Abstract
Background
Group B Streptococcus (GBS) remains a major cause of neonatal sepsis and is also associated with invasive and noninvasive infections in pregnant women and non-pregnant adults, elderly and patients with underlying medical conditions. Ten capsular serotypes have been recognized, and determination of their distribution within a specific population or geographical region is important as they are major targets for the development of vaccine strategies. We have evaluated the characteristics of GBS isolates recovered from individuals with infections or colonization by this microorganism, living in different geographic regions of Brazil.
Methods
A total of 434 isolates were identified and serotyped by conventional phenotypic tests. The determination of antimicrobial susceptibility was performed by the disk diffusion method. Genes associated with resistance to erythromycin (ermA, ermB, mefA) and tetracycline (tetK, tetL, tetM, tetO) as well as virulence-associated genes (bac, bca, lmb, scpB) were investigated using PCR. Pulsed-field gel electrophoresis (PFGE) was used to examine the genetic diversity of macrolide-resistant and of a number of selected macrolide-susceptible isolates.
Results
Overall, serotypes Ia (27.6%), II (19.1%), Ib (18.7%) and V (13.6%) were the most predominant, followed by serotypes IV (8.1%) and III (6.7%). All the isolates were susceptible to the beta-lactam antimicrobials tested and 97% were resistant to tetracycline. Resistance to erythromycin and clindamycin were found in 4.1% and 3% of the isolates, respectively. Among the resistance genes investigated, tetM (99.3%) and tetO (1.8%) were detected among tetracycline-resistant isolates and ermA (39%) and ermB (27.6%) were found among macrolide-resistant isolates. The lmb and scpB virulence genes were detected in all isolates, while bac and bca were detected in 57 (13.1%) and 237 (54.6%) isolates, respectively. Molecular typing by PFGE showed that resistance to erythromycin was associated with a variety of clones.
Conclusion
These findings indicate that GBS isolates circulating in Brazil have a variety of phenotypic and genotypic characteristics, and suggest that macrolide-resistant isolates may arise by both clonal spread and independent acquisition of resistance genes.
Background
Streptococcus agalactiae (Group B Streptococcus, GBS) is a leading cause of neonatal morbidity and mortality [1], and is also an important opportunistic agent of infections in pregnant women, as well as non-pregnant adults, especially the elderly or those with underlying medical conditions [2]. This microorganism is commonly found as a colonizer of the genital and the gastrointestinal tracts of both men and women, and vertical transmission from a colonized mother to her newborn during labor can result in life threatening infections.
GBS virulence is complex and multifactorial. Several virulence determinants are involved in the adhesion to and invasion of host cells, as well as in the immune system evasion. Surface components, including a polysaccharide capsule and proteins, such as Cα, Cβ, Rib and the laminin binding protein (LMB), and a number of enzymes (like the C5a peptidase) and toxins/cytolysins, are produced and have been associated with GBS virulence [3].
Capsular polysaccharides are recognized as playing a key role as virulence factors and are important targets for the development of vaccine strategies. GBS capsular polysaccharides have chemical and antigenic differences that enable the subdivision of this species into ten serotypes, denominated Ia, Ib, II-IX [4]. Vaccines against GBS infections must include the more common serotypes associated with disease in different populations and capsular polysaccharide-protein conjugate vaccines are in clinical trials [5]. The epidemiological distribution of these serotypes can vary according to several aspects, including the geographical region, the profile of the population being studied, and the source of the bacterial isolate [6]. Four serotypes (Ia, II, III and V) are usually the most frequently isolated in the United States and in some European countries [6-8], while serotypes VI to IX are rarely described [4,9]. In Brazil, the occurrence of serotypes Ia, Ib, II, III, IV and V has been described in a few studies conducted with isolates originated from the South and the Southeast regions [10-15].
S. agalactiae is still uniformly sensitive to penicillin, although isolates with reduced susceptibility to penicillin have been reported since 2008 [16-18]. Clindamycin or erythromycin is recommended for GBS intrapartum prophylaxis for penicillin-allergic women with high risk of anaphylaxis or when therapeutic failure is suspected [19]. However, increasing rates of resistance to these antibiotics have been detected in several regions of the world, including Europe [8,20], Asia [21], North America [22-24], and South America [11,13,15,25].
Despite the clinical and epidemiological impact of GBS infections and the trends to increasing occurrence of antimicrobial resistance, information about isolates from Brazil is still fragmentary and usually restricted to a few areas of this large country. There is no Brazilian study that provides a national overview of the phenotypic and genotypic characteristics of circulating isolates of GBS. In the present report we describe the serotype distribution, antimicrobial susceptibility, presence of virulence-related and antimicrobial resistance genes among GBS isolates collected from the five geographical regions in Brazil. Also, the genetic diversity of macrolide-resistant isolates was assessed and compared to a representative fraction of the macrolide-susceptible isolates, providing a national overview of the characteristics of strains circulating in the country.
Methods
Bacteria
Four hundred thirty four S. agalactiae isolates were included in this study. They were obtained during March 2005 to December 2009 from patients living in the five Brazilian geographical regions: South region [Paraná (35 isolates), Rio Grande do Sul (59 isolates)]; Southeast region [São Paulo (100 isolates), Rio de Janeiro (92 isolates), Minas Gerais (18 isolates)]; Mid-West region [Mato Grosso (16 isolates); Brasília (25 isolates)]; Northeast region [Bahia (17 isolates), Maranhão (27 isolates), Pernambuco (40 isolates)]; and North region [Amazonas (five isolates)]. The sources of the isolates included two groups of specimens: (i) from colonized patients [vaginal and perianal secretions (249 isolates)], (ii) from symptomatic adults [Table 1 (185 isolates)].
Table 1.
Serotype distribution of the 185 Streptococcus agalactiae isolates recovered from symptomatic adults included in the present study, according to the type of clinical specimen and geographical origin
| Clinical specimen (number of isolates) | Geographical region (number of isolates) | Serotype (number of isolates) b |
|---|---|---|
| Urine (167) |
South (83) |
Ia (35); Ib (11); II (12); III (6); IV (7); V (9); NT (3) |
| Southeast (57) |
Ia (13); Ib (10); II (9); III (6); IV (6); V (6); NT (7) |
|
| Mid-west (27) |
Ia (12); Ib (2); II (3); III (2); IV (3); V (2); NT (3) |
|
| Blood and other sterile fluids (10) |
Southeast (10) |
Ia (3); Ib (3); II (3); IV (1) |
| Male genital tract discharges (3) |
Mid-west (2) |
Ia (1); II (1) |
| Southeast (1) |
II (1) |
|
| Othera (5) |
Southeast (5) |
Ia (1); Ib (3); II (1) |
| Total (185) | All above (185) | Ia (65); Ib (29); II (30); III (14); IV (17); V (17); NT (13) |
aOther clinical specimens included abscesses and ear secretions.
bNT, nontypeable by using antisera against Ia, Ib, II-VIII serotypes.
Conventional methods were used for culturing procedures and identification of the isolates as GBS [19]. Serological grouping was performed using a commercial latex agglutination test, according to the manufacturer's instructions (Slidex Strepto Kit, bioMerieux, France). All of the strains were stored in Todd-Hewitt broth (Difco Laboratories, Detroit, Michigan, USA) with glycerol at -20°C.
The project was approved by the Research Ethics Committee of the Hospital Universitário of Universidade Federal do Rio de Janeiro (protocol number 21905), and written informed consents were obtained from all participants of the study.
Serotyping
All the isolates were serotyped by using the HCl extraction method and double immunodiffusion tests or capillary precipitation tests with typing antisera against nine of the capsular polysaccharides (Ia, Ib and II-VIII), prepared in house according to standardized methods [10]. Serotype IX was not investigated. Nontypeable isolates were designated as NT.
Antimicrobial susceptibility testing
All the 434 isolates were tested for ampicillin, cefotaxime, clindamycin, chloramphenicol, erythromycin, levofloxacin, and tetracycline (Oxoid) susceptibility by disk diffusion according to the CLSI guidelines [26]. The erythromycin-clindamycin double-disk test was used to determine the resistance phenotypes.
Detection of antimicrobial resistance genes
Macrolide and/or tetracycline-resistant isolates, as defined by phenotypic methods, were tested for the presence of the ermA, ermB, mefA, tetK, tetL, tetM and tetO genes by PCR using previously described primers [27,28].
Detection of virulence-associated genes
The presence of the bac, bca, lmb and scpB genes, which encode for alpha and beta proteins, laminin binding protein, and C5a peptidase, respectively, was evaluated in all 434 isolates by PCR using previously described primers [29,30].
Molecular typing by PFGE
All the erythromycin-resistant isolates (total of 18 strains) and 25 randomly selected erythromycin-susceptible isolates were characterized by PFGE. Chromosomal DNA was prepared in agarose plugs as previously described [31] and treated with 12U of SmaI (Invitrogen, San Diego, CA) for 18–24 h at 25°C. The fragments were separated by PFGE in 1.2% agarose gels in a CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA) with pulse times of 2 to 30 s for 23 h at 11.3°C and 6 V/cm. The restriction profiles were analyzed by using the BioNumerics software version 6.6 (Applied Maths, Ghent, East Flanders, Belgium). The Dice similarity coefficient was used to determine the similarity between each banding profiles, and a dendrogram was constructed using the unweighted-pair group method with arithmetic averages (UPGMA) with a tolerance coefficient of 1.0%. Isolates with similarities of ≥ 70% were considered as closely related and were clustered in a given clonal complex (CC). The clonal complexes (CCs) were assigned with capital letters (A to I). Isolates with similarities of < 70% were considered genetically unrelated.
Statistical analysis
The statistical analyses were performed using the SPSS program (“Statistical Package for the Social Sciences” version 19.0, IBM Brazil, São Paulo, SP, Brazil). The chi-square test was used to examine differences in serotype distribution between isolates recovered from infection and colonization specimens, as well as to evaluate correlation between erythromycin-resistance phenotypes and genotypes. Good evidence against the null hypothesis was considered with p values of <0.05.
The genetic diversity, as revealed by using PFGE, was calculated using the Simpson’s Index of Diversity (SID). The 95% confidence intervals (95% CI) were also calculated [32,33].
Results
Overall, serotypes Ia (120 isolates; 27.6% ± 4.2, 95% CI), II (83 isolates; 19.1% ± 3.7), Ib (81 isolates; 18.7% ± 3.7) and V (59 isolates; 13.6% ± 3.2) were the most predominant among the 434 GBS isolates included in the present study, followed by serotypes IV (35 isolates; 8.1% ± 2.6) and III (29 isolates; 6.7% ± 2.3). The sum of the four most common serotypes (Ia, Ib, II and V) constituted 79% (±3.8) of the isolates while percentages above 90% were obtained with the addition of serotypes III and IV. Serotypes VI, VII, and VIII were not found and 6.2% ± 2.3 (27 isolates) of the isolates were nontypeable.
Serotypes Ia, II and Ib were the most frequent among isolates recovered from infections, which were represented by the South, Southeast and Mid-West regions (Table 1). Although absolute numbers of colonization isolates were small in some regions, particularly the Mid-West, serotypes Ia and II were the most frequently observed in the South, Southeast and Mid-West regions, whereas serotypes Ib and V were the most common in the North/Northeast region (Figure 1).
Figure 1.
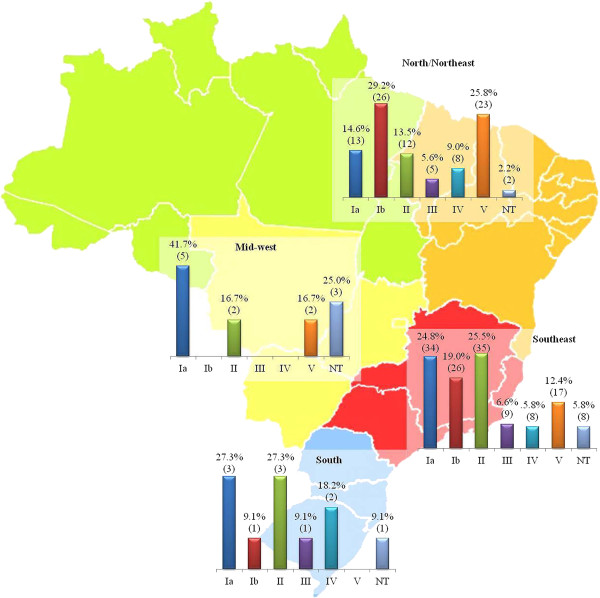
Serotype distribution among the 249 Streptococcus agalactiae isolates recovered from colonization included in the present study, according to geographical region. NT, nontypeable by using antisera against Ia, Ib, II-VIII serotypes.
Tests for detection of the virulence genes showed that all the 434 GBS isolates harbored both the lmb and scpB genes, whereas the bca and bac genes were detected in 54.4% ± 4.7 (236 isolates) and 13.1% ± 3.2 (57 isolates) of the isolates, respectively. The majority of isolates from infections (101 isolates; 54.6% ± 4.7) had neither bac or bca genes, while almost half of the colonization strains (122 isolates; 49% ± 4.7) were bca-positive (Table 2).
Table 2.
Distribution of bac and bca virulence-associated genes according to serotype and clinical origin of the 434 Streptococcus agalactiae isolates included in the present study
| Serotype (number of isolates) | Clinical origin (number of isolates) | Number (%) of bac (+) isolates | Number (%) of bca (+) isolates | Number (%) of bac (+)/ bca (+) isolates | Number (%) of bac (-)/ bca (-) isolates |
|---|---|---|---|---|---|
| Ia (120) |
Colonization (55) |
1 (1.8%) |
31 (56.4%) |
6 (10.9%) |
17 (30.9%) |
| Infection (65) |
2 (3.1%) |
26 (40%) |
3 (4.6%) |
34 (52.3%) |
|
| Ib (81) |
Colonization (53) |
1 (1.9%) |
23 (43.4%) |
10 (18.9%) |
19 (35.8%) |
| Infection (28) |
0 |
10 (35.7%) |
8 (28.6%) |
10 (35.7%) |
|
| II (83) |
Colonization (52) |
2 (3.8%) |
26 (50%) |
8 (15.4%) |
16 (30.8%) |
| Infection (31) |
0 |
12 (38.7%) |
2 (6.5%) |
17 (54.8%) |
|
| III (29) |
Colonization (15) |
2 (13.3%) |
7 (46.7%) |
0 |
6 (40%) |
| Infection (14) |
0 |
3 (21.4%) |
0 |
11 (78.6%) |
|
| IV (35) |
Colonization (18) |
0 |
11 (61.1%) |
2 (11.1%) |
5 (27.8%) |
| Infection (17) |
0 |
9 (52.9%) |
1 (5.9%) |
7 (41.2%) |
|
| V (59) |
Colonization (42) |
1 (2.4%) |
18 (42.8%) |
2 (4.8%) |
21 (50%) |
| Infection (17) |
0 |
3 (17.6%) |
1 (5.9%) |
13 (76.5%) |
|
| NTa (27) |
Colonization (14) |
1 (7.1%) |
6 (42.9%) |
4 (28.6%) |
3 (21.4%) |
| Infection (13) |
0 |
4 (30.8%) |
0 |
9 (69.2%) |
|
| Total (434) | Colonization (249) |
8 (3.2%) |
122 (49%) |
32 (12.9%) |
87 (34.9%) |
| Infection (185) | 2 (1.1%) | 67 (36.2%) | 15 (8.1%) | 101 (54.6%) |
aNT, nontypeable by using antisera against Ia, Ib, II-VIII serotypes.
All the 434 isolates, regardless serotype, were positive for the scpB and lmb virulence-associated genes.
Uniform susceptibility was detected to ampicillin, levofloxacin, cefotaxime, and chloramphenicol, while most (97% ± 1.6) of the isolates were resistant to tetracycline. Resistance to erythromycin and clindamycin was found in 4.1% ± 1.8 (18 isolates) and 3% ± 1.6 (13 isolates) of the isolates, respectively. Among the erythromycin-resistant (EryR) isolates, the constitutive macrolide-lincosamide-streptogramin B (cMLSB) phenotype was the predominant phenotype (13/18 isolates) followed by the M (3/18 isolates) and the inducible MLSB (2/18 isolates) phenotypes. The relationship among resistance phenotypes and genotypes was significant (p = 0.028). All the isolates with iMLSB and M phenotypes and 2 of the 13 isolates with cMLSB phenotype harbored the ermA gene, whereas the ermB gene was detected in five isolates expressing the cMLSB phenotype. Six isolates belonging to the cMLSB phenotype did not show the presence of the erm or mef genes, even after several attempts. Erythromycin-resistant strains were represented by multiple serotypes (Table 3).
Table 3.
Distribution of phenotypic and genotypic characteristics among the 18 macrolide-resistant Streptococcus agalactiae isolates included in the present study
| Isolate | City of origin a | Serotype | Phenotype b |
Genotype
c
|
||
|---|---|---|---|---|---|---|
| erm A | erm B | mef A | ||||
| 4676 |
Rio de Janeiro |
Ia |
iMLSB |
+ |
- |
- |
| 4677 |
Rio de Janeiro |
Ia |
M |
+ |
- |
- |
| 4583 |
Rio de Janeiro |
NTd |
cMLSB |
- |
+ |
- |
| 4740 |
Rio de Janeiro |
III |
cMLSB |
- |
- |
- |
| 4714 |
Rio de Janeiro |
II |
cMLSB |
- |
- |
- |
| 4835 |
Rio de Janeiro |
II |
M |
+ |
- |
- |
| 5008 |
Rio de Janeiro |
III |
cMLSB |
- |
+ |
- |
| 4619 |
Rio de Janeiro |
NTd |
cMLSB |
- |
- |
- |
| 282 |
Rio de Janeiro |
II |
cMLSB |
- |
- |
- |
| 4971 |
Rio de Janeiro |
Ia |
cMLSB |
- |
+ |
- |
| 907 |
Rio de Janeiro |
II |
cMLSB |
- |
- |
- |
| 1196 |
Rio de Janeiro |
Ib |
cMLSB |
- |
- |
- |
| 4501 |
Porto Alegre |
III |
M |
+ |
- |
- |
| 4555 |
Porto Alegre |
Ia |
cMLSB |
+ |
- |
- |
| 4732 |
Brasília |
Ia |
cMLSB |
- |
+ |
- |
| 4736 |
Brasília |
Ia |
cMLSB |
- |
+ |
- |
| 6717 |
Recife |
V |
iMLSB |
+ |
- |
- |
| 8134 | Manaus | Ib | cMLSB | + | - | - |
aRio de Janeiro is located in the Southeast region; Porto Alegre is located in the South region; Brasilia is located in the Mid-West; Recife is located in the Northeast and Manaus is located in the North region.
bcMLSB: constitutive macrolide-lincosamide-streptogramin B resistance phenotype; iMLSB: inducible macrolide-lincosamide-streptogramin B resistance phenotype; M: macrolide resistance phenotype.
cerm: erythromycin ribosome methylation; mef: macrolide efflux.
dNT, nontypeable by using antisera against Ia, Ib, II-VIII serotypes.
Two tetracycline resistance genes, tetM and tetO, were detected in 99.3% ± 0.8 and 1.8% ± 1.2 of the total GBS isolates, respectively, and five of them harbored both genes. The tetK and tetL genes were not detected.
One EryR strain could not be typed by PFGE, despite several attempts, due to resistance to SmaI digestion. Among the remaining 17 isolates, 5 CCs and 16 different PFGE profiles were identified, and the Simpson’s Index of Diversity (SID) for this subgroup of isolates was 0.897 (95% CI, 0.807-0.987). Among the 25 erythromycin-susceptible strains, 9 CCs and 25 different profiles were observed, with a SID of 0.960 (95% CI, 0.940-0.980). Overall, 9 CCs and 40 different PFGE profiles were identified among the 42 isolates (Figure 2), with a SID of 0.935 (95% CI, 0.891-0.979). Five of the CCs comprised both EryR and susceptible isolates. Among the resistant isolates, those harboring different resistance genes shared the same CC. Also, isolates belonging to different serotypes or recovered from different clinical sources were clustered in the same CC.
Figure 2.
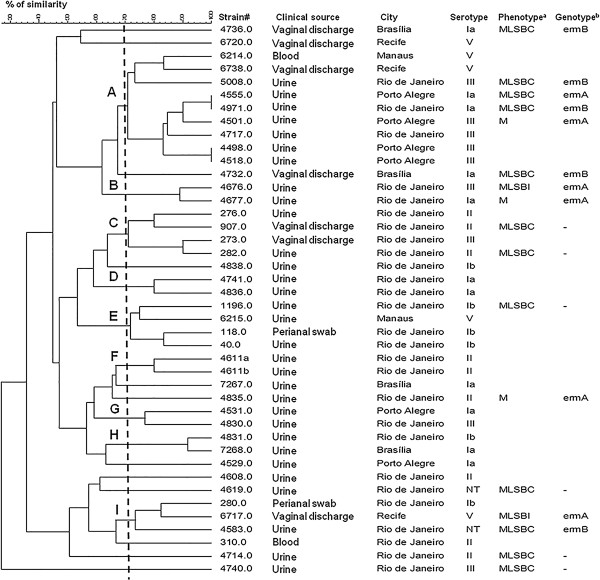
Dendrogram constructed by similarity and clustering analysis using the Dice coefficient and UPGMA of the digitalized PFGE profiles of 42 Streptococcus agalactiae isolates included in the present study. A total of 17 erythromycin-resistant isolates and 25 erythromycin-susceptible isolates were included. A tolerance of 1% was applied. The vertical line indicates the 70% level of similarity. The upper-case letters indicate the clonal complexes. cMLSB: constitutive resistance to macrolide-lincosamide-streptogramin B; iMLSB: inducible resistance to macrolide-lincosamide-streptogramin B; M: resistance to erythromycin only. (-): indicates the absence of resistance-associated genes.
Discussion
S. agalactiae is recognized as a frequent colonizing agent in pregnant women and is an important cause of neonatal sepsis and meningitis. Nevertheless, in the past decade GBS has been increasingly associated with invasive disease in non-pregnant adults [1,2].
The aim of this study was to phenotypically and genotypically characterize 434 GBS isolates obtained from different regions of Brazil, a continental country, with a large ethnic, climatic, social and economic diversity. Most of the previous studies performed in Brazil were limited to isolates from the South and Southeast regions [10-15]. This is the first study in Brazil encompassing the characterization of GBS isolates from 15 different locations representative of the five Brazilian geographical regions. Therefore, although sampling each geographic region and checking for variations among them was not a primary objective of the present study, it seemed useful and informative to display our results according to the geographical region. Nevertheless, our data might not fully and accurately represent each one of the five geographical regions.
Serotype determination has been traditionally applied in epidemiological studies on GBS and is an important aid in the development of broadly protective vaccines containing capsular polysaccharides or polysaccharides conjugated to protein [5]. The serotype distribution varies geographically and studies performed in European countries, United States and in Latin America have shown that serotypes Ia, Ib, III or V are usually the most frequently found [13,34]. In this study, serotypes Ia, Ib, II and V were the most commonly detected, in frequencies varying according to the region. Serotypes VI to VIII were not found, and these serotypes are still rarely reported from other locations [9]. Our results are in agreement with previous data obtained in earlier Brazilian studies with isolates obtained from the South and Southeast regions [10,15,35], as well as with data published in several other countries [24,25].
Serotypes were found in similar frequencies among isolates obtained from colonization as well as infections (p = 0.1392), suggesting that the capsule, despite being an essential factor for the virulence in GBS is not the only bacterial determinant responsible for the pathogenicity. Other studies have described the predominance of serotypes III and V among cases of infection in neonates and in non-pregnant adults, respectively [25,34] while serotype Ia was found to predominate among GBS causing invasive infections in non-pregnant adults in Portugal [36]. In Brazil, previous studies showed that serotypes Ia, II, III and V were found in 68.2% of isolates predominantly from vaginal specimens of asymptomatic pregnant woman [14]. On the other hand, serotype IV was detected in 13.1% of the isolates from Curitiba city, Paraná state, Brazil, being most of them from infectious cases [13], and serotypes Ib (34.9%) and Ia (25.6%) were predominant among a cohort of HIV-infected pregnant women in Rio de Janeiro city [35].
The occurrence of a nontypeable isolates rate of 6.2% in this study is consistent with the previously reported data [13]. Nevertheless, it should be pointed out that serotype IX was not investigated in the present study due to the lack of specific antiserum and that the detection of a serotype IX GBS isolate has been recently reported in the South region of Brazil [37]. Therefore, it is possible that the group of NT isolates may include representatives of this serotype.
The uniform susceptibility of GBS to beta-lactam antibiotics detected in this study is consistent with previously published data on S. agalactiae isolates in Brazil [10-15,30], as well as in several regions in the world [8,25]. However, an extensive network national surveillance should be implemented since isolates with reduced susceptibility to penicillin have been described in various regions of the world [16-18]. Although susceptibility of GBS to beta-lactam antibiotics was observed in the present study, reduced susceptibility has been reported elsewhere, underscoring the importance of an ongoing surveillance to monitor emergence of increased penicillin MICs among GBS isolates.
The low rates of resistance to erythromycin (4.1%) and clindamycin (3.0%) found in this study, are similar to that obtained in other studies conducted in Brazil, and some other countries in Latin America [11,13,15,25]. On the other hand, higher resistance rates have been observed in studies conducted in Asia, Europe, United States and Canada [6,22,23]. No correlation was found among erythromycin resistance and serotype, as previously observed [13]. Nonetheless, the predominance of serotype V among isolates with MLSB resistance has been reported [8]. Erythromycin-resistant isolates in the present study were obtained from colonization or non-invasive infections, as previously described [13,15]. These data suggest that the use of erythromycin and clindamycin as second-choice drugs in individuals with penicillin allergy should be accompanied by routine susceptibility testing to ensure proper therapy. If this is not possible, then treatment with vancomycin is recommended by CDC [19].
Among the EryR isolates, the ermA gene was the most frequent. Isolates presenting the cMLSB phenotype were mainly associated with the ermB gene, while those with iMLSB or M phenotypes harbored the ermA gene. Other authors have shown similar results [38,39]. The mefA gene was not found in this study, as previously described in Brazil [11,13], in contrast with the results obtained in other studies, where the mefA gene was detected in the isolates expressing the M phenotype [6,38]. The causes for the absence of resistance genes in six isolates with MLSB phenotype might reside in the fact that a great variety of genetic mechanisms can be associated with a single resistance phenotype. Here, we investigated the presence of the genes most commonly reported among GBS strains, however those 6 isolates could harbor less frequent determinants, such as ermC, ermF, ermQ, ermT, msrA or msrD; all of them already described among streptococci [40]. Also, mutations in 23S rRNA or in the genes encoding ribossomal proteins can also be speculated as possible causes of erythromycin-resistance in such strains.
Erythromycin-resistant isolates showed a slightly higher homogeneity by PFGE when compared to the susceptible strains analyzed. Nevertheless, a considerable genetic diversity was observed among the 42 isolates typed by PFGE, which clustered EryR and susceptible isolates in the same CC, suggesting that resistance to erythromycin is not necessarily a clonal characteristic among GBS strains included in the present study. On the other hand, the majority (64.7%) of the 17 EriR isolates were distributed among five of the nine CCs detected, suggesting that multiple clusters may have arisen simultaneously, mostly by independent acquisition of resistance genes by different strains. Similar results were previously shown in other studies [13,15,41]. Likewise, serotype does not seem to have a correlation with the classification of a GBS isolate in a clonal group. Relationships between the serotype and the source of isolation or between the serotypes and PFGE profiles were also not detected in another Brazilian study [14]. This fact could be explained by the occurrence of capsule switching between GBS isolates by horizontal gene transfer, as a result of pressures by immune response of the host [7]. One EryR isolate could not be typed by PFGE due to resistance to SmaI restriction. This phenomenon is not rare, and has been reported by other authors from different parts of the world [8,42,43]. Nevertheless, PFGE still represents an important molecular typing tool for GBS strains in Brazil, where, to our knowledge, other molecular methods such as MLST and MLVA have not or have rarely [37] been employed.
Resistance to tetracycline was detected in most of the isolates of this study, and the tetM gene was widely disseminated as previously reported in Brazil as well as in other countries [6,8,11,15,25]. All the isolates resistant to macrolides independent from the phenotypic and genotypic mechanism of resistance also had the tetM gene.
Conclusions
The present study provides an overview of the distribution of serotypes and on antimicrobial susceptibility of GBS isolates from Brazil, the largest country in Latin America. The findings suggest that GBS isolates circulating in Brazil have a wide phenotypic and genotypic diversity and that macrolide-resistant isolates may arise by both clonal spread and independent acquisition of resistance genes. These data are important to help in designing prevention and treatment strategies for GBS infections in the region.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VGD, LMT and SELF conceived the study, participated in the design and coordination, collaborated in the capsular typing, performed the antimicrobial susceptibility testing and drafted the manuscript. VMNA, ANO, CAGD, AFAB, ELTA, MABS, RS, PCSR, CFF, MA, KM, AAA, LFP, CM, EFCM, PCFJ, MSGR, PSM, RBGB, JKP, LMDC participated in data collection and presumptive identification of isolates obtained in 5 Brazilian regions. TCAP performed the PFGE analysis and the PCR reactions. ACNB participated in the capsular typing, performed the antimicrobial susceptibility testing, performed the PCR reactions and maintained GBS isolate collections. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Vanusa G Dutra, Email: vanusagd@oi.com.br.
Valéria MN Alves, Email: vmonteironeto@yahoo.com.br.
André N Olendzki, Email: andreolendzki@hotmail.com.
Cicero AG Dias, Email: cicero@ufspa.edu.br.
Alessandra FA de Bastos, Email: alessandra@sabinonline.com.br.
Gianni O Santos, Email: gianni@sabin.com.br.
Efigênia LT de Amorin, Email: efigenia@sergiofranco.com.brl.
Meireille ÂB Sousa, Email: meireille.sousa@hermespardine.com.br.
Rosemary Santos, Email: rosemarysantos@vivax.com.br.
Patricia CS Ribeiro, Email: waspatty@ig.com.br.
Cleuber F Fontes, Email: cleuber@ufba.br.
Marco Andrey, Email: mcoandrey@terra.com.br.
Kedma Magalhães, Email: kedma.biom@gmail.com.
Ana A Araujo, Email: ana.albertina@nkb.com.br.
Lilian F Paffadore, Email: slc_micro@hu.usp.br.
Camila Marconi, Email: camilamarconi@uol.com.br.
Eddie FC Murta, Email: eddiemurta@mednet.com.br.
Paulo C Fernandes Jr, Email: pcfarmaco@yahoo.com.br.
Maria SG Raddi, Email: stellaraddi@uol.com.br.
Penélope S Marinho, Email: penelopesaldanha@uol.com.br.
Rita BG Bornia, Email: rgbornia@terra.com.br.
Jussara K Palmeiro, Email: jussarakp@ig.com.br.
Libera M Dalla-Costa, Email: jussarakp@ig.com.br.
Tatiana CA Pinto, Email: tati.micro@gmail.com.
Ana Caroline N Botelho, Email: anabotelho25@gmail.com.
Lúcia M Teixeira, Email: lmt2@micro.ufrj.br.
Sérgio Eduardo L Fracalanzza, Email: fracalanzza@micro.ufrj.br.
Acknowledgements
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). We thank Dr. Ana Paula Colombo from the Universidade Federal do Rio de Janeiro, for the statistical analysis.
References
- Schuchat A. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin Microbiol Rev. 1998;14:497–513. doi: 10.1128/cmr.11.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoff TH, Farley MM, Petit S. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990–2007. Clin Infect Dis. 2009;14:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- Maisey HC, Doran KS, Nizet V. Recent advances in understanding the molecular basis of group B Streptococcus virulence. Expert Rev Mol Med. 2008;14:1–16. doi: 10.1017/S1462399408000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. Serotype IX, a proposed new Streptococcus agalactiae serotype. J Clin Microbiol. 2007;14:2929–2936. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CJ, Edwards MS. Group B streptococcal conjugate vaccines. Arch Dis Child. 2003;14:375–378. doi: 10.1136/adc.88.5.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan B, Schukken YH, Santisteban C, Boor KJ. Distribuition of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J Clin Microbiol. 2005;14:5899–5906. doi: 10.1128/JCM.43.12.5899-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S-L, Granlund M, Sellin M, Lagergard T, Spratt BG, Norgren M. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J Clin Microbiol. 2005;14:3727–3733. doi: 10.1128/JCM.43.8.3727-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi G, Imperi M, Baldassarri L, Pataracchia M, Alfarone G, Recchia S, Orefici G, Dicuonzo G, Creti R. Molecular epidemiology and distribuition of serotypes, surface proteins, and antibiotic resistance among Group B streptococci in Italy. J Clin Microbiol. 2007;14:2909–2916. doi: 10.1128/JCM.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Hans CS, Hans VN, Marcit SK, Helle BK. Emergence of invasive serotype VIII group B streptococcal infections in Denmark. J Clin Microbiol. 2003;14:4442–4444. doi: 10.1128/JCM.41.9.4442-4444.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchetrit LC, Fracalanzza SEL, Peregrino H, Camelo AA, Sanches LALR. Carriage of Streptococcus agalactiae in women and neonates and distribution of serological types: a study in Brazil. J Clin Microbiol. 1982;14:787–790. doi: 10.1128/jcm.15.5.787-790.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte RS, Bellei BC, Miranda OP, Brito MA, Teixeira LM. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob Agents Chemother. 2005;14:97–103. doi: 10.1128/AAC.49.1.97-103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões JA, Alves VM, Fracalanzza SEL, Camargo RP, Mathias HM, Brolazo EM. Phenotypical characteristics of group B Streptococcus in parturients. Braz J Infect Dis. 2007;14:261–266. doi: 10.1590/S1413-86702007000200019. [DOI] [PubMed] [Google Scholar]
- Palmeiro JK, Dalla-Costa LM, Fracalanzza SEL, Botelho ACN, Nogueira KS, Scheffer MC, Torres ARLS, Carvalho NS, Cogo LL, Madeira HMF. Phenotypic and genotypic characterization of group B streptococcal isolates in southern Brazil. J Clin Microbiol. 2010;14:4397–4403. doi: 10.1128/JCM.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa ABA, Silva GL, Pinto TCA, Oliveira ICM, Fernandes FG, Costa NS, Mattos MC, Fracalanzza SEL, Benchetrit LC. The genetic diversity and phenotypic characterisation of Streptococcus agalactiae isolates from Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2011;14:1002–1006. doi: 10.1590/S0074-02762011000800017. [DOI] [PubMed] [Google Scholar]
- Pinto TCA, Costa NS, Souza ARV, Silva LG, Corrêa ABA, Fernandes FG, Oliveira ICM, Mattos MC, Rosado AS, Benchetrit LC. Distribution of serotypes and evaluation of antimicrobial susceptibility among humam and bovine Streptococcus agalactiae strains isolated in Brazil between 1980 and 2006. Braz J Infect Dis. 2013;14:131–136. doi: 10.1016/j.bjid.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Suzuki S, Wachino J, Kurokawa H, Yamane K, Shibata N, Nagano N, Kato H, Shibayana K, Arakawa Y. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob Agents Chemother. 2008;14:2890–2897. doi: 10.1128/AAC.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahesh S. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob Agents Chemother. 2008;14:2915–2918. doi: 10.1128/AAC.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano N, Nagano Y, Toyama M, Kimura K, Tamura T, Shibayama K, Arakawa Y. Nosocomial spread of multidrug-resistant group B streptococci with reduced penicillin susceptibility belonging to clonal complex 1. J Antimicrob Chemother. 2012;14:849–856. doi: 10.1093/jac/dkr546. [DOI] [PubMed] [Google Scholar]
- CDC- Centers for Disease Control and Prevention. Prevention of perinatal group B streptococcal disease: revised guidelines from CDC. MMWR Recomm Rep. 2010;14(RR-10):1–31. [PubMed] [Google Scholar]
- Gonzalez JJ, Andreu A. The spanish group for the study of perinatal infection from the spanish society for clinical microbiology and infectious diseases. Multicenter study of the mechanisms of resistance and clonal relationships of Streptococcus agalactiae isolates resistant to macrolides, lincosamides, and ketolides in Spain. Antimicrob Agents Chemother. 2005;14:2525–2527. doi: 10.1128/AAC.49.6.2525-2527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsush PR, Teng LJ, Lee LN, HO SW, Yang PC, Luh KT. High incidence of erythromycin resistance among clinical isolates of Streptococcus agalactiae in Taiwan. Antimicrob Agents Chemother. 2001;14:3205–3208. doi: 10.1128/AAC.45.11.3205-3208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M, Delgaty KL, Ramotar K, Seetaram C, Toye B. Prevalence and mechanisms of erythromycin resistance in group A and group B Streptococcus implications for reporting susceptibility results. J Clin Microbiol. 2004;14:5620–5623. doi: 10.1128/JCM.42.12.5620-5623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt SM, Debusscher JH, Tallman PA, Manning SD, Marrs CF, Kurzynski TA, Foxman B. Frequency of antimicrobial resistance among invasive and colonizing Group B streptococcal isolates. BMC Infect Dis. 2006;14:57. doi: 10.1186/1471-2334-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phares CR, Lynfield R, Farley MM, Mohle-Boetani LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA. 2008;14:2056–2065. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- Martinez MA, Ovalle SA, Duran C, Reid I, Urriola G, Garay B, Cifuents M. Sorotipos y susceptibilidad antimicrobiana de Streptococcus agalactiae. Rev Méd Chil. 2004;14:549–555. doi: 10.4067/s0034-98872004000500003. [DOI] [PubMed] [Google Scholar]
- CLSI. 20th informational supplement Wayne. PA: CLSI; 2010. Performance standards for antimicrobial susceptibility testing: approved standard. M100-S20U. [Google Scholar]
- Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;14:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000;14:2778–2781. doi: 10.1093/jac/45.6.763. [DOI] [PubMed] [Google Scholar]
- Dmitriev A, Shakleina E, Tkacikova M, Mikula I, Totolian. Genetic heterogeneity of the pathogenic potentials of human and bovine group B streptococci. Folia Microbiol. 2002;14:291–295. doi: 10.1007/BF02817655. [DOI] [PubMed] [Google Scholar]
- Corrêa ABA, Oliveira ICM, Pinto TCA, Mattos MC, Benchetrit LC. Pulsed-field gel electrophoresis, virulence determinants and antimicrobial susceptibility profiles of type Ia group B streptococci isolated from humans in Brazil. Mem Inst Oswaldo Cruz. 2009;14:599–603. doi: 10.1590/S0074-02762009000400011. [DOI] [PubMed] [Google Scholar]
- Teixeira LM, Carvalho MGC, Merquior VLC, Steigerwalt AG, Brenner DJ, Facklam RR. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolates from human source. J Clin Microbiol. 1997;14:2778–2781. doi: 10.1128/jcm.35.11.2778-2781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;14:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001;14:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JL, Diekema DJ, Hunter SK, Rhomberg PR, Pfaller MA, Jones RN, Doern GV. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the Western Hemisphere. Am J Obstet Gynecol. 2000;14:859–862. doi: 10.1067/mob.2000.108839. [DOI] [PubMed] [Google Scholar]
- João EC, Gouvea MI, Menezes JA, Matos HJ, Cruz MLS, Rodrigues CAS, Caio ASR, de Souza MJ, Fracalanzza SEL, Botelho ACN, Calvet GA, Grinsztejn BGJ. Group B Streptococcus in a cohort of HIV-infected pregnant women: Prevalence of colonization, identification and antimicrobial susceptibility profile. Scand J Infect Dis. 2011;14:742–746. doi: 10.3109/00365548.2011.585178. [DOI] [PubMed] [Google Scholar]
- Martins ER, Melo-Cristino J, Ramirez M. Dominance of serotype Ia among group B streptococci causing invasive infections in nonpregnant adults in Portugal. J Clin Microbiol. 2012;14:1219–1227. doi: 10.1128/JCM.05488-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaguiri ES, Morguette AEB, Tavares ER, Santos PMMC, Morey AT, Cardoso JD, Perugini MRE, Yamauchi LM, Yamada-Ogatta SF. Commensal Streptococcus agalactiae isolated from patients seen at University Hospital of Londrina, Paraná, Brazil: capsular types, genotyping, antimicrobial susceptibility and virulence determinants. BMC Microbiol. 2013;14:297–305. doi: 10.1186/1471-2180-13-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura PAM, Schuab RBB, Neves FPG, Pereira CFA, de Paula GR, Barros RR. Antimicrobial resistance profiles and genetic characterisation of macrolide resistant isolates of Streptococcus agalactiae. Mem Inst Oswaldo Cruz. 2011;14:119–122. doi: 10.1590/S0074-02762011000200001. [DOI] [PubMed] [Google Scholar]
- Culebras E, Rodriguez-Avial I, Betriu C, Redondo M, Picazo J. Macrolide and tetracycline resistance and molecular relationships of clinical strains of Streptococcus agalactiae. Antimicrob Agents Chemother. 2002;14:1574–1576. doi: 10.1128/AAC.46.5.1574-1576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2008;14:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Souza VC, Kegele FC, Souza SR, Neves FP, de Paula GR, Barros RR. Antimicrobial susceptibility and genetic diversity of Streptococcus agalactiae recovered from newborns and pregnant women in Brazil. Scand J Infect Dis. 2013;14:780–785. doi: 10.3109/00365548.2013.810814. [DOI] [PubMed] [Google Scholar]
- Pillai P, Srinivasan U, Zhang L, Borchardt SM, Debusscher J, Marrs CF, Foxman B. Streptococcus agalactiae pulsed-field gel electrophoresis patterns cross capsular types. Epidemiol Infect. 2009;14:1420–1425. doi: 10.1017/S0950268809002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins ER, Andreu A, Correia P, Juncosa T, Bosch J, Ramirez M, Melo-Cristino J. Catalan Society for Clinical Microbiology and Infectious Diseases. Group B streptococci causing neonatal infections in Barcelona are a stable clonal population: 18-year surveillance. J Clin Microbiol. 2011;14:2911–2918. doi: 10.1128/JCM.00271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


