Abstract
Rationale
Increasing evidence has shown that proper control of mitochondrial dynamics (fusion and fission) is required for high capacity ATP production in heart. The transcriptional coactivators, peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1) α and β have been shown to regulate mitochondrial biogenesis in heart at the time of birth. The function of the PGC-1 coactivators in heart after birth is incompletely understood.
Objective
To assess the role of the PGC-1 coactivators during postnatal cardiac development and in the adult heart in mice.
Methods and Results
Conditional gene targeting was used in mice to explore the role of the PGC-1 coactivators during postnatal cardiac development and in adult heart. Marked mitochondrial structural derangements were observed in hearts of PGC-1α/β-deficient mice during postnatal growth, including fragmentation and elongation, associated with the development of a lethal cardiomyopathy. The expression of genes involved in mitochondrial fusion [mitofusin 1 (Mfn1), optic atrophy 1 (Opa1)] and fission [dynamin-related protein 1 (Drp1), fission protein 1 (Fis1)] was altered in hearts of PGC-1α/β-deficient mice. PGC-lα was shown to directly regulate Mfn1 gene transcription by coactivating the estrogen-related receptor α (ERRα) upon a conserved DNA element. Surprisingly, PGC-1α/β deficiency in the adult heart did not result in evidence of abnormal mitochondrial dynamics or heart failure. However, transcriptional profiling demonstrated that the PGC-1 coactivators are required for high level expression of nuclear- and mitochondrial-encoded genes involved in mitochondrial dynamics and energy transduction in adult heart.
Conclusion
These results reveal distinct developmental stage-specific programs involved in cardiac mitochondrial dynamics.
Keywords: PGC-1 coactivators, mitochondrial fusion, cardiac energy metabolism, cardiomyopathy, mitofusin
INTRODUCTION
The high energy demands of the postnatal heart are largely satisfied by adenosine triphosphate (ATP) generated via mitochondrial oxidative phosphorylation (OXPHOS). Accordingly, cardiac myocytes require a specialized, high capacity mitochondrial system. The importance of mitochondrial respiration for proper heart function is demonstrated by the tight linear relationship between cardiac oxygen consumption and work.1 Evidence is emerging that in many forms of heart failure, cardiac mitochondria revert to prenatal levels of function and morphology, leading to energy starvation, contributing to a vicious pathological cycle.2,3
During cardiac development, mitochondria undergo a maturation process. Mitochondrial maturation can be divided into three main developmental stages: prenatal, perinatal, and postnatal. In the prenatal stage, the early embryonic heart relies largely on non-mitochondrial energy sources (i.e. anaerobic glycolysis).4 During the transition from late fetal to postnatal periods (perinatal stage), the mitochondrial functional capacity of the heart increases dramatically, supporting a switch to reliance on fatty acids (FA) as the chief energy substrate.4-6 This increase in mitochondrial oxidative capacity is triggered by a burst of mitochondrial biogenesis at birth.7-9 The third stage of mitochondrial maturation occurs during the postnatal period when the cardiac myocyte undergoes growth, elongation, and assembly of adult sarcomeres. This final stage involves an increase in mitochondrial size and function, together with redistribution of the organelles throughout the myocyte.6 This intracellular redistribution results in the mitochondria being tightly packed between the longitudinally-oriented myofibrils with the development of contacts between mitochondria, myofibrils, and the sarcoplasmic reticulum.6 The mitochondrial architectural arrangement of the adult myocyte facilitates efficient high energy phosphate transfer between the mitochondria and key ATPases such as the myosin-ATPase and the sarcoplasmic reticulum ATPase.10 Recent evidence suggests that postnatal mitochondrial maturation in the mammalian heart involves coordinated mitochondrial dynamics (fusion and fission).11
The mechanisms regulating mitochondrial maturation are incompletely understood. Peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) and PGC-1β, are inducible, developmentally-regulated, transcriptional coregulators of cellular energy processes including OXPHOS and FA oxidation in mitochondrial-rich tissues such as heart, skeletal muscle, and brown adipose tissue.12-15 Using conditional gene targeting strategies in mice, we have shown that PGC-1α and PGC-1β serve overlapping roles in the surge of mitochondrial biogenesis that occurs in heart immediately following birth.12 Combined disruption of the PGC-1α and PGC-1β genes during the embryonic period resulted in a complete arrest of cardiac perinatal mitochondrial biogenesis leading to a lethal cardiomyopathy soon after birth.12 Conversely, overexpression of PGC-1α in the immediate postnatal period drives an exuberant mitochondrial biogenic response.16
The PGC-1 loss-of-function studies to date have not defined the role of these transcriptional coactivators during postnatal development or in the adult heart due to the lethal phenotype of PGC-1α/PGC-1β-deficient mice shortly after birth. The role of the PGC-1 coactivators after birth is an important question given the potential link of altered PGC-1 signaling to the development of heart failure.17,18 To address this question, we took advantage of a conditional gene targeting system in mice that resulted in deletion of the PGC-1β gene in both heart and skeletal muscle via the actions of Cre recombinase driven by the muscle creatine kinase (MCK) promoter (MCK-Cre) on a generalized PGC-1α null background (PGC-1α−/−βf/f/MCK-Cre).19,20 MCK-Cre is not fully active in the heart until after the perinatal mitochondrial biogenic surge. We found that loss of both PGC-1α and PGC-1β (but not either alone) during postnatal cardiac development resulted in dramatic defects in mitochondrial maturation and structure associated with reduced expression of genes involved in mitochondrial fusion and fission, leading to a progressive lethal cardiomyopathy. In striking contrast, disruption of the PGC-1 genes in the adult heart did not result in evidence of altered mitochondrial dynamics. These results reveal the importance of the PGC-1 coactivators for postnatal mitochondrial dynamics, maturation, and cardiac function. Our findings also suggest that the upstream pathways involved in the control of cardiac mitochondrial dynamics are developmental stage-specific.
METHODS
A Detailed Methods Section is available in the Online Data Supplement. Animal studies were conducted in strict accordance with the NIH guidelines for humane treatment of animals and approved by the IACUC committee at the Sanford-Burnham Medical Research Institute at Lake Nona. The gene array data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE43798.
RESULTS
PGC-1 coactivators are required for normal postnatal cardiac development
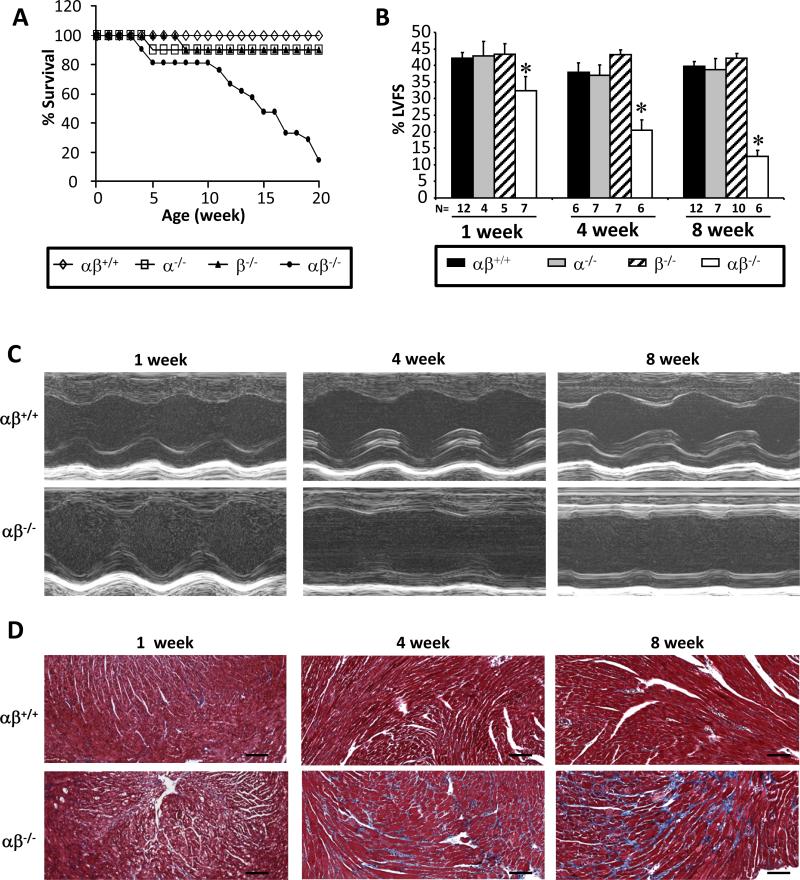
Mice deficient for both PGC-1α and PGC-1β in heart and skeletal muscle (PGC-1α−/−βf/f/MCK-Cre mice) were generated by crossing mice with skeletal muscle- and heart-specific disruption of the PGC-1β gene via Cre recombinase driven by an MCK promoter (PGC-1βf/f/MCK-Cre mice) to whole body PGC-1α-deficient mice.19,20 MCK promoter expression is activated during the late fetal stages and, to a greater extent, the early postnatal period.21 Accordingly, cardiac levels of PGC-1β mRNA in PGC-1α−/−βf/f/MCK-Cre were 30% of control at time of birth with progressive reduction during the postnatal period (Online Figure I). In contrast to the early postnatal lethality of mice with complete prenatal targeting of the PGC-1 genes,12 the PGC-1α−/−βf/f/MCK-Cre mice survive the perinatal period. However, the PGC-1α−/−βf/f/MCK-Cre mice exhibit a lethal phenotype beginning at 5 weeks of age with only 14% survival by 20 weeks (Figure 1A).
Figure 1. PGC-1α−/−βf/f/MCK-Cre mice develop progressive postnatal cardiomyopathy.
(A) Male and female mice were monitored for 20 weeks [PGC-1βf/f (αβ+/+, n=12), PGC-1α−/−βf/f (α−/−, n=10), PGC-1βf/f/MCK-Cre (β−/−, n=10), and PGC-1α−/−βf/f/MCK-Cre (αβ−/−, n=21)]. Graph denotes survival rates as a function of age (p<0.0001 based on Log-rank test). There was no significant difference in the longevity of male vs. female αβ−/− mice. (B) Left ventricular (LV) fractional shortening (FS) was determined using echocardiography for 1, 4, and 8 week-old mice. 1 week-old data represents male and female mice, n=4-12 mice per genotype. 4 and 8 week-old data represent female mice, n=6-12 mice per genotype. *p<0.05 relative to αβ+/+. (C) Representative M-mode echocardiographic images from 1, 4, and 8 week-old αβ+/+ and αβ−/− mice are shown. (D) Representative images of Mason’s trichrome staining of myocardial left ventricular sections from 1, 4, and 8 week-old PGC-1αβ+/+ and PGC-1αβ−/− mice are shown. Scale bars are 100 μm.
To investigate the cause of death in the PGC-1α−/−βf/f/MCK-Cre line, echocardiography was performed across several postnatal timepoints. Compared to wild-type (αβ+/+) and single PGC-1α- or PGC-1β-deficient mice, the PGC-1α−/−βf/f/MCK-Cre mice exhibited a progressive cardiomyopathy beginning at 1 week of age (Figures 1B, C). At 8 weeks of age, the mean percent LV fractional shortening (%LVFS) and ejection fraction (EF) of the PGC-1α−/−βf/f/MCK-Cre hearts were decreased by 68% and 67%, respectively, compared to αβ+/+ control animals (Figure 1B, Online Table II). Notably, significant cardiac remodeling, as reflected by expansion of chamber dimensions and relative wall thickness (RWT), did not occur until 8 weeks of age in the PGC-1α−/−βf/f/MCK-Cre mice (Online Table II). Only minimal fibrosis was noted in the PGC-1α−/−βf/f/MCK-Cre hearts at 4 weeks of age with a significant increase thereafter (Figure 1D, Online Figure II). There was no evidence of fibrosis in single PGC-1α or PGC-1β knockout animals at any timepoint (data not shown). In addition, caspase immunostaining was negative at all timepoints, in all genotypes, suggesting that apoptosis is an unlikely major primary cause of the cardiomyopathy (Online Figure III).
Derangements in mitochondrial structure and function in PGC-1α−/−βf/f/MCK-Cre hearts
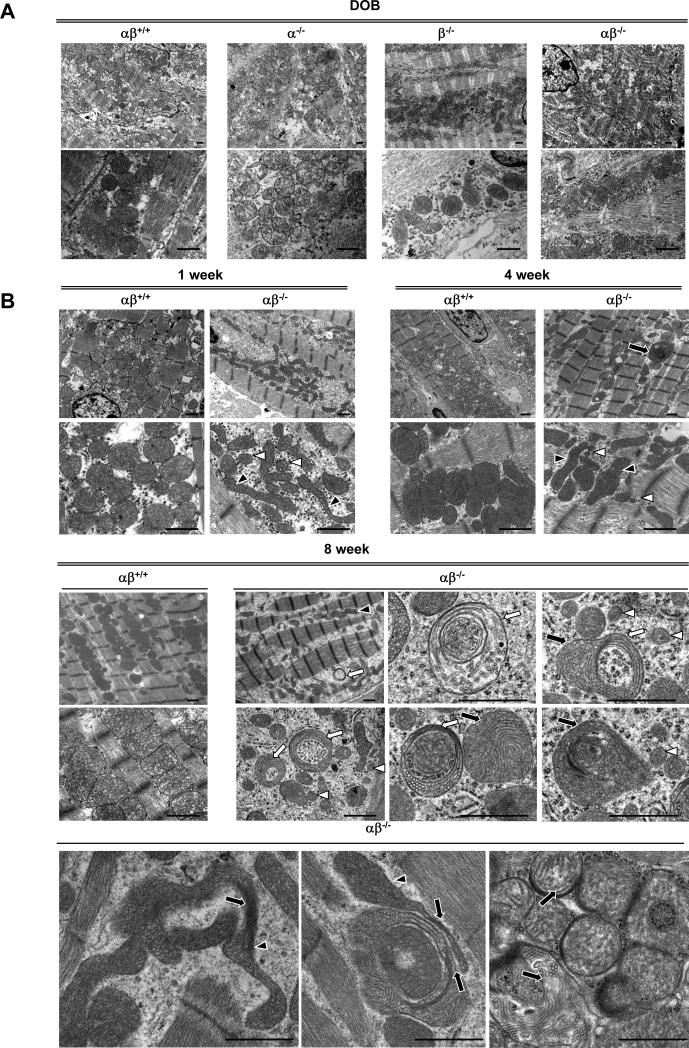
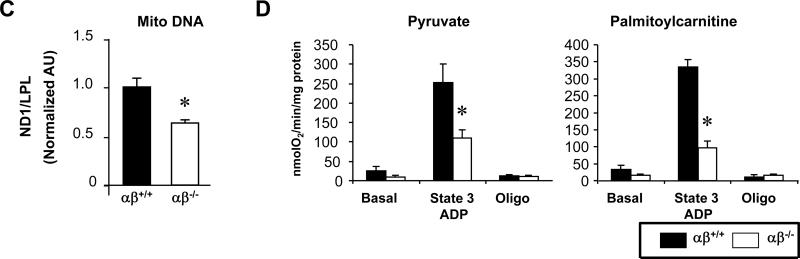
As an initial step to determine the basis for the cardiomyopathy of the PGC-1α−/−βf/f/MCK-Cre mice, EM assessment of the myocardium was conducted across postnatal timepoints. Consistent with the survival of the PGC-1α−/−βf/f/MCK-Cre mice at birth, the PGC-1α−/−βf/f/MCK-Cre hearts had normal mitochondrial density and structure on postnatal day 1, indicating that the mitochondrial biogenic surge at birth remained largely intact (Figure 2A). However, striking mitochondrial morphologic abnormalities were noted in the PGC-1α−/−βf/f/MCK-Cre hearts by 1 week of age, including fragmented and elongated mitochondria and a reduction in direct contacts between mitochondria and the sarcomeres (Figure 2B). Mitochondria of 4 week-old PGC-1α−/−βf/f/MCK-Cre mice displayed increased heterogeneity in size and shape (Figure 2B). By 8 weeks of age, further mitochondrial derangements were present including collapsed cristae and ring-like or “donut”-shaped mitochondria, in addition to a decrease in mitochondrial density and more pronounced loss of contacts between mitochondria and the sarcomeres (Figure 2B). Mitophagosomes were not observed. The observed decrease in mitochondrial density by EM at 8 weeks was associated with a 35% decrease in mitochondrial DNA (Figure 2C) and a decrease in levels of Complex I (8 kd subunit) and IV (COX IV) proteins. In contrast, Complex II (SDHA) protein levels were unchanged (Online Figure IV). Abnormalities in mitochondrial density and structure were not detected in the hearts of single PGC-1 knockout animals (data not shown) consistent with what we have reported previously.20
Figure 2. Progressive cardiac mitochondrial structural and functional derangements develop in PGC-1α−/−βf/f/MCK-Cre mice.
(A) Representative electron micrograph images taken from the LV free wall of animals on day of birth (DOB). Low magnification images were used to evaluate mitochondrial density, and high magnification images were used to determine mitochondrial structure. Images from 4 animals of each genotype were examined. Scale bars are 1μm. (B) Representative EMs of LV free wall of 1 week-old mice, and papillary muscle of 4 and 8 week-old mice from the genotypes indicated are shown. Arrows indicate structurally abnormal mitochondria including mitochondria with abnormal cristae structure (black arrows), elongated (black arrowheads), “donut-shaped” (white arrows), and small fragmented mitochondria (white arrowheads). (C) PCR of DNA from heart of PGC-1βf/f (αβ+/+) vs. PGC-1α−/−βf/f/MCK-Cre (αβ−/−) mice was performed to quantify mitochondrial DNA using primers for NADH dehydrogenase (ND1) normalized to genomic DNA using primers for lipoprotein lipase (LPL). The ND1 levels were normalized to genomic DNA content. Bars represent mean ± SEM. *p< 0.05. (D) Bars represent mean (± SEM) mitochondrial respiration rates determined on mitochondria isolated from the entire heart ventricle of 4 week-old female mice from the indicated genotypes. Rates were measured using a fluorescence-based oxygen sensing probe (FOXY, Ocean Optics) with either pyruvate or palmitoylcarnitine as substrate under the following conditions: basal, state 3 (ADP stimulated), and oligomycin-induced state 4 (oligo). *p<0.05 relative to αβ+/+.
Respiration rates were determined on mitochondria isolated from PGC-1α−/−βf/f/MCK-Cre mice compared to age-matched controls. Mitochondria from cardiac ventricles of 4 week-old PGC-1α−/−βf/f/MCK-Cre hearts exhibited a severe reduction in state 3 respiration rates using either pyruvate or palmitoylcarnitine as substrates (Figure 2D). Single PGC-1α knockout animals displayed a modest reduction in respiration rates relative to control mice (data not shown), consistent with the original description of this line.20
Evidence for regulation of cardiac mitochondrial fusion and fission genes by PGC-1 coactivators during postnatal development
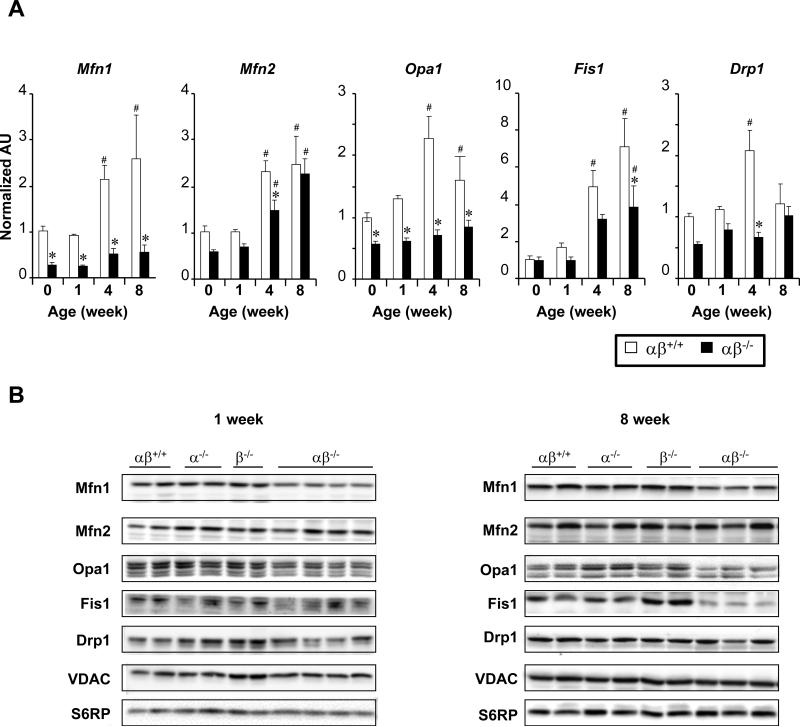
The mitochondrial structural abnormalities identified in the hearts of PGC-1α−/−βf/f/MCK-Cre mice suggested derangements in mitochondrial fusion and fission (dynamics). Indeed, fragmented, elongated, and “donut-shaped” forms have been described in mouse models and humans with genetic defects in the mitochondrial fusion machinery such as Charcot-Marie-Tooth disease, an inherited motor neuronopathy.22-26 The expression of a panel of genes encoding proteins known to be involved in mitochondrial fusion (Mfn1, Mfn2, Opa1) and fission (Fis1, and Dnm1l or Drp1) was assessed in hearts from PGC-1α−/−βf/f/MCK-Cre mice at birth (DOB), and 1, 4, and 8 weeks of age. The expression of this group of genes was induced during the postnatal period in αβ+/+ control animals (Figure 3A, white bars). This pattern of induction was markedly altered, in a gene-specific manner, in PGC-1α−/−βf/f/MCK-Cre hearts (Figure 3A, black bars). Notably, Mfn1 mRNA and protein levels were significantly decreased across all timepoints in the PGC-1α−/−βf/f/MCK-Cre hearts compared to controls (Figures 3A, B). This was also true for Opa1 (Figures 3A, B). A progressive decrease in Fis1 protein levels was also noted during the late postnatal period. In contrast, Mfn2 mRNA levels were only modestly reduced and Mfn2 protein levels were not different than control levels at any timepoint.
Figure 3. Reduced expression of genes involved in mitochondrial dynamics in PGC-1α−/−βf/f/MCK-Cre (αβ−/−) heart.
(A) Results of qRT-PCR analysis performed on RNA isolated from hearts of PGC-1βf/f (αβ+/+) vs. PGC-1α−/−βf/f/MCK-Cre (αβ −/−) mice from birth (0 weeks) to 8 weeks of age [mitofusin 1(Mfn1), mitofusin 2 (Mfn2), dynamin-related protein 1 (Drp1), optic atrophy 1(Opa1)]. The average of results from 4 animals per group is shown. *p<0.05 relative to corresponding αβ+/+, #p<0.05 relative to the week 0 timepoint of the corresponding genotype. (B) Representative Western blot analyses performed using protein extracts prepared from 1 and 8 week-old mouse heart ventricle tissue homogenate from genotypes denoted at the top. Voltage-dependent anion channel (VDAC) and S6 ribosomal protein (S6RP) were used as loading controls.
To determine whether reduced expression of Opa1 and Mfn1 is sufficient to cause the mitochondrial morphological changes seen in the PGC-1α−/−βf/f/MCK-Cre hearts, microRNAs were used to KD Opa1 and Mfn1 in C2C12 myocytes. In differentiated C2C12 myotubes, KD of Mfn1 or Opa1 alone had no effect on mitochondrial morphology; however, combined KD of Opa1 and Mfn1 caused mitochondrial fragmentation (Online Figure V). Interestingly, KD of either Mfn1 or Opa1 alone caused fragmentation in C2C12 myoblasts (data not shown). These results suggest that decreased expression of Opa1 and Mfn1, such as occurs in PGC-1α−/−βf/f/MCK-Cre heart, is sufficient to cause a mitochondrial fragmentation phenotype.
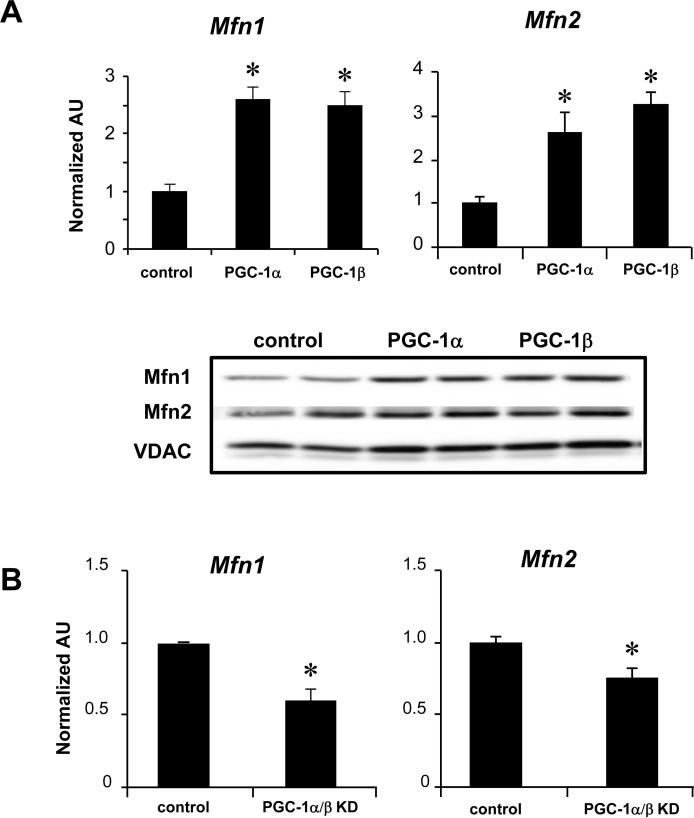
The abnormal expression of the Mfn1 gene in PGC-1α−/−βf/f/MCK-Cre hearts was of particular interest given recent evidence demonstrating the importance of Mfn1 and Mfn2 for mitochondrial fusion and maturation during embryonic27 and postnatal cardiac development.11 To explore the link between PGC-1 signaling and the expression of Mfn1 and Mfn2, the effect of adenoviral-mediated overexpression of PGC-1α was assessed in NRCM in culture. Overexpression of PGC-1α or PGC-1β increased expression of Mfn1 and Mfn2 (Figure 4A). Conversely, shRNA-mediated knockdown (KD) of PGC-1α and β resulted in a modest but significant decrease in Mfn1 and Mfn2 mRNA levels (Figure 4B). KD of either PGC-1α or PGC-1β, independently, did not decrease Mfn1 or Mfn2 mRNA levels (data not shown). Taken together, these results suggest that PGC-1α and PGC-1β regulate the expression of Mfn1 and Mfn2, but that in the PGC-1α−/−βf/f/MCK-Cre heart, independent regulatory pathways maintain Mfn2 levels.
Figure 4. Induction of Mfn1 and Mfn2 expression by PGC-1α and PGC-1β.
(A, top) qRT-PCR analysis was performed to quantify the Mfn1 and Mfn2 transcripts in NRCM after adenoviral-mediated overexpression of PGC-1α, PGC-1β, or GFP control. Bars represent the mean (±SEM) of 3 independent experiments, *p<0.05 relative to control. (bottom) Representative Western blot analyses using whole cell protein extracts after adenoviral-mediated overexpression of PGC-1α or β in H9c2 myotubes. (B) Results of qRT-PCR analysis to quantify Mfn1 and Mfn2 transcripts in NRCM after shRNA-mediated knockdown (KD) of PGC-1α and β [the shRNAs resulted in significant KD of PGC-1α (84%) and PGC-1β (67%)]. Bars represent the mean (±SEM) of 3 independent experiments, *p<0.05.
PGC-1 activates Mfn1 gene transcription via ERRα
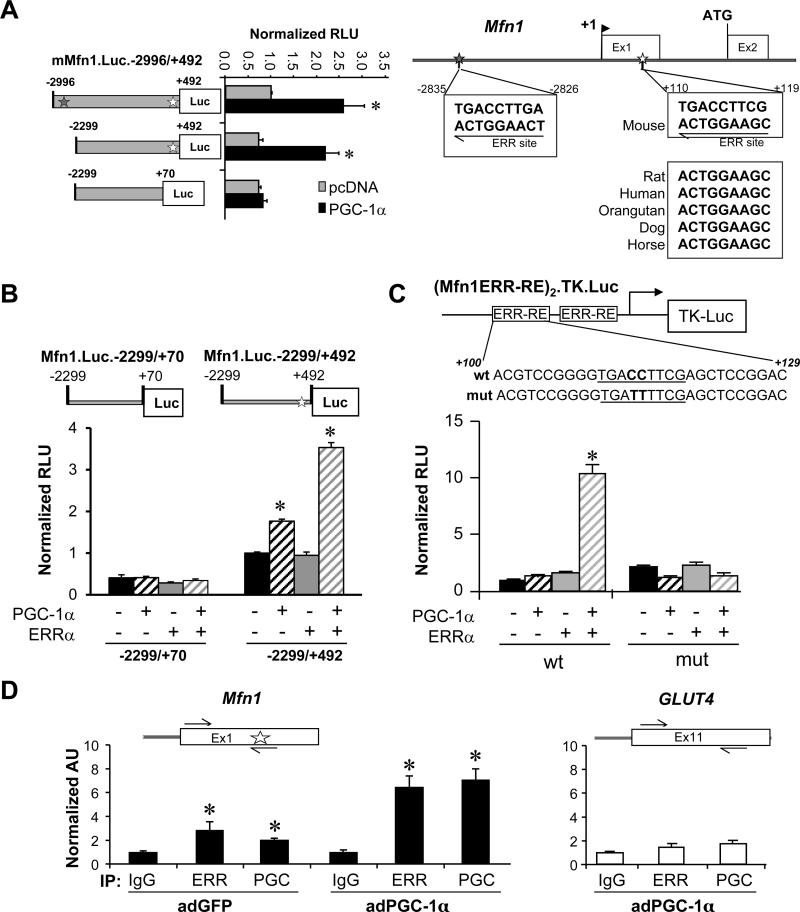
We next sought to determine whether PGC-1α directly activates Mfn1 gene transcription. To this end, Mfn1 promoter-reporter studies were conducted in C2C12 myotubes. Co-transfection studies were performed using a PGC-1α expression vector and a luciferase reporter containing a ~3.5kb fragment of the mouse Mfn1 promoter region (mMfn1.Luc.-2996/+492). PGC-1α significantly activated mMfn1.Luc.-2996/+492 (Figure 5A, left). Analysis of the DNA sequence of the Mfn1 promoter region revealed two consensus binding sites for ERR, a known target of PGC-1α,28-30 located 2826 bp upstream and 110 bp downstream of the transcription start site (Figure 5A, right). The latter, which is present in an untranslated exon 1, is highly conserved across species (Figure 5A, right). Deletion of the region containing the upstream putative ERR binding site (mMfn1.Luc.-2299/+492) had no effect on the PGC-1-mediated induction of the promoter (Figure 5A, left). However, removal of a region containing the conserved ERR binding site (mMfn1.Luc.-2299/+70) abolished the PGC-1-mediated activation (Figure 5A, left). Similar results were obtained with PGC-1β (Online Figure VI).
Figure 5. PGC-1α coactivates ERRα to increase transcription of the Mfn1 gene.
(A, left) Serial deletion reporter constructs were transfected into C2C12 myotubes in the presence of overexpressed PGC-1α (black bars) or vector backbone control (pcDNA3.1; gray bars). The bars represent mean luciferase activity (normalized relative light units, RLU) ±SEM. Gray (-2826) or white (+110) stars denote the position of putative ERR sites. *p<0.05. (right) Putative ERRα binding consensus site sequences. The positions of the putative ERR binding sites are denoted by stars. DNA sequence conservation between species is shown in the boxes. (B) Bars denote RLU of the Mfn1.Luc.-2299/+70 or the Mfn1.Luc.-2299/+492 reporter in the presence or absence of expressed PGC-1α and/or ERRα in C3H10T1/2 cells. *p<0.05 compared to reporter alone. (C) C3H10T1/2 cells were transfected with a reporter construct containing two copies of the short nucleotide fragment containing the conserved ERRα site (or a mutated version of the site) upstream of the TK promoter in the presence or absence of overexpressed PGC-1α and/or ERRα. Bars represent mean normalized RLU ±SEM for 3 independent experiments. *p<0.05. (D) Quantification of ChIP assays performed with H9c2 myotubes following infection by adPGC-1α or control virus using anti-PGC-1α, anti-ERRα, or IgG (negative control) as shown at the bottom. Schematics above the graphs indicate the relative positions of primers used for amplification (black arrows), and the relative position of the conserved ERR-RE (white star). Bars represent SYBR green quantification of 3 independent chromatin isolations and immunoprecipitations (arbitrary units ± SEM) normalized to the value for the IgG control (taken as 1.0). *p< 0.05 compared to IgG control.
To further evaluate the functionality of the conserved putative ERRα binding site, Mfn1 promoter-reporter experiments were performed in undifferentiated 10T1/2 cells, which express very low levels of endogenous ERRα and PGC-1α (data not shown). Forced expression of PGC-1 alone conferred a modest activating effect on mMfn1.Luc.-2299/+492 (Figure 5B) whereas ERRα alone had no effect, consistent with its dependence on PGC-1α for its transcriptional activating function (Figure 5B). Cotransfection of both PGC-1α and ERRα activated mMfn1.Luc.-2299/+492 in a synergistic manner (Figure 5B). The cooperative effect of PGC-1α and ERRα was not observed with a reporter that lacks the conserved +110/+119 ERRα binding site (mMfn1.Luc.-2299/+70, Figure 5B). Next, a reporter containing two copies of a Mfn1 promoter fragment containing the conserved +110/+119 putative ERRα binding site upstream of the HSV thymidine kinase (TK) promoter (Mfn1ERR-RE2.TK.Luc) was assessed. Whereas neither PGC-1α nor ERRα alone had a significant effect on Mfn1ERR-RE2.TK.Luc activity, together they activated the reporter over 10-fold (Figure 5C). In contrast, a reporter construct containing a mutated version of the ERR element (Mfn1ERR-RE2mut.TK.Luc) did not confer activation by PGC-1α/ERRα (Figure 5C).
ChIP studies were performed to assess occupation of PGC-1α and ERRα on the Mfn1 promoter region using chromatin isolated from H9c2 myotubes following infection with an adenoviral expression vector for PGC-1α and GFP (adPGC-1α or control virus (adGFP). Antibodies specific to ERRα and PGC-1α significantly enriched for the endogenous Mfn1 promoter region (Figure 5D) in conditions in which PGC-1 was not overexpressed (adGFP). The enrichment was further increased in the adPGC-1α condition (Figure 5D). In contrast, neither ERRα nor PGC-1α antibodies enriched a distal (control) region of the Slc24a (GLUT4) gene (Figure 5D) demonstrating the specificity of the IP results. Taken together, these results confirm that PGC-1α and ERRα occupy the ERRα-responsive region of the Mfn1 promoter to increase its transcription.
The hearts of adult PGC-1α/β-deficient mice do not exhibit evidence of altered mitochondrial dynamics
To determine if PGC-1 coactivators are also important for maintenance of mitochondrial density and structure in the adult heart, an inducible PGC-1 gene targeting strategy was undertaken. To this end, transgenic mice with the tamoxifen-inducible Cre recombinase-driven (MerCreMer) by the cardiac-specific MHC promoter31 were crossed to PGC-1βf/f mice and, subsequently, with whole body PGC-1α-deficient mice20 to generate PGC-1α−/−βf/f/MerCre mice. Adult cardiac PGC-1α/β-deficient mice were generated by injection of tamoxifen into 2-3 month old PGC-1α−/−βf/f/MerCre mice. One month after tamoxifen injection, only 3% of PGC-1β expression remained relative to age-matched control PGC-1α−/− animals (Online Figure VIIA). Marked reduction in cardiac PGC-1β gene expression was observed as early as 72 hours after tamoxifen injection (7% of expression remaining compared to control levels, Online Figure VIIA). The level of reduction in PGC-1β expression observed in the tamoxifen-treated adult PGC-1α−/−βf/f/MerCre mice was similar to that of the PGC-1α−/−βf/f/MCK-Cre animals.
The tamoxifen-treated PGC-1α−/−βf/f/MerCre mice appeared grossly normal and their survival was no different than controls (data not shown). Heart size, wall thickness, and function were normal in adult PGC-1α/β-deficient mice one and two months following tamoxifen injection (Online Figure VIIB and data not shown). Overall mitochondrial distribution, size, and density in the PGC-1α−/−βf/f/MerCre hearts were not different than the normal age-matched controls 1 month after tamoxifen injection (Online Figure VIIB, Figure 6A). However, increased numbers of lipid droplets were noted compared to the controls (Figure 6A). In addition, some cristae appeared dense and “collapsed” in a subset of the mitochondria (Figure 6A). The significance of this is unknown. The levels of several known PGC-1α isoforms were assessed using qRT-PCR in hearts of adult, tamoxifen-treated, PGC-1α−/−βf/f/MerCre mice to ensure that upregulation of these transcripts did not occur in the context of markedly reduced PGC-1β expression in the adult heart. The levels of transcripts encoding full-length PGC-1α and two shorter alternatively spliced forms of PGC-1α (NT-PGC-1α32 and PGC-1α433) were markedly reduced in the hearts of the adult PGC-1α/β-deficient mice relative to age-matched controls (Online Figure VIIC).
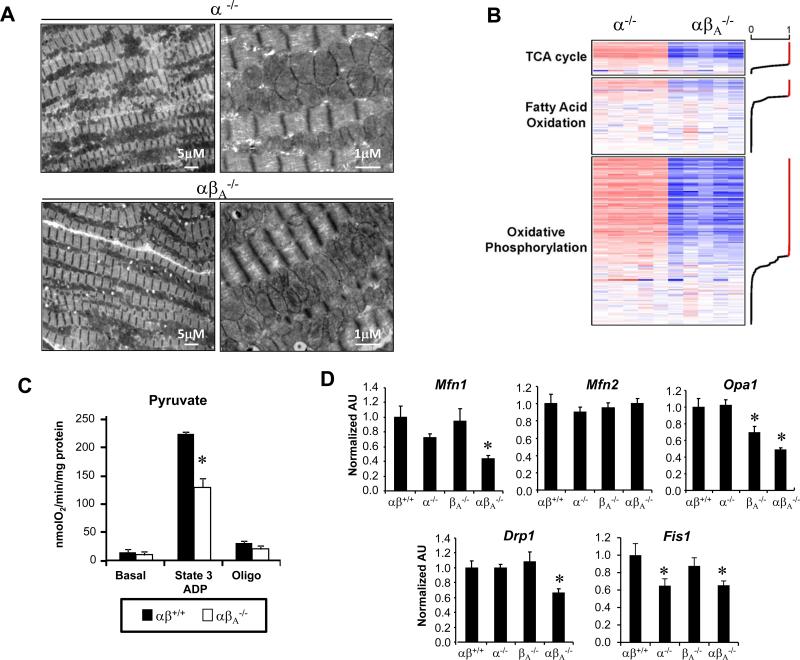
Figure 6. Global downregulation of genes involved in mitochondrial pathways in hearts of adult PGC-1α−/−βf/f/MerCre mice.
(A) Representative EMs of adult (A) PGC-1α−/−βf/f/MerCre (α−/−) or PGC-1α−/−βf/f/MerCre (αβA−/−) hearts taken from the papillary muscle of 4 month old female mice, 2 months following IP injection of tamoxifen or control. Images are representative of 3 animals per group. Scale bars are 5μm or 1mm, as indicated. (B) Heat map of Affymetrix Gene Chip profiling data representing genes within regulated mitochondrial pathways from cardiac tissue of 2-3 month old PGC-1α−/−βf/f/MerCre mice, 1 month after vehicle (α−/−) or tamoxifen injection (αβA−/−). Genes are ordered by the posterior probability (PP) of differential expression (DE) between α−/− and αβA−/−. Red indicates upregulation and blue indicates downregulation relative to the α−/− control animals. The line graph on the right margin shows the PP values for each gene; red indicates PP>0.949. (C) Bars represent mean (± SEM) mitochondrial respiration rates determined on mitochondria isolated from the entire heart ventricle of 2-3 month old control PGC-1αβ+/+ (αβ+/+) mice vs. PGC-1α−/−βf/f/MerCre (αβA−/−) mice, 1 month after injection with vehicle or tamoxifen (depending on the genotype). Rates were measured using Seahorse Bioscience XF96 Analyzer with pyruvate as a substrate under the following conditions: basal, state 3 (ADP stimulated), and oligomycin-induced state 4 (oligo). *p<0.05 relative to αβ+/+. (D) qRT-PCR analysis was performed to quantify levels of mRNA encoding fission/fusion proteins in cardiac tissue in 2-3 month old control PGC-1αβ+/+ (αβ+/+), PGC-1βf/f/MerCre (βA−/−), PGC-1α−/− (α−/−), PGC-1α−/− (α−/−), PGC-1α−/−βf/f/MerCre (αβA−/−) mice, 1 month after injection with vehicle or tamoxifen (depending on the genotype). The bars represent mean ± SEM values. *p<0.05 relative to αβ+/+.
To further explore the impact of PGC-1α/β deficiency in adult heart, transcriptional profiling was conducted on 12 week-old, tamoxifen-treated, PGC-1α−/−βf/f/MerCre mice. For these experiments, vehicle-injected PGC-1α−/−βf/f/MerCre mice (PGC-1α−/−) were used as controls in order to utilize littermate comparisons. The transcriptome of hearts from tamoxifen-injected PGC-1α−/−βf/f/MerCre mice revealed a dramatic global decrease in the levels of transcripts encoding genes of multiple mitochondrial pathways (Online Table III). Of particular interest was downregulation of genes involved in mitochondrial energy transduction and ATP-generating pathways including the citric acid cycle, FA metabolism, and OXPHOS pathways (Online Table IV, Figure 6B). Consistent with the transcriptional profiling results, state 3 respiration rates were reduced in mitochondria isolated from the tamoxifen-injected PGC-1α−/−βf/f/MerCre hearts (Figure 6C). Importantly, qRT-PCR (Figure 6D) and protein immunoblotting (Online Figure VIID) confirmed that expression of Mfn1 and the other fusion/fission genes were dysregulated in adult, tamoxifen-treated PGC-1α−/−βf/f/MerCre mice, in a pattern similar to that found during the postnatal period in the PGC-1α−/−βMCK-Cre mice. Taken together, these results strongly suggest that PGC-1 coactivators are required for mitochondrial dynamics during postnatal cardiac growth, but not in the normal adult heart.
DISCUSSION
Emerging evidence indicates that mitochondrial dynamics (fusion and fission) serves a critical function in the cellular distribution, maturation, and quality control of mitochondria.11,34 The results herein unveil a role for the transcriptional coactivators, PGC-1α and PGC-1β, as key upstream regulators of the mitochondrial dynamics machinery during postnatal growth of the heart. Our results support the following conclusions: 1) PGC-l coactivators are necessary for normal mitochondrial maturation, dynamics, and cardiac function during postnatal development; 2) PGC-1 coactivators regulate expression of a subset of cardiac genes involved in mitochondrial fusion and fission; 3) PGC-lα directly stimulates transcription of the Mfn1 gene by coactivating the orphan nuclear receptor ERRα; and 4) PGC-1 coactivators are not necessary for maintenance of mitochondria in the adult heart, but are required for high level expression of nuclear- and mitochondrial-encoded genes involved in mitochondrial energy transduction and OXPHOS pathways, and for full respiratory capacity.
The postnatal hearts of the PGC-1α−/−βf/f/MCK-Cre mice exhibit dramatic evidence of derangements in mitochondrial dynamics including fragmented, elongated, and “donut-shaped” mitochondria. Consistent with these findings, the expression of genes encoding proteins involved in mitochondrial fusion (Mfn1 and Opa1) and fission (Drp1 and Fis1) was reduced in the hearts of PGC-1α−/−βf/f/MCK-Cre mice. Among these genes, we identified Mfn1 as a direct PGC-1 target. PGC-1α was shown to coactivate the nuclear receptor ERRα on a conserved element in the Mfn1 gene. However, ERRα sites were not identified in the upstream regions of the other downregulated fusion/fission genes, although a functional non-classical PGC-1-responsive ERR site has been described previously in the Mfn2 gene promoter.35,36 Indeed, our cell culture overexpression studies (Figure 4A) demonstrated that PGC-1α and PGC-1β are capable of inducing both Mfn1 and Mfn2 expression. Surprisingly however, levels of Mfn2 mRNA and protein were not significantly downregulated in the PGC-1α−/−βf/f/MCK-Cre hearts. These results indicate that full activity of both mitofusins is necessary to support mitochondrial dynamics during the postnatal growth phase of heart development. Our findings also suggest that upstream regulatory pathways in addition to PGC-1 signaling serve to maintain Mfn2 expression during the postnatal growth phase. To this point, the transcription factor Sp1 has also been implicated in the transcriptional control of Mfn2.37 It should also be noted that a subset of mitochondrial respiratory complexes (I and IV) and mtDNA levels were decreased in the PGC-1α−/−βf/f/MCK-Cre hearts. Taken together, these results suggest that the mitochondrial and cardiac phenotypes are driven by a combination of altered dynamics and biogenesis.
In striking contrast to the postnatal cardiac phenotype, loss of PGC-1 coactivator function did not result in evidence of mitochondrial fragmentation and other features of altered fusion/fission in the adult mouse heart. This observation was surprising, given that Mfn1 mRNA and protein levels were reduced to a similar extent in both models. These results suggest that the activity of mitochondrial dynamics and its control are developmental stage-specific in heart. Consistent with this notion, rates of mitochondrial fusion and fission have been shown to be very low in adult cardiac myocytes38,39 compared to that of neonatal cardiac myocytes.40,41 Thus, loss-of-function of a single mitofusin gene could have a greater impact during periods of intense mitochondrial fusion and fission such as occurs during postnatal myocyte growth and maturation. Accordingly, the full activity of both Mfn1 and Mfn2, driven in part by the “boosting” activity of the PGC-1 coactivators, are likely required during the cardiac postnatal stage, but not in the adult. Consistent with this possibility, the mitochondrial phenotypes of single Mfn1 and Mfn2 mouse knockouts in adult heart and skeletal muscle are mild compared to the severe consequences of combined loss-of-function.11,38,42-45 A second possibility is that the mitofusins serve a subset of distinct biological roles. For example, high rates of mitochondrial fusion, such as occur during the postnatal period, may be directed largely by Mfn1 downstream of the “boosting” function of the PGC-1 coactivators. In contrast, other functions linked to the fusion/fission cycle, such as quality control via sorting for mitophagy,34,46 may be carried out in the adult heart by Mfn2.
Evidence is emerging that mitochondrial dynamics serve important roles in the maintenance of cardiac function and adaptation to stressful perturbations such as ischemia/reperfusion.47-49 Targeted disruption of the Drosophila ortholog of mammalian mitofusins (mitochondrial assembly regulatory factor)45 and of Mfn1 and Mfn2 in mice11,38 cause cardiomyopathy associated with altered mitochondrial structure. Given the potential pathogenic role of reduced PGC-1/ERR signaling and mitochondrial dysfunction in the development of heart failure,46,50,51 these results suggest that altered mitochondrial dynamics could contribute to pathologic remodeling leading to some forms of heart failure. For example, the results shown herein also raise the possibility that derangements in mitochondrial dynamics should be considered in the pathogenesis of childhood cardiomyopathies.
The lack of an obvious cardiac functional phenotype in adult PGC-1α−/−βf/f/MerCre mice was surprising. The PGC-1α−/−βf/f/MerCre hearts exhibited a global downregulation of genes involved in mitochondrial energy transduction and OXPHOS pathways along with reduced state 3 respiration rates. However, despite widespread alterations in genes involved in mitochondrial ATP production and reduced capacity for respiration, cardiac function was maintained in the adult PGC-α/β-deficient mice. These results suggest that the mitochondrial system of the adult mammalian heart has tremendous reserve for ATP production under basal, non-stressed, conditions. It is possible that processes such as energy transfer rather than ATP production capacity per se serve as the limiting factor for energy homeostasis in heart. Importantly, the adult PGC-1α−/−βf/f/MerCre heart did not exhibit morphological evidence of significant alterations in mitochondrial dynamics. It is tempting to speculate that mitochondrial structural organization, including appropriate juxtaposition with the myofilaments which was maintained in the adult but not the postnatal PGC-1α/β−/− hearts, is a critical determinant for maintaining adequate ATP production and transfer to the contractile apparatus.
Supplementary Material
Novelty and Significance.
What Is Known?
The energy demands of the heart are met by a high-capacity, dynamic mitochondrial system.
The development of the cardiac mitochondrial network involves a dramatic mitochondrial biogenic response at birth, followed by maturation and distribution of the organelles between the sarcomeres of the cardiac myocyte.
The regulatory circuitry driving the perinatal mitochondrial biogenic response was recently delineated but the mechanisms controlling mitochondrial maturation and maintenance in the postnatal heart is unknown.
What New Information Does This Article Contribute?
The transcriptional coregulators PPARγ coactivator 1 (PGC-1) α and β are required for the mitochondrial biogenic surge at birth in heart. Using conditional PGC-1α/β-deficient mice, we delineate the role of these factors during postnatal cardiac growth and in the adult heart.
During the postnatal period, PGC-1α/β-deficient mice develop a cardiomyopathy that is associated with mitochondrial fragmentation and elongation, indicative of altered fusion and fission (dynamics).
PGC-1 coactivators were then shown to control the expression of genes involved in mitochondrial fusion in cardiac myocytes.
Surprisingly, in contrast to the results during the postnatal growth stage, disruption of the PGC-1α/β genes in the adult heart did not result in cardiomyopathy or evidence of abnormalities alteration in mitochondrial dynamics.
The regulatory circuitry controlling mitochondrial biogenesis and function in heart is the subject of intense investigation. The transcriptional coregulators PGC-1α and β play a critical role in the surge of mitochondrial biogenesis that occurs around birth, equipping the heart with a high capacity mitochondrial system. In this study, conditional gene targeting was used to define the role of the PGC-1 coactivators in the postnatal and adult mouse heart. PGC-1α/β-deficiency in the postnatal growth stages resulted in lethal cardiomyopathy associated with dramatic mitochondrial fragmentation and elongation, consistent with alterations in mitochondrial fusion and fission (dynamics). Indeed, PGC-1 coactivators were shown to control transcription of the gene encoding mitofusin 1 (Mfn1), a protein involved in mitochondrial fusion. Surprisingly, when PGC-1 signaling was targeted in the adult heart, there was no evidence for alterations in mitochondrial dynamics and cardiac function was preserved. These findings emphasize the importance of mitochondrial dynamics during the postnatal growth phase of cardiac development and the importance of PGC-1 coactivators in this process. These results pave the way for future studies aimed at exploring this regulatory pathway for novel therapeutics aimed at the prevention or treatment of heart failure in both children and adults.
ACKNOWLEDGEMENTS
We wish to thank Rick Vega for critical reading, Lorenzo Thomas for assistance in preparation of the manuscript, and Jian-Liang Li at Sanford-Burnham Medical Research Institute’s (SBMRI) Applied Bioinformatics Core at Lake Nona, Florida for expert bioinformatic analysis. Special thanks to the Cardiometabolic Phenotyping, Histology, and Analytical Genomics Cores at SBMRI at Lake Nona, and to Juliet Zechner, Deanna Collia, Corin Riggs, Beatrice Alvarado, and Lauren Ashley Gabriel for assistance with the animal studies.
SOURCES OF FUNDING
This work was supported by NIH grants R01 DK045416 (D.P.K.), R01 HL58493 (D.P.K.), and R01 HL101189 (D.P.K., A.D.A., and D.M.M.).
Nonstandard Abbreviations and Acronyms
- ATP
Adenosine triphosphate
- AU
Arbitrary units
- ChIP
Chromatin immunoprecipitation
- CO
Cardiac output
- DOB
Day of birth
- Drp1 or Dnm1l
Dynamin-related protein 1
- EF
Ejection fraction
- EM
Electron microscopic/microscopy
- ERR
Estrogen-related receptor
- FA
Fatty acids
- Fis1
Fission protein 1
- HR
Heart rate
- KD
Knockdown
- LV
Left ventricular
- LVFS
Left ventricular fractional shortening
- LVIDd
Left ventricular internal dimension, diastole
- LVIDs
Left ventricular internal dimension, systole
- LVM
Left ventricular mass
- LVPWd
Left ventricular posterior wall, diastole
- LVPWs
Left ventricular posterior wall, systole
- MCK
Muscle creatine kinase
- MCK-Cre
Muscle creatine kinase promoter-driven Cre recombinase
- MerCreMer
Mutant estrogen receptor-driven Cre recombinase
- Mfn1
Mitofusin 1
- Mfn2
Mitofusin 2
- MHC
Alpha-myosin heavy chain
- NRCM
Neonatal rat cardiac myocytes
- Opa1
Optic atrophy 1
- OXPHOS
Oxidative phosphorylation
- PGC-1
Peroxisome proliferator-activated receptor gamma coactivator 1
- RLU
Relative light units
- RWT
Relative wall thickness (LVPWd+IVSd/LVIDd)
- S6RP
S6 ribosomal protein
- SEM
Standard error of the mean
- VDAC
Voltage-dependent anion channel
Footnotes
DISCLOSURES
The authors have no conflicts.
REFERENCES
- 1.Ventura-Clapier R, Garnier A, Veksler V, Joubert F. Bioenergetics of the failing heart. Biochim Biophys Acta. 2011;1813:1360–1372. doi: 10.1016/j.bbamcr.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Kelly DP, Scarpulla RC. Transcriptional control of striated muscle mitochondrial biogenesis and function. In: Hill JA, Olson EN, editors. Muscle: Fundamental biology and mechanisms of disease. Elsevier Academic Press; London: 2012. pp. 203–215. [Google Scholar]
- 3.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 4.Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 5.Warshaw JB, Terry ML. Cellular energy metabolism during fetal development. II. Fatty acid oxidation by the developing heart. J Cell Biol. 1970;44:354–360. doi: 10.1083/jcb.44.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter GA, Jr., Hom J, Hoffman D, Quintanilla R, de Mesy Bentley K, Sheu SS. Bioenergetics, mitochondria, and cardiac myocyte differentiation. Prog Pediatr Cardiol. 2011;31:75–81. doi: 10.1016/j.ppedcard.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallman M. Changes in mitochondrial respiratory chain proteins during perinatal development. Evidence of the importance of environmental oxygen tension. Biochim Biophys Acta. 1971;253:360–372. doi: 10.1016/0005-2728(71)90040-5. [DOI] [PubMed] [Google Scholar]
- 8.Smolich JJ, Walker AM, Campbell GR, Adamson TM. Left and right ventricular myocardial morphometry in fetal, neonatal, and adult sheep. Am J Physiol. 1989;257:H1–9. doi: 10.1152/ajpheart.1989.257.1.H1. [DOI] [PubMed] [Google Scholar]
- 9.Marin-Garcia J, Ananthakrishnan R, Goldenthal MJ. Heart mitochondrial DNA and enzyme changes during early human development. Mol Cell Biochem. 2000;210:47–52. doi: 10.1023/a:1007031919298. [DOI] [PubMed] [Google Scholar]
- 10.Piquereau J, Novotova M, Fortin D, Garnier A, Ventura-Clapier R, Veksler V, Joubert F. Postnatal development of mouse heart: formation of energetic microdomains. J Physiol. 2010;588:2443–2454. doi: 10.1113/jphysiol.2010.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papanicolaou KN, Kikuchi R, Ngoh GA, Coughlan KA, Dominguez I, Stanley WC, Walsh K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ Res. 2012;111:1012–1026. doi: 10.1161/CIRCRESAHA.112.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 16.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sihag S, Cresci S, Li AY, Sucharov CC, Lehman JJ. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci USA. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zechner C, Lai L, Zechner JF, Geng T, Yan Z, Rumsey JW, Collia D, Chen Z, Wozniak DF, Leone TC, Kelly DP. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franco D, Lamers WH, Moorman AF. Patterns of expression in the developing myocardium: towards a morphologically integrated transcriptional model. Cardiovasc Res. 1998;38:25–53. doi: 10.1016/s0008-6363(97)00321-0. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 24.Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhoeven K, Claeys KG, Zuchner S, et al. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129:2093–2102. doi: 10.1093/brain/awl126. [DOI] [PubMed] [Google Scholar]
- 27.Kasahara A, Cipolat S, Chen Y, Dorn GW, 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and notch signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- 28.Mootha VK, Handschin C, Arlow D, et al. ERRalpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 30.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Huypens P, Adamson AW, Chang JS, Henagan TM, Boudreau A, Lenard NR, Burk D, Klein J, Perwitz N, Shin J, Fasshauer M, Kralli A, Gettys TW. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J Biol Chem. 2009;284:32813–32826. doi: 10.1074/jbc.M109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruas JL, White JP, Rao RR, et al. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano FX, Liesa M, Bach D, Chan DC, Palacin M, Zorzano A. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes. 2006;55:1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 36.Liesa M, Borda-d'Agua B, Medina-Gomez G, Lelliott CJ, Paz JC, Rojo M, Palacin M, Vidal-Puig A, Zorzano A. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS One. 2008;3:e3613. doi: 10.1371/journal.pone.0003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorianello E, Soriano FX, Fernandez-Pascual S, Sancho A, Naon D, Vila-Caballer M, Gonzalez-Navarro H, Portugal J, Andres V, Palacin M, Zorzano A. The promoter activity of human Mfn2 depends on Sp1 in vascular smooth muscle cells. Cardiovasc Res. 2012;94:38–47. doi: 10.1093/cvr/cvs006. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Liu Y, Dorn GW., 2nd. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X, Sun L, Ji S, Zhao T, Zhang W, Xu J, Zhang J, Wang Y, Wang X, Franzini-Armstrong C, Zheng M, Cheng H. Kissing and nanotunneling mediate intermitochondrial communication in the heart. Proc Natl Acad Sci USA. 2013;110:2846–2851. doi: 10.1073/pnas.1300741110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Twig G, Liu X, Liesa M, Wikstrom JD, Molina AJ, Las G, Yaniv G, Hajnoczky G, Shirihai OS. Biophysical properties of mitochondrial fusion events in pancreatic beta-cells and cardiac cells unravel potential control mechanisms of its selectivity. Am J Physiol Cell Physiol. 2010;299:C477–487. doi: 10.1152/ajpcell.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beraud N, Pelloux S, Usson Y, Kuznetsov AV, Ronot X, Tourneur Y, Saks V. Mitochondrial dynamics in heart cells: very low amplitude high frequency fluctuations in adult cardiomyocytes and flow motion in non beating Hl-1 cells. J Bioenerg Biomembr. 2009;41:195–214. doi: 10.1007/s10863-009-9214-x. [DOI] [PubMed] [Google Scholar]
- 42.Papanicolaou KN, Khairallah RJ, Ngoh GA, Chikando A, Luptak I, O'Shea KM, Riley DD, Lugus JJ, Colucci WS, Lederer WJ, Stanley WC, Walsh K. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol Cell Biol. 2011;31:1309–1328. doi: 10.1128/MCB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papanicolaou KN, Ngoh GA, Dabkowski ER, O'Connell KA, Ribeiro RF, Jr., Stanley WC, Walsh K. Cardiomyocyte deletion of mitofusin-1 leads to mitochondrial fragmentation and improves tolerance to ROS-induced mitochondrial dysfunction and cell death. Am J Physiol Heart Circ Physiol. 2012;302:H167–179. doi: 10.1152/ajpheart.00833.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorn GW, 2nd, Clark CF, Eschenbacher WH, Kang MY, Engelhard JT, Warner SJ, Matkovich SJ, Jowdy CC. MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ Res. 2011;108:12–17. doi: 10.1161/CIRCRESAHA.110.236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Dorn GW., 2nd. Pink1-phosphorylated mitofusin 2 is a parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Hajnoczky G. Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death and Differentiation. 2011;18:1561–1572. doi: 10.1038/cdd.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 49.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial opa1, apoptosis, and heart failure. Cardiovasc Res. 2009;84:91–99. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosca MG, Tandler B, Hoppel CL. Mitochondria in cardiac hypertrophy and heart failure. J Mol Cell Cardiol. 2013;55:31–41. doi: 10.1016/j.yjmcc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aubert G, Vega RB, Kelly DP. Perturbations in the gene regulatory pathways controlling mitochondrial energy production in the failing heart. Biochim Biophys Acta. 2013;1833:840–847. doi: 10.1016/j.bbamcr.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.