzed, clones used in the libraries were randomly generated and often contain several genes. Although this may appear to be a weakness of the libraries, small gene duplications up to 100 kb rarely cause phenotypes (Venken et al., 2010) and larger clones are more likely to include all the necessary regulatory elements of a gene. The functionality of the three libraries was tested by integrating numerous clones into attP docking sites and showing that they rescue phenotypes associated with known mutations in the corresponding region of interest.
The amplifiable nature of P[acman] and FlyFos clones greatly facilitates recombineering. Prior to amplification, tags and mutations are introduced at low-copy number and fixed within a bacterial colony. These methods have already been used with P[acman] and FlyFos to (1) introduce deletions of regulatory elements for structure/function analysis (Pepple et al., 2008; Perry et al., 2010; Enneking et al., 2013; Cassidy et al., 2013), (2) incorporate protein tags for visualization of expression or acute protein inactivation (Venken et al., 2008; Venken et al., 2009; Ejsmont et al., 2009; Negre et al., 2011; Verstreken et al., 2009; Verstreken et al., 2005), and (3) integrate binary factors (e.g. GAL4 or QF) for cellular labeling (Stowers, 2011). Hence, tagged clones can be used to determine the expression patterns of numerous uncharacterized genes or label specific cell populations.
4.3 Transgenic Duplication Kit
The X chromosome of Drosophila melanogaster contains 22 Mb of euchromatic DNA, encompassing approximately 2,300 protein-coding genes (Adams et al., 2000). Mutations in essential genes or those involved in male fertility are often tedious to map unless a duplication is present on another chromosome that can be brought in through simple genetic crosses. Recently, two groups independently generated a set of duplications to expedite the mapping of X chromosome mutations (see also Section 2.6) (Figure 5B). One group made use of the available P[acman] libraries (Venken et al., 2010). In doing so, a minimal tiling path of 408 overlapping clones was integrated into a single attP docking site located on the 3rd chromosome (Groth et al., 2004; Bischof et al., 2007). Currently, 96% of the X chromosome is covered by the P[acman] duplications and contained in 382 fly strains. Approximately 100 clones were tested for rescue, and 92% rescued existing mutations as well as entire deficiencies previously generated with the FLP/FRT system (Parks et al., 2004; Ryder et al., 2004; Ryder et al., 2007). Interestingly, this group demonstrated that most genes are tolerated at twice the normal dosage, and the duplication collection enabled more precise mapping of two regions involved in diplolethality. The second group to generate an X chromosome duplication kit did so by genetic means, incorporating relatively large duplications of the X chromosome onto the Y (attached-XY). Due to the size of the duplications, this kit allows quick but rough mapping and currently covers 90% of the X chromosome(Cook et al., 2010b). Both duplication collections are available at the BDSC: http://flystocks.bio.indiana.edu/Browse/dp/BDSC-Dps.php.
4.4 Genome-Wide RNAi
There are currently two genome-wide RNAi libraries for Drosophila (http://stockcenter.vdrc.at/)—both created in the lab of Barry Dickson. These libraries use RNAi technology to knock down genes, aiding functional analysis. The first library, the GD library, was generated using P element transposons (Dietzl et al., 2007). However, since P element insertion sites cannot be controlled, a second library—KK—was created using the ΦC31 system. Other genome-wide RNAi resources are currently being generated to complement these two libraries. These include an independent collection based on long dsRNA as well as a collection based on short hairpins using a miRNA backbone (Ni et al., 2009; Ni et al., 2011). These and other RNAi reagents are discussed in detail in a subsequent chapter (Mohr et al., 2014).
4.5 Genome-wide Enhancer Bashing
The GAL4/UAS system has been extremely useful for interrogation of regulatory gene elements (Brand and Perrimon, 1993). Notably, many of the driver lines used in GAL4/UAS experiments were isolated by hopping P element enhancer detector elements (O′Kane and Gehring, 1987) carrying the binary GAL4 transactivator (Brand and Perrimon, 1993). Since P elements tend to integrate into the regulatory elements of genes, the neutral promoter driving GAL4 or lacZ expression comes under the control of endogenous enhancers (Bellen et al., 1989; Wilson et al., 1989; Bier et al., 1989; Brand and Perrimon, 1993). As such, many enhancer detector insertion strains display expression patterns that correspond to that of a nearby gene. However, expression can often not be limited to a small subset of cells or cell populations. This lack of cellular specificity is an issue if one desires to remove, functionally inactivate, or manipulate a few cells in a complex circuit (e.g. neurons).
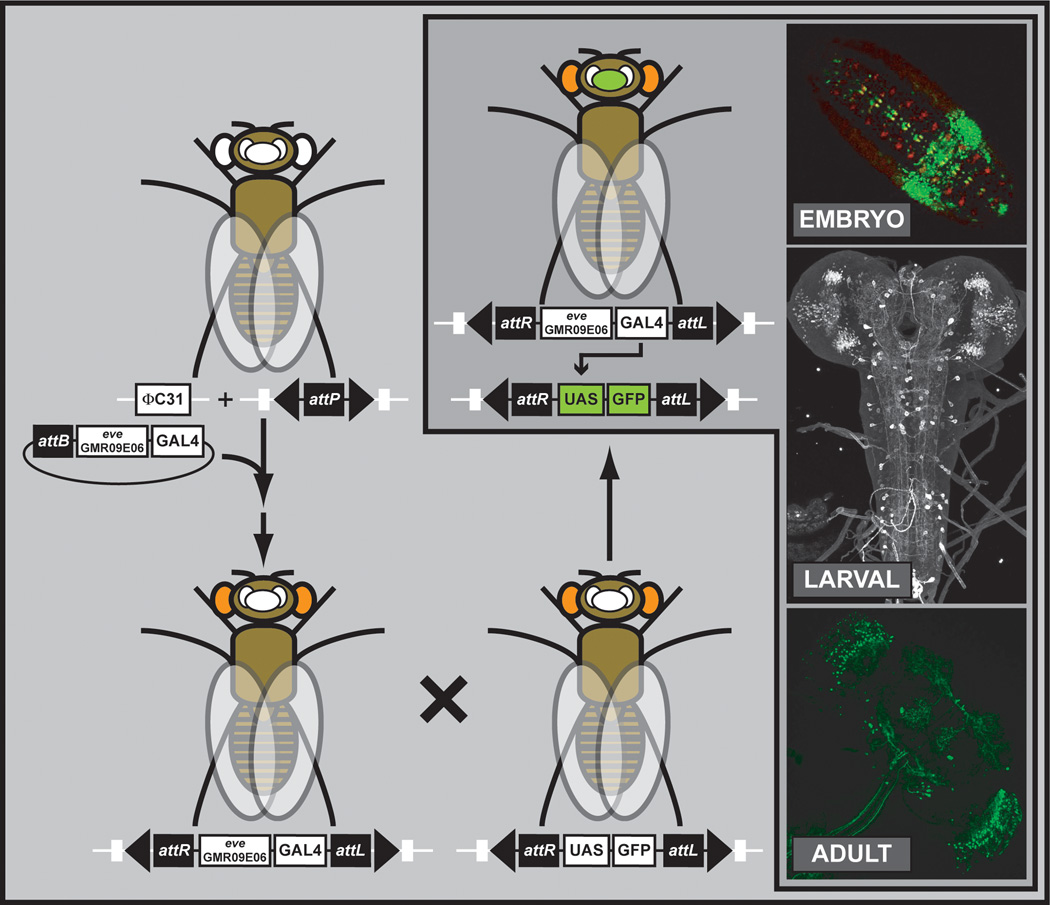
To isolate the individual components of a gene’s complex regulatory environment, it typically suffices to isolate smaller DNA fragments and bring them in front of a neutral promoter—the so-called enhancer bashing strategy. As position effects may influence the analysis of different regulatory regions, it is advantageous to integrate the different constructs into the same “neutral” site. Hence, a large set of ΦC31 attP docking sites were analyzed for position effects (Markstein et al., 2008), and one, attP2, with virtually no leaky expression, was selected to integrate the majority of the transgenic GAL4 library (Pfeiffer et al., 2008) (Figure 15).
Figure 15. Genome-wide Enhancer Bashing.
Thousand of enhancer fragments were fused to GAL4 and assayed for gene expression patterns. The GMR09E06 enhancer fragment of the even skipped (eve) is shown as an example. Expression patterns are shown for the embryo, larval brain and ventral nerve cord, and the adult brain. Images were obtained from the Janelia Farm Research Campus website summarizing the project (http://www.janelia.org/gal4-gen1).
The initial GAL4 collection is biased towards genes expressed in the nervous system (Pfeiffer et al., 2008), and the resulting fly strains (~ 7,000) have been screened for expression in a variety of tissues. Expression patterns were first established in the adult brain and ventral nerve cord (Jenett et al., 2012), where novel neuronal cell types were identified and brain asymmetry revealed. Interestingly, many lines expressed GAL4 in a pattern broader than anticipated, especially since regulatory elements of only 3 kb were used. In a second study, a subset of 5,000 transgenic lines was used to document expression patterns in the embryonic central nervous system at two different embryonic stages (i.e. stages 9–11, and stage 16) (Manning et al., 2012). The expression data will be valuable for determining the morphology and function of all neurons in the embryonic CNS. Finally, the lines were tested for expression patterns in third-instar imaginal discs, specifically the eye, antenna, leg, wing, haltere, and genital discs (Jory et al., 2012). In this study, DNA sequence motifs provided information about candidate transcription factors that may regulate enhancers. All the expression data obtained from these studies is available through Janelia Farm Research Campus (http://flweb.janelia.org/cgi-bin/flew.cgi). A second collection GAL4 collection, the VT collection, has also been generated using the same technology (Pfeiffer et al., 2008). Expression data for these lines have been obtained for embryos (Kvon et al., 2012) and the adult brain (http://stockcenter.vdrc.at/control/vtlibrary). To perform independent manipulations of different cell types, several of the fragments have been fused to an orthogonal binary activation system that consists of the transcriptional activator LexA that binds to LexO operators. Numerous LexA driven lines and reporters have been generated (Yagi et al., 2010; Diegelmann et al., 2008; Pfeiffer et al., 2010; Shearin et al., 2013; Gohl et al., 2011).
One of the driving goals of the GAL4 collection was to identify small subsets of labeled neurons. This was only partly achieved, and the same libraries have since been fused to the recently optimized split GAL4 system (Luan et al., 2006; Pfeiffer et al., 2010). In the split GAL4 system, the GAL4 transcription factor is split into two hemidrivers, one containing the DNA binding domain and the other containing the activation domain. Each hemidriver is driven by separate regulatory elements. Since each hemidriver cannot activate expression of the reporter, expression is not observed. However, the location where the expression domains overlap, the two halves of GAL4 can heterodimerize, and reconstitute a functional activator that can activate the expression of a reporter gene. Thus, expression patterns can be narrowed to a small overlapping cell population shared between two much larger expression domains. Other intersectional strategies follow along a similar vein and involve combining two sets of overlapping hemi-drivers, e.g. the xplit LexA system (Ting et al., 2011).
4.6 Genome-Wide Overexpression and In Vivo Transgene Upgrading
A final set of genome-wide reagents based on the ΦC31 integrase was recently initiated—the UAS ORFeome, a transgene collection aimed to represent at least one cDNA isoform of each Drosophila gene (Bischof et al., 2013). This library, which represents a large collection of genes regulating proliferation, cell cycle, and cell size, greatly facilitates large scale overexpression studies by using any GAL4 driver of interest to drive expression of the UAS-linked open reading frames of target genes. By using misexpression of genes, problems arising from genetic redundancy are prevented and novel gene functions can be uncovered. To date, 1,149 UAS-ORF fly lines have been created. As proof of principle, researchers cloned 655 growth-regulating genes in two variants—one with the open reading frame and a native stop codon, the other an open reading frame with a C-terminal 3xHA tag prior to the stop codon for affinity purification studies. Interestingly, to streamline the creation of transgenic flies, pools of plasmids were injected into embryos. Each plasmid, in addition to a single cDNA, contains a unique barcode serving as the plasmid identifier after retrieval of transgenic flies from the pooled injections.
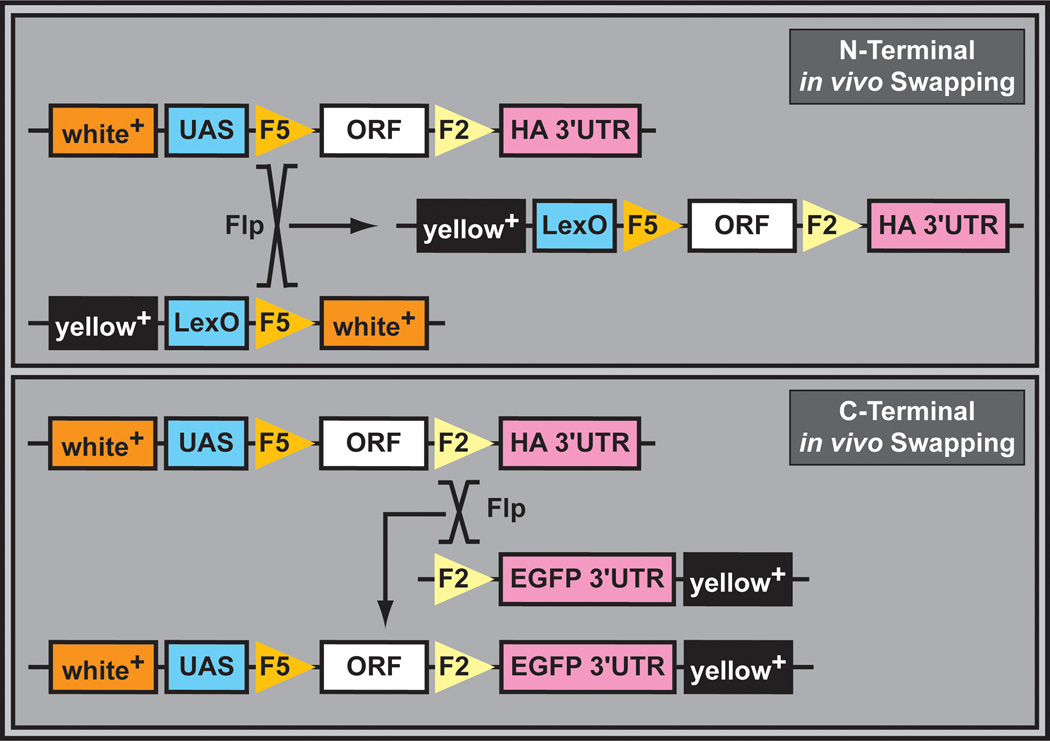
The versatility of the ORF collection was greatly expanded with the development of an in vivo swapping technique (Figure 16). Based on mitotic recombination (i.e. using the FLP recombinase), this system enables the rapid exchange of promoters and epitope tags through simple crosses. Promoters, N- or C-terminal tags, and polyadenylation signals can be exchanged at will. In vivo swapping was validated by changing promoters (e.g. UAS into LexO) and C-terminal protein tags (e.g. 3xHA into EGFP). To confirm the usefulness of this library for screening purposes, Wingless (Wg) pathway components were screened for a variety of phenotypic outputs (Schertel et al., 2013). Three serine/threonine kinases were identified as regulators of Wg signaling. One of these, Nek2, optimizes the Wg pathway response by direct phosphorylation of Dishevelled. While the ORF collection is likely to be a very valuable resource for studying growth regulation in vivo, similar caveats associated with EP and EY screens should be kept in mind (See also Section 3.4).
Figure 16. In vivo transgene upgrading through mitotic recombination.
N-terminal in vivo swapping, catalyzed by the FLP recombinase, results in exchange of the UAS promoter with novel promoter elements (e, g, LexO). Desired recombination events can be identified by exchange of the white+ marker with the yellow+ marker. C-terminal in vivo swapping, catalyzed by the FLP recombinase, results in exchange of the HA tag and the 3’UTR with the fluorescent EGFP tag and potentially a different 3’UTR. Desired recombination events can be screened by the simultaneous occurrence of the white+ and yellow+ markers.
5. Future Directions
One of the striking features of technology development is that relatively few methods are truly innovative and powerful on their own. Although exceptions such as the development of P element transgenesis (Rubin and Spradling, 1982; Spradling and Rubin, 1982), PCR (Saiki et al., 1988) and monoclonal antibodies (Kohler and Milstein, 1975) come to mind, very often seemingly less important technical innovations can be combined to create very powerful toolsets. For example, P element-mediated enhancer detection (O′Kane and Gehring, 1987) was shown to be a powerful methodology to identify new genes and their expression patterns. Having a “neutral and weak” promoter controlling LacZ expression in a transposon permitted the detection of innumerable gene expression patterns (Bellen et al., 1989; Bier et al., 1989; Wilson et al., 1989). However, it only became a major forerunner when combined with GAL4/UAS technology. This binary system from yeast had been shown to be efficient in flies (Fischer et al., 1988) and mice (Ornitz et al., 1991), but only when the LacZ of the enhancer detector system was replaced with GAL4 and a UAS-LacZ transgene was inserted in the genome, did both technologies (enhancer detection and GAL4/UAS binary system) develop their full potential. The GAL4/UAS methodology (Brand and Perrimon, 1993) ultimately is the most used technology in flies.
Although numerous methods are available to tackle scientific questions, often too little time is spent in weighing the quality, depth, and caveats of various approaches. Very often quick technological paradigms are available but they often have major limitations that are ignored. For example, RNAi against a given gene may be determined to cause a phenotype, but the data should be corroborated with mutations that are already publicly available. In addition, effeciency and specificity of RNAi should be tested. Similarly, when new mutations are isolated with chemical mutagenesis screens, it is important to either isolate another allele in the same complementation group or to characterize RNAis that cause the same or very similar phenotypes. Alternatively, a genomic rescue construct carrying the gene of interested should be tested. Indeed, the number of polymorphisms in some stocks is staggering, and many stocks that we tested carry mutations in two essential genes (Venken et al., 2010). Although these second site mutations can easily be outcrossed, this often seems to be a concern solely to/ behavioral biologists. Given the arsenal of tools that are available (Venken et al., 2011b; St Johnston D., 2013), this trend should be reversed.
In summary, we believe that given the very broad tool box that is currently available in the Drosophila community, and in particular the innumerable combinations and permutations that can be engineered, the future of Drosophila to study novel and well established biological processes is extremely promising. However, we also feel that the toolsets need to be used with caution and that the users need to know the strengths and weaknesses of these resources. This chapter should provide a starting point for researchers using these tools.
Acknowledgements
We apologize to those whose work we did not cite because of our focus and space limitations. We would like to thank Pavel Tomancak for help with appropriate representation of Figure 13, and Kristi Hoffman, Audrey Christiansen and Karen Schulze for comments on the manuscript. KJTV is supported by startup funds kindly provided by Baylor College of Medicine and the McNair Medical Institute, and grants from the Cancer Prevention and Research Institute of Texas, and the National Institutes of Health (1R21HG006726). HJB is supported by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhães TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, ndrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de PB, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook and Manual. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Azad P, Zhou D, Zarndt R, Haddad GG. Identification of genes underlying hypoxia tolerance in Drosophila by a P-element screen. G3 (Bethesda.) 2012;2:1169–1178. doi: 10.1534/g3.112.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-Specific Transformation of Drosophila via ϕC31 Integrase-Mediated Cassette Exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, Hoskins RA, Spradling A. The Drosophila Gene Disruption Project: Progress Using Transposons With Distinctive Site-Specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, O′Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Tong C, Tsuda H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 2010;11:514–522. doi: 10.1038/nrn2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. U. S. A. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Bischof J, Bjorklund M, Furger E, Schertel C, Taipale J, Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific ΦC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokel C. EMS screens : from mutagenesis to screening and mapping. Methods Mol. Biol. 2008;420:119–138. doi: 10.1007/978-1-59745-583-1_7. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJ, Ji J, Jiang H, Bellen HJ, White KP, Carthew RW. miR-9a Minimizes the Phenotypic Impact of Genomic Diversity by Buffering a Transcription Factor. Cell. 2013;155:1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro JP, Carareto CM. Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica. 2004;121:107–118. doi: 10.1023/b:gene.0000040382.48039.a2. [DOI] [PubMed] [Google Scholar]
- Cook KR, Parks AL, Jacobus LM, Kaufman TC, Matthews KA. New research resources at the Bloomington Drosophila Stock Center. Fly. 2010a;4:88–91. doi: 10.4161/fly.4.1.11230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Deal ME, Deal JA, Garton RD, Brown CA, Ward ME, Andrade RS, Spana EP, Kaufman TC, Cook KR. A new resource for characterizing X-linked genes in Drosophila melanogaster: systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics. 2010b;186:1095–1109. doi: 10.1534/genetics.110.123265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diantonio A, Parfitt KD, Schwarz TL. Synaptic transmission persists in synaptotagmin mutants of Drosophila. Cell. 1993;73:1281–1290. doi: 10.1016/0092-8674(93)90356-u. [DOI] [PubMed] [Google Scholar]
- Diegelmann S, Bate M, Landgraf M. Gateway cloning vectors for the LexA-based binary expression system in Drosophila. Fly. (Austin) 2008;2:236–239. doi: 10.4161/fly.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dudai Y, Jan YN, Byers D, Quinn WG, Benzer S. dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. U. S. A. 1976;73:1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsmont RK, Sarov M, Winkler S, Lipinsk KA, Tomancak P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods. 2009;6:435–437. doi: 10.1038/nmeth.1334. [DOI] [PubMed] [Google Scholar]
- Enneking EM, Kudumala SR, Moreno E, Stephan R, Boerner J, Godenschwege TA, Pielage J. Transsynaptic coordination of synaptic growth, function, and stability by the L1-type CAM Neuroglian. PLoS. Biol. 2013;11:e1001537. doi: 10.1371/journal.pbio.1001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Golic MM. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics. 1996;144:1693–1711. doi: 10.1093/genetics/144.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O′Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage ΦC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Hendrix D, Trang V, Levine M. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM, Harrell RA. Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol. Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- Horn C, Offen N, Nystedt S, Hacker U, Wimmer EA. piggyBac-based H sertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics. 2003;163:647–661. doi: 10.1093/genetics/163.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev. Genes Evol. 2000;210:630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, Tabor PE, Bailey MR, DeShazo DR, Catanese J, Mammoser A, Osoegawa K, de Jong PJ, Celniker SE, Gibbs RA, Rubin GM, Scherer SE. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science. 2000;287:2271–2274. doi: 10.1126/science.287.5461.2271. [DOI] [PubMed] [Google Scholar]
- Hotta Y, Benzer S. Genetic dissection of the Drosophila nervous system by means of mosaics. Proc. Natl. Acad. Sci. U. S. A. 1970;67:1156–1163. doi: 10.1073/pnas.67.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Klambt C. P-element mutagenesis. Methods Mol. Biol. 2008;420:97–117. doi: 10.1007/978-1-59745-583-1_6. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev. Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SC, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jory A, Estella C, Giorgianni MW, Slattery M, Laverty TR, Rubin GM, Mann RS. A survey of 6, 300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Rep. 2012;2:1014–1024. doi: 10.1016/j.celrep.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kania A, Salzberg A, Bhat M, D’Evelyn D, He Y, Kiss I, Bellen HJ. P-element mutations affecting embryonic peripheral nervous system development in Drosophila melanogaster. Genetics. 1995;139:1663–1678. doi: 10.1093/genetics/139.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Koana T, Hotta Y. Isolation and characterization of flightless mutants in Drosophila melanogaster. J. Embryol. Exp. Morphol. 1978;45:123–143. [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kondo S, Booker M, Perrimon N. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics. 2009;183:1165–1173. doi: 10.1534/genetics.109.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Stampfel G, Yanez-Cuna JO, Dickson BJ, Stark A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012;26:908–913. doi: 10.1101/gad.188052.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafave MC, Sekelsky J. Transcription initiation from within P elements generates hypomorphic mutations in Drosophila melanogaster. Genetics. 2011;188:749–752. doi: 10.1534/genetics.111.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB, Bacher F. Methods of feeding ethyl methane sulfonate (EMS) to Drosophila males. Drosophila Information Service. 1968;43 [Google Scholar]
- Liebl FL, Werner KM, Sheng Q, Karr JE, McCabe BD, Featherstone DE. Genome-wide P-element screen for Drosophila synaptogenesis mutants. J. Neurobiol. 2006;66:332–347. doi: 10.1002/neu.20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- Littleton JT, Bellen HJ. Genetic and phenotypic analysis of thirteen essential genes in cytological interval 22F1-2; 23B1-2 reveals novel genes required for neural development in Drosophila. Genetics. 1994;138:111–123. doi: 10.1093/genetics/138.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Stern M, Perin M, Bellen HJ. Calcium dependence of neurotransmitter release and rate of spontaneous vesicle fusions are altered in Drosophila synaptotagmin mutants. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10888–10892. doi: 10.1073/pnas.91.23.10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, Deng WM, Jiao R. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics. 2012;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Loukeris TG, Arca B, Livadaras I, Dialektaki G, Savakis C. Introduction of the transposable element Minos into the germ line of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9485–9489. doi: 10.1073/pnas.92.21.9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich T, Asztalos Z, Awano W, Baba K, Kondo S, Niwa S, Yamamoto D. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics. 2001;157:727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning L, Heckscher ES, Purice MD, Roberts J, Bennett AL, Kroll JR, Pollard JL, Strader ME, Lupton JR, Dyukareva AV, Doan PN, Bauer DM, Wilbur AN, Tanner S, Kelly JJ, Lai SL, Tran KD, Kohwi M, Laverty TR, Pearson JC, Crews ST, Rubin GM, Doe CQ. A resource for manipulating gene expression and analyzing cis-regulatory modules in the Drosophila CNS. Cell Rep. 2012;2:1002–1013. doi: 10.1016/j.celrep.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SQ, Hiesinger PR, Beronja S, Zhai RG, Schulze KL, Verstreken P, Cao Y, Zhou Y, Tepass U, Crair MC, Bellen HJ. Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron. 2005;46:219–232. doi: 10.1016/j.neuron.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley. Interdiscip. Rev. RNA. 2012;3:145–158. doi: 10.1002/wrna.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar C, Lopez-Varea A, Hernandez R, de Celis JF. A gain-of-function screen identifying genes required for vein formation in the Drosophila melanogaster wing. Genetics. 2006;174:1635–1659. doi: 10.1534/genetics.106.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz K, Zipperlen P, Dearolf C, Basler K, Hafen E. A reverse genetic screen in Drosophila using a deletion-inducing mutagen. Genome Biol. 2004;5:R83. doi: 10.1186/gb-2004-5-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba J, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Wirtz-Peitz F, Lee S, Kwon Y, Buckner M, Hoskins RA, Venken KJ, Bellen HJ, Mohr SE, Perrimon N. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190:931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norga KK, Gurganus MC, Dilda CL, Yamamoto A, Lyman RF, Patel PH, Rubin GM, Hoskins RA, Mackay TF, Bellen HJ. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 2003;13:1388–1396. doi: 10.1016/s0960-9822(03)00546-3. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O′Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc. Natl. Acad. Sci. U. S. A. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak WL, Grossfield J, Arnold KS. Mutants of the visual pathway of Drosophila melanogaster. Nature. 1970;227:518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, al-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Patton JS, Gomes XV, Geyer PK. Position-independent germline transformation in Drosophila using a cuticle pigmentation gene as a selectable marker. Nucleic Acids Res. 1992;20:5859–5860. doi: 10.1093/nar/20.21.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepple KL, Atkins M, Venken K, Wellnitz K, Harding M, Frankfort B, Mardon G. Two-step selection of a single R8 photoreceptor: a bistable loop between senseless and rough locks in R8 fate. Development. 2008;135:4071–4079. doi: 10.1242/dev.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- feiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS. ONE. 2010;5:e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, Wang S, Castiblanco C, Buszczak M, Hoskins RA, Cooley L. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman RR, Swan JM, Geyer PK. A Drosophila insulator protein facilitates dosage compensation of the X chromosome min-white gene located at autosomal insertion sites. Development. 1995;121:3573–3582. doi: 10.1242/dev.121.11.3573. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, Reuter G, Baisch H, Apelt C, Kauk A, Rudolph T, Kube M, Klimm M, Nickel C, Szidonya J, Maroy P, Pal M, Rasmuson-Lestander A, Ekstrom K, Stocker H, Hugentobler C, Hafen E, Gubb D, Pflugfelder G, Dorner C, Mechler B, Schenkel H, Marhold J, Serras F, Corominas M, Punset A, Roote J, Russell S. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, O′Kane CJ, Huen D, Sharma P, Asztalos Z, Baisch H, Schulze J, Kube M, Kittlaus K, Reuter G, Maroy P, Szidonya J, Rasmuson-Lestander A, Ekstrom K, Dickson B, Hugentobler C, Stocker H, Hafen E, Lepesant JA, Pflugfelder G, Heisenberg M, Mechler B, Serras F, Corominas M, Schneuwly S, Preat T, Roote J, Russell S. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Russell S. Transposable elements as tools for genomics and genetics in Drosophila. Brief. Funct. Genomic. Proteomic. 2003;2:57–71. doi: 10.1093/bfgp/2.1.57. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schertel C, Huang D, Bjorklund M, Bischof J, Yin D, Li R, Wu Y, Zeng R, Wu J, Taipale J, Song H, Basler K. Systematic screening of a Drosophila ORF library in vivo uncovers Wnt/Wg pathway components. Dev. Cell. 2013;25:207–219. doi: 10.1016/j.devcel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin HK, Dvarishkis AR, Kozeluh CD, Stowers RS. Expansion of the gateway multisite recombination cloning toolkit. PLoS ONE. 2013;8:e77724. doi: 10.1371/journal.pone.0077724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Bellen HJ, Hoskins RA. Drosophila P elements preferentially transpose to replication origins. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15948–15953. doi: 10.1073/pnas.1112960108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. Using mutants, knockdowns, and transgenesis to investigate gene function in Drosophila. Wiley. Interdiscip. Rev. Dev. Biol. 2013;2:587–613. doi: 10.1002/wdev.101. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- Staudt N, Molitor A, Somogyi K, Mata J, Curado S, Eulenberg K, Meise M, Siegmund T, Hader T, Hilfiker A, Bronner G, Ephrussi A, Rorth P, Cohen SM, Fellert S, Chung HR, Piepenburg O, Schafer U, Jackle H, Vorbruggen G. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS. Genet. 2005;1:e55. doi: 10.1371/journal.pgen.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS. An efficient method for recombineering GAL4 and QF drivers. Fly. (Austin.) 2011;5 doi: 10.4161/fly.5.4.17560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabad J, Bellen HJ, Venken KJ. An assay to detect in vivo Y chromosome loss in Drosophila wing disc cells. G3 (Bethesda) 2012;2:1095–1102. doi: 10.1534/g3.112.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Tien AC, Rajan A, Schulze KL, Ryoo HD, Acar M, Steller H, Bellen HJ. Ero1L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J. Cell Biol. 2008;182:1113–1125. doi: 10.1083/jcb.200805001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Gu S, Guttikonda S, Lin TY, White BH, Lee CH. Focusing transgene expression in Drosophila by coupling Gal4 with a novel split-LexA expression system. Genetics. 2011;188:229–233. doi: 10.1534/genetics.110.126193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunstall NE, Herr A, de BM, Warr CG. A screen for genes expressed in the olfactory organs of Drosophila melanogaster identifies genes involved in olfactory behaviour. PLoS. ONE. 2012;7:e35641. doi: 10.1371/journal.pone.0035641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A, Mito T, Noji S, Ueda R, Kawakami K. Transposition of the vertebrate Tol2 transposable element in Drosophila melanogaster. Gene. 2008;425:64–68. doi: 10.1016/j.gene.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 2005;6:167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134:3571–3584. doi: 10.1242/dev.005686. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ. Genome-Wide Manipulations of Drosophila melanogaster with Transposons, Flp Recombinase, and PhiC31 Integrase. Methods Mol. Biol. 2012;859:203–228. doi: 10.1007/978-1-61779-603-6_12. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Kasprowicz J, Kuenen S, Yan J, Hassan BA, Verstreken P. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 2008;36:e114. doi: 10.1093/nar/gkn486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics. 2010;186:1111–1125. doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, Bellen HJ. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011a;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011b;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Ohyama T, Haueter C, Habets RL, Lin YQ, Swan LE, Ly CV, Venken KJ, De CP, Bellen HJ. Tweek, an evolutionarily conserved protein, is required for synaptic vesicle recycling. Neuron. 2009;63:203–215. doi: 10.1016/j.neuron.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Pearson RK, Bellen HJ, O′Kane CJ, Grossniklaus U, Gehring WJ. P-element-mediated enhancer detection: an efficient method for isolating and (Tcharacterizing developmentally regulated genes in Drosophila. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- Witsell A, Kane DP, Rubin S, McVey M. Removal of the bloom syndrome DNA helicase extends the utility of imprecise transposon excision for making null mutations in Drosophila. Genetics. 2009;183:1187–1193. doi: 10.1534/genetics.109.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff RC, Ashburner M. The genetics of a small autosomal region of Drosophila melanogaster containing the structural gene for alcohol dehydrogenase. I. Characterization of deficiencies and mapping of ADH and visible mutations. Genetics. 1979a;92:117–132. doi: 10.1093/genetics/92.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff RC, Ashburner M. The genetics of a small autosomal region of Drosophila melanogaster containing the structural gene for alcohol dehydrogenase. II. Lethal mutations in the region. Genetics. 1979b;92:133–149. doi: 10.1093/genetics/92.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yagi R, Mayer F, Basler K. Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16166–16171. doi: 10.1073/pnas.1005957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Charng WL, Rana NA, Kakuda S, Jaiswal M, Bayat V, Xiong B, Zhang K, Sandoval H, David G, Wang H, Haltiwanger RS, Bellen HJ. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science. 2012;338:1229–1232. doi: 10.1126/science.1228745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, Greenbaum MP, Bellen HJ. Mapping Drosophila mutations with molecularly defined P element insertions. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Michelson Y, Freger V, Avraham Z, Venken KJ, Bellen HJ, Justice MJ, Wides R. Drosophila Ten-m and filamin affect motor neuron growth cone guidance. PLoS. ONE. 2011;6:e22956. doi: 10.1371/journal.pone.0022956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K, Parnas D, Fetter RD, Isacoff EY, Goodman CS. Watching a synapse grow: noninvasive confocal imaging of synaptic growth in Drosophila. Neuron. 1999;22:719–729. doi: 10.1016/s0896-6273(00)80731-x. [DOI] [PubMed] [Google Scholar]